Abstract
We have generated an FLT3/ITD knock-in mouse model in which mice with an FLT3/ITD mutation develop myeloproliferative disease (MPD) and a block in early B-lymphocyte development. To elucidate the role of FLT3/ITD signaling in B-cell development, we studied VDJ recombination in the pro-B cells of FLT3/ITD mice and discovered an increased frequency of DNA double strand breaks (DSBs) introduced by the VDJ recombinase. Early pro-B cells from FLT3/ITD mice were found to have a lower efficiency and decreased accuracy of DSB repair by nonhomologous end joining (NHEJ), which is required for rejoining DSBs during VDJ recombination. Reduced NHEJ repair probably results from reduced expression of Ku86, a key component of the classic DNA-PK-dependent NHEJ pathway. In compensation, early pro-B cells from FLT3/ITD cells mice show increased levels of the alternative, and highly error-prone, NHEJ pathway protein PARP1, explaining the increase in repair errors. These data suggest that, in early pro-B cells from FLT3/ITD mice, impairment of classic NHEJ decreases the ability of cells to complete postcleavage DSB ligation, resulting in failure to complete VDJ recombination and subsequent block of B-lymphocyte maturation. These findings might explain the poor prognosis of leukemia patients with constitutive activation of FLT3 signaling.
Introduction
In mouse, Fms-like tyrosine kinase 3 ligand (FLT3) is mainly expressed in normal hematopoietic stem/progenitor cells (HSPCs) and early B-cell progenitors.1–4 Its expression appears to be required for the initiation of B lymphopoiesis, and mice deficient in either FLT3 receptor or its ligand display a marked decrease in the B-cell compartment, in particular the earliest B precursors.5,6 Activating mutations of FLT3, either in the form of internal tandem duplication (ITD) mutations in the juxtamembrane domain or point mutations in the kinase domain, are frequently reported in acute myeloid leukemia and less frequently in acute lymphoblastic leukemia.7–9 The mechanism through which constitutively activated FLT3 contributes to leukemic transformation of HSPCs is not fully understood.
One essential characteristic of lymphocyte development is VDJ recombination, through which the somatic assembly of germline VDJ gene segments of T-cell receptor or immunoglobulin (Ig) gene loci occurs to produce genes encoding a unique receptor or Ig structure on each T or B lymphocyte, respectively.10 This process can further be dissected into 2 steps: site-specific cleavage of DNA and rejoining of broken DNA ends. The cleavage step is initiated by site-specific RAG1/RAG2 endonucleases, which introduce DNA double-strand breaks (DSBs) between participating gene segments,11,12 whereas the rejoining of broken DNA is completed by the nonhomologous end joining (NHEJ) pathway.10,13
Mammalian cells use several major pathways that function in different but complementary manners to repair DSBs. The classic DNA-PK-dependent nonhomologous end joining (C-NHEJ) pathway is the pathway cells use to repair the majority of DSBs, including those generated by VDJ recombination. Several of the components participating in this pathway have been identified: the heterodimer of Ku70 and Ku86 forms a complex with the DNA-dependent protein kinase catalytic subunit (DNA-PKcs), which bridges DNA ends and phosphorylates Artemis to activate its DNA end-processing activities.14–17 It also provides a platform for the ligation complex, consisting of the catalytic subunit DNA ligase IV and its cofactor XRCC4, to perform the ligation of DNA ends.18,19 In the presence of nonligatable DNA ends, XLF (XRCC4-like factor), also known as Cernunnos, interacts with DNA ligase IV/XRCC4 and stimulates the joining of mismatched DNA ends.20 The joining of DSBs by C-NHEJ results in the loss or addition of a few nucleotides at the break site. The presence of short microhomologies at the break site contributes to the alignment of the DNA ends.21
Accumulating evidence suggests that alternative backup NHEJ pathways play important roles in DSB repair.22–25 For example, rare aberrant VDJ coding joins are found in Ku or DNA-PKcs-absent lymphocytes.26,27 Chromosomal abnormalities, including c-myc/IgH translocations, are observed in the absence of either Ku or DNA ligase IV/XRCC4.26,28 These alternative pathways are slower and less efficient, featured by larger deletions and insertions and longer repair junctions with DNA sequence microhomologies.23,24 They are directly implicated in genomic instability and the development of cancer.22–25 Several DNA repair proteins, including DNA ligase III/XRCC1, poly(ADP) ribose polymerase 1 (PARP1), MRN, and WRN, have been implicated in these pathways.23,24 However, the regulation and underlying mechanisms are poorly understood.
Recently, we have generated an FLT3/ITD knock-in mouse model in which mice heterozygous for an FLT3/ITD mutation develop myeloproliferative disease and a block in B-lymphocyte development at the early pro-B stage.29 We found that, during the process of D-JH recombination in early pro-B cells from FLT3/ITD mice, impairment of the C-NHEJ pathway decreases the ability of cells to complete postcleavage DSB ligation, resulting in failure to complete D-JH recombination with a subsequent block of pro-B cell maturation. With the C-NHEJ pathway impaired, BM cells from FLT3/ITD mice appear to repair DSBs through the slow, less efficient, and highly error-prone alternative (ALT)–NHEJ pathways. Increased genomic instability caused by the abnormal repair of DSBs might contribute to the leukemic transformation of normal HSPCs by FLT3/ITD mutations. This could also explain the poor prognosis associated with FLT3/ITD mutations in leukemia patients.
Methods
Mice
FLT3/ITD knock-in mice were generated as previously reported.29 The mice were maintained on a C57BL/6 background and housed in microisolator cages in a pathogen-free animal facility. Two-month-old mice were used for all experiments. All animal experiments were reviewed and approved by the Johns Hopkins Animal Care and Use Committee.
