Abstract
Several genes including the cagA in the cag pathogenicity island (cag PAI) of Helicobacter pylori are thought to be associated with the gastroduodenal diseases and hence variation in the genetic structure of the cag PAI might be responsible for different clinical outcomes. Our study was undertaken to characterize the cag PAI of H. pylori strains from duodenal ulcer (DU) patients and asymptomatic or non-ulcer dyspepsia (NUD/AV) subjects from Kolkata, India. Strains isolated from 52 individuals (30 DU and 22 NUD/AV) were analyzed by PCR using 83 different primers for the entire cag PAI and also by dot-blot hybridization. Unlike H. pylori strains isolated from other parts of India, 82.6% of the strains used in this study had intact cag PAI, 9.6% had partially deleted cag PAI, and 7.7% of the strains lacked the entire cag PAI. Dot-blot hybridization yielded positive signals in 100% and 93.8% of PCR-negative strains for HP0522-523 and HP0532-HP0534 genes, respectively. An intact cagA promoter region was also detected in all cagA-positive strains. Furthermore, the expression of cagA mRNA was confirmed by RT-PCR for the representative strains from both DU and NUD/AV subjects indicating the active cagA promoter regions of these strains. A total of 66.7% of Kolkata strains produced a ~390-bp shorter amplicon than the standard strain 26695 for the HP0527 gene, homologue of virB10. However, sequence analyses confirmed that the deletion did not alter the reading frame of the gene, and mRNA transcripts were detected by RT-PCR analysis. The strains isolated from DU and NUD/AV express CagA protein and possess a functional type IV secretion system, as revealed by Western blot analyses. Interestingly, no significant differences in cag PAI genetic structure were found between DU and NUD/AV individuals suggesting that other bacterial virulence factors, host susceptibility, and environmental determinants also influence the disease outcome at least in certain geographical locations.
Keywords: Helicobacter pylori, cag PAI, Duodenal ulcer, Disease association
Introduction
Helicobacter pylori, a Gram-negative microaerophilic bacterium, chronically infects the gastric epithelium, and infection is associated with several gastroduodenal diseases such as chronic gastritis, peptic ulcer, and gastric cancer (Correa et al., 1992; NIH, 1994; Parsonnet, 1999). Although more than half of the human population carries the infection, only ~10–15% of the infected individuals develop such gastroduodenal diseases and hence, strain-specific genetic traits could be involved in H. pylori-related pathogenesis. The cagA gene, which encodes a protein of ~128 kDa (CagA), the CagA protein is one of the most well-studied virulence markers of H. pylori (Covacci et al., 1993; Tummuru et al., 1993). cagA along with several other virulence-associated genes constitute the ~40-kb cag pathogenicity island (cag PAI) and is present in ~50–70% and ~90% of the western and Asian H. pylori strains, respectively (Covacci et al., 1993; Ito et al., 1997; van Doorn et al., 1999). Vacuolating cytotoxin (VacA), a protein that can cause severe cytotoxicity in cell lines as well as in gastric mucosa, is coded by the vacA gene, which could be present in several allelic combinations (Cover et al., 1994; Atherton et al., 1995). H. pylori strains that carry s1m1-allelic combination are significantly more cytotoxic than strains that carry s1m2-allelic combination while strains that carry s2m2-allelic combination are a non-vacuolating form of VacA (Atherton et al., 1995, 1997). Interestingly, strains that carry cag PAI (cag+) are more likely to carry the s1m1-allelic combination of the vacA gene as compared to strains that lack cag PAI (cag−) (Atherton et al., 1995, 1997). In western countries, strains that carry s1m1cagA are significantly associated with H. pylori-related gastroduodenal diseases, although such association is not apparent in the Indian context (Cover et al., 1994; Atherton et al., 1997; Mukhopadhyay et al., 2000; Chattopadhyay et al., 2002; Datta et al., 2003). Moreover, expression of babA gene product, which governs adherence to Lewisb (histo-blood group antigen) on gastric epithelial cells and expression of Lewis antigens as part of their lipopolysaccharide, is also strongly associated with cag+ strains (Ilver et al., 1998). Therefore, it appears that cag+ and cag− strains probably have different requirements for their colonization in gastric epithelium.
