Abstract
The basal ganglia support learning to exploit decisions that have yielded positive outcomes in the past. In contrast, limited evidence implicates the prefrontal cortex for making strategic exploratory decisions when the magnitude of potential outcomes is unknown. Here we examine neurogenetic contributions to individual differences in these distinct aspects of motivated human behavior, employing a temporal decision making task and computational analysis. We show that genes controlling striatal dopamine function (DARPP-32 and DRD2) are associated with exploitative learning to incrementally adjust response times as a function of positive and negative decision outcomes. In contrast, a gene primarily controlling prefrontal dopamine function (COMT) is associated with a particular type of “directed exploration”, in which exploratory decisions are made in proportion to Bayesian uncertainty about whether other choices might produce outcomes that are better than the status quo. Quantitative model fits reveal that genetic factors modulate independent parameters of a reinforcement learning system.
Individuals differ in their choices and neural responses when confronted with decision uncertainty [1, 2]. Some people are motivated by having achieved desirable outcomes and are driven to work harder to attain even better ones, whereas others are primarily motivated to avoid negative outcomes [3]. However, often one doesn't know which outcomes should be considered positive until they compare them to those obtained from other decision strategies (e.g., do you choose to return to the same fail-safe sushi restaurant, or to try a new one because it might be even better? ). This classic problem of whether to sample other options or maintain the current strategy for maximizing reward is known as the exploration/exploitation dilemma [4, 5, 6, 7]. Here we examine neurogenetic contributions to exploitative and exploratory behavior.
In part, individual differences in personality variables are thought to reflect different parameters within the dopaminergic motivational system [8]. Dopaminergic genetic components that alter function in the striatum (and indirectly, its interactions with frontal cortex; [9]) differentiate between individuals who are more adept at learning from positive versus negative decision outcomes, via modulation of striatum and its interactions with frontal cortex [9, 10, 11]. Specifically, a functional polymorphism within the PPP1R1B gene coding for DARPP-32 [9] is predictive of “Go learning” to reproduce behaviors that yield positive outcomes [10]. DARPP-32 is a protein highly concentrated in the striatum, is phosphorylated by D1 receptor stimulation, and is required for striatal D1-receptor mediated synaptic plasticity and behavioral reward learning [12, 13, 14]. While DARPP-32 is also present in D2-containing neurons, D2 receptor stimulation de-phosphorylates DARPP-32 and does not mediate its effects on reward learning [13]. Conversely, polymorphisms within the DRD2 gene predictive of striatal D2 receptor density are associated with “NoGo learning” to avoid behaviors that yield negative outcomes [10, 11]. These findings converge with the notion that dopamine plays a key role in reinforcement learning [15], and in particular, that dopamine acts in the striatum to support learning from positive and negative outcomes via D1 and D2 receptors in separate neuronal striatonigral and striatopallidal populations [16, 17]. They also converge with rodent data showing that the transition to exploitative behavior is associated with the development of highly stabilized striatal firing patterns [18].
Whereas the role of striatal dopamine in reinforcement exploitation is relatively well established, the neurobiological correlates of exploration are far less developed. Computational considerations suggest that an adaptive heuristic is to explore in proportion to one's uncertainty about the consequent outcomes [4, 6, 19, 7]. Such computations might depend on neuromodulation within the prefrontal cortex (PFC) [7]. Functional neuroimaging evidence implicates anterior and orbital PFC in computations of uncertainty [20, 2], and in making exploratory decisions in a reinforcement learning environment [6]. Further, models and experimental data suggest that orbital PFC represents reward magnitudes, required to compute the expected value of decisions, especially over delays [21, 22, 6, 23]. At the genetic level, a gene coding for catechol-O-methyltransferase (COMT), substantially affects PFC dopamine levels, and in turn, PFC-dependent cognitive function [24]. COMT is an enzyme that breaks down dopamine, with the val allele associated with greater enzymatic efficacy and therefore lower PFC dopamine levels. It plays a comparatively minor role in striatum, due to its relatively sparse expression, and the presence of potent dopamine transporters and autoreceptors [25, 26, 24, 27].
We assessed these motivational components, including exploitation, exploration, and probability vs. magnitude learning, within a single “temporal utility integration task” [28]. We hypothesized that DARPP-32 and DRD2 genes, as markers of individual differences in striatal dopaminergic function, would be predictive of response time adaptation to maximize rewards. In contrast, we hypothesized that the COMT gene, as a proxy for prefrontal dopaminergic function, would be predictive of uncertainty-based exploration and enhanced representation of reward magnitudes.
Results
Temporal Integration of Expected Value
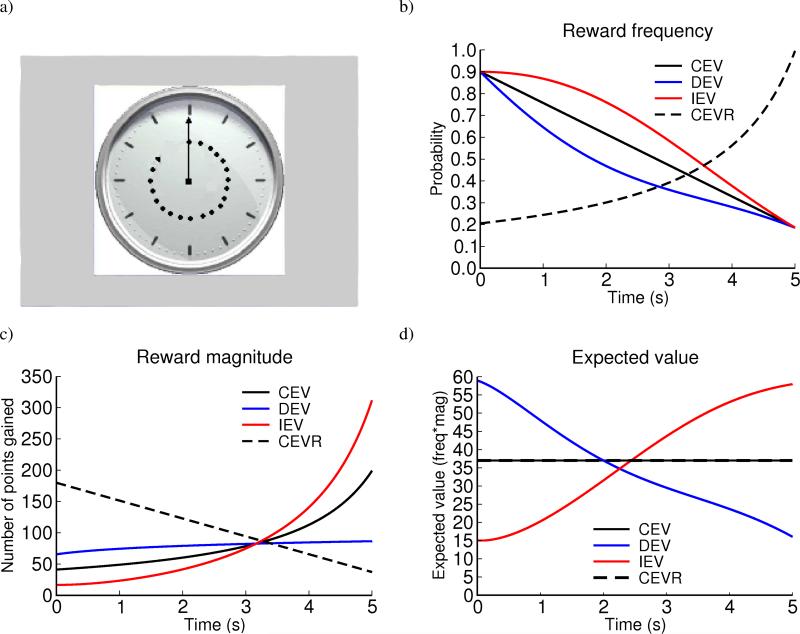
Participants observed a clock arm which completed a revolution over 5 seconds, and could stop the clock with a key press in an attempt to win points. Rewards were delivered with a probability and magnitude that varied as a function of response time (RT, Figure 1). The functions were designed such that the expected value (EV; probability*magnitude) increased, decreased, or remained constant (IEV, DEV, or CEV) with increasing response times (Figure 1). Thus in the DEV condition, faster RTs yield more points on average, such that performance benefits from “Go learning” to produce further speeded RTs. In contrast, fast RTs in the IEV condition yield below average outcomes, such that performance benefits from “NoGo learning” to produce adaptively slower responding. The CEV condition was included for a within-subject baseline RT measure for comparison with IEV and DEV. Because all RTs are equivalently rewarding in the CEV condition, participants’ RT in this condition controls for individual differences in overall motor responding. Given this baseline, an ability to adaptively integrate expected value would be indicated by relatively faster responding in the DEV condition and slower responding in the IEV condition. Dopaminergic manipulations in Parkinson's patients have opposite effects on these measures, likely via modulation of striatal dopamine [28].
Figure 1.
Task conditions: decreasing expected value (DEV), constant expected value (CEV), increasing expected value (IEV), and constant expected value - reverse (CEVR). The x axis corresponds to the time after onset of the clock stimulus at which the response is made. The functions are designed such that the expected value at the beginning in DEV is equal to that at the end in IEV so that at optimal performance, subjects should obtain the same average reward in both IEV and DEV. Faster responses were accompanied by longer inter-trial intervals so that reward-rate is roughly equalized across conditions. a) Example clock-face stimulus. Each trial ended when the subject made a response or otherwise when the 5 s duration elapsed. The number of points won on the current trial was displayed. b) Probability of reward occurring as a function of response time; c) Reward magnitude (contingent on probability in b); d) Expected value across trials for each time point. Note that CEV and CEVR have the same EV.