Flow cytometric analysis and cell sorting
Flow cytometric analysis was performed as described previously29 using a BD FACSCalibur or LSRII. Cell sorting was performed on a FACSAria (BD Biosciences). Phenotypic definitions used to define the compartments are: common lymphoid progenitors (CLPs): Lin−IL7Rα+Sca-1lowc-KITlow; pre-pro-B: B220+CD43+CD93+CD24−CD19−IgM−; early pro-B: B220+CD43+CD93+CD24+CD19−IgM−; late pro-B: B220+CD43+CD93+CD24+CD19+IgM−; pre-B: B220+CD43−CD24+CD19+IgM−; and B: B220+CD43−CD24+CD19+IgM+. Data were analyzed by FACSDIVA (BD Biosciences) or FlowJo Version 8.8.6 analysis software (TreeStar). A detailed record of antibodies used is available in the supplemental data (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
VDJ recombination assays
The D-JH rearrangement assay was performed as previously described.30 Genomic DNA extracted from sorted cells were amplified using primers DH: 5′-GGAATTCG(A/C)TTTTTGT(C/G)AAGGGATCTACTACTGTG-3′ and J3: 5′-GTCTAGATTCTCACAAGAGTCCGATAGACCCTGG-3′. Products were detected by hybridization to 32P-labeled probe JH3 (5′-AGACAGTGACCAGAGTCCCTTGG-3′). The ligation-mediated PCR (LM-PCR) assay for signal end breaks was performed as previously described31 using linkers and locus-specific primers.31 PCR products were detected by hybridization to 32P-labeled probe 5′ of JH locus. DNA band intensities were measured using QuantityOne Version 4.5.0 densitometry analysis software (Bio-Rad).
Immunocytochemistry
Immunocytochemistry analysis was performed as previously described. Flow cytometric analysis of γH2AX was performed according to Huang and Darzynkiewicz.32 Cells were fixed in 1% formaldehyde, permeabilized with 70% ethanol, stained with FITC-conjugated antiphospho-H2AX antibody and propidium iodide (Sigma-Aldrich), and then subjected to flow cytometry acquisition and analysis.
In vivo DNA repair assay
The in vivo NHEJ assay was performed as previously described.33 Repair efficiency was assessed as the total number of blue (correctly repaired) and white (incorrectly repaired) bacterial colonies produced. Misrepair errors were calculated as the frequency of white versus total colonies produced. Plasmid DNA from randomly selected white bacterial colonies was prepared and sequenced using either pUC18-F or pUC18-R primer. The BLAST program from National Center for Biotechnology Information Website was used for sequence alignment.
Quantitative RT-PCR analysis
Quantitative RT-PCR was performed using an iCycler iQ multicolor real-time PCR system (Bio-Rad) as described previously.29 A detailed list of primers used is available in the supplemental data.
Western blotting analysis
Nuclear extract was prepared as previously described,33 separated by SDS-PAGE, transferred onto PVDF membrane, and probed with the indicated primary and secondary antibodies. Primary antibodies used are goat anti-Ku86, PARP1, Ku70, DNA-PKcs, Lamin B, RAG1, and RAG2 (Santa Cruz Biotechnology) and rabbit anti–DNA ligase IIIα (Sigma-Aldrich). Relative levels of protein expression were measured using QuantityOne Version 4.5.0 densitometry analysis software (Bio-Rad).
In vitro B-cell culture
Sorted early B-cell progenitors (B220+CD43+CD24−/lowCD93+CD19−IgM−) were plated in Iscove modified Dulbecco medium supplemented with 10 ng/mL IL-7 (PeproTech) and lestaurtinib (LC Laboratories) in the presence of OP9 stromal cells (ATCC). Cultured cells were collected 48 hours later and subjected to subsequent analysis.
In vivo treatment with sorafenib
Sorafenib (LC Laboratories) was resuspended in a liquid vehicle composed of 30% (weight/volume) Cremophor EL, 30% (weight/volume) polyethylene glycol, 10% ethanol, and 10% glucose (all from Sigma-Aldrich) and administered into the mice with a dosage of 10 mg/kg daily for 3 days via oral gavages. Control mice were administered with vehicle only. BM were collected 12 hours after the last administration and subjected to subsequent analysis.
Lentiviral transduction and BM transplantation
pWCC-Ku86 plasmid was constructed by inserting Ku86 cDNA (a kind gift from Dr David Chen at University of Texas Southwestern Medical Center, Dallas, TX) into pWCC lentiviral vector under control of the EF1α promoter (supplemental Figure 1). Lentiviral particles were produced and concentrated as described previously.34 A total of 5 × 105 lineage-depleted BM cells were transduced and injected into the lateral tail veins of lethally irradiated (9 Gy) B6-Ly5.2 recipient mice (National Cancer Institute, Frederick, MD).
Results
FLT3/ITD expression blocks B-cell differentiation at the early pro-B cell stage
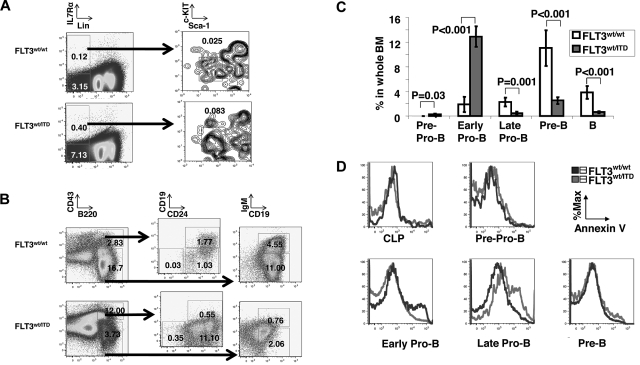
ITD mutations cause constitutive activation of FLT3 that deregulates hematopoietic development. We have previously reported that mice with a knock-in of an FLT3/ITD mutation in the FLT3 locus show a significant expansion of myeloid cells and a marked reduction of B cells in both the BM and spleen.29 To define the stage(s) at which B-cell development is blocked, we analyzed the cell surface expression of markers of B-cell progenitors in BM from young (2-month-old) heterozygous FLT3/ITD mice. In mice, FLT3 is reported to be expressed in both CLPs and early B-cell progenitors.1–4 Consistent with this, flow cytometric analysis showed that the frequencies of CLPs (0.065% ± 0.026% in the FLT3/ITD group vs 0.028% ± 0.008% in the wild-type group, P = .04, n = 5), pre-pro-B cells (Hardy fraction A, 0.29% ± 0.08% vs 0.03% ± 0.01%, P = .03, n = 5), and early pro-B cells (Hardy fraction B, 12.90% ± 1.67% vs 1.89% ± 1.21%, P < .001, n = 5) are significantly increased in the BM from FLT3/ITD mice compared with their wild-type littermate controls (Figure 1A-C). This suggests that constitutive FLT3 activation results in expansion of the earliest B precursors, including CLPs, pre-Pro-B, and early pro-B cells. However, the fraction of late pro-B cells (Hardy fraction C) were greatly reduced in BM from FLT3/ITD mice (0.48% ± 0.25% vs 2.29% ± 0.73%, P = .001, n = 5). Similar reductions in the factions of subsequent pre-B cells (Hardy fraction D; 2.52% ± 0.54% vs 11.05% ± 2.52%, P < .001, n = 5) and B cells (Hardy fraction E; 0.63% ± 0.14% vs 3.82% ± 1.04%, P < .001, n = 5) were also observed in BM from FLT3/ITD mice (Figure 1B-C). These data indicate that, in mice with the FLT3/ITD mutation, B-cell development is blocked at the early pro-B to late pro-B stage transition.