The cag PAI, which contains a different GC content than the H. pylori genome, probably entered the genome after the bacterium had evolved as a species (Tomb et al., 1998). It contains 27 genes, 6 of which are thought to encode a putative type IV secretion system, responsible for the translocation of the CagA into the host cell (Covacci et al., 1999; Stein et al., 2002). The CagA, after being translocated to the host cell, becomes phosphorylated on tyrosine residues by Src family kinases, and the phosphorylated CagA interacts with the SH2 domain of the SHP-2 (Higashi et al., 2002a, 2002b). This interaction leads to an altered cellular morphology and may eventually lead to gastric carcinoma (Asahi et al., 2000; Segal et al., 1999; Odenbreit et al., 2000; Stein et al., 2000; Higashi et al., 2002a).
An intact cag PAI may be responsible for the proinflammatory nature of H. pylori leading to gastroduodenal diseases like duodenal ulcer, gastric atrophy, and gastric cancer. The presence of intact cag PAI strains was found more frequently in patients with severe gastroduodenal disease (Nilsson et al., 2003). Partial deletions of the cag PAI appear to be sufficient to render the organism less pathogenic (Ali et al., 2005; Nilsson et al., 2003). The cag PAI is involved in the induction of interleukin-8 (IL-8) secretion, which is implicated in the inflammatory response of the gastric mucosa to H. pylori infection. However, the existence of strains inducing IL-8 secretion regardless of the cag PAI structure suggests that this region is not the only prerequisite for the IL-8 secretion (Audibert et al., 2001; Hsu et al., 2002). Furthermore, the cag PAI status did not affect the attachment of the bacterium to the gastric epithelial cells. In some populations, cagA-related genes are associated with an abrogated apoptotic response, whereas other studies showed that apoptosis was increased in the antrum and body (of the stomach) only in patients with cagA-positive H. pylori strains (Peek et al., 1997; Moss et al., 2001). Moreover, cag PAI-positive H. pylori strains induce apoptosis more rapidly than cag PAI-negative mutant strains, suggesting that the H. pylori binding and subsequent apoptosis are differentially regulated with regard to bacterial properties (Minohara et al., 2007). The cagA sequences of H. pylori strains isolated from Kolkata, India, are clustered with the cagA sequences of strains isolated from western countries and differed significantly from cagA sequences of strains isolated from East Asia (China or Japan) (Mukhopadhyay et al., 2000; Datta et al., 2003; Chattopadhyay et al., 2004). There is also a distinct polymorphic site at the right end of the cag PAI of Kolkata H. pylori strains (Kersulyte et al., 2000). It has also been observed that the presence of the IS605 element both in cagA+ and cagA− strains did not systematically modify the severity of associated disease in the study population (Owen et al., 2001).
Studies concerning the variation within the cag PAI of H. pylori infection associated with a variety of outcomes ranging from seemingly asymptomatic coexistence to peptic ulcer disease and gastric cancer showed variable results in different geographical populations (Yakoob, 2009). One study from southern India reported that intact cag PAI is present in only 12% of the population which correlated well with the data that 15% of the infected patients are symptomatic (Kauser et al., 2004). Another study claimed that the presence of an intact cag PAI correlated with the development of more severe pathology, and such strains were found more frequently in patients with severe gastroduodenal disease (Ali et al., 2005).
These considerations and our interest in the dynamics of genetic traits associated with H. pylori infection and disease association motivated us to conduct the present study (i) whether the Bengali population, which is different from the south Indian population, has a similarly low percentage of intact cag PAI carrying H. pylori strains, and (ii) whether the presence of intact cag PAI is correlated with the development of a more severe pathology to understand the disease process and pathogen–host interaction.
