Abstract
Background
Cell-to-cell communication (quorum sensing (QS)) co-ordinates bacterial behaviour at a population level. Consequently the behaviour of a natural multi-species community is likely to depend at least in part on co-existing QS and quorum quenching (QQ) activities. Here we sought to discover novel N-acylhomoserine lactone (AHL)-dependent QS and QQ strains by investigating a bacterial community associated with the rhizosphere of ginger (Zingiber officinale) growing in the Malaysian rainforest.
Results
By using a basal growth medium containing N-(3-oxohexanoyl)homoserine lactone (3-oxo-C6-HSL) as the sole source of carbon and nitrogen, the ginger rhizosphere associated bacteria were enriched for strains with AHL-degrading capabilities. Three isolates belonging to the genera Acinetobacter (GG2), Burkholderia (GG4) and Klebsiella (Se14) were identified and selected for further study. Strains GG2 and Se14 exhibited the broadest spectrum of AHL-degrading activities via lactonolysis while GG4 reduced 3-oxo-AHLs to the corresponding 3-hydroxy compounds. In GG2 and GG4, QQ was found to co-exist with AHL-dependent QS and GG2 was shown to inactivate both self-generated and exogenously supplied AHLs. GG2, GG4 and Se14 were each able to attenuate virulence factor production in both human and plant pathogens.
Conclusions
Collectively our data show that ginger rhizosphere bacteria which make and degrade a wide range of AHLs are likely to play a collective role in determining the QS-dependent phenotype of a polymicrobial community.
Background
Bacteria employ sophisticated cell-to-cell communication networks which instigate population-wide behavioural changes in response to environment stimuli. Such population-dependent adaptive behaviour results in altered gene expression in response to the production and sensing of chemical information in the form of diffusible signal molecules, commonly referred to as autoinducers. The process, whereby an increase in the concentration of signal molecule(s) in the extracellular milieu reflects cell population density is called 'quorum sensing' (QS). At a threshold concentration of the QS signal molecule (when the population is considered to be 'quorate'), the target genes are induced or repressed. In different bacterial genera, these may include genes which code for the production of secondary metabolites, plasmid transfer, motility, virulence, and biofilm development (for reviews see [1,2]).
In many Gram-negative bacteria, QS depends on the actions of N-acylhomoserine lactone (AHL) signal molecules [1,2]. These consist of a homoserine lactone ring linked via a saturated or unsaturated acyl chain (generally between 4 and 18 carbons) and without or with a keto or hydroxy substituent at the C3-position (for reviews see [1,2]). AHL biosynthesis primarily depends on the actions of enzymes belonging to the LuxI or LuxM protein families while the response to an AHL is usually driven by the interaction between the signal molecule and a member of the LuxR protein family of response regulators [1,2].
Since QS controls a range of biological functions associated with virulence and as the emergence of multi-antibiotic resistant bacterial strains is in the ascendency, there is increasing pressure to discover novel therapeutic approaches to combat bacterial infections [3,4]. Interrupting QS may represent one such method which has the added advantage that the targets are not normally essential for bacterial survival and therefore are not subject to the same selective pressures observed for conventional growth inhibitory antimicrobials [3-6].
Quorum quenching (QQ) refers to the process in which QS is disrupted. QQ can be achieved in several ways such as through the enzymatic destruction of QS signal molecules, the development of antibodies to QS signal molecules or via agents which block QS. In this context both the QS signal synthase and sensor or response regulator proteins are the primary targets [3-6].
Under alkaline conditions AHLs are rapidly inactivated by pH-dependent lactonolysis in which the homoserine lactone ring is hydrolysed to the ring open form (i.e. the corresponding N-acylhomoserine compound) in a reaction which can be reversed by acidification [7,8]. This reaction can also be driven enzymatically by lactonases such as AiiA, AttM, AiiB [9,10] and AhlD [11]. There is also a second class of AHL-degrading enzymes which are amidases/acylases such as AiiD [12] and PvdQ [13] which cleave the AHL amide bond releasing homoserine lactone and the corresponding fatty acid.
The ability to inactivate AHLs enzymatically is shared by diverse bacteria belonging to the α-Proteobacteria including Agrobacterium, Sphingomonas, Sphingopyxis and Bosea, the β-Proteobacteria such as Variovorax, Ralstonia and Comamonas, the γ-Proteobacteria including Pseudomonas and Acinetobacter, Firmicutes such as Bacillus and Actinobacteria such as Rhodococcus as well as the Streptomyces sp. (reviewed by Uroz et al [6]).
Since QS often controls virulence in both plant and animal pathogens [1,2], QQ bacteria have potential as biocontrol agents which protect plants from pathogens while novel AHL-inactivating enzymes may have utility as therapeutic agents [6]. Consequently, we have been exploring novel rhizosphere environments for bacterial communities displaying both AHL-dependent QS and AHL-degrading activities. Since both beneficial rhizosphere bacteria and pathogens may use the same or very similar AHLs, it is important that QQ directed toward the latter do not perturb the former [6]. Hence the identification of strains expressing highly specific QQ enzymes would have significant utility. Here we focus on the AHL-inactivating activities of a community of bacteria associated with the roots of Zingiber officinale (ginger) growing in the Malaysian rainforest. Three AHL-inactivating bacteria belonging to the genera Acinetobacter, Burkholderia and Klebsiella were identified and isolated using an enrichment assay employing N-(3-oxohexanoyl)homoserine lactone (3-oxo-C6-HSL) as the sole nitrogen and carbon source. While the Acinetobacter and Klebsiella strains both exhibited broad spectrum lactonase activity, the Burkholderia strain reduced 3-oxo-AHLs to the corresponding 3-hydroxy compounds. The Acinetobacter and Burkholderia strains both possessed AHL-producing and AHL-inactivating activities and in co-culture experiments we show that all three ginger rhizosphere isolates are capable of reducing the production of key P. aeruginosa QS-dependent virulence determinants and Erwinia carotovora-mediated tissue damage in a potato tuber infection model.
Results
Selection of QQ bacteria from ginger rhizosphere
To enrich for rhizosphere-associated bacteria with AHL-degrading capabilities, a ginger rhizosphere suspension was used to inoculate a basal medium containing 3-oxo-C6-HSL as the sole source of carbon and nitrogen [14]. Bacterial growth was evident within 48 h but only in the samples containing 3-oxo-C6-HSL (data not shown). The enrichment culture was plated onto solidified basal KG medium [14] containing 3-oxo-C6-HSL which was passaged for single colonies which were subcultured on LB agar. Seven ginger rhizosphere-associated bacteria with four distinctive morphotypes (GG1, GG2, GG3, GG4, GG5, GGp and Se14) were chosen for further study.
The ginger rhizosphere strains were identified by 16S rDNA sequencing and analysis of the aligned sequences (1498 nucleotides) was performed by web-based similarity searches against the GenBank database. The strains were identified as Acinetobacter spp. (GG2 and GG3), Burkholderia spp. (GG1 and GG4), Klebsiella sp. (GG5 and Se14) and Microbacterium sp. (GGp). Since the 16S rDNA sequence data indicated that GG1, GG3 and GG5 are very closely related to GG4, GG2 and Se14 respectively, we chose to focus on GG2, GG4 and Se14. GGp was also omitted from further investigation. The GG2 16S rDNA sequence showed 99% identity with Acinetobacter spp. and clustered phylogenetically with Acinetobacter calcoaceticus [GenBank Accession Number EF432578] and a poorly characterized Acinetobacter sp. [GenBank Accession Number DQ366106]). The GG4 16S rDNA shared 99% sequence identity with Burkholderia cepacia PRE5 [GenBank Accession Number AY946011) while Se14 is most closely related to Klebsiella species PN2 [GenBank Accession Number AY946011]. The accession numbers for the 16S rDNA sequences of Acinetobacter sp. (GG2) [GenBank: GQ245971], Burkholderia sp. (GG4) [GenBank: HQ728437] and Klebsiella sp. (Se14) [GenBank: HQ728438] have been deposited with GenBank.