Figure 1.
FLT3/ITD blocks B-lineage differentiation at the early pro-B stage. Flow cytometry analysis of the fraction of CLPs (A) and early B-cell differentiation (B) in whole BM from 2-month-old FLT3/ITD mice and wild-type littermate controls. (A) Far left panels: Cells are whole BM cells. (B) Cells are CD93+ cells in whole BM. Numbers shown on plots are percentages in whole BM. (C) The graph summarizes the differences in the fraction of CLPs and B-cell progenitors. Data are mean ± SEM (error bars) (n = 5). (D) Flow cytometry analysis for annexin V staining of B-cell compartments demonstrates increased apoptosis in late pro-B cells from FLT3/ITD mice. Data are representative of 3 independent experiments.
During B-lymphoid development, D to JH recombination initially occurs in early pro-B cells. Pro-B cells that fail to assemble a functional heavy chain gene are growth arrested and eventually are eliminated by apoptosis. We analyzed the frequency of early apoptotic cells within B-cell progenitors using annexin V staining. No differences were found in the percentage of apoptotic cells in CLPs, pre-pro-B, or early pro-B populations between FLT3/ITD and wild-type control mice. However, an increased fraction of annexin V-positive cells was detected in the late pro-B compartment from FLT3/ITD mice compared with their wild-type littermates (Figure 1D). The increased apoptotic rate in late pro-B cells coincides with the developmental block at this stage, suggesting that FLT3 activation affects the differentiation and survival of late pro-B cells.
We did not observe significant changes in thymocyte subsets in either the thymus or BM, as evidenced by expression of CD90, CD25, CD44, CD3, CD4, and CD8a (data not shown). This is consistent with the absence of FLT3 expression in T lymphocytes and the previous report that treatment with FLT3 ligand greatly reduces the fraction of B220+CD19+ cells in mice without affecting T-cell subsets.35
VDJ recombination is impaired in early pro-B cells with FLT3/ITD mutation
Because we have demonstrated here that B-cell development is blocked at the early pro-B stage in FLT3/ITD mice, we are interested in understanding the changes in the VDJ recombination activity of developing B cells from FLT3/ITD mice.
To assess VDJ recombination activity in early pro-B cells from FLT3/ITD mice, we performed a D-JH rearrangement assay. We observed greatly reduced D-JH rearrangements in early pro-B cells (Figure 2A), indicating defective D to JH recombination in these cells.
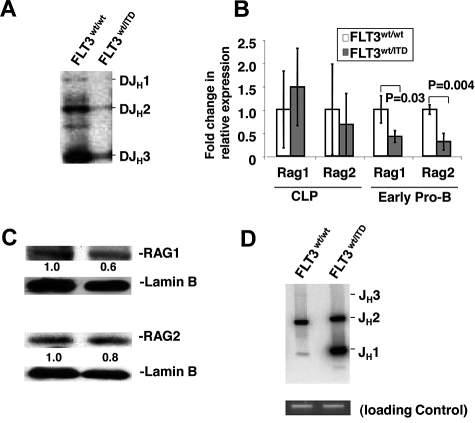
Figure 2.
VDJ recombination is impaired in early pro-B cells with FLT3/ITD mutation. (A) Early pro-B cells from FLT3/ITD mice demonstrate significantly decreased D-JH rearrangement. Densitometry analysis shows that recombination of D-JH1, D-JH2, and D-JH3 in FLT3/ITD+ early pro-B cells was less than 0.05-, 0.1-, and 0.1-fold, respectively, of that in wild-type cells. Completed D-JH rearrangements were assayed by PCR of sorted early pro-B cells from FLT3/ITD or wild-type littermate controls. (B) Quantitative RT-PCR analysis shows changes in relative expression levels of Rag1 and Rag2 in CLPs and early pro-B cells (n = 3). (C) Western blotting analysis shows expression of RAG1 and RAG2 in early pro-B cells. Values below the gel image indicate relative fold changes of protein levels normalized to lamin B. (D) Broken signal ends at the JH locus accumulate in early pro-B cells from FLT3/ITD mice. Densitometry analysis shows that recombination intermediates for JH1 and JH2 in FLT3/ITD+ early pro-B cells were 20- and 1.8-fold of that in wild-type cells, respectively. Sorted early pro-B cells were subjected to LM-PCR assays for JH-associated broken ends. The expected gene arrangements (DJH1-3 in panel A) or broken JH fragments (JH1-3 in panel D) are indicated. PCR amplification with primers for a nonrearranging locus (CD14) was used as a quantity control for the linker-ligated DNA samples. Data are representative of 3 independent experiments.
The VDJ recombination process can be further divided into 2 steps: site-specific DNA cleavage and the joining of broken DNA ends. The former is carried out by RAG1 and RAG2 to recognize and induce DSBs at the recombination signal sequences, whereas the latter is completed by NHEJ pathways.10–13 Reduced D to JH rearrangements could result from either reduced cleavage of genomic DNA or impaired DSB end joining. We first analyzed expression of Rag1 and Rag2 in CLPs and early pro-B fraction by quantitative RT-PCR. Our results showed that Rag1 and Rag2 expression in CLPs from FLT3/ITD mice was comparable with the wild-type control group. In contrast, expression in early pro-B cells from FLT3/ITD mice was reduced to 42.0% ± 12.5% (P = .03, n = 5) and 30.6% ± 17.9% (P = .004, n = 5) compared with the levels expressed in the wild-type control group (Figure 2B). Western blotting analysis also demonstrated slightly reduced levels of RAG1 and RAG2 in early pro-B cells from FLT3/ITD mice (Figure 2C). Reduced RAG1/RAG2 expression in FLT3/ITD mice indicates that DNA cleavage is reduced in early pro-B cells in these mice, which might be caused by deregulated B-lineage transcription regulation as a result of constitutively activated FLT3 signaling because early activation and later repression of FLT3 signaling play an important role in B-cell specification and commitment, respectively.1–4
To further evaluate end-joining ability after DNA cleavage, we assayed the signal end recombination intermediates, an indicator of ongoing VDJ recombination, by LM-PCR. Based on the finding that RAG1 and RAG2 expression was reduced in early pro-B cells from FLT3/ITD mice, we expected to detect reduced levels of double-strand recombination intermediates. To our surprise, double-strand DNA breaks at the JH recombination signal, indicating ongoing D to JH rearrangement, were more abundant in early pro-B cells from FLT3/ITD mice than in the cells from their wild-type littermates (Figure 2D).
