Abstract
The NR4A subfamily of nuclear receptors (NR4A1, NR4A2, and NR4A3) function as transcription factors that transduce diverse extracellular signals into altered gene transcription to coordinate apoptosis, proliferation, cell cycle arrest, and DNA repair. We previously discovered that 2 of these receptors, NR4A1 and NR4A3, are potent tumor suppressors of acute myeloid leukemia (AML); they are silenced in human AML, and abrogation of both genes in mice leads to rapid postnatal development of AML. Reduced expression of NR4As is also a common feature of myelodysplastic syndromes (MDSs). Here we show that reduced gene dosage of NR4A1 and NR4A3 in hypoallelic (NR4A1+/−NR4A3−/− or NR4A1−/−NR4A3+/−) mice below a critical threshold leads to a chronic myeloid malignancy that closely recapitulates the pathologic features of mixed myelodysplastic/myeloproliferative neoplasms (MDS/MPNs) with progression to AML in rare cases. Enhanced proliferation and excessive apoptosis of hematopoietic stem cells and myeloid progenitors, together with elevated DNA damage, contribute to MDS/MPN disease. We identify the myeloid tumor suppressor genes Egr1 and JunB and the DNA damage checkpoint kinase, polo-like kinase 2 (Plk2) as deregulated genes whose disrupted signaling probably contributes to MDS/MPN. These mice provide a novel model to elucidate the molecular pathogenesis of MDS/MPN and for therapeutic evaluation.
Introduction
Chronic myeloid malignancies represent a heterogeneous group of diseases that have been classified by the most recent (2008) World Health Organization classification system into 3 major categories. They include: myeloproliferative neoplasms (MPNs), characterized by increased nonlymphoid hematopoietic cells in the spleen or bone marrow (BM); myelodysplastic syndromes (MDSs), characterized by ineffective hematopoiesis, cytopenias, and hypercellular BM with dysplastic cell myeloid morphology; and MDS/MPN with overlapping myeloproliferative and myelodysplastic features.1 In the latter category of neoplasms, cytopenias and dysplastic changes are often seen in conjunction with features more commonly associated with MPN, including elevated white blood cell counts, thrombocytosis, and organomegaly.2 MDS/MPNs in adults include BCR-ABL1–negative atypical chronic myeloid leukemia, chronic myelomonocytic leukemia, and a rare unclassifiable MDS/MPN subgroup with relatively poor prognosis. Although MDS and MPNs are considered separate entities, subgroups of patients display partially overlapping cytogenetic abnormalities, such as gain of chromosome 8 (trisomy 8), loss of chromosome 7, and deletions of 5q or 20q, suggesting that common molecular mechanisms may contribute to both disorders.3,4 Despite progress in identification of cytogenetic abnormalities associated with a subset of MDS/MPNs, more than half of patients present with normal karyotypes, and the molecular basis of their disease pathogenesis remains poorly understood.3,4 The extensive morphologic and genetic heterogeneity of MDS/MPNs, together with a lack of molecular targets and limited availability of mouse models to mimic MDS/MPN, have hampered development of targeted therapies to treat these diseases. The propensity of MDS/MPN diseases to transform to acute myeloid leukemia (AML) and the relatively poor prognosis of MDS/MPN patients underscore the need for a better understanding of the molecular mechanisms underlying the development of these diseases.
The NR4A subclass of nuclear receptor transcription factors compose 3 highly homologous proteins (NR4A1, NR4A2, and NR4A3).5–8 Through their conserved DNA-binding domains, they interact with common cis-acting DNA elements to regulate transcription of overlapping target genes.7,9 Cellular levels of NR4As are tightly controlled under normal physiologic conditions, and they are transiently induced and activated by a diverse variety of extracellular signals, including genotoxic cellular stress, hormones, inflammatory cytokines, and metabolic, mitogenic, and apoptotic signals.10–18 On receipt of these diverse stimuli, they function as intracellular relay stations to interpret and transduce distinct signaling information into activation or repression of distinct functional clusters of target genes. NR4A activation is generally short lived, and the cellular outcome is stimulus- and cell context–dependent differential activation of NR4A target genes that regulate proliferation, cell cycle arrest, differentiation, apoptosis, and/or DNA repair.19 NR4A1 and NR4A3 play a central role in negative selection of T-lymphocytes,20 as well as surface IgM-mediated and viral-induced apoptosis of B cells.21,22 NR4As have also been implicated in the regulation of proliferation, apoptosis, and cell cycle arrest of cancer cells and play a major role in apoptotic responses of epithelial cancer cells, including sensitivity to antineoplastic agents.23–26 Furthermore, NR4A3 is a target for chromosomal translocations involving the Ewing sarcoma gene leading to aberrant regulation of NR4A3-dependent target genes and development of extraskeletal myxoid sarcoma.27 Together, these findings suggest that deregulated NR4A function may play a central role in susceptibility to tumorigenesis.
We recently uncovered an essential function of NR4A1 and NR4A3 as novel tumor suppressors of AML. Targeted deletion of both genes (NR4A1/3-null) in mice causes extremely rapid postnatal development of transplantable AML, leading to pup death within 3 to 4 weeks after birth.28 We also demonstrated that NR4A1 and NR4A3 expression is silenced in leukemic blasts from AML patients regardless of their cytogenetic background or French-American-British classification.28 Further, a recent genome-wide comparison of transcriptional profiles of highly purified normal human BM hematopoietic stem cells (HSCs) and leukemia stem cells from AML patients with distinct cytogenetic backgrounds has confirmed down-regulation of all 3 NR4As in all AML leukemia stem cells relative to normal HSCs.29 Interestingly, reduced expression of NR4As also appears to be a common feature of human MDS. Genome-wide comparisons of the gene transcript profiles of normal CD34+ progenitor cells and CD34+ progenitor cells from MDS patients with distinct disease classifications have revealed down-regulated expression of all NR4A transcripts by 40% to 80% in MDS relative to normal CD34+ stem and progenitor cells.30,31 Whereas these studies suggest that reduced NR4A expression/function may contribute to the pathogenesis of MDS, the contribution of NR4A dysregulation to development of chronic myeloid malignancies is unknown.
Because human MDS is associated with reduced NR4A expression and because reduced transcription factor function in HSCs and progenitor cells is a common feature of myeloproliferative and myelodysplastic diseases in addition to AMLs,32–34 we asked whether reduction of NR4A1 or NR4A3 levels, rather than complete abrogation of NR4A gene expression, can also contribute to chronic myeloid malignancies. Here we show that reduction of NR4A1/3 gene dosage beyond a critical threshold in mice is sufficient to cause MDS/MPN with progression to AML.
Methods
Mice
NR4A1−/−NR4A3+/− and NR4A1+/−NR4A3−/− mice were generated on a C57BL/6 and 129SvJ hybrid background by breeding previously generated NR4A1+/+NR4A3−/− and NR4A1−/−NR4A3+/+ mice. Nonlymphoid hematopoietic neoplasms were characterized according to guidelines of the Mouse Models of Human Cancers Consortium (www.emice.nci.nih.gov/aam/mouse).35 Mice were monitored for onset of disease by performing complete blood counts (Advia 120; Bayer-Siemens) with automated and manual differentials. Animals were killed when they became moribund (indicated by hunched posture, lethargy, and difficulty breathing). BM cellularity was determined by manual counts. Blood smears, BM, and spleen cytospins were stained with Wright-Giemsa stain. All mouse experiments were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee (protocol #AN-1861).