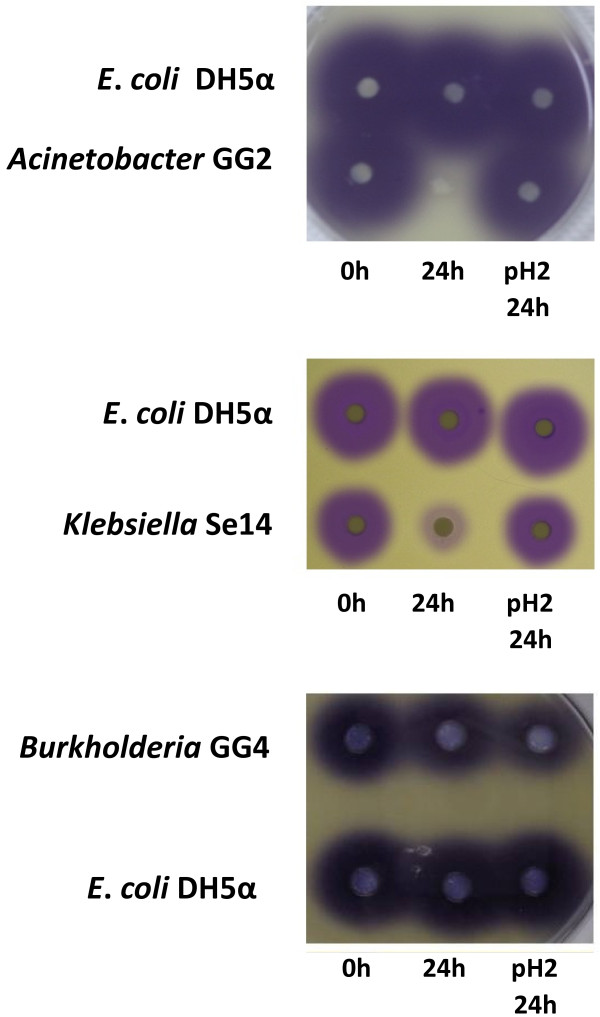
The 3-oxo-C6-HSL-inactivating activity of each strain was assessed, and Figure 1 shows the lack of any residual 3-oxo-C6-HSL after incubation with GG2 or with Se14 for 24 h. However, 3-oxo-C6-HSL was still detected after incubation with GG4 cells for 24 h (Figure 1).
Figure 1.
3-oxo-C6-HSL degradation by Acinetobacter GG2, Burkholderia GG4 and Klebsiella Se14 quorum quenching bacteria isolated from the ginger rhizosphere. Each rhizosphere bacterium or E. coli DH5α was incubated with 3-oxo-C6-HSL for 0, 24 h after which the cell culture supernatants were either spotted directly onto paper disks or acidified to pH 2 for 24 h to recyclize any ring opened 3-oxo-C6-HSL before spotting onto paper disks. These were placed on an agar plate overlaid with C. violaceum CV026 and incubated. A purple halo indicates the presence of 3-oxo-C6-HSL.
Characterization of QQ Activities of Acinetobacter GG2, Burkholderia GG4 and Klebsiella Se14
To determine the range of AHLs inactivated by each of the three ginger rhizosphere isolates, whole cells resuspended in PBS buffer were incubated for up to 96 h with a range of AHLs differing in acyl chain length (C4-C14), the presence or absence of a C3 substituent (oxo or hydroxy) or with a series of 3-hydroxy-C14-HSLs with a double bond at either C9, C10, C11 or C13 (Table 1). After incubation, any remaining AHLs were detected using the appropriate AHL biosensor as described in the Methods section and compared with Escherichia coli DH5α and PBS as negative controls. The data obtained are summarized in Table 1. Using biosensor assays Klebsiella Se14 inactivated all of the AHLs tested while Acinetobacter GG2 showed broad activity but was most effective against the long chain unsubstituted or 3-hydroxy substituted saturated or unsaturated acyl chain-AHLs (Table 1). Burkholderia GG4 exhibited no apparent activity against the AHLs using these biosensor assays (data not shown).
Table 1.
AHLs degraded by GG2 and Se14
| Types of AHL tested | AHL-degradation pattern | |
|---|---|---|
| GG2 | Se14 | |
| C4-HSL | + | + + + |
| C5-HSL | + + | + + + |
| C6-HSL | + + | + + + |
| C7-HSL | + + | + + + |
| C8-HSL | + + | + + + |
| C9-HSL | + + + | + + + |
| C10-HSL | + + + | + + + |
| C11-HSL | + + + | + + + |
| C12-HSL | + + + | + + + |
| C14-HSL | + + + | + + + |
| 3-hydroxy-C4-HSL | + + | + + + |
| 3-hydroxy-C6-HSL | + + | + + + |
| 3-hydroxy-C8-HSL | + + | + + + |
| 3-hydroxy-C10-HSL | + + + | + + + |
| 3-hydroxy-C12-HSL | + + + | + + + |
| 3-hydroxy-C14-HSL | + + + | + + + |
| 3-oxo-C8-HSL | + + | + + + |
| 3-oxo-C10-HSL | + + | + + + |
| 3-oxo-C12-HSL | + + | + + + |
| 3-oxo-C14-HSL | + + | + + + |
| Δ9-3-hydroxy-C14-HSL | + + + | + + + |
| Δ10-3-hydroxy-C14-HSL | + + + | + + + |
| Δ11-3-hydroxy-C14-HSL | + + + | + + + |
| Δ13-3-hydroxy-C14-HSL | + + + | + + + |
Degradation of AHLs by GG2 and Se14. Degradation of AHL: +  : weak; ++
: weak; ++  : moderate; + + +
: moderate; + + +  : significant. Insets denote the digital image of AHLs detected using the biosensors E. coli [pSB401] and/or E. coli [pSB1075]; evaluated according to the reduction in bioluminescence. All experiments took into account the detection limit of the biosensors used for each AHL-degradation assay.
: significant. Insets denote the digital image of AHLs detected using the biosensors E. coli [pSB401] and/or E. coli [pSB1075]; evaluated according to the reduction in bioluminescence. All experiments took into account the detection limit of the biosensors used for each AHL-degradation assay.
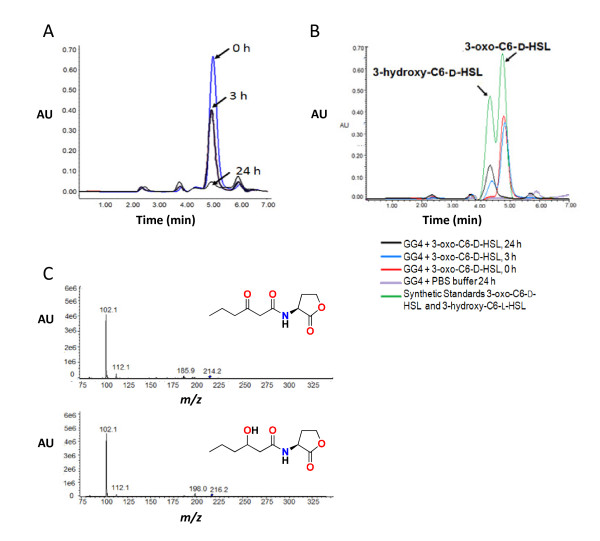
Since natural AHLs are in the L-configuration, we sought to determine whether the AHL inactivating activities observed were stereospecific. After incubation of GG2 and Se14 whole cells with the D-isomer of 3-oxo-C6-HSL (3-oxo-C6-D-HSL), the reaction mixture was extracted and analysed by HPLC rather than using the AHL biosensors which do not respond to D-isomers. For GG2 and Se14 the peak corresponding to 3-oxo-C6-D-HSL was reduced after 3 h incubation and effectively absent after 24 h. The data for Acinetobacter strain GG2 are shown in Figure 2A. Similar results were obtained for Se14 (data not shown) indicating that AHL inactivation by these two ginger rhizosphere bacteria is not stereospecific. Although the biosensor assays did not demonstrate any AHL inactivation for Burkholderia GG4, HPLC analysis revealed that the peak corresponding to 3-oxo-C6-D-HSL was reduced with the concomitant appearance of a new peak (Figure 2B) suggesting that GG4 was capable of modifying AHLs.
Figure 2.