The observed reduced D-JH rearrangements and the accumulation of signal ends generated during D-JH recombination suggest that early pro-B cells from FLT3/ITD mice are impaired in their ability to ligate broken DNA ends.
Early pro-B cells with FLT3/ITD mutation accumulate DSBs and have reduced ability and impaired fidelity to repair DSBs
In mammalian cells, H2AX, a histone H2A variant, is phosphorylated on serine 139 to form γH2AX at the site of DNA DSBs.36 γH2AX is an established marker for the presence of DSBs. Immunocytochemistry results demonstrated an increased number of γH2AX “foci” in early pro-B cells from FLT3/ITD mice compared with the wild-type controls (9.3 ± 2.5 vs 2.8 ± 1.4, P < .001, n = 12; Figure 3A-B), supporting the finding that D-JH intermediates accumulate in FLT3/ITD+ early pro-B cells. Flow cytometric analysis confirmed this result, demonstrating increased levels of γH2AX in early pro-B cells from FLT3/ITD mice, with an increased mean fluorescence index for the FLT3/ITD cells in G0/G1 phase compared with the wild-type group in the same cell cycling phase (Figure 3C-D).
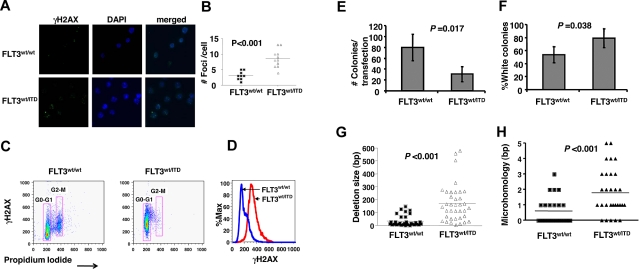
Figure 3.
Early pro-B cells from FLT3/ITD mice accumulate DSBs and are defective for DSB repair. (A-B) γH2AX foci are increased in early B-cell progenitors from FLT3/ITD mice compared with wild-type littermate controls. Images were acquired at room temperature using a Nikon TE 2000-E microscope system (Nikon) with a Nikon Plan APO VC 100×/1.40 oil objective (original magnification ×1000) and Nikon EZ-C1 Version 3.5 software. Flow cytometric analysis shows increased phosphorylation levels of H2AX in early pro-B cells from FLT3/ITD mice (C), with an increased mean fluorescence for cells in G0/G1 phase (D), compared with their wild-type controls. Early pro-B cells from FLT3/ITD mice demonstrate decreased NHEJ efficiency (E) and increased errors (F) after repair of DSBs. Repair efficiency is assessed as the number of total bacterial colonies obtained from the transfection of 5 × 106 sorted early pro-B cells. Misrepair rate is calculated as the fraction of white in total (blue and white) colonies. Data are expressed as mean ± SEM (error bars). Early pro-B cells from FLT3/ITD mice demonstrate an increased frequency of larger deletions (G) and the use of longer microhomologous sequences (H) for the repair of DNA. Data are representative of 3 independent experiments.
To more quantitatively assess the effects of FLT3/ITD mutations on the repair of DSBs during VDJ recombination in B-cell development, we performed an in vivo plasmid end-joining assay that relies on the repair of a DSB within the LacZα gene of PUC18 transfected into cells, followed by transformation of repaired plasmids in Escherichia coli. The results showed that early pro-B cells from FLT3/ITD mice produce a significantly reduced number of total bacterial colonies. For every 5 × 106 transfected early pro-B cells, 79 ± 24 bacterial colonies were generated in the wild-type group, whereas only 31 ± 14 colonies were generated in the FLT3/ITD group (P < .05, n = 4). Early pro-B cells from FLT3/ITD mice also appeared to generate a higher frequency of misrepaired bacterial colonies compared with their littermates (79% ± 14% for FLT3/ITD vs 53% ± 12% for the wild-type group, P = .038, n = 4; Figure 3E-F). Moreover, sequencing of the region surrounding the DSB in misrepaired plasmids showed larger deletions in the FLT3/ITD cells, compared with results from the wild-type control group (Figure 3G). Detailed sequencing analysis also revealed that 73.8% (31 of 44) of the DSBs in FLT3/ITD+ early pro-B cells were repaired using microhomologous sequences compared with 36% (9 of 25) in wild-type control group (P < .001). It is also notable that the average sizes of microhomologous sequences appear to be larger in FLT3/ITD+ early pro-B cells compared with wild-type control (2.1 ± 1.3 vs 1.5 ± 0.7 bp, P < .001, Figure 3H). Interestingly, in one of the samples from FLT3/ITD group, a 370-bp fragment of genomic DNA from mouse chromosome 5 was found to be inserted into the repaired plasmid (supplemental Figure 2). The translocation of mouse chromosomal DNA to the repaired plasmid suggests increased genomic instability in early pro-B cells from FLT3/ITD mice.
These results suggest that FLT3/ITD+ early pro-B cells have a reduced ability to repair DSBs and that this repair is accompanied by an increased error rate, an increase in larger deletions, and larger microhomologies. It is unlike the classic DNA-PK and Ku-dependent NHEJ where errors typically involve the deletion of fewer basepairs. These data provide additional evidence that FLT3/ITD+ pro-B cells recruit ALT end-joining pathway to repair DSBs.