Materials and methods
Patient samples
A total of 73 adult participants [duodenal ulcer (DU) patients and non-ulcer dyspepsia (NUD) or asymptomatic volunteers (AV)] of both sexes (aged between 20 and 65 years) underwent a non-sedated upper gastrointestinal endoscopy (GIF XQ 30, Olympus optical company, Japan) under topical lignocaine anesthesia at the hospital of the Institute of Post Graduate Medical Education and Research, Kolkata, India, during the years 2002–2004. Among the 40 DU cases (17 females and 23 males), the mean age difference was 46±11.7 years vs. 43.7±9.2 years and among 33 NUD or AV cases (10 females and 23 males), the mean age difference was 31.4±6.5 years vs. 33.4±6.7 years, respectively. Diagnosis of duodenal ulcer was based on visual mucosal examination of the stomach and duodenum during endoscopy and also on any patient history of previous peptic ulcer. A detailed history was taken, and a physical examination of each subject was carried out prior to endoscopy. The objective of the study was explained to every individual. Informed consents were obtained from each individual under protocols approved by the institutional ethical committees of the Post Graduate Medical Education and Research and National Institute of Cholera and Enteric Diseases, Kolkata, West Bengal, India. Exclusion criteria were: use of antibiotics, antihistamins, and proton pump inhibitors 3 months prior to this study. During endoscopy, 2 biopsies, one from the antrum and the other from the corpus of the stomach, were obtained from each subject. Biopsies taken in 0.6 ml of Brucella broth (Difco Laboratories, Detroit, MI) with 15% glycerol were transported to the Bacteriology Division of the National Institute of Cholera and Enteric Diseases in ice-cold condition and were stored at − 70°C until culture.
H. pylori culture
In the laboratory, Brucella broth containing the specimen was vortexed for 2 min and 200 µl of the mixture was streaked on Petri plates containing brain heart infusion (BHI) agar (Difco Laboratories) supplemented with 7% sheep blood, 0.4% IsoVitaleX, amphotericin B (8 µg/ml) (Sigma Chemicals Co., St. Louis, MO), trimethoprim (5 µg/ml) (Sigma Chemicals Co.), and vancomycin (Sigma Chemicals Co.) (6 µg/ml). Plates were incubated at 37°C in a double gas incubator (Heraeus Instruments, Germany), which maintains an atmosphere of 5% O2, 10% CO2, and 85% N2, for 3–6 days. H. pylori colonies, which appeared as translucent water droplets, were identified based on their typical colony morphology and positive urease, oxidase, and catalase tests. The H. pylori cells were preserved in sterile BHI broth with 20% glycerol at −70°C.
Characterization by PCR-based assay
H. pylori genomic DNA was extracted by the CTAB (hexadecyltrimethyl ammonium bromide) method (Ausubel et al., 1993) from 24-h grown confluent lawn of bacterial culture on brain heart infusion agar (BHIA; Difco Laboratories) plates. Specific PCR was carried out in 20-µl volume containing 10 ng of bacterial genomic DNA, 20 pM of each primer, 0.25 mM of each dNTP (Takara, Shuzo, Japan), 1 U of Taq DNA polymerase (Takara) in standard PCR buffer (Takara) containing 1.5 mM MgCl2, and the products were amplified in 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min (1 min/kb). Primers used for PCR, generating probes for hybridization, and nucleotide sequencing are listed in Table 1a and 1b. All strains underwent multiple PCR assays using 83 primers in various combinations. The strains were first amplified with Primer set A, and those strains which failed to amplify with the standard primer set A for a gene were further amplified by PCR using primers within and outside the gene designated as either primer set B or primer set C or both as shown in Table 2.
Table 1.
Sequences and locations of the oligonucleotide primer sets used in this study.