HPLC analysis of the degradation of 3-oxo-C6-D-HSL after incubation with Acinetobacter GG2 and Burkholderia GG4. (A) The D-isomer of 3-oxo-C6-HSL was incubated for 0- (blue line), 3- (black line) and 24 h (grey line) with GG2, the culture supernatant extracted with ethyl acetate and subjected to HPLC analysis. The data show the disappearance of the AHL peak at 5.0 min after 24 h incubation. (B) When incubated with GG4 over a period from 0- (red line), 3- (blue line) and 24 h (black line), the 3-oxo-C6-D-HSL peak is replaced by a new peak at about 4.3 min which co-migrates with 3-hydroxy-C6-HSL. The controls used were synthetic 3-oxo-C6-D-HSL, 3-hydroxy-C6-D-HSL (green line) and PBS buffer incubated with GG4 for 24 h to ensure no 3-hydroxy-C6-HSL production by GG4 (purple line). (C) MS showing the presence of 3-oxo-C6-HSL at 0 h (upper panel; m/z 214.2 [M+H]) and 3-hydroxy-C6-HSL after 24 h (lower panel; m/z 216.2 [M+H]) when 3-oxo-C6-L-HSL was incubated with GG4.
Identification of the AHL degradation products
To determine whether Acinetobacter strain GG2 inactivated AHLs through cleavage of the acyl chain or via lactonolysis or both, 3-oxo-C6-HSL was first incubated with GG2 cells for 24 h. The cells were removed and the supernatant was collected, acidified to pH 2 and incubated for a further 24 h. This results in the pH-mediated re-cyclization of any ring opened compound present [8] which was subsequently detected using the C. violaceum CV026 AHL biosensor [15]. Figure 1 shows that while no 3-oxo-C6-HSL was detected in the supernatant after 24 h incubation with GG2, it could be recovered by acidification indicating that GG2 possesses lactonase activity. To investigate whether GG2 also exhibits amidase activity a cell-free GG2 24 h culture supernatant grown in the presence of 3-oxo-C6-HSL was treated with dansyl chloride which reacts with the exposed free amine of the homoserine lactone ring following release of the AHL acyl chain [16]. No dansylated homoserine lactone was detected indicating that GG2 does not exhibit acylase activity (data not shown).
Similar acidification experiments to those described above for Acinetobacter GG2 were carried out for Klebsiella Se14. These showed that Se14 also possesses a lactonase. Since Klebsiella pneumoniae has previously been reported [11] to possess a homologue of the Arthrobacter lactonase gene ahlD termed ahlK, we used primers based on ahlK to determine whether the gene was also present in Se14. A single PCR product was obtained and sequenced and found to be identical to the ahlK gene (data not shown). When Se14 ahlK was expressed in E. coli DH5α, AHL biosensor assays revealed that there was a reduction in bioluminescence consistent with a decrease in 3-oxo-C6-HSL when compared to the vector control. Upon acidification of the supernatant AHL biosensor activity could be restored thus confirming that AhlK has lactonase activity (data not shown).
When Burkholderia strain GG4 was incubated with 3-oxo-C6-D-HSL for 3 h and examined by HPLC, we noted the appearance of a new peak (retention time 4.3 min) that was absent when either GG2 or Se14 was incubated with the same D-isomer (retention time 4.8 min) (Figure 2B). Similar results were obtained following incubation of the natural L-isomer of 3-oxo-C6-HSL with GG4 and the new peak was found to co-migrate with the L-isomer of 3-hydroxy-C6-HSL (data not shown) suggesting that GG4 has oxido-reductase activity towards 3-oxo-AHLs. To confirm the oxido-reductase activity of GG4, 3-oxo-C6-L-HSL incubated with GG4 for 0 h and 24 h was analysed by mass spectrometry and the appearance of 3-hydroxy-C6-HSL was confirmed by detection of the precursor ion (m/z 216.2 ([M+H])) and fragment ions of m/z 198.0 ([M+H-H2O]) and 102.0 (Figure 2C) which are characteristic of 3-hydroxy-AHLs which readily lose a water molecule and the homoserine lactone moiety respectively [17,18].
Similar results were obtained on incubation of GG4 with the L-isomers of 3-oxo-C4-HSL or 3-oxo-C8-HSL in that new HPLC peaks co-eluting with 3-hydroxy-C4-HSL and 3-hydroxy-C8-HSL synthetic standards, respectively, were observed after incubation for 6 h (data not shown). This oxido-reductase activity was only apparent when 3-oxo-AHLs were incubated with GG4 whole cells but not cell lysates (data not shown).
Acinetobacter GG2 and Burkholderia GG4 produce AHLs
Since some but not all Acinetobacter and Burkholderia strains have previously been reported to produce AHLs, we wondered whether QQ and QS activities co-exist in GG2, GG4 and Se14. To determine whether any of the three ginger rhizosphere strains produced AHLs, they were first cross-streaked against each of three different AHL biosensors namely C. violaceum CV026, E. coli [pSB401] and E. coli [pSB1075], and the plates examined over time for the induction of violacein or bioluminescence (data not shown). GG2 induced bioluminescence in E. coli [pSB1075] indicating that it was producing long chain AHLs, GG4 activated both CV026 and E. coli [pSB401] indicative of short chain AHL production while Se14 failed to activate any of the three AHL biosensors.
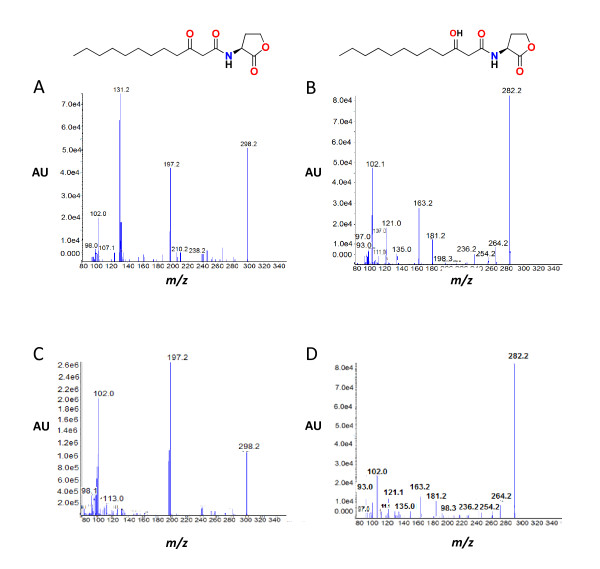
To identify unequivocally the AHLs produced by GG2, spent culture supernatant extracts were analysed by liquid chromatography (LC) coupled to hybrid quadruple-linear ion trap mass spectrometry (LC-MS/MS), which revealed the presence of 3-oxo-C12-HSL and 3-hydroxy-C12-HSL by comparison of their retention times, precursor and principal fragment ions with synthetic standards. Figure 3 shows the fragmentation patterns for 3-oxo-C12-HSL (precursor ion m/z 298.2 [M+H]; fragment ions m/z 197.2, 102.0 (Figure 3A and Figure 3C) and 3-hydroxy-C12-HSL (precursor ion m/z 282.2 [M-H2O]; fragment ions m/z 181.2, 102.1) (Figure 3B and Figure 3D). The mass spectra of the extracted AHLs were similar to the corresponding synthetic compounds. Quantitative analysis by LC-MS/MS of the AHLs produced by GG2 over a 24 h period revealed that 3-hydroxy-C12-HSL was the most abundant AHL produced by GG2 which attains a maximum level after 12 h growth, but is almost undetectable after 24 h (data not shown).
Figure 3.
Mass spectra of the AHLs produced by GG2. Extracts from spent culture supernatants of GG2 were analysed by LC-MS/MS. The fragment ion at m/z 102 is characteristic of the homoserine lactone ring (A and B). By comparison with the corresponding synthetic AHL standards (C and D) the precursor ion of m/z 298.2 and fragment ion of m/z 197.2 demonstrate the presence of 3-oxo-C12-HSL (A) whereas the precursor ion of m/z 282.2 (which corresponds to [M-H2O]) and fragment ion of m/z 181.2 are characteristic for 3-hydroxy-C12-HSL (B). AU: Absorbance unit.
LC-MS/MS analysis of GG4 supernatants confirmed the presence of 3-oxo-C6-HSL (precursor ion m/z 214.2 [M+H]; fragment ions m/z 113.0, 102.0); C8-HSL (precursor ion m/z 228.2 [M+H]; fragment ions m/z 109.1, 102.0), 3-hydroxy-C8-HSL (precursor ion m/z 226.2 [M-H2O]; fragment ions m/z 125.1, 102.0) and C9-HSL (precursor ion m/z 242.2 [M-H2O]; fragment ions m/z 142.2, 102.1) (Additional File 1). The mass spectra of the extracted AHLs were indistinguishable from the corresponding synthetic compounds (Additional File 1).