We also included a fourth condition (constant expected value - reverse, CEVR) in which reward probability increased while magnitude decreased. This condition serves two purposes: First, because both CEV and CEVR have equal expected values across time, any difference in RT in these two conditions can be attributed to a participants’ potential bias to learn more about reward probability than about magnitude or vice-versa. Second, CEVR provides another measure of avoidance learning. That is, despite the constant expected value, a bias to learn from negative outcomes will produce slowed responses due to their high probability of occurrence at early response times.
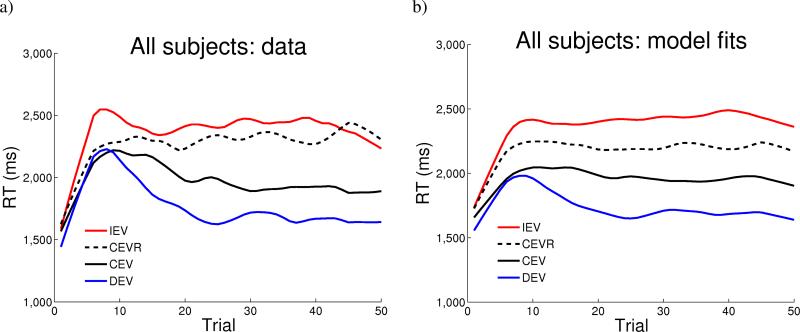
Overall, participants exhibited robust learning (Figure 2a; see Figure S5 for RTs for each genotype). Compared to the baseline CEV condition, RTs in the IEV condition were significantly slower (F(1,67) = 28.5, p < 0.0001), whereas those in the DEV condition were significantly faster (F(1,67) = 6.7 , p = 0.01).
Figure 2.
Response times as a function of trial number, smoothed (with weighted linear least squares fit) over a 10 trial window, in a) all 69 participants, b) computational model.
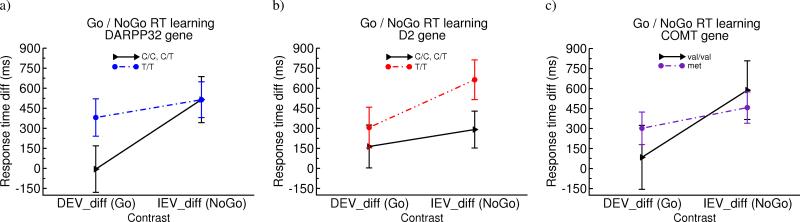
There were no effects of any gene on baseline RTs in the CEV condition, or on overall response time (all p's > 0.25). Nevertheless, within-subject RT modulations due to reward structure were predictably altered by striatal genotype (Figure 3). DARPP-32 T/T carriers showed enhanced “Go learning”, with faster RTs in the last block of DEV condition (F[1,64] = 4.4, p = 0.039), and, marginally, relative to CEV (DEVdiff; F[1,64] = 3.1, p = 0.08, an effect that was significant across all trials; p < 0.05). There was no DARPP-32 effect on “NoGo learning” (IEV RT's, or IEVdiff; p's > 0.8). Conversely, DRD2 T/T carriers, who have the highest striatal D2 receptor density [29, 10], showed marginally slower RTs in IEV, indicative of enhanced NoGo learning (F[1,66] = 3.3, p = 0.07 for both IEV and IEVdiff), with no effect on Go learning (p's > 0.3). Modeling results reported below, together with CEVR performance, more strongly support the conclusion that DARPP-32 and DRD2 genes modulate learning to speed and slow RTs from positive and negative outcomes. Finally, there was no effect of COMT on any of these measures (p's > 0.35). This constellation of genetic effects converge with those found previously [10], but extend them to a completely different task context, dependent measure, and sample. Moreover, these same response time adaptations due to reward structure are sensitive to dopaminergic manipulation in Parkinson's disease [28].
Figure 3.
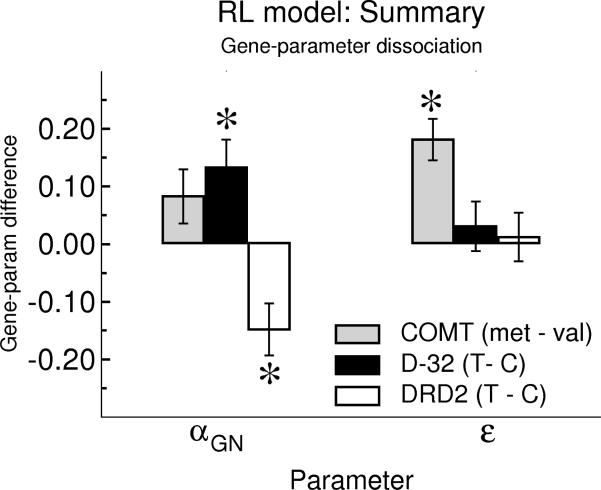
Relative within-subjects biases to speed RTs in DEV relative to CEV (DEVdiff = CEV – DEV) and to slow RTs in IEV (IEVdiff IEV = IEV – CEV). Values represent mean (standard error) in the last quarter of trials in each condition. a) DARPP-32 gene, b) DRD2 gene, c) COMT gene.
Further analysis revealed genetic contributions to learning from probability relative to magnitude of reinforcement, as assessed by comparing RTs in the CEVR condition (alone and relative to CEV; p = 0.02, Supplement). Specifically, those with enhanced D2 function showed significantly greater sensitivity to frequent negative outcomes in CEVR, again consistent with enhanced NoGo learning. There was also some evidence for COMT met allele carriers to be more sensitive to reward magnitudes (Figure S1).
Trial-to-Trial RT adaptation: Exploration?
Although on average participants incrementally changed response times dependent on reward structure, single subject data revealed large RT swings from one trial to the next (Figure ). These swings did not reflect adaptive changes following rewards or lack thereof [28]. Instead, preliminary analyses indicated that RT swings simply reflected a regression to the mean, whereby faster than average responses were more likely to be followed by relatively slower responses and vice-versa (p < 0.0001; Supplement). As will be clear, however, these RT swings reflect more than just a statistical necessity, and likely represent participants’ tendency to explore the space of responses to determine the reward structure. We investigate this effect in the mathematical reinforcement learning (RL) model developed below.
Computational Model
We previously simulated performance in this task using an a priori neural network model of the basal ganglia [28]. The model simulates interactive neural dynamics among corticostriatal circuits and accounts for various effects of dopaminergic manipulation on action selection and reinforcement learning [16, 30, 31, 32]. Simulated dopamine (DA) medications induce speeded RTs in the DEV condition as a result of D1-dependent Go learning in striatonigral cells. However, the same increased DA release impedes the ability to slow down in IEV, due to excessive D2 receptor stimulation on striatopallidal cells, and concomitant impairments in NoGo learning. Simulated DA depletion produces the opposite result: less speeding in DEV but better slowing in IEV and CEVR, mirroring Parkinson's patients’ performance in the task [28].
Here we develop an abstract mathematical model designed to quantitatively fit individual participants’ response times on a trial-to-trial basis. The purpose of this modeling is threefold: (i) to demonstrate the core computational principles by which the more complex neural model captures the incremental RT changes as a function of reward prediction error; (ii) to augment the model to capture strategic exploratory behavior as a function of reward uncertainty; and (iii) to determine whether best-fitting model parameters for both exploitative and exploratory decisions are predictably modulated as a function of genotype [10].
The point of departure for the model is the central assumption common with virtually all reinforcement models, namely that participants develop an expected value V(t) for the reward they expect to gain in a given trial t. This value is updated as a function of each reward experience using a simple delta rule:
where α is a learning rate that modifies the extent to which values are updated from one trial to the next, and δ is the reward prediction error reported by DA neurons [33, 15], which is simply the reward outcome minus the prior expected value:
This value integration is posited to be computed by brain areas upstream of dopamine neurons comprising the “critic”, which learns as a function of prediction errors to faithfully represent expected value [5, 34, 35]. Our model further shares the assumption that these same prediction error signals train the “actor” in striatum [34]. This process can occur in at least two ways. First, we model a simple, likely implicit process, whereby accumulated positive prediction errors translate into approach-related speeded responses (“Go learning”), whereas accumulated negative prediction errors produce relative avoidance and slowed responses (“NoGo learning”) [28, 32]. These processes are posited to rely on D1 and D2 receptor mechanisms in separate populations of striatonigral and striatopallidal cells [16, 28, 32, 36]. Because of these differential learning mechanisms, we use different learning rates and for each:
where αG controls D1-dependent speeding from positive prediction errors (δ+) and αN controls D2-dependent slowing from negative prediction errors (δ−), for action a and clock-face state s. On each trial RTs were predicted to speed/slow according to differences between current Go and NoGo values.