Early pro-B cells from FLT3/ITD mice show decreased expression of Ku86 and increased expression of PARP1
In mammalian cells, the majority of DSBs are repaired through the Ku and DNA-PK-dependent C-NHEJ pathway. Recently, ALT NHEJ pathways have been reported to repair DSBs when C-NHEJ is down-regulated or defective.22–25 Importantly, this pathway has been implicated in the generation of large deletions and translocations in cancer.23,24 To determine whether transcription of C-NHEJ genes were potentially decreased in FLT3/ITD pro-B cells, we examined mRNA levels of C-NHEJ genes, including DNA-PKcs, Ku70, Ku86, and DNA ligase IV, using quantitative RT-PCR. We detected an approximately 5-fold decrease in Ku86 expression in early pro-B cells from FLT3/ITD mice compared with the wild-type controls (Figure 4Ai). No significant differences in RNA expression of Ku70, DNA-PKcs, DNA ligase IV, or XRCC4 were detected. Western blotting of proteins in nuclear extracts from the early pro-B cells of FLT3/ITD mice confirmed decreased expression of Ku86 compared with wild-type mice (Figure 4B-i).
Figure 4.
Early pro-B and lineage-negative BM cells from FLT3/ITD mice show decreased expression of Ku86 and increased expression of PARP1. (A) RNA was extracted from sorted early pro-B cells, reverse transcribed, and subjected to quantitative RT-PCR assays. Data are representative of 5 independent experiments. (B) Western blotting analysis using nuclear extracts from sorted early pro-B cells (i-ii) demonstrates decreased expression of Ku86 and increased expression of PARP1 in samples from FLT3/ITD mice. Lamin B was used as a loading control. (C) Transduction of Ku86 into FLT3/ITD Lin− BM partially overcomes the block of B-lymphocyte development in recipients. Representative flow cytometry analyses of recipient PB and BM are shown. GFP+B220+CD19− cells from recipients transplanted with Ku86-transduced FLT3/ITD Lin− cells demonstrate increased repair efficiency (D) and the use of less and shorter microhomologous sequences (E) for the repair of DNA. (F) Quantitative RT-PCR (i) and Western blotting assay (ii) of lineage-depleted BM cells from FLT3/ITD mice have significantly decreased Ku86 and increased PARP1 expression compared with control mice. Data are mean ± SEM (error bars). Data are representative of 3 independent experiments. Values below the gel image indicate relative fold changes of protein levels normalized to lamin-B.
To confirm the suppressive effect of FLT3/ITD on Ku86, lineage-depleted BM from FLT3/ITD mice were lentivirally transduced with Ku86 and transplanted into lethally irradiated recipients. Twelve weeks after transplantation, analysis of peripheral blood from recipients transplanted with Ku86-transduced FLT3/ITD+ Lin− cells showed an increased CD19+ fraction in total GFP+ cells compared with the group that received vector alone-transduced FLT3/ITD+ Lin− cells (23.5% ± 5.2% vs 15.6% ± 3.6%, P = .01, n = 5 in each group, Figure 4C). Similar results were found in the BM of recipients transplanted with Ku86-transduced FLT3/ITD+ Lin− cells (12.2% ± 2.9% vs 7.7% ± 0.8%, P = .06, Figure 4C). Thus, it appears that exogenous Ku86 partially overcomes the block of B-cell development caused by FLT3/ITD expression.
The PUC18-based in vivo DNA repair assay of the GFP+B220+CD19− cells from recipient mice demonstrated that the Ku86-transduced FLT3/ITD+ cells showed an improved DNA repair efficiency compared with the group transduced with vector (92 ± 8 vs 56 ± 9 per 5 × 106 transfected cells, P = .008, n = 3; Figure 4D). Further sequencing of the misrepaired colonies showed slightly less use of microhomologies in DSB repair in Ku86-transduced FLT3/ITD cells compared with the vector-transduced cells (68% vs 78%). The microhomologous sequences also appeared to be shorter in size in Ku86-transduced FLT3/ITD cells (1.7 ± 1.3 vs 0.9 ± 0.8 bp, P = .02, n = 22, Figure 4E). These results indicate that Ku86-transduced FLT3/ITD cells use less alternative NHEJ pathways to repair DSBs. These data support the finding that FLT3/ITD mutations exert a suppressive role on Ku86 expression.
In mammalian cells, Ku70 and Ku86 form a heterodimer, which plays an essential role in the NHEJ pathway by recruiting DNA-PKcs, Artemis, and ligase IV/XRCC4 complex for the processing and joining of the DNA ends.37 Reduced expression of Ku86 suggests that C-NHEJ pathways are impaired in FLT3/ITD pro-B cells.
In the event of defective C-NHEJ, DNA end joining would need to be carried out by alternative pathways.22–25 We next determined whether, in compensation for the decrease in C-NHEJ, some components of the ALT-NHEJ pathway, such as PARP1 or DNA ligase III, might be up-regulated in early pro-B cells from FLT3/ITD mice. Interestingly, quantitative RT-PCR analysis demonstrated a 2-fold increase in PARP-1 expression. Western blotting analysis of protein nuclear extracts from FLT3/ITD B-cells confirmed that PARP1 was up-regulated, compared with wild-type controls (Figure 4Aii,Bii). We did not find elevated levels of DNA ligase IIIα expression in FLT3/ITD mice by either quantitative RT-PCR or Western blotting analysis. In mouse, FLT3 is mainly expressed in hematopoietic stem/progenitors. Interestingly, by quantitative RT-PCR and Western blotting analysis, we also observed decreased expression of Ku86 and increased expression of PARP1 in the lineage-depleted BM, which contains most of the hematopoietic progenitors expressing FLT3, from FLT3/ITD mice (Figure 4Fi-ii).
These results suggest that both early pro-B cells and the lineage-negative BM cells from FLT3/ITD mice are defective in C-NHEJ, the major pathway for DSB repair. Instead, they probably use the slow and less efficient ALT-NHEJ pathway for the joining of DSBs cleaved by RAG1/RAG2 activity during VDJ recombination in the case of pro-B cells or by other DNA damage events in the case of lineage-negative cells.
Treatment of B cells from FLT3/ITD mice with FLT3 tyrosine kinase inhibitors overcomes the block of B-cell development
To further evaluate the effect of constitutively activated FLT3 signaling on B-cell development, we sorted early pro-B cells and used them in an in vitro differentiation assay. In this assay, 1 × 106 sorted cells were cultured with IL-7 in the presence or absence of different concentrations of lestaurtinib, an FLT3 inhibitor. After 2 days of culture without the inhibitor, 79% of the early pro-B cells from wild-type control mice were still viable (as shown by trypan blue exclusion) and readily generated 28.0% ± 5.2% B220+CD19+ cells. In contrast, whereas 76% of the cultured early pro-B cells from FLT3/ITD mice were viable, only 13.9% ± 2.5% of the viable cells were B220+CD19+ when cultured under the same conditions (Figure 5A-B).