QQ biocontrol activity of the ginger rhizosphere isolates
To determine whether any of the three ginger rhizosphere bacterial isolates were capable of quenching virulence factor production in human (P. aeruginosa) and plant (Er. carotovora) pathogens which utilize different AHLs, we undertook co-culture experiments. Figure 4A shows that Acinetobacter GG2 and Burkholderia GG4 both reduced elastase production approximately two-fold when compared to the P. aeruginosa PAO1 control whereas the Klebsiella strain Se14 was the most effective, reducing elastase levels about 16-fold. None of the QQ bacteria inhibited the growth of P. aeruginosa which reached a similar viable count in co-culture as was attained in monoculture (data not shown). GG2 and Se14 both effectively reduced the expression of lecA in P. aeruginosa although GG4 had comparatively little effect (Figure 4B).
Figure 4.
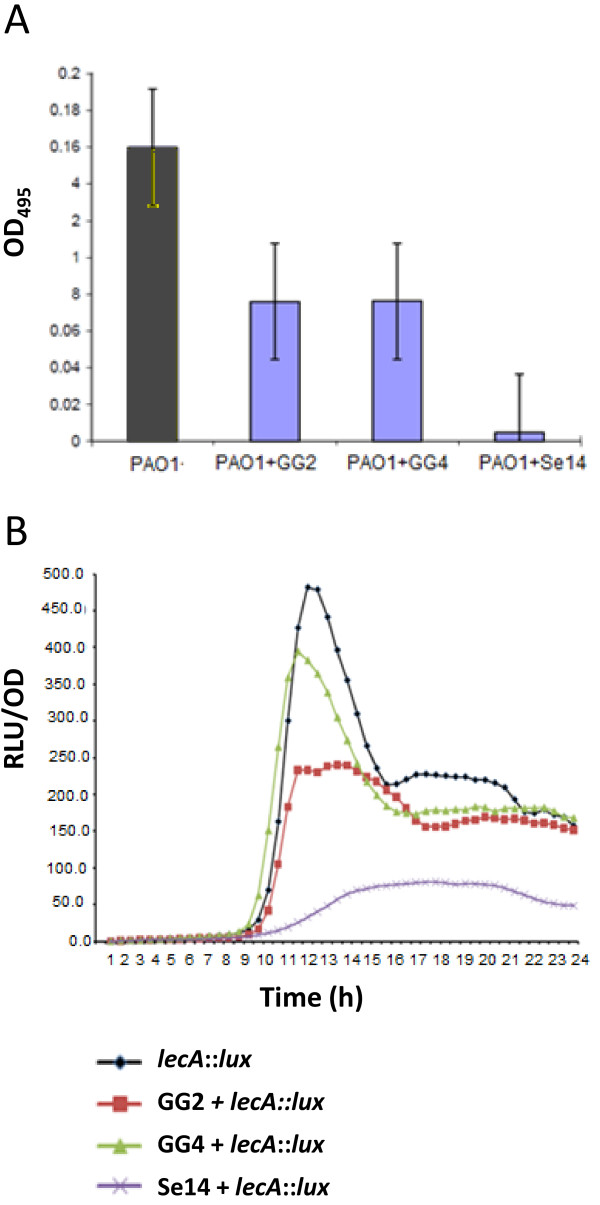
Quenching of elastase production and lecA expression in P. aeruginosa by ginger rhizosphere strains. (A) Elastase production by P. aeruginosa following monoculture (PAO1) or in co-culture with GG2 (PAO1+GG2), GG4 (PAO1+GG4) or Se14 (PAO1+Se14) at a starting inoculum ratio of 1:1 for 24 h. (B) Expression of a lecA::lux fusion following monoculture or co-culture of P. aeruginosa PAO1 with GG2, GG4 or Se14 at a starting inoculum ratio of 1:1 for 24 h. The data are presented as RLU/OD to account for any differences in growth.
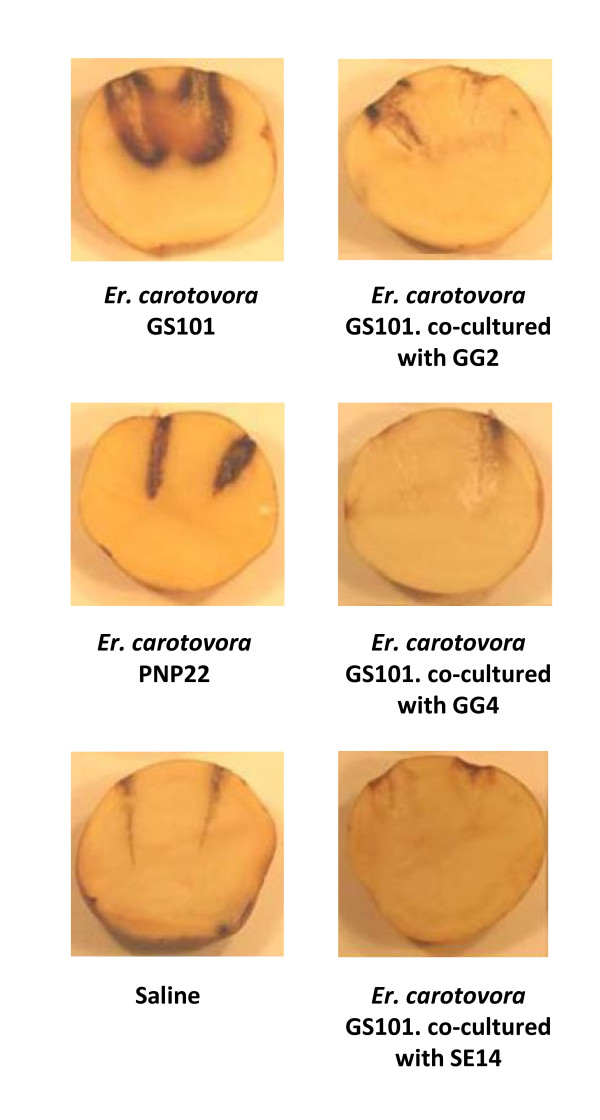
The QQ potential of GG2, GG4 and Se14 for attenuating the 3-oxo-C6-HSL-dependent pectinolytic activity of Er. carotovora was assessed in planta using a potato tuber infection model after viable count experiments confirmed that none of the strains affected the growth of Er. carotovora in co-culture experiments (data not shown). Er. carotovora causes substantial tissue necrosis when injected into the potato tuber but when co-cultured with any of the three ginger rhizosphere isolates, maceration of the potato tissue by the phytopathogen was greatly reduced (Figure 5).
Figure 5.
Quenching of the pectinolytic activity of Er. carotovora by ginger rhizosphere strains. The ability of GG2, GG4 and Se14 to reduce Er. carotovora-mediated soft rot in potato tuber tissues in co-culture was compared with the parent Erwinia strain in monoculture and, as negative controls, either the AHL-negative Er. carotovora mutant PNP22 or saline.
Discussion
In the present work, four bacterial morphotypes from the same ginger rhizosphere bacterial community were isolated and identified as a consequence of their ability to grow on an enrichment medium [14] containing 3-oxo-C6-HSL as the sole carbon and nitrogen source. BLAST search analyses of the 16S rDNA sequences identified the strains as belonging to the genera Acinetobacter, Burkholderia, Klebsiella and Microbacterium. In semi-quantitative whole cell assays, we evaluated the AHL-inactivating spectrum of the three Gram-negative isolates. The broadest range of activity was noted for Klebsiella strain Se14 which inactivated each of the 24 structurally diverse AHLs evaluated including the D-isomer of 3-oxo-C6-HSL. Similarly Acinetobacter strain GG2 exhibited a broad spectrum of activity but was less effective against short chain AHLs. In contrast, Burkholderia GG4 was inactive against the unsubstituted AHLs but was active against the 3-oxo-AHLs. Although AHL-degrading activity has not previously been characterized in the genus Burkholderia, a soil isolate from this genus capable of growing on AHLs as the sole nitrogen but not carbon source was reported by Yang et al [19]. This differs from Burkholderia strain GG4 which did not grow on 3-oxo-C6-HSL as a source of both carbon and nitrogen and probably came through the enrichment process as a consequence of AHL turnover by the other bacteria in the ginger rhizosphere community. Nevertheless, when GG4 was incubated with 3-oxo-C6-HSL in PBS buffer, GG4 reduced this AHL to the corresponding 3-hydroxy compound. Similar results were obtained for 3-oxo-C4-HSL and 3-oxo-C8-HSL as well as the D-isomer of 3-oxo-C6-HSL indicating that the activity was not AHL chain length dependent or stereospecific. This simple reduction of a 3-oxo-AHL to the corresponding 3-hydroxy compound is likely to impact on QQ. For example, in Er. carotovora where carbapenem antibiotic biosynthesis and exoenzyme production are regulated by 3-oxo-C6-HSL, the corresponding 3-hydroxy compound has only 1% of the activity of the 3-oxo-AHL [20]. For P. aeruginosa, the 3-hydroxy-C12-HSL was approximately 8-fold less active than the 3-oxo compound [21]. These data suggest that simple modification of the 3-oxo moiety is likely to substantially reduce the activity of 3-oxo-AHLs and to contribute to the QQ activity within a bacterial community. A similar oxido-reductase activity has been observed for a strain of Rhodococcus erythropolis isolated from the tobacco rhizosphere [22]. In contrast to Burkholderia strain GG4, this Gram positive bacterium (R. erythropolis) was unable to reduce 3-oxo-C6-HSL and required an AHL acyl chain of at least eight carbons [22]. However in common with GG4, the activity was only observed on incubation of 3-oxo-AHLs with whole, live bacterial cells as cell lysates were inactive [22].