In addition to this implicit process capturing putative striatal contributions to approach/avoidance, we also model a more strategic process in which participants separately track of reward structure for different (“fast” and “slow”) responses (Supplement). With these action representations, participants need only adapt RTs in proportion to the difference between their expected reward values. This would allow, for example, participants to delay responding when slow RTs yield larger rewards on average (as in IEV), or to speed up if they don't. We model this process using Bayesian integration, assuming subjects represent the prior distributions of reward prediction errors separately for fast and slow responses, and update them as a function of experience via Bayes’ rule:
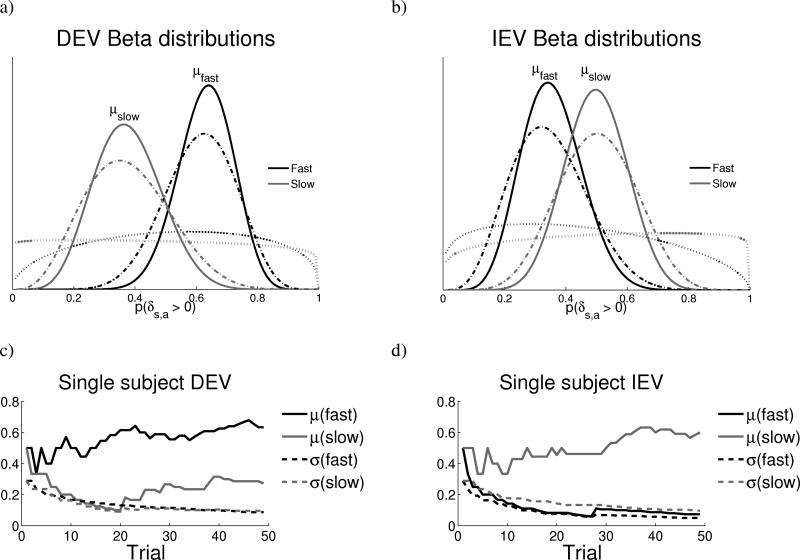
where θ reflects the parameters governing the belief distribution about the reward prediction errors for each response, and δ1...δn are the prediction errors observed thus far (on trials 1 to n). Simply stated, Bayes’ rule implies that the degree to which each outcome modifies participants’ beliefs about obtainable rewards depends on their prior experience and, given this prior, the likelihood that the outcome would occur. As experience is gathered, the means of the posterior distributions accurately represent reward structure in each condition (Figure 6).
Figure 6.
Evolution of action-value distributions. a), b) Beta probability density distributions representing the belief about the likelihood of reward prediction errors following fast and slow responses, averaged across all subjects’ data. The x axis is the probability of a positive prediction error and the y-axis represents the belief in each probability, with the mean value μ representing the best guess. Dotted lines reflect distributions after a single trial; dashed lines after 25 trials; solid lines, after 50 trials. (See supplemental animation #1 for dynamic changes in these distributions across all trials for a single subject). Differences between the μfast and μslow were used to adjust RTs to maximize reward likelihood. The standard deviation σ was taken as an index of uncertainty. Exploration was predicted to modulate RT in direction of greater uncertainty about whether outcomes might be better than the status quo. c), d) Trajectory of means and standard deviations for a single subject in DEV and IEV conditions. Uncertainties σ decrease with experience. Corresponding Beta hyperparameters η, β are shown in the supplement.
We considered that participants either track the probability of a reward prediction error (i.e., the probability that a dopamine burst occurs), using beta distributions Beta(η,β), or the magnitude of expected rewards, represented by Normal distributions N(μ, σ2). We focus here on the beta distribution implementation, which provided a better fit to the behavioral data. Nevertheless all genetic results presented below held when using Normal distributions and a Kalman filter (Supplement). In either case, RTs were predicted to adapt in proportion to the difference between the best estimates of reward structure for fast and slow responses, i.e., the following term was added to the RT prediction: ρ[σslow(s,t) – σfast(s,t)], where ρ is a free parameter.
We also modeled other parameters that contribute to RT in this task, including simple baseline response speed (irrespective of reward), captured by free parameter K, autocorrelation between the current and previous RT (λ) regardless of reward, and a tendency to adapt RTs toward the single largest reward experienced thus far (“going for gold”, parameter ν). Finally, we posited that exploratory strategies would contribute to participants’ RT adjustments, as participants sampled the outcomes available to determine which response is most adaptive. This process is modeled as a dynamic Explore process depending on Bayesian uncertainty, elaborated further below, and hypothesized to rely on prefrontal-dependent processes. The complete RT update is thus as follows:
For each subject, a single set of best fitting parameters was derived across all conditions. The model captures the qualitative pattern of results, with predicted RT changing as a function of reward structure (Figure 2b; see Figure S6 for model fits for each genotype). Positive prediction errors are most prevalent for early responses in DEV, and accordingly model RTs are fastest in this condition. Negative prediction errors are most prevalent in IEV and CEVR, leading to slowed model responses.
We hypothesized that these relative learning rate parameters for determining exploitative responses would be modulated by striatal genotype. Indeed, DARPP-32 T/T carriers, who should have increased striatal D1-dependent learning [10, 13, 14] had relatively larger αG than αN than did C carriers, suggesting relatively greater sensitivity to positive than negative prediction errors (Figure 5; F(1,65) = 4.0, p = 0.05). Conversely, DRD2 T/T carriers, with relatively greater D2 receptor density [29], showed relatively greater learning from negative prediction errors (F(1,66) = 5.3, p = 0.02). Relative learning rates were not modulated by COMT genotype (p > 0.2), and other than the Explore parameter, no other parameters differed as a function of any genotype (all p's > 0.2).
Figure 5.
Genetic effects on reinforcement model parameters. DARPP-32 T/T carriers showed relatively greater learning rates from gains than losses (αGN = αG – αN) compared to C carriers. DRD2 T/T carriers showed the opposite pattern. The COMT gene did not affect learning rates, but met carriers had significantly higher uncertainty-based explore parameter (ε) values (which are divided by 104 to be displayed on the same scale) than did val/val participants. Error bars reflect standard error.
Uncertainty-Based Exploration
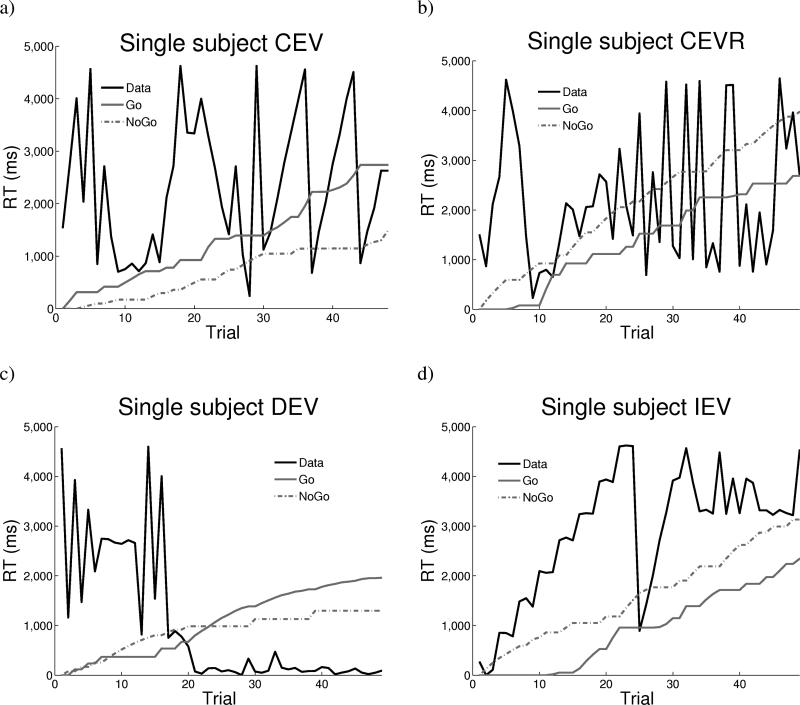
The above model provides an account of incremental RT changes as a function of reward prediction error, and provides evidence for the mechanisms posited to mediate these effects in neural networks [28]. Nevertheless, inspection of individual subject data reveals more complex dynamics than those observed in the averaged data (Figure 4). These plots show RTs across trials for an arbitrary single participant, along with model Go and NoGo terms. Asymptotically, the participant converges on a faster RT in DEV, and slower RT in IEV, relative to CEV. However, at the more fine-grained scale, there are often large RT swings from one trial to the next which are not captured by model learning mechanisms.