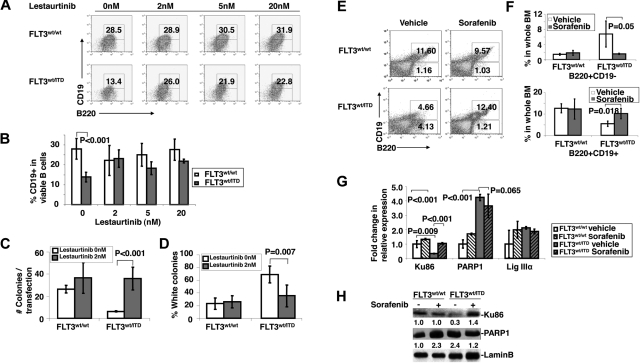
Figure 5.
Treatment of FLT3/ITD cells with FLT3 inhibitors overcomes the block of B-cell development. (A-B) In vitro culture of early pro-B cells from FLT3/ITD mice with lestaurtinib generates a higher frequency of the more differentiated B220+CD19+ cells compared with the wild-type control group. A total of 20 000 events were acquired using FACSCalibur (BD Biosciences). Analysis was based on gated viable populations according to forward/side scatter. (C-D) Treatment with 2nM lestaurtinib improves DNA repair efficiency and reduces repair errors in B220+ cells generated from in vitro culture of early pro-B cells with FLT3/ITD mutation but not from wild-type cells. Results of PUC18-based in vivo DNA repair assay are shown. Graphs show the repair efficiency (C) and misrepair errors (D) (n = 3). (E-F) BM from sorafenib-treated FLT3/ITD mice shows a higher frequency of B220+CD19+ cells compared with the vehicle-treated group. Data are representative of 5 independent experiments. Data are expressed as mean ± SEM (error bars). Quantitative RT-PCR (G) and Western blotting analysis (H) demonstrate that treatment with sorafenib increases expression of Ku86 in B220+ cells from FLT3/ITD mice. Data are mean ± SEM (error bars); n = 3. Values below the gel image indicate relative fold changes of protein levels normalized to lamin-B.
After culture in the presence of 20nM lestaurtunib for 2 days, 51% of the cells from wild-type BM and 53% from FLT3/ITD BM were still viable. Treatment of the early pro-B cells from FLT3/ITD BM with lestaurtinib significantly increased the frequency of B220+CD19+cells (from 13.9% ± 2.5% to 21.8% ± 0.1%, P = .006, n = 3) while having little to no effect on the frequency of the wild-type group (from 28.0% ± 5.2% to 27.6% ± 5.3%, P = .93, n = 3; Figure 5A-B). These data suggest that constitutively activated FLT3 specifically blocks B-cell differentiation, which can be overcome by selective inhibition of FLT3 tyrosine kinase activity.
Our data thus far suggest that constitutively activated FLT3 impairs C-NHEJ during VDJ recombination and subsequently blocks B-cell development. We next determined whether inhibition of FLT3 signaling would affect DNA repair efficiency in the B-cell compartment. We thus performed in vivo DNA repair assays on B220+ cells collected from the cultured early pro-B cells before and after treatment with lestaurtinib. B220+ cells from FLT3/ITD mice demonstrate a decrease in DNA repair efficiency when cultured in the absence of lestaurtinib, compared with cells from the wild-type group (Figure 5C). In the PUC18-based in vivo DNA repair assay, treatment of FLT3/ITD+ early pro-B cells with 2nM lestaurtinib resulted in increased DNA repair efficiency and decreased repair errors to levels comparable with that demonstrated in wild-type pro-B cells (Figure 5 C-D).
To further evaluate the effects of constitutive FLT3 activation on NHEJ pathways and B-cell development, we administered sorafenib, a small molecule inhibitor targeting the tyrosine kinase activity of FLT3, to FLT3/ITD mice. Similar to what has been observed in the treatment of early pro-B cells with lestaurtinib in vitro, BM obtained from sorafenib-treated FLT3/ITD mice showed significantly higher fraction of B220+CD19+ cells compared with the vehicle-treated FLT3/ITD mice. No significant difference was observed in response to treatment of the wild-type groups (Figure 5E-F). Western blotting analysis of B cells showed increased expression of Ku86 in sorafenib-treated FLT3/ITD mice compared with those with vehicle treatment, whereas expression of PARP1 in the FLT3/ITD mice was slightly decreased after treatment with sorafenib (Figure 5G-H).
These results indicate that suppression of constitutive activation of FLT3 by selective FLT3 tyrosine kinase inhibitors up-regulates expression of Ku86 therefore partly restores the C-NHEJ pathway. Treatment with FLT3 tyrosine kinase inhibitors overcomes the block of B-cell development in FLT3/ITD mice.
In summary, our results suggest that, in pro-B cells, the FLT3/ITD mutation impairs the classic NHEJ pathway, which causes defects in joining DSBs generated during VDJ recombination. Impaired VDJ recombination partially explains the block of B-cell development observed in mice expressing an FLT3/ITD mutation. Pro-B cells from FLT3/ITD mice repair DSBs through the highly error-prone ALT-NHEJ, which might lead to an increase in genomic instability.
Discussion
Recent studies have revealed that activation of FLT3 contributes to B-lineage specification (induction of B lineage-specific gene-expression program), although its repression is required for further B-lineage development (repression of alternative gene-expression programs).38 Forced expression of FLT3 significantly reduces the percentage of B cells (B220+CD19+) in a transplantation model.2 In vivo treatment with FLT3 ligand also reduces the proportion of B220+CD19+ cells in mice, with myeloid and T-cell progenitor activity largely preserved.35 These data suggest that properly regulated activation of FLT3 is essential for early B-cell development.38 Our results here support the idea that constitutive activation of FLT3 results in a block at an early stage of B-cell development, and suppression of FLT3 activity is essential for further B-lineage commitment.