For Klebsiella and Acinetobacter, AHL-inactivating activity has previously been noted by Park et al [11] and Kang et al [23], respectively. For the former, an AHL-degrading enzyme (AhlK) related to AhlD from Arthrobacter has been cloned and sequenced and by homology suggested to be a lactonase [11]. Here we have shown that the same gene is conserved in the Klebsiella ginger rhizosphere isolate Se14 and have demonstrated that the recombinant enzyme expressed in E. coli is indeed a lactonase with very broad AHL-inactivating activity including both short and long chain AHLs (with saturated or unsaturated acyl side chains of 4 to 14 carbons). These include N-(3-hydroxy-7-cis-tetradecanoyl)homoserine lactone (3-hydroxy-C14:1-HSL), an AHL which was originally termed the Rhizobium small bacteriocin [24] because it inhibits the growth of Rhizobium leguminosarum strains which carry a 'sensitivity locus' on Sym plasmids such as pRLJ1 [24]. 3-hydroxy-C14:1-HSL is also produced by soil bacteria such as Pseudomonas fluorescens [17].
Acinetobacter GG2 also degraded a wide range of short and long chain AHLs via a lactonase activity although we were unable to identify the gene involved. Although the mechanism of AHL degradation has not previously been determined in this genus, an Acinetobacter strain isolated from cucumber rhizosphere has been reported to degrade both C6-HSL and N-octadecanoyl homoserine lactone (C18-HSL) as well as the AHLs produced by a biocontrol strain of Pseudomonas chlororaphis and a phytopathogenic strain of Burkholderia glumae [23]. Interestingly, Acinetobacter GG2 not only degrades AHLs but also produces AHLs which we identified as 3-hydroxy-C12-HSL (major) and C12-HSL (minor). Previously Niu et al [25] showed that the human nosocomial pathogen, Acinetobacter baumannii, produces 3-hydroxy-C12-HSL and C12-HSL via the LuxI synthase, AbaI, the expression of which is AHL dependent. In A. baumannii, AHL-dependent QS appears to contribute to biofilm development since abaI mutants were less biofilm proficient than the parent strain [25].
Acinetobacter strains isolated from contact lenses have been reported not to produce any AHLs [26] whereas a study of 43 Acinetobacter strains isolated from both hospital patients and the environment and assayed using AHL biosensors showed that most of the strains examined produced AHLs although these were not chemically characterized [27]. Furthermore a comparative genome analysis of three different Acinetobacter strains from three different environments revealed the presence of a luxIR-type locus in a multidrug resistant clinical A. baumannii isolate which was disrupted by an insertion element in a sensitive strain isolated from human body lice but completely absent from a soil isolate [28].
In Acinetobacter GG2, 3-hydroxy-C12-HSL accumulated in the growth medium reaching a maximal level after 12 h before rapidly being degraded. This indicates GG2 tightly controls its own AHL production and turnover and suggests that sustained expression (or repression) of the QS target genes is not required in stationary phase. The coupling of AHL synthesis and degradation in the same bacterium has previously been noted for Agrobacterium tumefaciens which produces and degrades 3-oxo-C8-HSL during early stationary phase via a lactonase encoded by attM which is activated by starvation signals and the stress alarmone (p)ppGpp [29,30]. Similarly, a marine Shewanella strain which produces AHLs in late exponential phase degraded its long chain AHLs in stationary phase via both lactonase and acylase/amidase activities [31]. In polymicrobial biofilms, this Shewanella isolate interfered with AHL production in other bacteria and as a consequence, their ability to enhance the settlement of algal zoospores was compromised [31]. Here, we also found that the ginger rhizosphere Burkholderia isolate GG4 is not only capable of interfering with QS by reducing 3-oxo-AHLs to the corresponding 3-hydroxy compounds but also produces AHLs including 3-oxo-C6-HSL, C9-HSL and 3-hydroxy-C8-HSL. While most Burkholderia strains synthesize C6-HSL and C8-HSL [32,33], 3-hydroxy-C8-HSL production has only been confirmed in the pathogen, Burkholderia mallei [32] and tentatively identified in the environmental non-pathogenic Burkholderia xenovorans [33]. In B. mallei, C8-HSL and 3-hydroxy-C8-HSL are produced by two different AHL synthases (BmaI1 and BmaI3) [32]. In Burkholderia GG4, it remains to be established whether 3-hydroxy-C8-HSL is produced directly via a LuxI-type synthase or is a consequence of the reduction of 3-oxo-C8-HSL.
Bacteria such as GG2, GG4 and Se14 which produce and/or modify/degrade QS signals are likely to have a major impact on the properties of polymicrobial bacterial communities. Here we have shown that the ginger rhizosphere isolates were each capable of reducing virulence factor production in both P. aeruginosa and Er. carotovora. However, GG4 was unable to down-regulate lecA (which codes for the cytotoxic galactophilic lectin A [34]) expression probably as a consequence of its inability to reduce C4-HSL [35] in contrast to elastase which is predominantly LasR/3-oxo-C12-HSL dependent [36].
Conclusions
Three different QQ bacteria associated with the ginger rhizosphere have been isolated and characterized as belonging to the genera Acinetobacter (GG2), Burkholderia (GG4) and Klebsiella (Se14). GG2 and Se14 exhibited the broadest spectrum of AHL degrading activity via lactonolysis while GG4 reduced 3-oxo-AHLs to the corresponding 3-hydroxy compounds. In GG2 and GG4, AHL-dependent QQ co-exists with AHL-dependent QS suggesting that these bacteria are likely to play a major role in determining the QS-dependent phenotype of the polymicrobial community from which they were isolated. This was confirmed by co-culture experiments in which all three rhizosphere bacteria attenuated virulence factor production in both a human and a plant pathogen without inhibiting growth of either pathogen.
Methods
Bacterial strains, growth media and culture conditions
The bacterial strains used in this study are listed in Table 2. Bacteria were routinely grown in Luria Bertani (LB) medium buffered when required with 50 mM 3-[N-morpholino] propanesulfonic acid (MOPS) to pH 6.8 to prevent alkaline hydrolysis of AHLs [8]. For the enrichment of QQ bacteria from the ginger rhizosphere, KG medium supplemented with 3-oxo-C6-HSL (500 μg/ml) was used [14]. C. violaceum CV026, Er. carotovora strains and the rhizosphere isolates were grown at 28°C, E. coli and P. aeruginosa strains at 37°C. When required, the E. coli growth medium was supplemented with ampicillin (100 μg/ml) and tetracycline (5 μg/ml). C. violaceum CV026 required kanamycin (30 μg/ml) and chloramphenicol (30 μg/ml).
Table 2.