Figure 4.
Trial-to-trial RT adjustments in a single subject in a) CEV, b) CEVR, c) DEV, and d) IEV. Model Go and NoGo terms (magnified by 4x) accumulate as a function of positive and negative prediction errors. Go dominates over NoGo in DEV and the reverse in IEV, but these incremental changes do not capture trial-by-trial dynamics. For this subject, = 0.63 and = 0.74 (ms/point).
We hypothesized that these RT swings are rational, in that they might reflect exploratory strategies to gather statistics of reward structure. Several solutions have been proposed to manage the exploration/exploitation tradeoff. If performance is unsatisfactory over extended periods, stochastic noise can simply be added to behavioral outputs, promoting random exploratory choices [7]. Alternatively, exploration can be strategically directed toward particular choices in proportion to the amount of information that would be gained, regardless of past performance [4, 37, 38, 6]. Our model embodies the assumption that exploratory decisions occur in proportion to the participant's relative uncertainty about whether responses other than those currently being exploited might yield better outcomes. This assumption builds on prior modeling in which exploration is encouraged by adding an “uncertainty bonus” to the value of decision options having uncertain outcomes [4, 37, 38, 6]. Here we posit that exploration occurs in proportion to uncertainty about the probability that the explored option will yield a positive reward prediction error (or, in alternative models, uncertainty about the expected value of such rewards or reward prediction errors; Supplement). The Bayesian framework for integrating reward statistics provides a natural index of uncertainty: the standard deviations of the prior distributions [39], which decrease after sampling a given action (albeit at a slower rate for more variable outcomes).
Initially, distributions representing belief about reward structure for each response category are wide, reflecting maximum uncertainty (Figure 6). As experience with each option is gathered, the distributions evolve to reflect the underlying reward structures, such that the mean belief is higher for fast responses in DEV and for slow responses in IEV. Moreover, the standard deviations, and hence uncertainties, decrease with experience. This process is analogous to estimating the odds of a coin flip resulting in heads or tails, with uncertainty about those odds decreasing with the number of observations. With these distributions, the relative uncertainties for fast and slow responses in a given trial can be used as a rational heuristic to drive exploration. In particular, the Explore term of the model is computed as follows:
where ε is a free parameter that scales exploration in proportion to relative uncertainty and σδ|s,a=Slow, σδ|s,a=Fast are the standard deviations quantifying uncertainty about reward prediction error likelihood given slow and fast responses, respectively. Thus, with sufficiently high ε, RT swings are predicted to occur in the direction of greater uncertainty about the likelihood that outcomes might be better than the status quo.
Overall, including this uncertainty-based exploration term provided a better fit to trial-by-trial choice than the base model without exploration (and penalizing the fit for the additional parameters; see Supplement). Although the model cannot deterministically predict RT swings (which reflect the output of multiple interacting processes, including those sensitive to previous reinforcement), there is nevertheless a reliable positive correlation between the model's uncertainty-based exploratory predictions and participants’ actual RT swings from on trial to the next (r(4214) = 0.31, p < 0.0001; Figure 7 and Figure S3).
Figure 7.
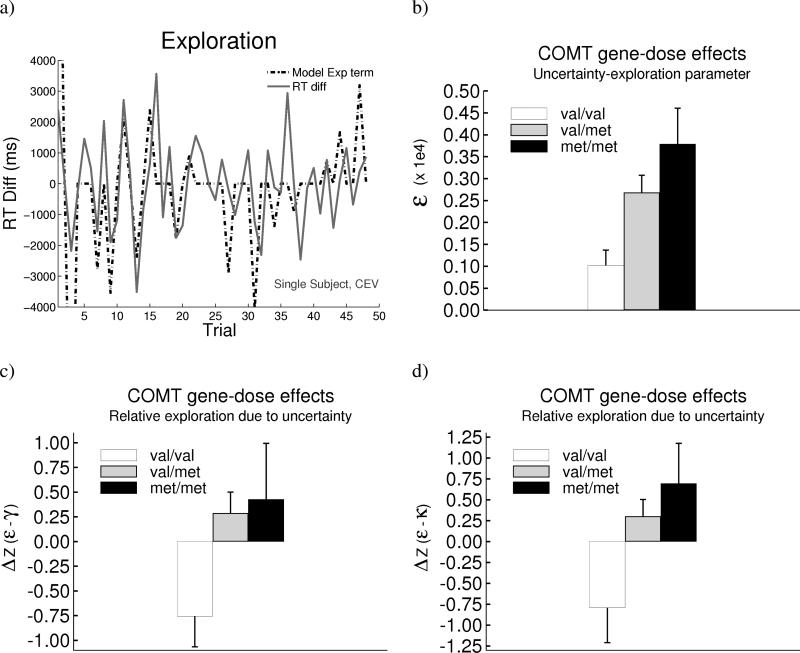
COMT gene predicts directed exploration toward uncertain responses. a) RT swings (change in RT from the previous trial) in a single met/met subject in the CEV condition, and the corresponding model uncertainty-based Explore term (amplified to be on the same RT scale). See supplemental animation #2 for this subject's evolution of beta distributions in CEV. b) COMT gene-dose effect on the uncertainty-based exploration parameter ε. Gene-dose effects were also observed when comparing relative contributions of ε compared with c) a reverse-momentum parameter γ, and d) a lose-switch parameter κ. Relative Z-scores are plotted here due to comparison of parameters scaling quantities of different magnitudes. Error bars reflect standard error.
Moreover, this relationship was particularly evident for COMT met allele carriers (Figure S3), supporting a role for PFC neuromodulatory control over exploration as a function of decision uncertainty. The ε parameter that scales exploration in proportion to uncertainty was significantly higher among met allele carriers (Figure 5; F(1,67) = 8.2, p = 0.006). Further, there was a monotonic gene-dose effect, with ε values largest in met/met participants, intermediate in val/met, and smallest in val/val carriers (Figure 7b; F[1,67] = 9.5, p = 0.003). No such effects on ε were observed for DARPP-32 or DRD2 genotypes (p's > 0.5).
Importantly, the COMT exploration effects appear to be specific to uncertainty. First, overall RT variability (in terms of standard deviation) did not differ as a function of genotype (p > 0.2). Second, a number of foil models attempting to account for RT swings without recourse to uncertainty confirmed that only the uncertainty-based exploration parameter can account for COMT effects (Supplement). For example, we included a “reverse-momentum” parameter γ, which predicted RT swings to counter a string of progressively speeded or slowed responses, regardless of uncertainty. While this model provided a reasonable fit to RT swings overall, the uncertainty model was superior only in COMT met allele carriers (Supplement). We also included a “lose-switch” parameter κ, which predicted RTs to adjust from fast to slow or vice-versa following a negative prediction error. Notably, there were COMT gene-dose effects not only on raw ε values but also their relative weighting compared to either γ or κ (p's < 0.004; Figure 7c,d). This result implies that the contribution of COMT to RT swings is specific to uncertainty.