Immunohistochemistry
Tissues were fixed in 4% paraformaldehyde for 12 to 16 hours at 4°C and washed in 70% ethanol. Tissues were embedded in paraffin and cut into 5-μm sections. Standard hematoxylin and eosin staining was performed. Antigen Unmasking Solution (Vector Laboratories) was used to perform antigen retrieval for primary antibodies to myeloperoxidase (Dako North America). A biotinylated goat anti–rabbit IgG (Vector Laboratories) secondary antibody was used. To detect myeloperoxidase antibody binding, sections were treated with the avidin-biotin-peroxidase complex detection system and VIP kit (Vector Laboratories) and then counterstained with either hematoxylin or 1% methyl green (Fisher Scientific). Pictures of stained sections were obtained using an Olympus BX41 microscrope and Olympus DP71 camera (Olympus). Images were acquired using the Olympus DP controller and all images were processed with Adobe Photoshop Version 10.0 (Adobe Systems).
γH2AX immunostaining
Cells were irradiated in vitro with 230 cGy of γ-radiation and allowed to recover at 37°C. Cytospins of BM cells were taken before irradiation and at 15 minutes and 3 hours after irradiation. Cells were fixed for 20 minutes in 4% paraformaldehyde and permeabilized with 0.01% Triton X-100 in phosphate-buffered saline for 10 minutes. Samples were blocked overnight (at 4°C) in 5% bovine serum albumin + 0.01% Triton X-100 and incubated for 1 hour overnight with primary antibody specific for γH2AX (1:250) (BioLegend). After washing with phosphate-buffered saline, slides were incubated for 1 hour with a rat-antimouse coupled to Alexa-488 (green) secondary antibody (1:1000; Invitrogen). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Vector Laboratories). A total of 200 cells were counted per animal.
Flow cytometry/cell cycle
BM cells were isolated from femurs and tibias by flushing the bones with 1× Hanks balanced salt solution with 5% fetal bovine serum. Red blood cells were lysed with ACK lysing buffer (Quality Biologicals). For multicolor analysis, cells from BM and spleen were incubated with single-color fluorochrome-conjugated antibodies (cKit, Sca1, CD11b, Gr-1, CD19, B220, CD71, and Ter119; eBioscience) or antibody cocktails/kits (lineage exclusion, annexin V; BD Biosciences) and analyzed on an LSR Fortesa cytometer (BD Biosciences). For sorting, cells were incubated with monoclonal antibodies conjugated to fluorochromes, and viable cells were sorted using a FACSAria (BD Biosciences). DNA content was analyzed by propidium iodide/RNase staining (BD Biosciences). Data were analyzed with FlowJo Version 8.7.3 (TreeStar) and Modfit Version 3.2.1 (Verity) software.
BrdU
Mice were injected intraperitoneally with 100 μL of a 10-mg/mL solution of bromodeoxyuridine (BrdU) 2 hours before death. BM cells were stained with surface antigen markers, fixed with formaldehyde, and stained with anti-BrdU and 7-amino-actinomycin D (BD Biosciences).
Real-time PCR
Total RNA was isolated using RNeasy with on-column DNase digestion (QIAGEN). cDNA was synthesized using a High Capacity cDNA Kit according to the manufacturer's instructions (Applied Biosystems). Primers and probes for detecting mouse NR4A1, NR4A2, Junb, cJun, Plk2, and B2m (encoding β2-microglobulin) were from TaqMan (Applied Biosystems). PCR amplifications were performed using the ABI Step One Plus Sequence Detection System (Applied Biosystems) under standard conditions. Transcript levels were determined using the standard curve method and normalized to corresponding B2m levels.
Spectral karyotype analysis
BM was cultured in RPMI 1640 containing 15% fetal calf serum, 1% l-glutamine, and 1% penicillin-streptomycin. The cultures were harvested using standard protocols. The metaphase spreads were aged at room temperature for 3 days for spectral karyotype (SKY) analysis. The cocktail of mouse chromosome paints was obtained from Applied Spectral Imaging (ASI). Hybridization and detection were carried out according to the manufacturer's protocol, with slight modifications. Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole. For each mouse, 5 to 10 metaphase spreads were analyzed by SKY. Images were acquired with a SD300-8A Spectra cube (ASI) mounted on a Zeiss Axioplan II microscope using a custom-designed optical filter (SKY-1; Chroma Technology) and analyzed using SKY View, Version 2.1 software (ASI).
Statistical analysis
Data are mean plus or minus SEM or SD as indicated. The significance of the differences between groups was determined using Student t test (2-tailed). A P value of less than .05 was considered significant for all analyses.
Results
Reduction of NR4A1 and NR4A3 gene dosage leads to MDS/MPN with progression to AML
We previously demonstrated that germline deletion of NR4A1 and NR4A3 in mice leads to rapid development of AML, resulting in postnatal death of NR4A1/3 null mice within 3 to 4 weeks of birth. To determine whether susceptibility to hematologic malignancy is affected by NR4A1/NR4A3 gene dosage, we monitored mice with reduced NR4A1 and NR4A3 gene dosage for up to 15 months. Whereas mice carrying a null mutation of either NR4A1 (NR4A1−/−) or NR4A3 (NR4A3−/−) alone displayed no overt hematologic abnormalities and had a normal life span, hypoallelic mice retaining only a single allele of either NR4A1/3 receptor (NR4A1−/−NR4A3+/− and NR4A1+/−NR4A3−/−) began to develop outward signs of disease, including lethargy, weight loss, ruffled fur, and hunched posture, and became moribund between 3 and 15 months of age (Figure 1A). Notably, disease penetrance among animals was significantly higher and latency of disease onset was shorter in NR4A1+/−NR4A3−/− relative to NR4A1−/−NR4A3+/− mice. On necropsy, moribund mice displayed severe splenomegaly, and a subset also displayed hepatomegaly (Figure 1B-C). Peripheral blood analysis at the time of death (Table 1; supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) revealed various degrees of leukocytosis with thrombocytopenia, mild to moderate anemia, decreased lymphocytes, and variable neutrophilia among animals. Serial complete blood count analysis showing absolute blood cell counts over time showed that the majority of hypoallelic mice develop pancytopenia and anemia at the moribund stage. Although some animals do not exhibit pancytopenia at death, they have markedly increased circulating white blood cells and neutrophils (supplemental Figure 1). Morphologic analysis of peripheral blood smears revealed increased myeloid cells (Figure 2A) with dysplastic features (Figure 2D) that consisted mainly of hypersegmented granulocytes. Occasional myeloid cells with Pseudo-Pelger-Huet-like hypolobation, dysplastic megakaryocytes, and dysplastic erythroid cells with inclusions resembling Howell-Jolly bodies, all frequent features of MDS, were also observed (Figure 2D). BM cellularity was variable from hypocellular to hypercellular among animals. Overproduction of myeloid cells at all stages of maturation was observed in BM (Figure 2B) with extensive dissemination of myeloid elements in spleen (Figure 2C). As in the peripheral blood, many of these cells were dysplastic (Figure 2D). Other than the hematopoietic phenotype, these mice displayed no other overt abnormalities in physiologic homeostasis. According to the Bethesda criteria,35 these disease features most closely resemble a mixed myelodysplastic/myeloproliferative neoplasm (MDS/MPN; supplemental Table 3), and we conclude that the mice probably die of organ failure because of extensive myeloid cell infiltration and/or pancytopenia in a subset of mice.