In this report, FLT3/ITD mice were observed to express 5-fold decreases in Ku86 levels in their early B progenitors. Consistent with the phenotype of the Ku86-deficient mice, which are severely defective for VDJ recombination,27 FLT3/ITD+ B progenitor cells also show defective VDJ recombination and a block in early B-cell development. The defect in Ku86 results in a decreased ability to initiate the C-NHEJ pathway to rejoin the RAG-cleaved DSBs during VDJ recombination.27 Ku is a heterodimer of Ku70 and Ku86 and plays an essential role in the NHEJ pathway. The mechanisms through which Ku proteins are regulated are not fully understood. It has been reported to be highly sensitive to oxidative damage.39,40 We have previously found increased reactive oxygen species in FLT3/ITD+ cells.33 It is possible that constitutively activated FLT3 in FLT3/ITD cells leads to constitutive elevation of reactive oxygen species, which might indirectly regulate Ku expression. Alternatively, the Ku promoter may be silenced through methylation of CpG islands.41 It has also been reported that, in Ku-null human somatic cells, DNA DSB repair activity is dominated by microhomology joining events, indicative of ALT-NHEJ.42 Nevertheless, Ku is the critical C-NHEJ factor that regulates the cell's choice of DNA DSB pathways. The availability of Ku proteins is the major decision point for the cells to repair DSBs by NHEJ.
PARP-1 has recently been reported to be recruited for DSBs only when the essential components of the C-NHEJ are compromised, particularly in the absence of or decreased levels of Ku.23,43 Consistent with this, we detected moderately elevated levels of PARP1 in FLT3/ITD+ early pro-B cells. Although we did not detect an increased expression of DNA ligase IIIα in early pro-B cells from heterozygous FLT3/ITD mice, significantly increased DNA ligase IIIα levels were detected in homozygous FLT3/ITD mice in another study.44 It is not clear whether constitutively activated FLT3 signaling directly activates ALT-NHEJ pathways. It appears more probable that, in FLT3/ITD+ cells, ALT-NHEJ pathways are activated as a consequence of repressed Ku86 activity and impaired C-NHEJ pathway.
Compared with the C-NHEJ pathway, the ALT-NHEJ pathway is slower and less efficient.24 More importantly, their catalyses of DSB repair are characterized by the use of microhomologies.22–25 Therefore, the ALT-NHEJ pathways are considered highly error-prone in nature. This in turn may cause genomic instability and cancer in the affected organism. For example, in B cells from XRCC4−/− or ligase IV−/− mice that are deficient for C-NHEJ, ALT-NHEJ is reported to support class-switch recombination and frequently joins IgH locus breaks to other chromosomes to generate translocations.22–25 Our findings that early pro-B cells from FLT3/ITD mice generate larger-size deletions and use microhomologies for the repair of DSBs support the idea that activated ALT-NHEJ pathways might be recruited for DSB repair in cells carrying FLT3/ITD mutations. The translocation of mouse genomic DNA into the plasmid sequence suggests increased genomic instability in these cells.
Clinically, constitutive activation of FLT3 by FLT3/ITD mutations in acute myeloid leukemia and acute lymphoblastic leukemia is associated with poor prognosis because of a higher relapse rate.7–9 We have previously observed increased levels of DSBs and misrepair errors in cell lines carrying FLT3/ITD mutations.32 We found that impaired C-NHEJ and subsequent enhanced ALT-NHEJ occur not only in early pro-B cells undergoing VDJ recombination, but also in lineage-negative bone marrow populations, which contain most of the hematopoietic progenitors expressing FLT3.44 Meanwhile, it has been reported that FLT3/ITD mutations are associated with an increased frequency of the more error-prone single-strand annealing DNA repair, a homology-directed DNA repair that involves the annealing of homologous single strands at repeated sequences and causes a deletion between repeats.45 During chemotherapy, with the C-NHEJ pathway impaired, cells may use ALT-NHEJ or single-strand annealing pathways to repair DSBs caused by chemotherapeutic agents to ensure their own survival. However, the more error-prone nature of these pathways may lead to further errors during DNA repair and result in increased genomic instability, which could eventually contribute to the aggressiveness and poor prognosis that characterize leukemia associated with constitutively activated FLT3 mutation.
In conclusion, our data suggest that, in pro-B cells from FLT3/ITD mice, impairment of the classic NHEJ pathway decreases the ability of cells to complete postcleavage DSB ligation initiated as part of the VDJ recombination process, resulting in failure to complete D-JH recombination and a subsequent block of pro-B-cell differentiation. It appears that early-pro-B cells from FLT3/ITD mice repair DSBs through the slow, inefficient, and highly error-prone ALT-NHEJ pathway, which probably promotes genomic instability. The increased genomic instability might underlie an important mechanism for the leukemic transformation of normal HSPCs by FLT3/ITD mutations and explain the poor prognosis of leukemia patients with this mutation.
Supplementary Material
Acknowledgments
The authors thank members of the Small, Rassool, and Desiderio laboratories for their insightful discussions and valuable comments on the manuscript.
This work was supported by the National Cancer Institute (grants CA90668, CA70970), the Leukemia & Lymphoma Society, and the Giant Food Pediatric Cancer Research Fund. D.S. is also supported by the Kyle Haydock Professorship.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: L.L. designed and performed experiments, analyzed data, and wrote the first draft; L.Z., J.F., and K.G. performed experiments; S.D. designed experiments; and F.V.R. and D.S. designed experiments, analyzed data, and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Donald Small, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins, CRB Rm 251, 1650 Orleans St, Baltimore, MD 21231; e-mail: donsmall@jhmi.edu.