Discussion
Individuals differ substantially in their motivational drives. The present findings demonstrate three distinct aspects of value-based decision making associated with independent genetic factors (see summary Figure 5). These genes modulate specific aspects of dopaminergic function in brain areas thought to support exploration and exploitation [10, 6, 7, 18]. Behaviorally, exploitative choices were manifest by RT differences between conditions in which rewards could on average be maximized by responding earlier (DEV) or later (IEV) in the trial, compared to baseline (CEV) conditions. Modeling showed that striatal genetic effects are accounted for by individual differences in learning rates from positive and negative prediction errors and their coupling with response speeding and slowing. This result is non-trivial: striatal genes could have affected exploitation by modulating the extent to which RTs are adjusted as a function of mean reward value estimates (i.e., the ρ parameter). Similarly, while trial-to-trial RT swings are readily viewable in single subject data (Figure 4), the specific components due to uncertainty-based exploration, and individual differences therein, were only extracted with the computational analysis.
Our observation that DARPP-32 and DRD2 modulate reinforcement learning in the temporal decision making domain is consistent with similar genetic effects in choice paradigms [10], and with data from Parkinson's patients on and off medication in this same task [28]. Recent rodent studies show direct support for the model's dual D1 and D2 mechanisms of synaptic plasticity [17, 16].
The present human genetic data provide support for the mechanisms posited in models of striatal dopamine, in which accumulated reward prediction errors over multiple trials produce speeded responses, whereas negative prediction errors slow responses [28, 40]. Our assumption that DARPP-32 genetic effects reflect striatal D1-receptor mediated “Go learning” is supported by evidence that the DARPP-32 protein is highly concentrated in the striatum [12] and is critical for D1- but not D2-dependent synaptic plasticity and behavioral reward learning [13, 14]. These data also converge with effects of pharmacological manipulation of striatal D1 receptors on appetitive approach and response speeding to obtain rewards in monkeys and rats [36, 41].
Similarly, our assumption that DRD2 genetic effects reflect primarily striatal D2-receptor mediated learning is supported by evidence that T/T homozygotes exhibit enhanced striatal D2 receptor density [29, 42]. Theoretically, striatal D2 receptors are thought to be necessary for learning in striatopallidal neurons when DA levels are low [16], as is the case during negative prediction errors [43, 44, 45], or as a result of Parkinson's disease [30, 28]. Indeed, synaptic potentiation in striatopallidal neurons is elevated under conditions of DA depletion [17]. Conversely, rats with reduced striatal D2 receptor density [46] are less sensitive to aversive outcomes, persisting to take addictive drugs even when followed by shocks [47].
Perhaps less clear is the precise neurobiological mechanism by which COMT modulates uncertainty-based exploration. Indeed, the mechanisms of exploration are understudied compared to those of exploitation. Nevertheless, neuroimaging studies reveal that in non-reinforcement learning contexts, anterior prefrontal cortical regions reflect Bayesian uncertainty [20], and that this same region is activated when participants make exploratory decisions in a RL environment [6]. Our findings provide the first evidence for exploratory decisions that occur in proportion to uncertainty about whether other responses might produce better outcomes than the status quo. This exploration strategy is strongly motivated by prior theoretical work [38, 6, 7], and appears to be highly dependent on prefrontal genetic function. Furthermore, our originally reported COMT effects on trial-to-trial “lose-shift” behavior in choice paradigms [10] might be more parsimoniously explained by uncertainty-based exploratory mechanisms. Indeed, in that study, met carriers exhibited greater propensity to shift only in the initial trials of the task when reward structure is most uncertain. Thus, these exploratory strategies may be viewed as an attempt to minimize uncertainty.
In contrast to the multiple extant neural models of exploitation, a dearth of models have investigated how neuronal populations can learn to represent quantities of uncertainty as a function of experience. Nevertheless, the sorts of Bayesian probability distributions required for the uncertainty computations used here are naturally coded in populations of spiking neurons [48, 49]. Thus future research should examine how such representations can be learned, and whether prefrontal DA supports the uncertainty computations per se, the active maintenance of relative uncertainties over time, or simply the final decision to over-ride exploitative strategies in order to explore when uncertainty is sufficiently high.
Methods
Sample
We tested 73 healthy participants recruited from the University of Arizona undergraduate psychology subject pool and who provided informed written consent. Two subjects declined genetic sampling, and are excluded from analysis. Failed genetic assays eliminated a further two COMT samples, two DRD2 samples, and three DARPP-32 samples. The remaining 69 subjects (46 female) had a mean age of 19 (SE = .2), and comprised 48 Caucasians, 14 Hispanics, 2 Asians, 1 African-American, and 4 subjects who categorized themselves as “Other”. The breakdown of COMT genotypes was 19:43:7 (val/val:val/met:met/met). The breakdown of DRD2 genotypes was 31:38 (C carriers:T/T). The breakdown of DARPP-32 genotypes was 38:29 (T/T:C carriers; note that in our prior report the T/T genotype was incorrectly referred to as A/A, and C carriers as G carriers, due to mislabeling the base-pair complement [10]. Thus the T/T subjects here reflect the same genotype previously associated with enhanced Go learning). Genetic effects were independent: there was no association between the distribution of any polymorphism and any other (e.g., DRD2 genotype was not predictive of COMT genotype, etc; Fisher's exact test, p > 0.3). All genotypes were in Hardy-Weinberg equilibrium (p's > 0.1), with the exception of COMT ([1] = 5.6, p < .05). This deviation is likely due to heterogeneity in the population; when analyzing Caucasians alone, Hardy-Weinberg equilibrium was not violated (p> 0.1).
Genotyping
Genotyping procedures were carried out in the Molecular Psychiatry Laboratory at the University of Arizona. DNA samples were extracted from saliva samples using Oragene DNA Collection Kits (DNAGenotek). Genomic DNA was amplified using standard polymerase chain reaction (PCR) protocols.
Dopamine- and adenosine-3’,5’-monophosphate (cAMP)-regulating phosphoprotein SNP (DARPP-32, rs907094)
Genomic DNA was amplified for the DARPP-32 (also called PPP1R1B) SNP using standard PCR protocol. Amplification of the 404 bp region was carried out using the sense primers DD-F 5’- GCATTGCTGAGTCTCACCTGCAGTCT- and anti-sense primers DD-R 3’5’-ATTGGGAGAGGGACTGAGCCAAGGATGG-3’ in a reaction volume of 25 μl consisting of 2.5 ng of DNA, .25 mM dNTP's, .25 μM sense and anti-sense primers, 1X QIAGEN PCR buffer and 1.5 U Taq DNA polymerase (QIAGEN). Thermocycling conditions consisted of an initial denaturation step of 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 72 °C for 60 s, and 72°C for 60 s, with a final extension step of 72 °C for 10 min. PCR products were sequenced using the ABI 3730XL DNA Analyzer ® (Applied Biosystems) and visualized using Chromas Vs. 2.13 (Technelysium).
COMT rs4680
Genomic DNA was amplified for the Comt4680 polymorphism using standard PCR protocol. Amplification of the 109 bp region was carried out using the sense primers Comt-F 5’-TCTCCACCTGTGCTCACCTC-3 ’ and anti-sense primers Comt-R 5 ’-GATGACCCTGGTGATAGTGG -3’ in a reaction volume of 25 μl consisting of 2.5 ng of DNA, 0.25 mM dNTP's, 0.25 μM sense and anti-sense primers, 1X QIAGEN PCR buffer and 1 U Taq DNA polymerase (QIAGEN). Thermocycling conditions consisted of an initial denaturation step of 95 °C for 5 min, followed by 35 cycles of 95 °C for 15 s, 54 °C for 20 s, and 72 °C for 30 s, with a final extension step of 72 °C for 5 min. The restriction enzyme Nla III (5 U New England Biolabs) was added to a 20 μl aliquot of the PCR product and digested for 2 hours at 37 °C. 5 μl of the digested PCR product was added to 4 μl of Orange G DNA loading buffer and loaded onto a 3% agarose gel. Images were captured via the Gel Doc XR System (BioRad, USA).
DRD2 rs6277
Optimization of tetra-primer ARMS PCR for the detection of the DRD2 polymorphism was performed empirically using primers designed by original software developed by the founders of the tetra-primer ARMS PCR method and available on the website: http://cedar.genetics.soton.ac.uk/public_html/primer1.html with a Tm optimized to 72°C and a GC content of 48.7%.