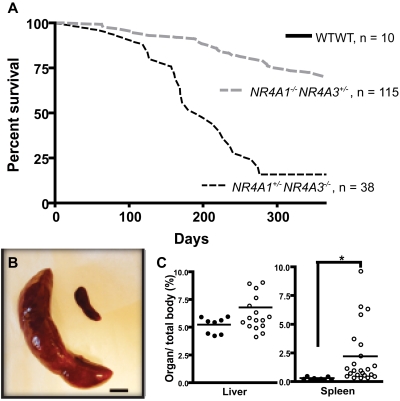
Figure 1.
Phenotype of NR4A1+/−NR4A3−/− and NR4A1−/−NR4A3+/− mice. (A) Kaplan-Meier curve shows survival of NR4A1+/−NR4A3−/− (n = 38) and NR4A1−/−NR4A3+/− (n = 115) mice compared with wild-type control mice (n = 10). (B) Massive splenomegaly in diseased mouse (left) compared with age-matched control (right). Image shown represents an 18-fold difference in weight. Scale bar represents 1 cm. (C) Quantification of hepatomegaly (left) and splenomegaly (right) in hypoallelic mice (○, n = 18 for liver and n = 23 for spleen) compared with healthy littermates (●, n = 8 for liver and n = 9 for spleen). Horizontal bars represent the mean. *P = .001. Both NR4A1−/−NR4A3+/− and NR4A1+/−NR4A3−/− mice were used for this assay; analysis was conducted when animals were moribund.
Table 1.
Peripheral blood analysis
| Group | n | WBCs, × 103/μL | RBCs, × 106/μL | Hematocrit, % | Platelets, × 103/μL | Neutrophils, % | Lymphocytes, % | Monocytes, % | Eosinophils, % | Basophils, % | Blasts, % |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild-type | |||||||||||
| NR4A1+/+NR4A3+/+ | 4 | 5.98 ± 0.9 | 9.57 ± 0.46 | 45.11 ± 1.19 | 1164 ± 155 | 10.0 ± 1.7 | 83.9 ± 1.6 | 1.5 ± 0.3 | 2.4 ± 0.4 | 0.1 ± 0.1 | 0.97 ± 0.8 |
| Disease-free | |||||||||||
| NR4A1−/−NR4A3+/+ | 8 | 4.36 ± 1.2 | 9.93 ± 0.41 | 44.40 ± 1.10 | 1148 ± 163 | 12.3 ± 1.1 | 83.1 ± 2.7 | 1.4 ± 0.3 | 2.9 ± 0.5 | 0.2 ± 0.1 | 1.13 ± 0.6 |
| NR4A1+/+NR4A3−/− | 9 | 5.2 ± 1.1 | 9.96 ± 0.38 | 44.35 ± 1.16 | 1161 ± 160 | 13.4 ± 2.2 | 83.3 ± 1.8 | 1.2 ± 0.4 | 2.5 ± 0.6 | 0.2 ± 0.1 | 1.30 ± 0.7 |
| MDS/MPN* | |||||||||||
| NR4A1+/−NR4A3−/− | 25 | 12.75 ± 14.2 | 8.16 ± 2.54 | 37.37 ± 11.8 | 744 ± 369 | 32.0 ± 13.1 | 53.3 ± 16.5 | 3.1 ± 2.4 | 4.2 ± 3.1 | 0.4 ± 0.1 | 5.9 ± 4.6 |
| NR4A1−/−NR4A3+/− | 20 | 10.83 ± 6.71 | 8.34 ± 1.52 | 42.28 ± 7.07 | 799 ± 476 | 31.7 ± 15.4 | 51.1 ± 19.7 | 4.2 ± 3.6 | 5.5 ± 3.3 | 0.9 ± 0.3 | 6.1 ± 5.1 |
| AML† | |||||||||||
| NR4A1+/−NR4A3−/− | 3 | 23.6 ± 20.5 | 6.33 ± 3.91 | 29.37 ± 18.4 | 235 ± 266 | 23.4 ± 3.2 | 40.3 ± 7.6 | 2.1 ± 0.4 | 2.9 ± 0.4 | 0.2 ± 0.2 | 31.0 ± 7.4 |
| NR4A1−/−NR4A3+/− | 2 | 8.05 ± 1.18 | 6.50 ± 1.70 | 22.80 ± 4.20 | 114 ± 32.6 | 15.2 ± 1.4 | 48.9 ± 7.1 | 1.9 ± 0.6 | 3.6 ± 1.3 | 0.3 ± 0.3 | 30.1 ± 5.3 |
Values are reported as mean ± SD. WBCs indicates white blood cells; and RBCs, red blood cells.
Mice were classified as MDS/MPN or AML based on the Bethesda criteria. In addition to abnormal blood counts, MDS/MPN mice had elevated blasts (but < 20%) in their bone marrow and spleen, and dysplastic granulocytes were present in the peripheral blood, bone marrow, and spleen. MDS/MPN mice also displayed splenomegaly and myeloid cell infiltration into spleen, liver, and lung.
AML mice had > 20% blasts in their peripheral blood and bone marrow and also displayed splenomegaly and myeloid cell infiltration into spleen, liver, and lung.
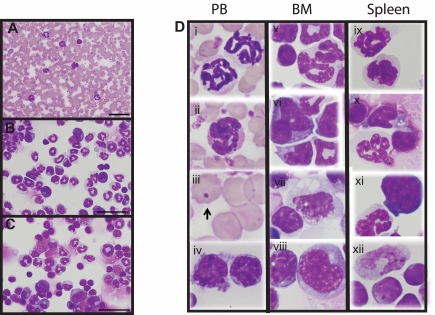
Figure 2.
Hypoallelic mice have a mixed MDS/MPN disease. Peripheral blood smear (A), BM cytospin (B), and spleen cytospin (C) showing increased myeloid cells (animal #3339 used for images). Scale bars represent 50 μm (100×/1.30 NA oil objective magnification). (D) Dysplastic cells observed in peripheral blood, BM, and spleen of hypoallelic mice include hypersegmented granulocytes (i, ii, v, ix, and x), pseudo-Pelger-Huet-like atypical white cells (vi and xi), Howell-Jolly bodies (iii, indicated by arrow), dysplastic megakaryocytes (vii), and increased immature white cells (iv, vii, viii, and xii). Both NR4A1−/−NR4A3+/− (n = 2, at 9-11 months) and NR4A1+/−NR4A3−/− (n = 4, at 4-7 months) mice were used for this assay.
A subset of hypoallelic mice also presented with markedly elevated immature blasts (> 20%; Table 1; supplemental Table 2). Analysis of BM and spleen cytospins, as well as peripheral smears from these mice, confirmed that they consisted predominantly of immature myeloid blasts (> 20% blasts; Figure 3) consistent with disease progression to AML. However, incidence of disease progression before death from MDS/MPN is rare, with 4 of 115 NR4A1−/−NR4A3+/− mice (at 8-12 months) and 3 of 38 NR4A1+/−NR4A3−/− mice (at 3-7 months) exhibiting more than 20% blasts.