Strains used in the study
| Strain | Description | Source/reference |
|---|---|---|
| E. coli | ||
| DH5α | recA endA1 hsdR17 supE4 gyrA96 relA1 Δ(lacZYA-argF)U169 (Φ80dlacZΔM15) | [37] |
| pSB1075 | lasRlasl' (P. aeruginosa PAO1)::luxCDABE (Photorhabdus luminescens [ATCC 29999]) fusion in pUC18 AmpR, AHL biosensor producing bioluminescence | [40] |
| pSB401 | luxRluxl' (Photobacterium fischeri [ATCC 7744])::luxCDABE (Photorhabdus luminescens [ATCC 29999]) fusion; pACYC184-derived, TetR, AHL biosensor producing bioluminescence | [40] |
| C. violaceum | ||
| CV026 | Double mini-Tn5 mutant derived from ATCC 31532, KanR, HgR, cviI::Tn5 xylE, plus spontaneous StrR AHL biosensor, produces violacein pigment only in the presence of exogenous AHL | [15] |
| Er. carotovora | ||
| GS101 | AHL producing Erwinia strain, pectinolytic positive | [44] |
| PNP22 | AHL-synthase mutant | [44] |
| P. aeruginosa | ||
| PAO1 | Prototroph | Lab collection |
| lecA::lux | lecA::luxCDABE genomic reporter fusion in PAO1 | [35] |
| Ginger rhizosphere-associated bacteria | ||
| Acinetobacter GG2 | Ginger rhizosphere-associated bacterium | This study |
| Burkholderia GG4 | Ginger rhizosphere-associated bacterium | This study |
| Klebsiella Se14 | Ginger rhizosphere-associated bacterium | This study |
Enrichment procedures for bacteria degrading AHL from ginger rhizosphere
Ginger roots were collected at the Rimba Ilmu, University of Malaya (Malaysia). Soil was removed by rinsing thoroughly with sterile distilled water after which 1 g of rhizosphere tissue was added to 10 ml of KG medium, vortexed vigorously for 10 min, sonicated for 1 min and finally vortexed for 1 min. After which, 2 ml of this suspension was briefly centrifuged to remove root debris, re-centrifuged at 13,000 × g (5 min) after which the pellet was washed and resuspended in 2 ml of KG medium to give the final rhizosphere suspension. Then, 100 μl of this suspension was inoculated into 3 ml of KG medium containing 3-oxo-C6-HSL (500 μg/ml) and the cells were grown at 28°C with shaking at 220 rpm. After 48 h, a 5% (v/v) transfer was made to fresh, sterile KG medium and subsequent transfers made at 48 h intervals. After six enrichments the appropriately diluted cell cultures were plated onto LB agar and KG medium supplemented with 3-oxo-C6-HSL (50 μM) solidified with 1.5% (w/v) Bacto-Agar to isolate individual colonies.
DNA manipulation
Genomic DNA and plasmid extraction, manipulation and competent cells were prepared using standard methods [37]. Treatment of PCR mixtures without DNA template was performed as previously described [38]. PCR mix (Promega, UK) was used to amplify 16S rDNA with the universal primers 27F and 1525R (Table 3). PCR conditions, cloning and sequencing of the PCR products were carried out as previously described [14]. DNA sequences were analysed with the Lasergene computer package (DNAstar) in combination with the BLAST programs available from NCBI http://www.ncbi.nlm.nih.gov/ while phylogenetic analyses were performed as previously described [14]. The ahlK gene was amplified from Klebsiella Se14 by PCR using the primers KF and KR (Table 3). A single band of 0.85 kb was amplified and ligated to pGEM-T Easy and introduced into E. coli DH5α. A positive clone exhibiting QQ activity was sequenced.
Table 3.
Oligonucleotide Primers
Synthesis of AHLs and related compounds
AHLs including the D-isomer of 3-oxo-C6-HSL were synthesized, purified and characterized as previously described [20,39].
AHL-inactivation assays
GG2, GG4 and Se14 were grown overnight at 28°C with shaking (220 rpm) in LB medium to approximately 109 cfu/ml, cells (100 ml) were collected by centrifugation, washed and resuspended in 100 ml of PBS (100 mM, pH 6.5). AHLs were evaporated to dryness in a suitable tube and rehydrated with cell suspension providing a final AHL concentration of 1 μM (for biosensor activation assays) or 50 μM (for HPLC analysis). The reaction mixture was incubated for up to 24 h at 28°C with gentle shaking. To stop the reaction, an equal volume of ethyl acetate was added, after which the AHLs were extracted with ethyl acetate. Any residual AHLs were detected using the lux-based biosensors E. coli [pSB401] or E. coli [pSB1075] for long and short chain AHLs, respectively, [40] using a microtitre plate bioassay based on that described by Reimmann et al [41]. Bioluminescence in the microtitre plate wells was visualized using Luminograph LB980 photon video camera (Berthold).
To determine whether AHLs were being inactivated by lactonolysis, i.e. by the formation of the corresponding N-acylhomoserine compound, the method described by Yates et al [8] was used. This is based on acidification of the reaction mixture to pH 2 with HCl (10 mM) to promote recyclization of the homoserine lactone ring.
HPLC analysis of AHLs and AHL-degradation products
HPLC analysis of AHLs and their degradation products was performed as described before [17,20] on an analytical C8 reverse-phase preparative HPLC column (Kromasil C8; 250 × 4.6 mm) attached to a photodiode array (PDA) system (Waters 996 PDA system operating with a Millennium 2010 Chromatography Manager, Waters, England) and eluted with acetonitrile/water isocratic or gradient combinations as described before [17].
Identification of AHLs
AHLs were unequivocally identified by LC-MS/MS as described before [17,42] using enhanced product trap experiments (EPI) triggered by precursor ion scanning between the m/z range 150-500 and in particular for the fragment ion m/z 102 which is characteristic for the homoserine lactone ring moiety. The EPI spectra (m/z range 80-400) containing a fragment ion at m/z 102 were compared for the retention time and spectral properties to a series of corresponding synthetic AHL standards. The 3-hydroxy-AHLs were identified by comparison with a synthetic standard based on the LC retention times, the MS-MS fragmentation product ions ([M+H-H2O] and m/z 102). 3-hydroxy-AHLs characteristically lose a water molecule during MS fragmentation generating a characteristic ion of [M-18] [17,43].
P. aeruginosa QQ co-culture assays
The ability of ginger rhizosphere isolates to attenuate P. aeruginosa QS-regulated virulence determinants (elastase and lectin A) were determined by growing cultures of P. aeruginosa PAO1, GG2, GG4 and Se14 separately at 28°C for 24 h with shaking (220 rpm), normalizing at an OD600 of 1.0 followed by co-culturing at a 1:1 ratio. Total viable cell counts were carried out to ensure that neither organism significantly reduced the growth of the other.
The elastolytic activity of P. aeruginosa was determined as described before using elastin-Congo red (ECR) as substrate. Briefly, 100 μl of cell free bacterial spent culture supernatants from both mono-culture and co-culture experiments were added separately to 900 μl ECR buffer (100 mM Tris [pH 7.5], 1 mM CaCl2) containing 20 mg of ECR and incubated with shaking at 37°C for 3 h. Insoluble ECR was removed by centrifugation at 7,000 × g for 5 min. The absorbance of the supernatant was determined at OD495. The expression of lecA was determined using a P. aeruginosa lecA::lux reporter strain [35] in a 96-well microtitre plate using an automated combined luminometer/spectrometer (Anthos Labtech LUCYI). Briefly, 200 μl of a 1:500 dilution of an overnight culture of the P. aeruginosa lecA::lux reporter and either GG2, GG4 or Se14 (100 μl from each culture in co-culture experiments, or 200 μl in monoculture experiments) were loaded into the plate wells and luminescence and OD495 were determined every 30 min for 24 h. Each co-culture experiment was carried out in triplicate and repeated at least twice.
Quenching of the pectinolytic activity of Er. carotovora
Inhibition of the pectinolytic activity of Er. carotovora was carried out with modification as described before [44,45] using potato tubers. Tubers were washed, sterilized with 70% v/v ethanol, then extensively rinsed with sterile water and finally dried under sterile conditions. Bacterial cells were grown overnight at 28°C in LB, washed, resuspended and diluted in sterile saline to OD600 of 1.0. Bacterial suspensions (Er. carotovora GS101 or the Er. carotovora AHL-synthase mutant PNP22 [44]) (negative controls), monocultures or co-cultured with GG2, GG4 or Se14 were introduced directly into the tubers using a 200-μl tip fitted on a micropipette. Tubers were incubated at 25°C, 90% humidity for 3 days. The results of the inoculation were assessed by visual inspection after slicing the tubers.