| (a) List of Helicobacter pylori cag pathogenicity island (PAI)-spanning primer sets designed from the strain 26695. | |||||
|---|---|---|---|---|---|
| Primer position in 26695 |
Designation (Tomb et al., 1997) |
PAI primer | Nucleotide sequence (5’-3’) | Primer set | Size of PCR product (bp) |
| 546899 | HP0519 | PAI-1S | ACACTGCCAAGCCCGATGCTGTA | ||
| 548162 | PAI-1AS | GATACAGCGGTTGCTAGT | PAI-1S- PAI-1AS | 1264 | |
| 547464 | HP0520 | PAI-2S | GTGTCTTGAGCGGTGCTATG | ||
| 548829 | PAI-2AS | ATCTCTTAGGGGCGAACACACTTC | PAI-2S- PAI-2AS | 1366 | |
| 547652 | HP0520AS1 | CAGTTGGTTCGTTGGTAAC | |||
| 547850 | HP0521S1 | GCTGTAAGGGCGTTTTAC | |||
| 549312 | HP0522AS3 | GATCAAAGTCCCCTCATAGC | |||
| 548547 | PAI-3S | CATCACAGGCTCATTAGAG | |||
| 549779 | PAI-3AS | CTGTTGTTCACCCTAGAGAG | PAI-3S- PAI-3AS | 1233 | |
| 549703 | HP0523 | PAI-4S | GGTTGCGACAATGAAGTG | ||
| 550007 | PAI-5S | GCGCTTACAATGGGGAATGAA | |||
| 550666 | PAI-6S | TCTTAGCGCCATTCCTACCATAACC | |||
| 551769 | PAI-6AS | CGTTCATTGGCTTGATTGCTCCTAC | PAI-6S- PAI-6AS | 1104 | |
| 552610 | HP0525 | PAI-8S | GGCCAAACGGATAAACGCTTCTTCA | ||
| 553630 | HP0526 | PAI-8AS | GTGGCGTTTCAGATCCTAGGGATAG | PAI-8S- PAI-8AS | 1021 |
| 553964 | PAI-new 1R | GCCTCTTCTTTAAAAGATAGCAAC | |||
| 558446 | PAI-14S | CAATCTAGCGCCACTTGAAC | |||
| 558925 | PAI-15S | CACGATAAGAACAGCGACTAC | |||
| 560058 | PAI-15AS | CTATGGTGAATTGGAGCGTGTG | PAI-15S-PAI-15AS | 1134 | |
| 560207 | HP0528 | PAI-17S | CAATGGCGGCATCAGTCATGCTCAA | ||
| 561183 | PAI-17AS | ACTTATCGTAGATGCGCCTGACC | PAI-17S-PAI-17AS | 977 | |
| 561843 | HP0529 | PAI-20S | TAGCAACAGAGGGCGTTATG | ||
| 562568 | PAI-19AS | TCAAAGGAGCGGACGCTGCTGTT | |||
| 562972 | PAI-20AS | CACAAGTTTAGCCGCTAGCA | PAI-20S-PAI-20AS | 1130 | |
| 563540 | HP0530 | PAI-22S | GATAGCTTCTGCTCGGACTT | ||
| 564932 | PAI-new 2F | CAAAGAAACAAGGCGGTGCA | |||
| 565031 | PAI-23AS | TGCACCGCCTTGTTTCTTTG | |||
| 564889 | PAI-24S | GTAGCACTAACGACAAGGTGCT | |||
| 565975 | PAI-24AS | TAACGCCCGTTGGCGTTTCTCT | PAI-24S-PAI-24AS | 1087 | |
| 565573 | PAI-25S | CTTGCATGGCTATGATGTGAG | |||
| 566555 | PAI-25AS | GCATACAAACAAGGGAGCGTTAG | PAI-25S-PAI-25AS | 983 | |
| 567109 | PAI-26AS | TCATCTTTCACGCAGAGC | |||
| 566816 | PAI-27S | CAGAGCGGTCATAATTCAAAGAGC | |||
| 567212 | HP0535S2 | CCAACCAAAGCAGATCCCATGT | |||
| 568157 | HP0536 | PAI-27AS | CTTATGGGGCAGGGGTGATTTTAG | PAI-27S-PAI-27AS | 1342 |
| 567426 | HP0535AS2 | TTGTTGGGTGGCGGAACAAA | |||
| 567409 | PAI-28S | TGTTCCGCCACCCAACAAAGAA | |||
| 568993 | HP0537 | PAI-30S | GAGGCTCTAGAGAAAGAGAC | ||
| 569303 | PAI-29 AS | GCTAATCGGCTCGCTTTT | |||
| 569724 | HP0538 | PAI-31 S | CGTAGATAGCGATCCTATG | ||
| 570078 | HP0538 | PAI-30AS | CTCTCAAAGCGTTAGTGG | PAI-30S-PAI-30AS | 1086 |
| 570169 | PAI-32S | AAGCGGCTAAGCACAAAG | |||