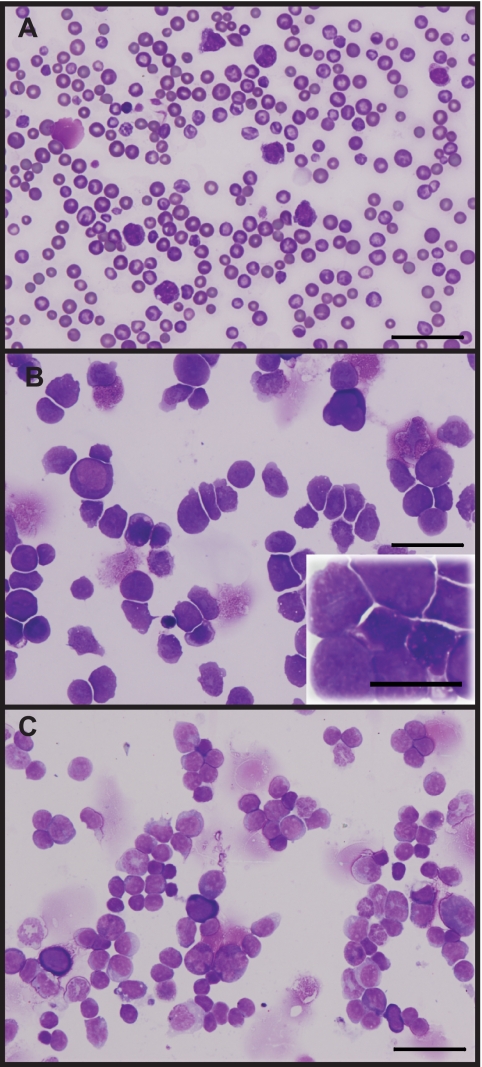
Figure 3.
A rare subset of hypoallelic mice transform to AML. Peripheral blood smear (A), BM cytospin (B), and spleen cytospin (C) showing more than 20% immature/blast cells. Scale bars represent 50 μm (60×/1.25 NA oil objective magnification, inset 100×/1.30 NA oil objective magnification). Images shown are from a representative AML stage NR4A1+/−NR4A3−/− mouse; analysis was conducted when animal was moribund (at 9 months).
Histologic analysis of spleen sections from MDS/MPN mice showed abnormal tissue architecture with a reduction and dispersion of the white pulp and increased red pulp resulting from extramedullary hematopoiesis (Figure 4B). Myeloperoxidase immunohistochemical analysis revealed that the disruption of lymphoid tissue in the spleen and the presence of abnormal perivascular infiltrations in the liver and lung were the result of the dissemination of myeloid cells (Figure 4F,H; and data not shown). We also analyzed both BM and spleen by flow cytometry and found the myeloid progenitor compartment (CD11b+/Gr1low/−) to be expanded in both NR4A1−/−NR4A3+/− and NR4A1+/−NR4A3−/− hypoallelic mice compared with control littermates (Figure 4I; supplemental Figure 2A). Furthermore, we also noted a decrease in lymphoid cells expressing B220 and CD19 (Figure 4J), consistent with the lymphopenia observed in peripheral blood. Analysis of CD71 and Ter119 showed no increase in erythropoiesis (supplemental Figure 2B). Finally, analysis of thymic architecture demonstrated complete effacement of the thymic cortex because of extensive infiltration of myeloid cells (supplemental Figure 2C-E), consistent with our previous findings of massively reduced T-cell development in NR4A1/3-null mice.28 Together, these data demarcate a clear shift in hematopoietic potential of hypoallelic BM toward myelopoiesis.
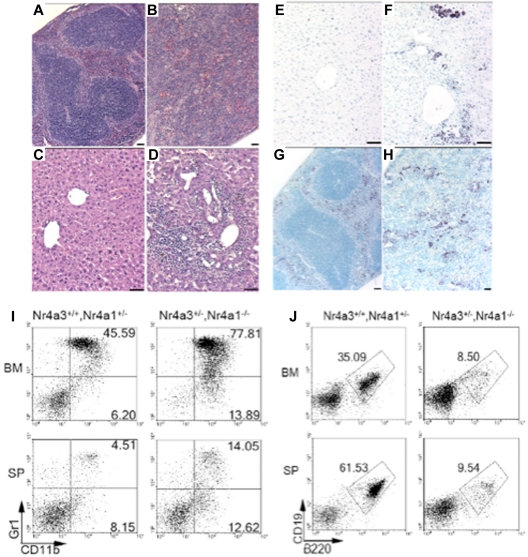
Figure 4.
Perivascular infiltrations and lymphoid tissue disruption resulting from abnormal myeloid dissemination and increased myelopoiesis in hypoallelic mice. Hematoxylin and eosin-stained paraffin sections of spleen (A-B) and liver (C-D). Spleen from a healthy littermate (A) and an MDS/MPN NR4A1+/−NR4A3−/− mouse (at 6 months; B) revealed loss of distinct splenic architecture, including well-defined red and white pulp. Histologic analysis of livers from a healthy littermate (C) and an MDS/MPN NR4A1+/−NR4A3−/− mouse (at 6 months) (D) showed large regions of perivascular infiltration. Scale bar represents 50 μm (60×/1.25 objective magnification). (E-H) Antimyeloperoxidase immunohistochemistry revealed myeloid contribution to infiltrates in the liver (F) and spleen (H). These infiltrates were not present in control liver (E) or spleen (G). Scale bars represent 50 μm. (I-J) Representative dot plots from flow cytometric analysis of BM and spleen (SP) demonstrate an increase in myeloid cells (CD11b+/Gr-1− and CD11b+/Gr-1+) in diseased mice compared with healthy littermates (I). A decrease in the percentage of B-lymphocytes (B220+/CD19+) was also noted in diseased mice compared with healthy littermates (J). Numbers shown are the average percentage of total cells for the associated region (averaged from n = 4 mice per genotype; age range, 9-12 months).
Collectively, these data indicate that reduction of NR4A1/NR4A3 gene dosage below a critical threshold is sufficient to cause development of a mixed MDS/MPN disease with progression to AML.