Abbreviations
AHL: N-acylhomoserine lactones; 3-oxo-C6-HSL: N-(3-oxo-hexanoyl)homoserine lactone; C4-HSL: N-butanoylhomoserine lactone; C5-HSL: N-pentanoylhomoserine lactone; C6-HSL: N-hexanoyl homoserine lactone; C7-HSL: N-heptanoylhomoserine lactone; C8-HSL: N-octanoylhomoserine lactone; C9-HSL: N-nonanoylhomoserine lactone; C10-HSL: N-decanoylhomoserine lactone; C11-HSL: N-undecanoylhomoserine lactone; C12-HSL: N-dodecanoylhomoserine lactone; C14-HSL: N-tetradecanoylhomoserine lactone; 3-hydroxy-C4-HSL: N-(3-hydroxybutanoyl)homoserine lactone; 3-hydroxy-C6-HSL: N-(3-hydroxyhexanoyl)homoserine lactone; 3-hydroxy-C8-HSL: N-(3-hydroxyoctanoyl)homoserine lactone; 3-hydroxy-C10-HSL: N-(3-hydroxydecanoyl)homoserine lactone; 3-hydroxy-C12-HSL: N-(3-hydroxydodecanoyl)homoserine lactone; 3-hydroxy-C14-HSL: N-(3-hydroxytetradecanoyl)homoserine lactone; Δ9-3-hydroxy-C14-HSL: N-(3-hydroxy-9-cis- tetradecanoyl)homoserine lactone; Δ10-3-hydroxy-C14-HSL: N-(3-hydroxy-10-cis-tetradecanoyl)homoserine lactone; Δ11-3-hydroxy-C14-HSL: N-(3-hydroxy-11-cis-tetradecanoyl)homoserine lactone; Δ13-3-hydroxy-C14-HSL: N-(3-hydroxy-13-cis-tetradecanoyl)homoserine lactone; CFU: colony forming units, QS: quorum sensing, QQ: quorum quenching;
Authors' contributions
KGC carried out the experiments other than LC-MS/MS. SA, KM helped draft the manuscript. SRC supervised the AHL syntheses and interpreted the MS spectra. MC established the HPLC method. CLK, CKS and PW conceived the study, helped in the biological interpretation, and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Mass spectra AHLs produced by GG4. Extracts from spent culture supernatants of GG4 were analysed by mass spectrometry. The peak ion at m/z 102 is characteristic of the homoserine lactone ring (A, B, E and F). By comparison with the corresponding synthetic standards (C, D, G and H) the precursor ion at m/z 214.2 and fragment ion at m/z 113.0 suggest the presence of 3-oxo-C6-HSL (A); the precursor ion at m/z 228.2 and fragment ion m/z 109.1 are indicative of C8-HSL (B); the precursor ion at m/z 226.2 [M-H2O] and fragment ion m/z 125.1 are indicative of 3-hydroxy-C8-HSL (E); the precursor ion at m/z 242.2 and fragment ion of m/z 142.2 are indicative of C9-HSL (F). AU: Absorbance unit.
Contributor Information
Kok-Gan Chan, Email: kokgan@um.edu.my.
Steve Atkinson, Email: steve.atkinson@nottingham.ac.uk.
Kalai Mathee, Email: kalai.mathee@fiu.edu.
Choon-Kook Sam, Email: choonkook.sam@nie.edu.sg.
Siri Ram Chhabra, Email: S.R.Chhabra@nottingham.ac.uk.
Miguel Cámara, Email: miguel.camara@nottingham.ac.uk.
Chong-Lek Koh, Email: chonglek.koh@nie.edu.sg.
Paul Williams, Email: paul.williams@nottingham.ac.uk.
Acknowledgements
KG Chan received a Commonwealth Split-site PhD Scholarship (Commonwealth Scholarship Commission, United Kingdom) and a PhD studentship from University of Malaya. The authors thank Alex Truman for AHL synthesis and Dr Catherine Ortori for LC-MS/MS analysis and Mavis Daykin for HPLC analysis. This work was supported by the grants from the University of Malaya namely HIR Grant (J-00000-73552), and partly supported by two Research University Grants (RG003-09BIO, TB013-2009C) to KG Chan.
References
- Williams P, Winzer K, Chan W, Cámara M. Look who's talking: communication and quorum sensing in the bacterial world. Phil Trans R Soc B. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology. 2007;153:3923–3938. doi: 10.1099/mic.0.2007/012856-0. [DOI] [PubMed] [Google Scholar]
- Williams P. Quorum sensing: an emerging target for antibacterial chemotherapy? Expert Opinion in Therapeutic Targets. 2002;6:257–274. doi: 10.1517/14728222.6.3.257. [DOI] [PubMed] [Google Scholar]
- Bjarnsholt T, Givskov M. Quorum sensing inhibitory drugs as next generation antimicrobials: worth the effort? Curr Infect Dis Rep. 2008;10:22–28. doi: 10.1007/s11908-008-0006-y. [DOI] [PubMed] [Google Scholar]
- Lowery CA, Salzameda NT, Sawada D, Kaufmann GF, Janda KD. Medicinal chemistry as a conduit for the modulation of quorum sensing. J Med Chem. 2010;53:7467–7489. doi: 10.1021/jm901742e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroz S, Dessaux Y, Oger P. Quorum sensing and quorum quenching: the yin and yang of bacterial communication. ChemBioChem. 2009;10:205–216. doi: 10.1002/cbic.200800521. [DOI] [PubMed] [Google Scholar]
- Byers JT, Lucas C, Salmond GPC, Welch M. Nonenzymatic turnover of an Erwinia carotovora quorum-sensing signalling molecule. J Bacteriol. 2002;184:1163–1171. doi: 10.1128/jb.184.4.1163-1171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, Goldner M, Dessaux Y, Cámara M, Smith H, Williams P. N-acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect Immun. 2002;70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong YH, Xu JL, Li XZ, Zhang LH. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc Natl Acad Sci USA. 2000;97:3526–3531. doi: 10.1073/pnas.060023897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier A, Uroz S, Smadja B, Fray R, Latour X, Dessaux Y, Faure D. The Ti plasmid of Agrobacterium tumefaciens harbors an attM- paralogous gene, aiiB, also encoding N-acylhomoserine lactonase activity. Appl Environ Microbiol. 2003;69:4989–4993. doi: 10.1128/AEM.69.8.4989-4993.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Lee SJ, Oh TK, Oh JW, Koo BT, Yum DY, Lee JK. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology. 2003;149:1541–1550. doi: 10.1099/mic.0.26269-0. [DOI] [PubMed] [Google Scholar]
- Lin YH, Xu JL, Hu JY, Wang LH, Ong SL, Leadbetter JR, Zhang LH. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol Microbiol. 2003;47:849–860. doi: 10.1046/j.1365-2958.2003.03351.x. [DOI] [PubMed] [Google Scholar]
- Sio CF, Otten LG, Cool RH, Diggle SP, Braun PG, Bos R, Daykin M, Cámara M, Williams P, Quax WJ. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infect Immun. 2006;74:1673–1682. doi: 10.1128/IAI.74.3.1673-1682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KG, Yin WF, Sam CK, Koh CL. A novel medium for the isolation of N-acylhomoserine lactone-degrading bacteria. J Ind Microbiol Biotechnol. 2009;36:247–251. doi: 10.1007/s10295-008-0491-x. [DOI] [PubMed] [Google Scholar]
- McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Cámara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GSAB, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- Uroz S, Chhabra SR, Cámara M, Williams P, Oger P, Dessaux Y. N-acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151:3313–3322. doi: 10.1099/mic.0.27961-0. [DOI] [PubMed] [Google Scholar]
- Laue BE, Jiang Y, Chhabra SR, Jacob S, Stewart GSAB, Hardman A, Downie JA, O'Gara F, Williams P. The biocontrol strain Pseudomonas fluorescens F113 produces the Rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecanoyl)homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology. 2000;146:2469–2480. doi: 10.1099/00221287-146-10-2469. [DOI] [PubMed] [Google Scholar]
- Chhabra SR, Philip B, Eberl L, Givskov M, Williams P, Cámara M. In: Topics in Current Chemistry. Schulz S, editor. Vol. 240. Springer-Verlag, Berlin Heidelberg; 2005. Extracellular communication in bacteria; pp. 279–315. [Google Scholar]
- Yang WW, Han JI, Leadbetter JR. Utilization of homoserine lactone as a sole source of carbon and energy by soil Arthrobacter and Burkholderia species. Arch Microbiol. 2006;185:47–54. doi: 10.1007/s00203-005-0065-5. [DOI] [PubMed] [Google Scholar]