References
- 1.Adolfsson J, Borge OJ, Bryder D, et al. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15(4):659–669. doi: 10.1016/s1074-7613(01)00220-5. [DOI] [PubMed] [Google Scholar]
- 2.Holmes ML, Carotta S, Corcoran LM, Nutt SL. Repression of Flt3 by Pax5 is crucial for B-cell lineage commitment. Genes Dev. 2006;20(8):933–938. doi: 10.1101/gad.1396206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogawa M, ten Boekel E, Melchers F. Identification of CD19(-)B220(+)c-Kit(+)Flt3/Flk-2(+)cells as early B lymphoid precursors before pre-B-I cells in juvenile mouse bone marrow. Int Immunol. 2000;12(3):313–324. doi: 10.1093/intimm/12.3.313. [DOI] [PubMed] [Google Scholar]
- 4.Wasserman R, Li YS, Hardy RR. Differential expression of the blk and ret tyrosine kinases during B lineage development is dependent on Ig rearrangement. J Immunol. 1995;155(2):644–651. [PubMed] [Google Scholar]
- 5.Mackarehtschian K, Hardin JD, Moore KA, Boast S, Goff SP, Lemischka IR. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 1995;3(1):147–161. doi: 10.1016/1074-7613(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 6.McKenna HJ, Stocking KL, Miller RE, et al. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95(11):3489–3497. [PubMed] [Google Scholar]
- 7.Armstrong SA, Mabon ME, Silverman LB, et al. FLT3 mutations in childhood acute lymphoblastic leukemia. Blood. 2004;103(9):3544–3546. doi: 10.1182/blood-2003-07-2441. [DOI] [PubMed] [Google Scholar]
- 8.Gilliland DG, Griffin JD. The roles of FLT3 in hematopoiesis and leukemia. Blood. 2002;100(5):1532–1542. doi: 10.1182/blood-2002-02-0492. [DOI] [PubMed] [Google Scholar]
- 9.Levis M, Small D. FLT3: ITDoes matter in leukemia. Leukemia. 2003;17(9):1738–1752. doi: 10.1038/sj.leu.2403099. [DOI] [PubMed] [Google Scholar]
- 10.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 11.McBlane JF, van Gent DC, Ramsden DA, et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83(3):387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 12.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 13.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 14.Falzon M, Fewell JW, Kuff EL. EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single- to double-strand transitions in DNA. J Biol Chem. 1993;268(4962):10546–10552. [PubMed] [Google Scholar]
- 15.Gottlieb TM, Jackson SP. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72(1):131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 16.Jeggo PA, Lobrich M. Artemis links ATM to double strand break rejoining. Cell Cycle. 2005;4(3):359–362. doi: 10.4161/cc.4.3.1527. [DOI] [PubMed] [Google Scholar]
- 17.Mimori T, Hardin JA. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261(22):10375–10379. [PubMed] [Google Scholar]
- 18.Grawunder U, Wilm M, Wu X, et al. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388(6441):492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 19.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412(6847):607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 20.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell. 2006;124(2):301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Roth DB, Wilson JH. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6(12):4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corneo B, Wendland RL, Deriano L, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449(7161):483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 23.Nussenzweig A, Nussenzweig MC. A backup DNA repair pathway moves to the forefront. Cell. 2007;131(2):223–225. doi: 10.1016/j.cell.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 24.Rassool FV, Tomkinson AE. Targeting abnormal DNA double strand break repair in cancer. Cell Mol Life Sci. 2010;67(21):3699–3710. doi: 10.1007/s00018-010-0493-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan CT, Boboila C, Souza EK, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449(7161):478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 26.Jankovic M, Nussenzweig A, Nussenzweig MC. Antigen receptor diversification and chromosome translocations. Nat Immunol. 2007;8(8):801–808. doi: 10.1038/ni1498. [DOI] [PubMed] [Google Scholar]
- 27.Zhu C, Bogue MA, Lim DS, Hasty P, Roth DB. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86(3):379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhu C, Mills KD, Ferguson DO, et al. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109(7):811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Piloto O, Nguyen HB, et al. Knock-in of an internal tandem duplication mutation into murine FLT3 confers myeloproliferative disease in a mouse model. Blood. 2008;111(7):3849–3858. doi: 10.1182/blood-2007-08-109942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tretter T, Ross AE, Dordai DI, Desiderio S. Mimicry of pre-B cell receptor signaling by activation of the tyrosine kinase Blk. J Exp Med. 2003;198(12):1863–1873. doi: 10.1084/jem.20030729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlissel MS, Corcoran LM, Baltimore D. Virus-transformed pre-B cells show ordered activation but not inactivation of immunoglobulin gene rearrangement and transcription. J Exp Med. 1991;173(3):711–720. doi: 10.1084/jem.173.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang X, Darzynkiewicz Z. Cytometric assessment of histone H2AX phosphorylation: a reporter of DNA damage. Methods Mol Biol. 2006;314:73–80. doi: 10.1385/1-59259-973-7:073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sallmyr A, Fan J, Datta K, et al. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111(6):3173–3182. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- 34.Li L, Piloto O, Kim KT, et al. FLT3/ITD expression increases expansion, survival and entry into cell cycle of human haematopoietic stem/progenitor cells. Br J Haematol. 2007;137(1):64–75. doi: 10.1111/j.1365-2141.2007.06525.x. [DOI] [PubMed] [Google Scholar]
- 35.Ceredig R, Rauch M, Balciunaite G, Rolink AG. Increasing Flt3L availability alters composition of a novel bone marrow lymphoid progenitor compartment. Blood. 2006;108(4):1216–1222. doi: 10.1182/blood-2005-10-006643. [DOI] [PubMed] [Google Scholar]
- 36.Rogakou EP, Pilch DR, Orr AH, et al. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 37.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005;4(6):639–648. doi: 10.1016/j.dnarep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 38.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26(6):715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Boldogh I, Roy G, Lee MS, et al. Reduced DNA double strand breaks in chlorambucil resistant cells are related to high DNA-PKcs activity and low oxidative stress. Toxicology. 2003;193(1):137–152. doi: 10.1016/j.tox.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 40.Andrews BJ, Lehman JA, Turchi JJ. Kinetic analysis of the Ku-DNA binding activity reveals a redox-dependent alteration in protein structure that stimulates dissociation of the Ku-DNA complex. J Biol Chem. 2006;281(19):13596–13603. doi: 10.1074/jbc.M512787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ludwig DL, Chen F, Peterson SR, Nussenzweig A, Li GC, Chen DJ. Ku80 gene expression is Sp1-dependent and sensitive to CpG methylation within a novel cis element. Gene. 1997;199(1):181–194. doi: 10.1016/s0378-1119(97)00366-1. [DOI] [PubMed] [Google Scholar]
- 42.Fattah F, Lee EH, Weisensel N, Wang Y, Lichter N, Hendrickson EA. Ku regulates the non-homologous end joining pathway choice of DNA double-strand break repair in human somatic cells. PLoS Genet. 2010;6(2):e1000855. doi: 10.1371/journal.pgen.1000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Audebert M, Salles B, Calsou P. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J Biol Chem. 2004;279(53):55117–55126. doi: 10.1074/jbc.M404524200. [DOI] [PubMed] [Google Scholar]
- 44.Fan J, Li L, Small D, Rassool F. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood. 2010;116(24):5298–5305. doi: 10.1182/blood-2010-03-272591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fernandes MS, Reddy MM, Gonneville JR, et al. BCR-ABL promotes the frequency of mutagenic single-strand annealing DNA repair. Blood. 2009;114(9):1813–1819. doi: 10.1182/blood-2008-07-172148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.