MDS/MPN is associated with increased proliferation and excessive apoptosis of the HSCs and myeloid progenitor-enriched compartments
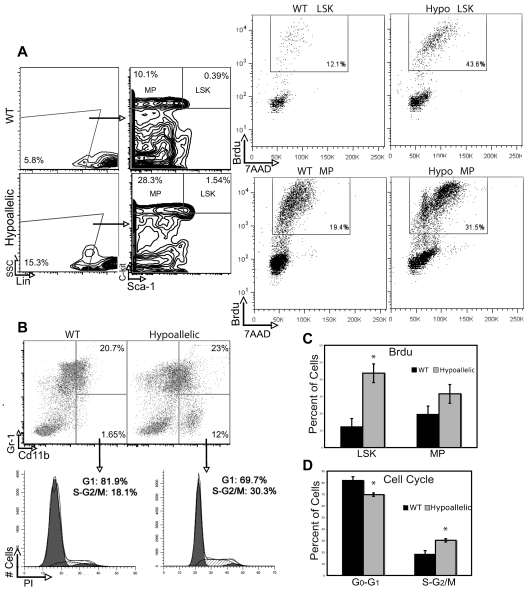
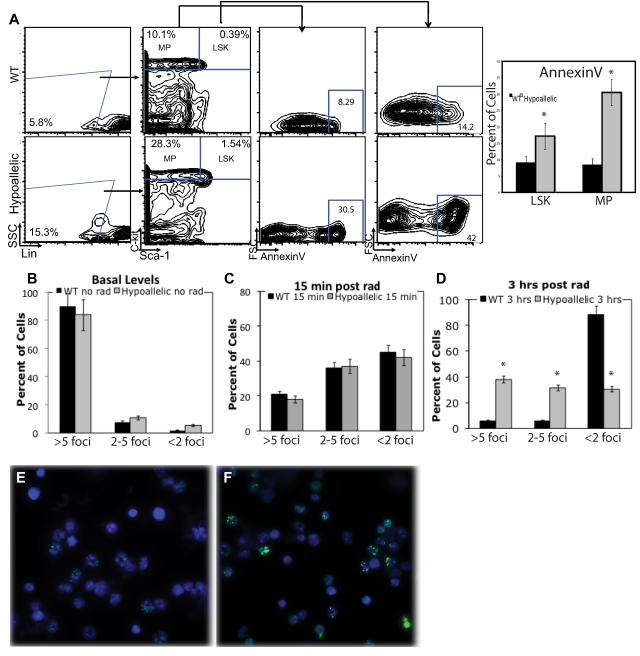
We next asked whether abnormal proliferation or apoptosis of HSCs and myeloid progenitor cells might contribute to the expansion of myeloid progenitors in hypoallelic BM. To measure proliferation rates, we injected mice with BrdU 2 hours before death and sorted Lin−Sca1+c-kit+ (LSK; HSCs and multipotent progenitors) as well as myeloid progenitor (MPs; Lin−Sca1−c-kit+) populations followed by fluorescence-activated cell sorter analysis of 7-amino-actinomycin D (7AAD) to measure DNA content and BrdU to label cells in S phase. We observed a significant increase (3.5-fold) in proliferating primitive LSK cells and a 1.5-fold increase in MPs in S phase in hypoallelic relative to WT mice (Figure 5A,C). Cell cycle analysis of more downstream myelomonocytic progenitors (CD11b+/Gr1low/−) using propidium iodide also confirmed a significant increase in cycling progenitors in S + G2/M phases (Figure 5B-C). Annexin V staining of BM LSK and MPs revealed a significant increase in numbers of apoptotic cells in both compartments in hypoallelic compared with wild-type mice (Figure 6A). Thus, both abnormal proliferation of HSCs and myeloid progenitor cells, coupled with excessive apoptosis, appear to contribute to the development of MDS/MPN disease in hypoallelic mice.
Figure 5.
Hypoallelic mice have increased proliferation and cell cycle defects in HSCs and myeloid progenitor-enriched compartments. (A) BrdU was administered to mark cells that entered S phase, and 7-amino-actinomycin D (7AAD) indicated DNA content. LSK and MP cells from MDS/MPN hypoallelic mice (backgate analysis shown at left) contain more dividing cells (43.6% and 31.5%, respectively) than age-matched wild-type controls (12.1% and 19.4%, respectively). Percentages shown represent averages of all samples; n = 3 in each group. Both NR4A1−/−NR4A3+/− (n = 1, at 11 months) and NR4A1+/−NR4A3−/− mice (n = 2, at 4-6 months) were used for this assay. (B) Propidium iodide incorporation revealed a larger number of proliferating cells (S + G2/M) in CD11b+/Gr-1low/− immature myeloid cells of MDS/MPN hypoallelic animals (30.3%) compared with age-matched controls (18.1%). Percentages shown represent averages of all samples; n = 4 for in each group. Both NR4A1−/−NR4A3+/− (n = 1, at 10.5 months) and NR4A1+/−NR4A3−/− mice (n = 3, at 3-5 months) were used for this assay. (C-D) Bar charts represent data shown in panels A and B, respectively. *P < .01. Error bars represent SEM.
Figure 6.
Increased apoptosis and DNA damage accumulation in stem and myeloid progenitor cells. (A) Annexin V was used to mark apoptotic cells. LSK and MP cells from MDS/MPN hypoallelic mice (backgate analysis shown at left) contain more apoptotic cells (30.5% and 42%, respectively) than age-matched wild-type controls (8.29% and 14.2%, respectively). Percentages shown represent averages of all samples; n = 3 in each group. Both NR4A1−/−NR4A3+/− (n = 1, at 11.5 months) and NR4A1+/−NR4A3−/− mice (n = 2, at 6-7 months) were used for this assay. *P < .01. Error bars represent SEM. (B) CD11b+/Gr-1low/− cells from MDS/MPN NR4A1+/−NR4A3−/− mice (n = 4, at 4-6 months) were analyzed for DNA damage. Bar chart of percentage γH2AX-positive cells in unirradiated samples shows that 3 of 4 hypoallelic mice analyzed have increased basal DNA damage. P = .07 and P = .055 for more than 5 foci and 2 to 5 foci, respectively. Error bars represent SEM. (C-F) CD11b+/Gr-1low/− cells from MDS/MPN hypoallelic mice were analyzed for DNA damage after in vitro exposure to γ-irradiation. (C) Bar chart of percentage of γH2AX positive cells 15 minutes after irradiation shows equal DNA damage induction. (D) Bar chart of percent γH2AX positive cells 3 hours after irradiation. N = 5 in each group. (E-F) At 3 hours after irradiation, anti-γH2AX immunofluorescence (green) showed a significantly higher amount of DNA damage remaining in diseased mice (F) compared to healthy controls (E; 100×/1.30 NA oil objective magnification). Both NR4A1−/−NR4A3+/− (n = 2, at 10-12 months) and NR4A1+/−NR4A3−/− mice (n = 3, at 5-8 months) were used for this assay. Error bars denote SEM.
Excessive DNA damage in myeloid cells from NR4A1/3 hypoallelic mice
To determine whether development of myelodysplastic features may be associated with increased DNA damage of myeloid cells, we examined the extent of DNA damage before and after in vitro exposure to γ-irradiation by analysis of the extent of double-strand break induction by γH2AX immunostaining to detect double-strand break foci. Quantitation of γH2AX-positive foci before irradiation showed a trend toward elevated levels of DNA damage (> 5 foci per cell) in myeloid cells (CD11b+/Gr1low/−) from 3 of 4 MDS/MPN hypoallelic mice tested relative to cells from wild-type mice, although this was not statistically significant (Figure 6B; supplemental Figure 3A). In addition, both wild-type and hypoallelic mice respond similarly to irradiation at an early time point (15 minutes; Figure 6C); yet although wild-type myeloid cells have repaired their DNA damage by 3 hours after irradiation, DNA damage remains relatively high in hypoallelic cells, as evidenced by a significantly higher percentage of γH2AX-positive cells (Figure 6D-F).
To determine whether dysplastic cellular changes were reflected in gross chromosomal abnormalities in BM of hypoallelic mice, we performed a 23-color SKY analysis of whole BM isolated from NR4A1−/−NRA3+/− mice with MDS/MPN (data not shown) as well as mice that had progressed to AML (supplemental Figure 4). SKY analysis confirmed that the normal karyotype is preserved even as animals progress to AML, confirming that no additional gross genetic alterations are required for AML transformation. Analysis of NR4A transcript levels showed that disease development was not associated with loss of heterozygosity of the remaining NR4A1 allele from NR4A1+/−NR4A3−/− mice (supplemental Figure 5A; supplemental Table 4), or silencing of the third NR4A family member, NR4A2 (supplemental Figure 5B). Thus, a threshold level of NR4A signaling is maintained in these mice, and reduced expression of NR4A1 and NR4A3 alone is sufficient to cause chronic disease.