- Chhabra SR, Stead P, Bainton NJ, Salmond GPC, Stewart GSAB, Williams P, Bycroft BW. Autoregulation of carbapenem biosynthesis in Erwinia carotovora ATCC 39048 by analogues of N-(3-oxohexanoyl)-L-homoserine lactone. J Antibiotics. 1993;46:441–454. doi: 10.7164/antibiotics.46.441. [DOI] [PubMed] [Google Scholar]
- Passador L, Tucker KD, Guertin KR, Journet MP, Kende AS, Iglewski BH. Functional analysis of the Pseudomonas aeruginosa autoinducer PAI. J Bacteriol. 1996;178:5995–6000. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroz S, Chhabra SR, Cámara M, Williams P, Oger P, Dessaux Y. N-acylhomoserine lactone quorum-sensing molecules are modified and degraded by Rhodococcus erythropolis W2 by both amidolytic and novel oxidoreductase activities. Microbiology. 2005;151:3313–3322. doi: 10.1099/mic.0.27961-0. [DOI] [PubMed] [Google Scholar]
- Kang BR, Lee JH, Ko SJ, Lee YH, Cha JS, Cho BH, Kim YC. Degradation of acyl-homoserine lactone molecules by Acinetobacter sp. strain C1010. Can J Microbiol. 2004;50:935–941. doi: 10.1139/w04-083. [DOI] [PubMed] [Google Scholar]
- Gray KM, Pearson JP, Downie JA, Boboye BE, Greenberg EP. Cell-to-cell signaling in the symbiotic nitrogen-fixing bacterium Rhizobium leguminosarum: autoinduction of stationary phase and rhizosphere-expressed genes. J Bacteriol. 1996;178:372–376. doi: 10.1128/jb.178.2.372-376.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu C, Clemmer KM, Bonomo RA, Rather PN. Isolation and characterization of an autoinducer synthase from Acinetobacter baumannii. J Bacteriol. 2008;190:3386–3392. doi: 10.1128/JB.01929-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Thuruthyil SJ, Willcox MD. Production of N-acylhomoserine lactones by Gram-negative bacteria isolated from contact lens wearers. Clin Exp Ophthalmol. 2001;29:150–152. doi: 10.1046/j.1442-9071.2001.00397.x. [DOI] [PubMed] [Google Scholar]
- González RH, Dijkshoorn L, Van den Barselaar M, Nudel C. Quorum sensing signal profile of Acinetobacter strains from nosocomial and environmental sources. Rev Argent Microbiol. 2009;41:73–78. [PubMed] [Google Scholar]
- Vallenet D, Nordmann P, Barbe V, Poirel L, Mangenot S, Bataille E, Dossat C, Gas S, Kreimeyer A, Lenoble P, Oztas S, Poulain J, Segurens B, Robert C, Abergel C, Claverie JC, Raoult D, Médigue C, Weissenbach J, Cruveiller S. Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS One. 2008;3:e1805. doi: 10.1371/journal.pone.0001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HB, Wang LH, Zhang LH. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2002;99:4638–4643. doi: 10.1073/pnas.022056699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HB, Wang C, Zhang LH. The quormone degradation system of Agrobacterium tumefaciens is regulated by starvation signal and stress alarmone (p)ppGpp. Mol Microbiol. 2004;52:1389–1401. doi: 10.1111/j.1365-2958.2004.04061.x. [DOI] [PubMed] [Google Scholar]
- Tait K, Williamson H, Atkinson S, Williams P, Cámara M, Joint I. Turnover of quorum sensing signal molecules modulates cross-kingdom signalling. Environ Microbiol. 2009;11:1792–1802. doi: 10.1111/j.1462-2920.2009.01904.x. [DOI] [PubMed] [Google Scholar]
- Duerkop BA, Herman JP, Ulrich RL, Churchill ME, Greenberg EP. The Burkholderia mallei BmaR3-BmaI3 quorum-sensing system produces and responds to N-(3-hydroxy-octanoyl)homoserine lactone. J Bacteriol. 2008;190:5137–5141. doi: 10.1128/JB.00246-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Moreno ZR, Devescovi G, Myers M, Hallack L, Mendonça-Previato L, Caballero-Mellado J, Venturi V. Commonalities and differences in regulation of N-acyl homoserine lactone quorum sensing in the beneficial plant-associated Burkholderia species cluster. Appl Environ Microbiol. 2010;76:4302–4317. doi: 10.1128/AEM.03086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Stacey RE, Dodd C, Cámara M, Williams P, Winzer K. The galactophilic lectin LecA contributes to biofilm development in Pseudomonas aeruginosa. Environ Microbiol. 2001;8:1095–1104. doi: 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- Winzer K, Falconer C, Garber NC, Diggle SP, Cámara M, Williams P. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J Bacteriol. 2000;182:6401–6411. doi: 10.1128/JB.182.22.6401-6411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M, Urbanowski ML, Greenberg EP. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc Natl Acad Sci USA. 2004;101:15833–15839. doi: 10.1073/pnas.0407229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York; 2003. [Google Scholar]
- Chan KG, Wong CS, Yin WF, Sam CK, Koh CL. Rapid degradation of N-3-oxo-acylhomoserine lactones by a Bacillus cereus isolate from Malaysian rainforest soil. Antonie van Leeuwenhoek. 2010;98:299–305. doi: 10.1007/s10482-010-9438-0. [DOI] [PubMed] [Google Scholar]
- Chhabra SR, Harty C, Hooi DSW, Daykin M, Williams P, Telford G, Pritchard DI, Bycroft BW. Synthetic analogues of bacterial quorum sensing molecules as immune modulators. J Med Chem. 2003;46:97–104. doi: 10.1021/jm020909n. [DOI] [PubMed] [Google Scholar]
- Winson MK, Swift S, Fish L, Throup JP, Jorgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GSAB. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol Lett. 1998;163:185–192. doi: 10.1111/j.1574-6968.1998.tb13044.x. [DOI] [PubMed] [Google Scholar]
- Reimmann C, Ginet N, Michel L, Keel C, Michaux P, Krishnapillai V, Zala M, Heurlier K, Triandafillu K, Harms H, Défago G, Haas D. Genetically programmed autoinducer destruction reduces virulence gene expression and swarming motility in Pseudomonas aeruginosa PAO1. Microbiology. 2002;148:923–932. doi: 10.1099/00221287-148-4-923. [DOI] [PubMed] [Google Scholar]
- Atkinson S, Chang CY, Sockett RE, Cámara M, Williams P. Quorum sensing in Yersinia enterocolitica controls swimming and swarming motility. J Bacteriol. 2006;188:1451–1461. doi: 10.1128/JB.188.4.1451-1461.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PD, Gao P, Daly SL, Cha C, Cronan JEM Jr, Rinehart KL, Farrand SK. Detecting and characterizing N-acylhomoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Yu B, Bainton NJ, Birdsall M, Bycroft BW, Chhabra SR, Cox, Golby P, Reeves PJ, Stephens S, Winson MK, Salmond GPC, Stewart GSAB, Williams P. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uroz S, Angelo-Picard C, Carlier A, Elasri M, Sicot C, Petit A, Oger P, Faure D, Dessaux Y. Novel bacteria degrading N-acylhomoserine lactones and their use as quenchers of quorum-sensing-regulated functions of plant-pathogenic bacteria. Microbiology. 2003;149:1981–1989. doi: 10.1099/mic.0.26375-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass spectra AHLs produced by GG4. Extracts from spent culture supernatants of GG4 were analysed by mass spectrometry. The peak ion at m/z 102 is characteristic of the homoserine lactone ring (A, B, E and F). By comparison with the corresponding synthetic standards (C, D, G and H) the precursor ion at m/z 214.2 and fragment ion at m/z 113.0 suggest the presence of 3-oxo-C6-HSL (A); the precursor ion at m/z 228.2 and fragment ion m/z 109.1 are indicative of C8-HSL (B); the precursor ion at m/z 226.2 [M-H2O] and fragment ion m/z 125.1 are indicative of 3-hydroxy-C8-HSL (E); the precursor ion at m/z 242.2 and fragment ion of m/z 142.2 are indicative of C9-HSL (F). AU: Absorbance unit.