Genomic DNA was amplified for the DRD2 polymorphism using tetra-primer ARMS PCR protocol as described [50]. Amplification of the total 295 bp region was carried out using the outer sense primers DRD2-F 5’- ACGGCTCATGGTCTTGAGGGAGGTCCGG- 3’ and outer anti-sense primers DRD-R 5’-CCAGAGCCCTCTGCCTCTGGTGCAGGAG -3’ as well as inner sense primers DRD-Fi 5’- ATTCTTCTCTGGTTTGGCGGGGCTGGCA -3’ and inner anti sense primers 5’-CGTCCCACCACGGTCTCCACAGCACTACC -3’in a reaction volume of 25 μl consisting of 2.5 ng of DNA, 0.25 mM dNTP's, 0.025 μM outer sense and anti sense primers, 0.25 μM inner sense and anti-sense primers, 1X QIAGEN PCR buffer and 2 U Taq DNA polymerase (QIAGEN). Thermocycling conditions consisted of an initial denaturation step of 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 72 °C for 60 s, and 72 °C for 60 s, with a final extension step of 72 °C for 10 min. Five microliters of the PCR product was added to 4 μl of Orange G DNA loading buffer and loaded onto a 3% agarose gel and run in 0.5TAE buffer for 20 min at 72 V. The gels were pre-stained with GelStar® Nucleic Acid Gel Stain and images were captured via the Gel Doc XR System (BioRad, USA).
Genotyping for DRD2 was carried in triplicate, and identification of each individual allele was conducted by three independent observers with 100% agreement.
Ethnicity
Because there was some heterogeneity in the sample (14 subjects were Hispanic) it is critical to establish whether genetic effects are not to due occult stratification. To this end we reanalyzed the data omitting the 14 Hispanics and found very similar patterns of results for each genotype. Similar results also were found when omitting all non-Caucasians. We also reanalyzed all the data and included an additional factor into the general linear model according to whether subjects were Hispanic or not. In this analysis, all genetic effects remained significant and there was no effect of ethnicity, nor an interaction between ethnicity and genotype (p's > 0.25). Again, similar findings were included if the factor coded whether subjects were Caucasian or not. Finally, Hardy Weinberg equilibrium data were also analyzed when excluding Hispanic and other non-Caucasians, and all genotype frequencies did not deviate from equilibrium.
Task Methods
Task instructions were as follows:
“You will see a clock face. Its arm will make a full turn over the course of 5 seconds. Press the ‘spacebar’ key to win points before the arm makes a full turn. Try to win as many points as you can!
“Sometimes you will win lots of points and sometimes you will win less. The time at which you respond affects in some way the number of points that you can win. If you don't respond by the end of the clock cycle, you will not win any points.
“Hint: Try to respond at different times along the clock cycle in order to learn how to make the most points. Note: The length of the experiment is constant and is not affected by when you respond.” This hint was meant to prevent participants from responding quickly simply to leave the experiment early, and in an attempt to equate reward rate (i.e., rewards per second) across conditions. In addition, earlier responses were associated with longer inter-trial intervals so that this was roughly the case. However, because subjects may be averse to waiting through long inter-trial intervals, and because we also wished to reduce the predictability of the onset of the next trial's clock face stimulus we set the inter-trial interval to (5000-RT)/2. Thus, faster responses were associated with longer wait times, but the onset of each trial was temporally unpredictable.
The order of condition (CEV, DEV, IEV, CEVR) was counterbalanced across participants. A rest break was given between each of the conditions (after every 50 trials). Subjects were instructed at the beginning of each condition to respond at different times in order to try to win the most points, but were not told about the different rules (e.g., IEV, DEV). Each condition was also associated with a different color clock face to facilitate encoding that they were in a new context, with the assignment of condition to color counterbalanced. Participants completed 50 trials of one condition before proceeding to the next, for a total of 200 trials.
To prevent participants’ from explicitly memorizing a particular value of reward feedback for a given response time, we also added a small amount of random uniform noise (+/− 5 points) to the reward magnitudes on each trial.
Analysis
General linear models were used for all statistical analysis. COMT gene dose effects were tested by entering the number of met alleles expressed by each subject as a continuous variable. Behavioral analyses, except where indicated, examined RTs in the last quarter (12 trials) of each condition, by which time participants were likely to have learned the reward structure of the particular clock face [28] . (While it is possible to compute learning from the first to last quarter of each condition, some participants learned to discriminate reward structure even in the first quarter, minimizing the difference across quarters. We therefore focus analyses on the last quarter in which performance is expected to stabilize. Further, the model-based analyses converge with those derived from these behavioral measures without confining analysis to any part of the learning curve.) In some analyses the degrees of freedom are one less than they should be because there was a computer crash for one subject who did not complete all conditions.
Model Methods
In all models, we used the Simplex method with multiple starting points to derive best fitting parameters for each individual participant that minimized the sum of squared error (SSE) between predicted and actual RTs across all trials. A single set of parameters was derived for each subject providing the best fit across all task conditions. Data were smoothed with a 5 trial moving average for fitting of sequential time series responses, although similar results were produced without such smoothing, just with larger overall SSE's for all models. Model fits were evaluated with Akaike's Information Criterion, which penalizes model fits for models with additional parameters:
where k is the number of parameters, n is the number of data points to be fit, and SSE is the sum of squared error between the model predictions and actual response times across all trials for each subject. The model with the lowest AIC value is determined to be the best fit.
Exploit Model
There are several ways in which RTs might be modeled in this task. Our first aim was to derive a simple model to approximate the mechanisms embodied within our a priori neural network model of the basal ganglia, which predicted the double dissociation between RTs in the DEV and IEV conditions dependent on dopaminergic medication status in Parkinson's disease [28]. Because that model is complex and involves multiple interacting brain areas, we sought to capture its core computations in abstract form, and to then fit free parameters of this reduced model to individual subject data, which in turn can be linked to striatal dopaminergic genes. A similar procedure was used in a choice rather than response time task [10].
We model the incremental RT changes in the different conditions via separate Go and NoGo parameters that learn from positive and negative prediction errors and serve to speed and slow RTs, respectively. These parameters correspond to D1 and D2-dependent learning in striatonigral and striatopallidal neurons. The terms “Go” and “NoGo” are shorthand descriptions of the functions of the two pathways in the neural model, whereby Go and NoGo activity separately report the learned probability that a given action in the current state would produce a positive and negative outcome, respectively. In choice paradigms, the probability that an action is taken is proportional to the relative (Go – NoGo) activity for that action, as compared to all other actions. Here, as far as the striatum is concerned in the model, there is only one action (“hit the spacebar”), and the relative (Go – NoGo) activity simply determines the speed at which that action is executed.
Positive and negative prediction errors are computed relative to current expected value V, which are then used to update V estimates for subsequent trials, and also to train the Go and NoGo striatal values. This scheme is reminiscent of “actor-critic” reinforcement learning models [5, 34], where the critic is the V system, the prediction errors of which are reflected in phasic dopaminergic signals, and the actor comprises Go and NoGo striatal neuronal populations [16, 28].
The expected value V was initialized to 0 at the beginning of the task. The final V value at the end of each condition was carried over to the beginning of the next, on the assumption that any rewards obtained at the beginning of a condition are compared relative to their best estimate of expected value in the task at large (e.g., 50 points might be interpreted as a positive prediction error if in the last block they had on average obtained 20 points, but would be a negative prediction error if their previous average point value was 100). Go and NoGo values were initialized to 0 and accumulated as a function of reward prediction errors for each state (clock face). [Although the Go and NoGo terms accumulate monotonically as a function of experience, in the neural model Go synapses are weakened following negative prediction errors and NoGo synapses are strengthened, preventing these values from saturating. Here the contributions of Go and NoGo terms were small enough for this to not be necessary; however adding a decay term to Go/NoGo values to prevent increases without bound did not change the basic pattern of results.] Finally, due to model degeneracy, α was held constant and was set to 0.1 to allow integration of history, allowing other Go/NoGo learning parameters to vary freely. This same critic learning rate was used in the neural network implementation [28].