Development of MDS/MPN is associated with disrupted expression of essential myeloid transcription factors Egr1 and JunB and the checkpoint polo-like kinase, Plk2
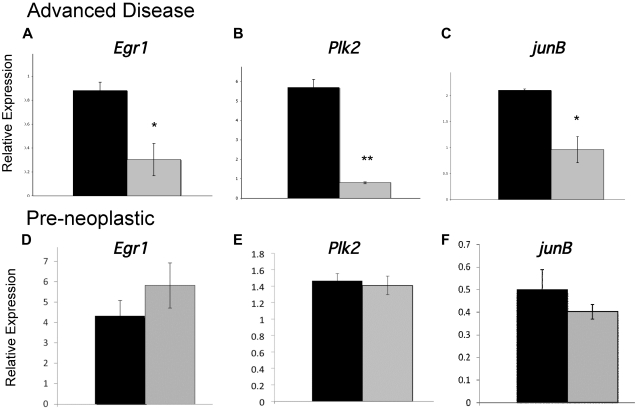
To identify genes whose dysregulation in hypoallelic mice may contribute to MDS/MPN development, we examined the expression of several candidate genes, which are critical for regulation of myeloid homeostasis, whose disruption has been implicated in chronic myeloproliferative disease or MDS and whose proximal promoters contain putative NR4A response elements.9 We identified 3 genes, Egr1, Plk2, and JunB, whose down-regulated expression is associated with MDS/MPN disease development in hypoallelic mice. Egr1 is a member of the WT1 family of transcription factors and a tumor suppressor gene that is essential for maintenance of hematopoietic stem cell quiescence36; it is localized to chromosome 5 in a region that is recurringly deleted in approximately 10% of MDS patients, and haploinsufficiency of Egr1 predisposes mice to MDS/MPN.33 The second gene, Plk2, is a serine/threonine kinase that is induced by p53 and regulates cell cycle checkpoints and cellular response to DNA damage.37 Recent evidence suggests that Plk2 is indirectly silenced in AML38 and this checkpoint kinase has been proposed to function as a tumor suppressor of hematologic malignancies.39,40 Finally, we have previously shown that the AP-1 transcription factor JunB, which is required for normal myeloid homeostasis and whose dysregulation is sufficient to cause chronic MPD in mice,41 is down-regulated in NR4A1/3-null mice.28 Analysis of the expression of all 3 transcripts in CD11b+/GR-1low/− myeloid cells showed significant down-regulation in MDS/MPN diseased hypoallelic mice (Figure 7A-C). However, expression was unaffected in hypoallelic mice before disease development (at 6 weeks of age; Figure 7D-F). These data suggest that reduced expression of these genes is a secondary consequence of disease development rather than the result of a direct loss of transcriptional control by NR4As in hypoallelic mice. Given their previously implicated roles myeloid malignancies, deregulation of Egr1, Plk2, and JunB signaling probably contributes to MDS/MPN disease but is an indirect consequence of NR4A loss.
Figure 7.
Development of MDS/MPN is associated with disrupted expression of essential myeloid transcription factors. (A-C) Real-time PCR analysis of indicated transcripts using RNA isolated from CD11b+/Gr-1low/− cells from MDS/MPN NR4A1+/−NR4A3−/− mice (n = 3, at 4-10 months, blast levels between 12% and 18%; gray columns) and control NR4A1+/−NR4A3+/+ mice (n = 7, at age 4-10 months; black columns). Animals with greater than 10% blasts were considered advanced diseased based on the criteria of Sokal et al and the International Bone Marrow Transplant Registry.42,43 (D-F) Real-time PCR analysis of indicated transcripts using RNA isolated from CD11b+/Gr-1low/− cells from 6-week-old preneoplastic NR4A1−/−NR4A3+/− mice (gray columns, n = 3) and 6-week-old NR4A1+/+NR4A3+/− control mice (black columns, n = 3). All results were normalized to B2m expression. *P < .05. **P < .02. Error bars represent SEM.
Discussion
We have generated a novel mouse model that recapitulates key pathologic features of mixed MDS/MPN disease observed in human patients with susceptibility to progress to AML. Reduction of expression levels of NR4A1 and NR4A3 nuclear receptor transcription factors leads to peripheral blood cytopenias with dysplastic myeloid morphology in the presence of a hypocellular or hypercellular BM. These MDS features appeared in conjunction with hepatosplenomegaly, variable leukocytosis, and overproduction of myeloid cells in BM, spleen, and peripheral blood. We noted that BM cellularity decreased as splenomegaly increased, consistent with increased extramedullary hematopoiesis. Although the majority of mice died from chronic MDS/MPN, a subset progressed to AML before they became moribund. In contrast to NR4A1/3 null mice, which develop AML extremely rapidly,28 the relatively long latency of development of MDS/MPN and transformation to AML on reduction of NR4A gene dosage suggests that cooperating mutations may be required for disease development. Reduced expression and function of lineage restricted transcription factors that promote myeloid cell differentiation has been shown to play a critical role in the development of AML.44,45 In addition, 2 recent genome-wide comparison studies have examined the transcriptional profiles of normal CD34+ progenitor cells and CD34+ progenitor cells from MDS patients with distinct disease classifications. Data from these studies indicate down-regulation of all 3 NR4As by 40% to 80% in MDS progenitors relative to normal progenitors.30,31 Our data indicate that coordinate control of myelopoiesis is highly sensitive to NR4A expression levels and that deregulated expression/function can lead to a spectrum of myeloid malignancies. Further, they reveal a common molecular mechanism that contributes to MDS/MPN and AML via disrupted NR4A signaling.
Both NR4A1 and NR4A3 are expressed in HSCs and myeloid progenitor cells.28 However, the latency of MDS/MPN development and penetrance among animals were markedly different between hypoallelic mice lacking NR4A3 relative to NR4A1. Despite relatively higher levels of NR4A1 expression in both compartments of normal mice,28 depletion of NR4A3 in a haploinsufficient NR4A1 background (NR4A1+/−NR4A3−/−) resulted in a shorter latency of MDS/MPN development with markedly increased disease penetrance (85%) relative to NR4A1−/−NR4A3+/− mice. This may suggest that the 2 receptors may function cooperatively rather than redundantly in hematopoietic cells to suppress myeloid leukemogenesis.
The cellular defects associated with MDS/MPN development lie at both the stem cell (HSC) and MP levels and involve increased proliferation of HSCs and MP at the expense of other blood lineages, as well as excessive apoptosis in both cellular compartments. These are common pathologic features observed in human patients with MDS, in contrast to AML.46 Interestingly, increased survival, not excessive apoptosis, was observed during early postnatal development of AML in our NR4A1/3 null model.28 Thus, apoptosis is a characteristic of chronic disease but not AML in NR4A mutant mice. Examination of the molecular mechanisms underlying these defects indicates that NR4As may function upstream of the myeloid tumor suppressors, Egr1 and JunB, to indirectly regulate their expression and prevent development of MDS/MPN. Haploinsufficiency of Egr1, a tumor suppressor located within a segment of chromosome 5 that is commonly deleted in human MDS patients, including those with therapy-related MDS/AML, is sufficient to predispose mice to MDS/MPN.33 The AP-1 tumor suppressor, JunB, is an essential regulator of myelopoiesis; down-regulated expression has been observed in both chronic myeloid leukemia and AML patients, and abrogation in mice leads to chronic myeloproliferative disease.47,48
Although no gross genomic abnormalities were detected in leukemic BM derived from NR4A hypoallelic mice, increased DNA damage in myeloid cells of these animals may contribute to the myelodysplastic phenotype and to increased genome instability, providing an extended opportunity to acquire additional mutations to facilitate leukemic progression to AML. NR4As have previously been implicated in cellular responses to genotoxic stress10 and in the repair of ultraviolet-induced DNA damage of melanocyte cells.12 However, the molecular mechanisms by which NR4As regulate cellular responses to DNA damage are unknown. Our finding that the DNA damage checkpoint kinase, Plk2, is down-regulated in hypoallelic mice provides a novel molecular link between NR4As and the DNA damage response in myeloid cells. Depletion of Plk2 leads to elevated replication stress-induced DNA damage, a dysfunctional S-phase checkpoint, and elevated apoptosis in lung cancer cells.49 These cellular defects closely resemble those observed in myeloid cells of NR4A1/3 hypoallelic mice. Further, microRNA-mediated silencing of Plk2 has recently been implicated in the development of human AML.38 Although the Plk2 promoter contains putative NR4A response elements, we did not observe changes in Plk2 expression in preneoplastic mice, suggesting that this gene is not a direct target of NR4As in myeloid cells. Because Plk2 is a transcriptional target of the tumor suppressor p53,37 which in turn is activated by Egr1 during irradiation induced cell cycle arrest,50 it is possible that NR4A-mediated regulation of Plk2 in myeloid cells may occur indirectly through induction of an Egr1/p53-dependent pathway. Future studies will be required to determine which of these tumor suppressors represent direct transcriptional targets of NR4As.