| 571054 | HP0539 | PAI31AS | CACAGACGCTTGTAGAAAG | ||
| 571040 | PAI33S | CTACAAGCGTCTGTGAAG | |||
| 571479 | HP0540 | PAI-32AS | AGAGACCAACCAACAAGTGC | PAI-32S-PAI-32AS | 1311 |
| 571729 | PAI-34S | GCGCGTTCAAATCTACTG | |||
| 572648 | HP0541 | HP0540AS3 | GCTTGAACCCGCCTTAAA | ||
| 573078 | PAI-34AS | CACTCCTGCATGCCCTATTG | PAI-34S-PAI-34AS | 1350 | |
| 574444 | HP0543 | PAI-37S | GCTTCAACGCTCATATCAG | ||
| 574614 | PAI-36AS | CATAAGCGAGGACATGCAGAAC | |||
| 574991 | HP0544 | PAI-38S | GAGCGGTAAGGTTTTGTTCGGTGAT | ||
| 575532 | PAI-37AS | GTCAGACTTGCGACTCAAAG | PAI-37S-PAI-37AS | 1089 | |
| 576094 | PAI-39S | GCCGCCCAAGCAAAAGGATTTA | |||
| 576584 | PAI-38AS | CCAACGCAGCGACTTTCTCTATG | |||
| 577589 | PAI-39AS | ATGGGGTGATCCTTACTAACAACTA | PAI-39S-PAI-39AS | 1498 | |
| 578305 | PAI-40AS | CCAACAAACAACGCTGCTTTC | |||
| 578096 | HP0545 | PAI-41S | GCCACAAACACCCCTCTCTTTA | ||
| 579016 | PAI-41AS | CAATCCTTTAATGGCGGTCACCAG | PAI-41S-PAI-41AS | 921 | |
| 578847 | HP0546 | PAI-42S | CAGTCGCCTGACCTCTTTTGAT | ||
| 580440 | PAI-42AS | CTGAAAAGATTGTTTGGCAGA | PAI-42S-PAI-42AS | 1594 | |
| 579910 | HP0547 | PAI-43S | AAGGAGAAACAATGACTAACGAAACTATTG | ||
| 581979 | PAI-45S | GAATTGTCTGATAAACTTGAAA | |||
| 583130 | PAI-45AS | GCGTATGTGGCTGTTAGTAGCG | PAI-45S-PAI-45AS | 1152 | |
| 584021 | PAI-46AS | GTTGATGCTCCCCTTCAACA | |||
| 579530 | HP0546AS1 | TTTGGTTTGTGTGTGTCATACT | |||
| (b) List of Helicobacter pylori cag pathogenicity island (PAI)-spanning oligonucleotide primer sets designed from the accession numbers U60176 and AC000108. | |||||
|---|---|---|---|---|---|
| Gene of DNA region amplified |
Primer | Primer sequence | Primer set | Amplicon size (bp) |
Reference |
| LEC | LecF1 | ACATTTTGGCTAAATAAACGCTG | Ikenoue et al. (2001) | ||
| LecR1 | TCTCCATGTTGCCATTATGCT | LecF1- LecR1 | 384 | Ikenoue et al. (2001) | |
| LecF2 | ATAGCGTTTTGTGCATAGAA | Ikenoue et al. (2001) | |||
| LecR2 | ATCTTTAGTCTCTTTAGCTT | LecF2- LecR2 | 877 | Ikenoue et al. (2001) | |
| cagE | cagE-F1 | GCGATTGTTATTGTGCTTGTAG | 329 | Ikenoue et al. (2001) | |
| cagE-R1 | GAAGTGGTTAAAAAATCAATGCCCC | cagE-F1- cagE-R1 | Ikenoue et al. (2001) | ||
| cagT | cagT-F1 | CCATGTTTATACGCCTGTGT | 301 | Ikenoue et al. (2001) | |
| cagT-R1 | CATCACCACACCCTTTTGAT | cagT-F1- cagT-R1 | Ikenoue et al. (2001) | ||
| cagA promoter | cagAP-F1 | GTGGGTAAAAATGTGAATCG | cagAP-F1-PAI-42AS | 730 | Ikenoue et al. (2001) |
| cagAP-F2 | CTACTTGTCCCAACCATTTT | cagAP-F2-PAI-42AS | 1181 | Ikenoue et al. (2001) | |
| cagA | cag 5cf | GTTGATAACGCTGTCGCTTCA | Chattopadhyay et al. (2003) | ||
| cag 3cr | GGGTTGTATGATATTTTCCATAA | Cag 5cf-cag 3cr | 350 | ||
| cag PAI empty site | Lunil 1 | ACATTTTGGCTAAATAAACGCTG | Mukhopadhyay et al. (2000) | ||
| R5280 | CCAACGTGCGTAAAAGGGAATTAG | Lunil 1-R5280 | 550 | ||
Table 2.