Bayesian integration of expected value
The Go and NoGo learning mechanisms capture a relatively automatic process in which the striatum speeds/slows responses after positive/negative prediction errors, independent of the RTs that produced those reinforcements. This mechanism may result from the architecture of the basal ganglia, which supports approach and avoidance behavior for positive and negative outcomes. This mechanism is also adaptive in the current task if participants’ initial responses are faster than the midpoint (as was typically the case), in which case positive prediction errors predominate in DEV and negative prediction errors predominate in IEV, leading to speeding and slowing respectively. The improved behavioral fit (including penalty for additional parameters) provided by including these mechanisms suggests that these tendencies capture some of the variance in this task. However, note that these mechanisms are not necessarily adaptive in all cases: for example, slow responses that produce positive prediction errors (e.g., in IEV) would lead to subsequent speeding according to this mechanism.
We posited that in addition to Go/NoGo learning, subjects would attempt to explicitly keep track of the rewards experienced for different responses and then produce those responses that had been rewarded most. It is unrealistic to assume that participants track reward structure for all possible response times. Instead, we employed a simplifying (and perhaps more plausible) assumption that participants simply track reward structure for responses categorized as “fast” or “slow”. Given that the reward functions are monotonic (and assuming subjects believe this to be the case), one only needs to track rewards separately for fast and slow responses to determine which has the highest expected value, and to respond faster or slower in proportion to the difference in these values.
We thus categorized each response depending on whether it was faster or slower than the participants local mean RTavg , which was itself tracked with the delta rule:
(This choice for tracking average RT was not critical; all results are similar even if simply defining fast and slow according to the first and second halves of the clock. However using an adaptive local mean RT is more general, and may prove useful if the reward functions are non-monotonic.)
We represented participants’ beliefs about reward structure for these two response categories in Bayesian terms, assuming participants represent not only a single value of each response but rather a distribution of such values, and crucially, the uncertainty about them [39]. In particular, we posited that participants would track the estimated likelihood of obtaining a positive reward prediction error for each response, or the magnitude of such prediction errors, as a function of past set of dopamine bursts reported by midbrain dopamine neurons. Any probability distribution in the exponential family of distributions can be represented in a population of spiking neurons [48, 49], so a priori it is not clear whether it is more plausible for participants to track simply the probability of a dopamine burst occurring at all, or to instead represent the magnitude of the typical prediction error. Model fits to data were clearly superior for probability simulations, which we focus on here; nevertheless, as reported below, all genetic findings hold when modeling reward magnitudes (or reward prediction error magnitudes), with a Kalman filter.
We represented the likelihood of reward prediction errors for each state s and fast or slow action a as beta distributions Beta(ηs,a,βs,a) (see below). The probability of a reward prediction error can be represented as a binomial process , and the beta distribution is the conjugate prior to the binomial distribution. This implies that the application of Bayes rule to update the prior distribution results in a posterior distribution that is itself also a beta distribution with new parameters. [Strictly speaking, a binomial process assumes that each observation is independent. This assumption is violated in the case of reward prediction errors because a given reward value may be interpreted as a positive or negative prediction error depending on prior reinforcement context. The beta distribution is nevertheless a simplifying assumption that provided a substantial improvement to behavioral fit. Furthermore, we also modeled a version in which we track the probability of obtaining a non-zero reward, rather than a reward prediction error. In this model, we also binarized responses such that “fast” and “slow” responses were categorized according to those that were in the first and second halves of the clock. In this case, each observation is indeed independent, and all core results continued to hold.]
The probability density function of the beta distribution is as follows:
where the integral in the denominator is the beta function B(η,β) and is a normalization factor that ensures that the area under the density function is always 1. The defining parameters of the posterior distribution for each state s are calculated after each outcome using Bayes’ rule:
Explore Model
Due to the conjugate prior relationship between binomial and beta distributions, this update is trivial without having to directly compute Bayes’ equation above. The η and β parameters are updated for each state/action by simply incrementing the prior η and β hyperparameters after each instance of a positive or negative prediction error, respectively (see Figure S4 for trajectories of hyperparameters for a single subject.)
The participant can then compare the means of each posterior distribution and adjust RTs so as to increase the probability of obtaining a reward prediction error. The mean of the beta distribution is simply μ = η / (η + β). Thus this component of the exploitation model predicts that subjects adjust RTs according to ρ[μslow(s,t) – μfast(s,t)], where ρ is a free parameter scaling the degree to which participants utilize these mean estimates in adapting their RTs.
In addition to the Go/NoGo learning and Bayesian integration mechanisms, model fits to data were also substantially improved by a mechanism in which participants adapted RTs toward that which had produced the single largest reward thus far (“going for gold”), regardless of the reward probability. This tendency was captured by free parameter ν, and was not associated with any genotype (nor was it required for the core results of the paper to hold, but may be useful for future studies of the neural and genetic mechanisms of this behavior). We modeled this by keeping track of the RT that yielded rewards that were at least one standard deviation greater than all rewards observed thus far in the block, and adapting all subsequent RTs toward this value. Further, participants’ response on one trial may be heavily influenced by that of the previous trial, independent of value. Accordingly we introduce a parameter λ to capture individual differences in this influence of previous responses.
Thus the full RT model is as follows:
The computations of the final Explore term is discussed next.
One of the central advantages of the Bayesian framework is that it provides an estimate not only of the “best guess” (the mean, or expected value μ of the beta distribution), but also the uncertainty about that mean, quantified by the standard deviation σ of that distribution. We attempted to predict RT swings from one trial to the next, hypothesizing that RT swings reflect exploration when participants are uncertain about whether they might obtain better outcomes. The standard deviation of the beta distributions for each state (clock-face) can be computed analytically in each trial as a measure of uncertainty:
The model Explore term was applied on each trial as a function of the relative differences in uncertainty about the likelihood of reward prediction errors given fast and slow responses:
In this way exploratory-based RT swings are predicted to occur in the direction of greater uncertainty (thereby acting to reduce this uncertainty). Note that for trials immediately following an exploratory RT swing, as it stands this implementation would roughly double-count exploration, because the λ parameter already reflects autocorrelation between the previous and current RT (where in this case the previous trial was an exploratory swing). To mitigate against this double-counting, we set the Explore term to 0 in trials immediately following an exploratory RT swing (defined as a change in RT that was in the same direction predicted by the uncertainty Explore term). The results were not sensitive to this particular implementation, however. [For example, similar findings were found without resetting Explore to 0, but instead including a parameter into the RT estimate that reflects the effects of previous RT swings from trial n-2 to n-1 (in addition to λ which accounts for the raw RT in trial n-1). This additional parameter was negative, such that a large RT swing in trial n-1 was predictive of a swing in the opposite direction in trial n. In this model, without resetting Explore, all genetic findings remained significant, including the COMT gene-dose Explore effect; p=.01.]
A number of models of RT swings were compared in an effort to determine whether COMT effects were specific to uncertainty.
Sutton (1990) Exploration Bonus
In this model, exploration is increasingly encouraged for options that had not been explored for several trials. Specifically, exploration is predicted to increase with the square-root of the number of trials since making that choice, scaled by free parameter ζ:
“Lose-Switch” model
In this model, RT swings are predicted to occur after negative prediction errors, such that participants switch to a slower response if the previous response was fast and vice-versa. The degree of adaptation was scaled by free parameter κ.
“Regression to the mean” model
Here responses are predicted to speed/slow as a function of whether the previous response was faster or slower than the local mean, regardless of the outcome. The degree of adaptation was scaled by free parameter ξ.
where RT'(s,t) is the new RT prediction including regression to the mean.
“Reverse-momentum” model
This model attempts to capture periodic changes in RT whereby subjects reverse the direction of their responses if they had progressively sped up or slowed down over the last number of trials. The degree of RT adjustment was predicted to linearly increase with the number of preceding responses that had been progressively speeded/slowed, and scaled by a free parameter γ. Further, this RT reversal was only predicted to occur if the number of progressively speeded/slowed responses exceeded a minimum threshold θ, also a free parameter (this parameter allows for variability in the period of RT swings and was required for the good fits described below).
Model comparison results are presented in the supplement.
Supplementary Material
Acknowledgments
We thank S. Williamson and E. Carter for help with DNA analysis and administering cognitive tasks to participants, and N. Daw, P. Dayan, and R. O'Reilly for helpful discussions. This research was supported by US National Institutes of Mental Health grant R01 MH080066-01.