In conclusion, we have generated a novel mouse model that closely resembles many of the pathologic features of human MDS/MPN. We show that reduced expression of NR4A1 and NR4A3, a common feature of human MDS, is sufficient to cause MDS/MPN disease in mice. The availability of this novel model and its propensity to progress to AML will enhance our understanding of the molecular pathogenesis of human MDS/MPN as well as the molecular mechanisms that contribute to transformation to acute leukemia.
Supplementary Material
Acknowledgments
The authors thank Jie Yang for technical support.
This work was supported by National Institutes of Health, National Cancer Institute grant RO1CA111411 (O.M.C.) and National Science Foundation Graduate Research Fellowship award 2009079140 (A.M.R.-H.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: A.M.R.-H. designed and conducted experiments and wrote the manuscript; S.E.M. and A.M.S. contributed to performance of experiments; and O.M.C. designed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.E.M. is Department of Medicine, Institute for Diabetes, Metabolism and Obesity, University of Pennsylvania School of Medicine, Philadelphia, PA.
Correspondence: Orla M. Conneely, Department of Molecular and Cellular Biology, M511 DeBakey Bldg, Baylor College of Medicine, 1 Baylor Plaza, Houston, TX 77030; e-mail: orlac@bcm.edu.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed. Geneva, Switzerland: IARC; 2008. [Google Scholar]
- 2.Orazi A, Germing U. The myelodysplastic/myeloproliferative neoplasms: myeloproliferative diseases with dysplastic features. Leukemia. 2008;22(7):1308–1319. doi: 10.1038/leu.2008.119. [DOI] [PubMed] [Google Scholar]
- 3.Nimer SD. Myelodysplastic syndromes. Blood. 2008;111(10):4841–4851. doi: 10.1182/blood-2007-08-078139. [DOI] [PubMed] [Google Scholar]
- 4.Bacher U, Schnittger S, Kern W, Weiss T, Haferlach T, Haferlach C. Distribution of cytogenetic abnormalities in myelodysplastic syndromes, Philadelphia negative myeloproliferative neoplasms, and the overlap MDS/MPN category. Ann Hematol. 2009;88(12):1207–1213. doi: 10.1007/s00277-009-0745-3. [DOI] [PubMed] [Google Scholar]
- 5.Milbrandt J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron. 1988;1(3):183–188. doi: 10.1016/0896-6273(88)90138-9. [DOI] [PubMed] [Google Scholar]
- 6.Law SW, Conneely OM, DeMayo FJ, O'Malley BW. Identification of a new brain-specific transcription factor, NURR1. Mol Endocrinol. 1992;6(12):2129–2135. doi: 10.1210/mend.6.12.1491694. [DOI] [PubMed] [Google Scholar]
- 7.Hedvat CV, Irving SG. The isolation and characterization of MINOR, a novel mitogen-inducible nuclear orphan receptor. Mol Endocrinol. 1995;9(12):1692–1700. doi: 10.1210/mend.9.12.8614405. [DOI] [PubMed] [Google Scholar]
- 8.Maruyama KTT, Ohkura N, Bandoh S, Hosono T, Yamaguchi K. The NGFI-B subfamily of the nuclear receptor superfamily [review]. Int J Oncol. 1998;12(6):1237–1243. doi: 10.3892/ijo.12.6.1237. [DOI] [PubMed] [Google Scholar]
- 9.Wilson TEFT, Johnston M, Milbrandt J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science. 1991;252:1296–1300. doi: 10.1126/science.1925541. [DOI] [PubMed] [Google Scholar]
- 10.Wansa KD, Harris JM, Yan G, Ordentlich P, Muscat GE. The AF-1 domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine antimetabolite 6-mercaptopurine. J Biol Chem. 2003;278(27):24776–24790. doi: 10.1074/jbc.M300088200. [DOI] [PubMed] [Google Scholar]
- 11.Wansa KD, Muscat GE. TRAP220 is modulated by the antineoplastic agent 6-mercaptopurine, and mediates the activation of the NR4A subgroup of nuclear receptors. J Mol Endocrinol. 2005;34(3):835–848. doi: 10.1677/jme.1.01739. [DOI] [PubMed] [Google Scholar]
- 12.Smith AG, Luk N, Newton RA, Roberts DW, Sturm RA, Muscat GE. Melanocortin-1 receptor signaling markedly induces the expression of the NR4A nuclear receptor subgroup in melanocytic cells. J Biol Chem. 2008;283(18):12564–12570. doi: 10.1074/jbc.M800480200. [DOI] [PubMed] [Google Scholar]
- 13.Davis IJ, Lau LF. Endocrine and neurogenic regulation of the orphan nuclear receptors Nur77 and Nurr-1 in the adrenal glands. Mol Cell Biol. 1994;14(5):3469–3483. doi: 10.1128/mcb.14.5.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassett MH, White PC, Rainey WE. A role for the NGFI-B family in adrenal zonation and adrenocortical disease. Endocr Res. 2004;30(4):567–574. doi: 10.1081/erc-200043715. [DOI] [PubMed] [Google Scholar]
- 15.Murphy EPMA, Conneely OM, Bresnihan B, FitzGerald O. Involvement of the nuclear orphan receptor NURR1 in the regulation of corticotropin-releasing hormone expression and actions in human inflammatory arthritis. Arthritis Rheum. 2001;44(4):782–793. doi: 10.1002/1529-0131(200104)44:4<782::AID-ANR134>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 16.Hazel TGND, Lau LF. A gene inducible by serum growth factors encodes a member of the steroid and thyroid hormone receptor superfamily. Proc Natl Acad Sci U S A. 1988;85(22):8444–8448. doi: 10.1073/pnas.85.22.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pei L, Castrillo A, Chen M, Hoffmann A, Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J Biol Chem. 2005;280(32):29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- 18.Pearen MA, Myers SA, Raichur S, Ryall JG, Lynch GS, Muscat GE. The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 2008;149(6):2853–2865. doi: 10.1210/en.2007-1202. [DOI] [PubMed] [Google Scholar]
- 19.Maxwell MA, Muscat GE. The NR4A subgroup: immediate early response genes with pleiotropic physiological roles. Nucl Recept Signal. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng LECF, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor-1 orphan steroid receptors in T-cell apoptosis. EMBO J. 1997;16(8):1865–1875. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mapara MY, Weinmann P, Bommert K, Daniel PT, Bargou R, Dorken B. Involvement of NAK-1, the human nur77 homologue, in surface IgM-mediated apoptosis in Burkitt lymphoma cell line BL41. Eur J Immunol. 1995;25(9):2506–2510. doi: 10.1002/eji.1830250915. [DOI] [PubMed] [Google Scholar]