Percentage of detection of various cag PAI genes using the different primer sets A, B, and C.
| Primer set | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Gene | Primer set A (n=48) | Detection (%) | Primer set B | Detection (%) |
Primer set C | Detection (%) |
(B+C) | (A+B+C) |
| 1 | HP0519, 520, 521 | PAI-1S- PAI-1AS | 47/48 (95.7%) | 47/48 (95.7%) | |||||
| 2 | HP0522, 523 | PAI-3S- PAI-3AS | 22/48 (45.7%) | PAI-3S- PAI-6AS | 10/26 (38.5%) | a | 38.5% | 32/48(66.6%) | |
| 3 | HP0524 | PAI-6S- PAI-6AS | 47/48 (97.8%) | 47/48 (97.8%) | |||||
| 4 | HP0525-526 | PAI-8S & PAI new1R | 44/48(87.36%) | PAI8S-PAI8AS | 5/6 (83.3%) | 83.3% | 47/48 (97.7%) | ||
| 5 | HP0527 | PAI14S-PAI15AS | 46/48 (95.7%) | 46/48 (95.7%) | |||||
| 6 | HP0528 | PAI-17S & PAI 17AS | 44/48 (91.5%) | PAI 16S - PAI 17AS | 3/4 (75%) | 75% | 47/48 (97.7%) | ||
| 7 | HP0529 | PAI-20S & PAI 20AS | 41/48 (85.2%) | PAI 20S-PAI 19AS | 5/7 (71.4%) | 71.4% | 47/48 (97.7%) | ||
| 8 | HP0530-531 | PAI22S- PAI 23AS | 37/48 (76.9%) | PAI 22S- PAI 25AS | 4/11 (36.4%) | PAI22SHP0535AS2 | 3/7 (42.9%) | 63.6% | 44/48 (91%) |
| 9 | HP0532-534 | PAInew2F- PAI 25AS | 3/48 (6.2%) | PAI 24S- PAI 25AS | 24/45 (53.3%) | PAI 24S- 26AS | 10/21(47.6%) | 75.5% | 37/48 (76.7%) |
| 10 | HP0535-536 | PAI27S - PAI 27AS | 16/48 (33.3%) | PAI27S-HP0535AS2 | 29/31 (93.5%) | HP0535S2-PAI27AS | 1/3 (33.3%) | 96.7% | 46/48 (95.6%) |
| 11 | HP0538-538 | PAI30S - PAI 30AS | 29/48 (60.3%) | PAI31S - PAI 30AS | 18/19 (94.7%) | PAI30S - PAI 29AS | 94.7% | 47/48 (97.7%) | |
| 12 | HP0538-539 | PAI32S - PAI 32AS | 44/48 (91.5%) | PAI 31S-PAI 32AS | 3/4 (75%) | PAI32S-PAI31AS | 75% | 47/48 (97.7%) | |
| 13 | HP0540-541 | PAI34S - PAI 34AS | 42/48 (87.4%) | PAI34S-HP0540AS3 | 6/6 (100%) | 100% | 48/48 (100%) | ||
| 14 | HP0543-544 | PAI37S - PAI 37AS | 45/48 (93.6%) | PAI37S - PAI 36AS | 3/3(100%) | PAI38S-PAI37AS | 100% | 48/48 (100%) | |
| 15 | HP0544 | PAI-39S- PAI 39AS | 42/48 (87.36%) | PAI37S - PAI 36AS | 2/4 (50%) | PAI38S-PAI37AS | 2/2 (100%) | 100% | 48/48 (100%) |
| 16 | HP0545-546 | PAI41S - PAI 41AS | 43/48 (89.4%) | PAI 41S-PAI 40AS | 3/5 (60%) | 60% | 46/48(95.7%) | ||
| 17 | HP0546-547 | PAI42S- PAI 42AS | 40/48 (83.2%) | PAI42S- PAI-41AS | 8/8 (100%) | PAI43S-PAI42AS | 100% | 48/48 (100%) | |
| 18 | HP0547 | PAI-45S -45AS | 38/48 (79%) | cag5cf-cag3cr | 18/18 (100%) | 100% | 48/48 (100%) | ||
| 19 | Left-end region of cag PAI | LecF1-LecR1 | 19/48 (39.5%) | LecF2-LecR2 | 27/29 (93.1%) | 93.1% | 46/48 (95.7%) | ||
| 20 | Promoter region of cag PAI | cagAPF1-PAI42AS | 33/48 (68.6%) | cagAPF2-PAI42AS | 15/15 (100%) | 100% | 48/48 (100%) | ||
| Mean | 71% | 77.9% | 56% | 84% | 95% | ||||
PCR assay by PAI-1S and PAI-6AS (designed from HP0519 and HP0524, respectively) yielded around 4.8-kb amplicon and indirectly indicated that the remaining 16 strains also contained HP0522-523 (Fig. 3C). As these primers do not fall into the HP0522-23 region, it was not included in the above table.