Footnotes
Author Contributions
M.J.F., B.B.D. and F.M. designed the study; M.J.F. conducted the modeling and analyzed the behavioral data; B.B.D. collected data; J.O.-T. and F.M. extracted the DNA and conducted genotyping; M.J.F., B.B.D. and F.M. wrote the manuscript.
References
- 1.Scheres A, Sanfey AG. Behavioral and brain functions. 2006;2:35. doi: 10.1186/1744-9081-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu M, Bhatt M, Adolphs R, Tranel D, Camerer CF. Science (New York, N. 2005;310:1680. doi: 10.1126/science.1115327. [DOI] [PubMed] [Google Scholar]
- 3.Frank MJ, Woroch BS, Curran T. Neuron. 2005;47:495. doi: 10.1016/j.neuron.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Gittins JC, Jones D. Progress in Statistics. North Holland: 1974. [Google Scholar]
- 5.Sutton RS, Barto AG. Reinforcement Learning: An Introduction. MIT Press; Cambridge, MA: 1998. [Google Scholar]
- 6.Daw ND, O'Doherty JP, Dayan P, Seymour B, Dolan RJ. Nature. 2006;441:876. doi: 10.1038/nature04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen JD, McClure SM, Yu AJ. Philos Trans R Soc Lond B Biol Sci. 2007;362:933. doi: 10.1098/rstb.2007.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Depue RA, Collins PF. The Behavioral and brain sciences. 2001;22:491. doi: 10.1017/s0140525x99002046. [DOI] [PubMed] [Google Scholar]
- 9.Meyer-Lindenberg A, et al. The Journal of clinical investigation. 2007;117:672. doi: 10.1172/JCI30413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank MJ, Moustafa AA, Haughey HM, Curran T, Hutchison KE. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16311. doi: 10.1073/pnas.0706111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein TA, et al. Science (New York, N. 2007;318:1642. [Google Scholar]
- 12.Ouimet CC, Miller PE, Hemmings HC, Walaas SI, Greengard P. The Journal of neuroscience. 1984;4:111. doi: 10.1523/JNEUROSCI.04-01-00111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stipanovich A, et al. Nature. 2008;453:879. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calabresi P, et al. The Journal of neuroscience. 2000;20:8443. doi: 10.1523/JNEUROSCI.20-22-08443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Montague PR, Dayan P, Sejnowski TJ. The Journal of Neuroscience. 1997;16:1936. doi: 10.1523/JNEUROSCI.16-05-01936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank MJ. Journal of Cognitive Neuroscience. 2005;17:51. doi: 10.1162/0898929052880093. [DOI] [PubMed] [Google Scholar]
- 17.Shen W, Flajolet M, Greengard P, Surmeier DJ. Science (New York, N. 2008;321:848. doi: 10.1126/science.1160575. 10.1126/science.1160575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graybiel AM. Annual review of neuroscience. 2008;31:563. doi: 10.1146/annurev.neuro.29.051605.112851. [DOI] [PubMed] [Google Scholar]
- 19.Kakade S, Dayan P. Neural Networks. 2002;15:549. doi: 10.1016/s0893-6080(02)00048-5. [DOI] [PubMed] [Google Scholar]
- 20.Yoshida W, Ishii S. Neuron. 2006;50:781. doi: 10.1016/j.neuron.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Frank MJ, Claus ED. Psychological review. 2006;113:300. doi: 10.1037/0033-295X.113.2.300. [DOI] [PubMed] [Google Scholar]
- 22.Roesch MR, Olson CR. Science (New York, N. 2004;304:307. doi: 10.1126/science.1093223. [DOI] [PubMed] [Google Scholar]
- 23.Rudebeck PH, Walton ME, Smyth AN, Bannerman DM, Rushworth MFS. Nature neuroscience. 2006;9:1161. doi: 10.1038/nn1756. [DOI] [PubMed] [Google Scholar]
- 24.Meyer-Lindenberg A, et al. Nature neuroscience. 2005;8:594. doi: 10.1038/nn1438. [DOI] [PubMed] [Google Scholar]
- 25.Slifstein M, et al. Molecular psychiatry. 2008;13:821. doi: 10.1038/mp.2008.19. [DOI] [PubMed] [Google Scholar]
- 26.Gogos JA, et al. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9991. doi: 10.1073/pnas.95.17.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forbes EE, et al. Molecular psychiatry. 2008;14 doi: 10.1038/sj.mp.4002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moustafa AA, Cohen MX, Sherman SJ, Frank MJ. Journal of Neuroscience. 2008;28:12294. doi: 10.1523/JNEUROSCI.3116-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirvonen M, Laakso K, abd Nagren A, Rinne J, Pohjalainen T, Hietala J. Molecular Psychiatry. 2005;10:889. doi: 10.1038/sj.mp.4001561. [DOI] [PubMed] [Google Scholar]
- 30.Frank MJ, Seeberger LC, O'Reilly RC. Science. 2004;306:1940. doi: 10.1126/science.1102941. [DOI] [PubMed] [Google Scholar]
- 31.Santesso D, Evins A, Frank M, Cowman E, Pizzagalli D. Human Brain Mapping. doi: 10.1002/hbm.20642. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiecki TV, Riedinger K, Meyerhofer A, Schmidt WJ, Frank MJ. Psychopharmacology. doi: 10.1007/s00213-008-1457-4. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayer HM, Glimcher PW. Neuron. 2005;47:129. doi: 10.1016/j.neuron.2005.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Doherty J, et al. Science (New York, N. 2004;304:452. doi: 10.1126/science.1094285. [DOI] [PubMed] [Google Scholar]
- 35.O'Reilly RC, Frank MJ, Hazy TE, Watz B. Behavioral Neuroscience. 2007;121:31. doi: 10.1037/0735-7044.121.1.31. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura K, Hikosaka O. The Journal of neuroscience. 2006;26:5360. doi: 10.1523/JNEUROSCI.4853-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutton RS. In: Porter BW, Mooney RJ, editors. Proceedings of the Seventh International Conference on Machine Learning; Morgan Kaufmann, Palo Alto, CA. 1990. pp. 216–224. [Google Scholar]
- 38.Dayan P, Sejnowski TJ. Machine Learning. 1996;25:5. [Google Scholar]
- 39.Daw ND, Niv Y, Dayan P. Nature neuroscience. 2005;8:1704. doi: 10.1038/nn1560. [DOI] [PubMed] [Google Scholar]
- 40.Niv Y, Daw ND, Joel D, Dayan P. Psychopharmacology. 2007;191:507. doi: 10.1007/s00213-006-0502-4. [DOI] [PubMed] [Google Scholar]
- 41.Dalley JW, et al. Proc Natl Acad Sci U S A. 2005;102:6189. doi: 10.1073/pnas.0502080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20552. doi: 10.1073/pnas.0707106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hollerman JR, Schultz W. Nature Neuroscience. 1998;1:304. doi: 10.1038/1124. [DOI] [PubMed] [Google Scholar]
- 44.Satoh T, Nakai S, Sato T, Kimura M. Journal of Neuroscience. 2003;23:9913. doi: 10.1523/JNEUROSCI.23-30-09913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bayer HM, Lau B, Glimcher PW. J Neurophysiol. 2007;98:1428. doi: 10.1152/jn.01140.2006. [DOI] [PubMed] [Google Scholar]
- 46.Dalley JW, et al. Science (New York, N. 2007;315:1267. [Google Scholar]
- 47.Belin D, Mar AC, Dalley JW, Robbins TW, Everitt BJ. Science (New York, N. 2008;320:1352. doi: 10.1126/science.1158136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zemel RS, Dayan P, Pouget A. Neural computation. 1998;10:403. doi: 10.1162/089976698300017818. [DOI] [PubMed] [Google Scholar]
- 49.Ma WJ, Beck JM, Latham PE, Pouget A. Nature neuroscience. 2006;9:1432. doi: 10.1038/nn1790. [DOI] [PubMed] [Google Scholar]
- 50.Ye S, Dhillon S, Ke X, Collins AR, Day IN. Nucleic acids research. 2001;29:e88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.