- 22.Lee JMLK, Weidner M, Osborne BA, Hayward SD. Epstein-Barr virus EBNA2 blocks Nur77-mediated apoptosis. Proc Natl Acad Sci U S A. 2002;99(18):11878–11883. doi: 10.1073/pnas.182552499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu QLS, Ye XF, Huang ZW, Su WJ. Dual roles of Nur77 in selective regulation of apoptosis and cell cycle by TPA and ATRA in gastric cancer cells. Carcinogenesis. 2002;23(10):1583–1592. doi: 10.1093/carcin/23.10.1583. [DOI] [PubMed] [Google Scholar]
- 24.Uemura HCC. Antisense TR3 orphan receptor can increase prostate cancer cell viability with etoposide treatment. Endocrinology. 1998;139(5):2329–2334. doi: 10.1210/endo.139.5.5969. [DOI] [PubMed] [Google Scholar]
- 25.Kolluri SK, Zhu X, Zhou X, et al. A short Nur77-derived peptide converts Bcl-2 from a protector to a killer. Cancer Cell. 2008;14(4):285–298. doi: 10.1016/j.ccr.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moll UM, Marchenko N, Zhang XK. p53 and Nur77/TR3-transcription factors that directly target mitochondria for cell death induction. Oncogene. 2006;25(34):4725–4743. doi: 10.1038/sj.onc.1209601. [DOI] [PubMed] [Google Scholar]
- 27.Labelle YZJ, Stenman G, Kindblom LG, et al. Oncogenic conversion of a novel orphan nuclear receptor by chromosome translocation. Hum Mol Genet. 1995;4(12):2219–2226. doi: 10.1093/hmg/4.12.2219. [DOI] [PubMed] [Google Scholar]
- 28.Mullican SE, Zhang S, Konopleva M, et al. Abrogation of nuclear receptors Nr4a3 and Nr4a1 leads to development of acute myeloid leukemia. Nat Med. 2007;13(6):730–735. doi: 10.1038/nm1579. [DOI] [PubMed] [Google Scholar]
- 29.Majeti R, Becker MW, Tian Q, et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci U S A. 2009;106(9):3396–3401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sternberg A, Killick S, Littlewood T, et al. Evidence for reduced B-cell progenitors in early (low-risk) myelodysplastic syndrome. Blood. 2005;106(9):2982–2991. doi: 10.1182/blood-2005-04-1543. [DOI] [PubMed] [Google Scholar]
- 31.Pellagatti A, Cazzola M, Giagounidis AA, et al. Gene expression profiles of CD34+ cells in myelodysplastic syndromes: involvement of interferon-stimulated genes and correlation to FAB subtype and karyotype. Blood. 2006;108(1):337–345. doi: 10.1182/blood-2005-12-4769. [DOI] [PubMed] [Google Scholar]
- 32.Sportoletti P, Grisendi S, Majid SM, et al. Npm1 is a haploinsufficient suppressor of myeloid and lymphoid malignancies in the mouse. Blood. 2008;111(7):3859–3862. doi: 10.1182/blood-2007-06-098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joslin JM, Fernald AA, Tennant TR, et al. Haploinsufficiency of EGR1, a candidate gene in the del(5q), leads to the development of myeloid disorders. Blood. 2007;110(2):719–726. doi: 10.1182/blood-2007-01-068809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenbauer F, Koschmieder S, Steidl U, Tenen DG. Effect of transcription-factor concentrations on leukemic stem cells. Blood. 2005;106(5):1519–1524. doi: 10.1182/blood-2005-02-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kogan SC, Ward JM, Anver MR, et al. Bethesda proposals for classification of nonlymphoid hematopoietic neoplasms in mice. Blood. 2002;100(1):238–245. doi: 10.1182/blood.v100.1.238. [DOI] [PubMed] [Google Scholar]
- 36.Min IM, Pietramaggiori G, Kim FS, Passegue E, Stevenson KE, Wagers AJ. The transcription factor EGR1 controls both the proliferation and localization of hematopoietic stem cells. Cell Stem Cell. 2008;2(4):380–391. doi: 10.1016/j.stem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Burns TF, Fei P, Scata KA, Dicker DT, El-Deiry WS. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23(16):5556–5571. doi: 10.1128/MCB.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Lu J, Sun M, et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc Natl Acad Sci U S A. 2008;105(40):15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Syed N, Smith P, Sullivan A, et al. Transcriptional silencing of Polo-like kinase 2 (SNK/PLK2) is a frequent event in B-cell malignancies. Blood. 2006;107(1):250–256. doi: 10.1182/blood-2005-03-1194. [DOI] [PubMed] [Google Scholar]
- 40.Smith P, Syed N, Crook T. Epigenetic inactivation implies a tumor suppressor function in hematologic malignancies for Polo-like kinase 2 but not Polo-like kinase 3. Cell Cycle. 2006;5(12):1262–1264. doi: 10.4161/cc.5.12.2813. [DOI] [PubMed] [Google Scholar]
- 41.Passegue E, Wagner EF, Weissman IL. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell. 2004;119(3):431–443. doi: 10.1016/j.cell.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 42.Sokal JE, Cox EB, Baccarani M, et al. Prognostic discrimination in “good-risk” chronic granulocytic leukemia. Blood. 1984;63(4):789–799. [PubMed] [Google Scholar]
- 43.Savage DG, Szydlo RM, Chase A, Apperley JF, Goldman JM. Bone marrow transplantation for chronic myeloid leukaemia: the effects of differing criteria for defining chronic phase on probabilities of survival and relapse. Br J Haematol. 1997;99(1):30–35. doi: 10.1046/j.1365-2141.1997.3453159.x. [DOI] [PubMed] [Google Scholar]
- 44.Rosenbauer F, Wagner K, Kutok JL, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36(6):624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 45.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat Rev Immunol. 2007;7(2):105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 46.Kerbauy DB, Deeg HJ. Apoptosis and antiapoptotic mechanisms in the progression of myelodysplastic syndrome. Exp Hematol. 2007;35(11):1739–1746. doi: 10.1016/j.exphem.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steidl U, Rosenbauer F, Verhaak RG, et al. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat Genet. 2006;38(11):1269–1277. doi: 10.1038/ng1898. [DOI] [PubMed] [Google Scholar]
- 48.Passegue E, Jochum W, Schorpp-Kistner M, Mohle-Steinlein U, Wagner EF. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell. 2001;104(1):21–32. doi: 10.1016/s0092-8674(01)00188-x. [DOI] [PubMed] [Google Scholar]
- 49.Matthew EM, Yen TJ, Dicker DT, et al. Replication stress, defective S-phase checkpoint and increased death in Plk2-deficient human cancer cells. Cell Cycle. 2007;6(20):2571–2578. doi: 10.4161/cc.6.20.5079. [DOI] [PubMed] [Google Scholar]
- 50.Krones-Herzig A, Adamson E, Mercola D. Early growth response 1 protein, an upstream gatekeeper of the p53 tumor suppressor, controls replicative senescence. Proc Natl Acad Sci U S A. 2003;100(6):3233–3238. doi: 10.1073/pnas.2628034100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.