Analysis by dot-blot hybridization and nucleotide sequencing
PCR products were purified with the QIA quick gel extraction kit (Qiagen Corporation, Chatsworth, CA) according to the manufacturer’s instruction and were directly sequenced using the BigDye terminator cycle sequencing kit (Perkin-Elmer, Applied Biosystems, Foster City, CA) on an automated DNA sequencer (ABI Prism 310). DNA sequence editing and analysis were performed with programs in the GCG package programs (Genetics Computer Group, Madison, WI). Dot-blot hybridization was performed by using Hybond-N1 nylon membranes and an ECL kit (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer’s instructions. Probes for various genes were generated by PCR using H. pylori strain 26695 genomic DNA as the template with specific primer pairs.
Gene expression assay by semiquantitative RT-PCR
Total RNA was extracted from H. pylori culture using the RNeasy Mini kit (Qiagen) according to the manufacturer’s protocol. RNA samples were electrophoresed in agarose gel, and the ethidium bromide-stained gel was inspected to check RNA quality. The absence of genomic DNA contamination was verified by PCR using taq DNA polymerase without reverse transcriptase. A total of 300 ng of RNA was reversely transcribed using the One-Step RTPCR Kit (Qiagen). The primers 5’- ACTCTAACGATCAAGAGATTATCAAAGG -3’ (sense) and 5’ - TGTATAAGGTTCTATTGGGATCGTCATT -3’ (antisense) were used to amplify 288 bp of cagY mRNA and Cag 5cf-Cag 3cr (Table 1b) for 350 bp of cagA mRNA. Ure B was amplified using the primers 5’- CGT CCG GCA ATA GCT GCC ATA GT -3’ (sense) and 5’- GTA GGT CCT GCT ACT GAA GCC TTA -3’ (antisense) to generate a 464-bp product. Data for cagY mRNA were normalized to data for urease. The results were analyzed with Quantity One software (Bio-Rad).
Infection of AGS cells with H. pylori strains and immunoblotting
Human gastric epithelial AGS cells were cultured in RPMI 1640 (GIBCO BRL, Grand Island, NY) containing 10% fetal bovine serum (Invitrogen Corp., USA) at 37°C in a 5% CO2–air humidified atmosphere. H. pylori (3×108 cells) was added to AGS cells (3×106 cells per 100-mm dish), which were then cultured in an antibiotic-free medium at a multiplicity of infection of 100. After incubation in a 5% CO2 atmosphere for 5 h, infected AGS cells were washed 3 times with 0.01 M phosphate buffered saline (PBS) (pH 7.5) containing 2 mM Na3VO4 and then lysed in ice-cold lysis buffer containing 50 mM Tris, pH 6.8, NP 40, 10% glycerol, 5 mM EDTA (pH 8.0), 25 mM NaF, 2 mM sodium orthovanadate, 5 mM sodium pyrophosphate, 200 mM NaCl, and protease inhibitor cocktail (Roche). The cell lysates were centrifuged at 5000 × g for 10 min at 4°C, and protein estimation was done.
Equal amounts of cell lysate were separated by SDS-polyacrylamide gel electrophoresis (7.5% polyacrylamide) and blotted onto a nitrocellulose membrane (Transblot, Bio-Rad Laboratories, USA) at 20 V for 0.5 h at room temperature in transfer buffer. The membranes were blocked with 3% (w/v) bovine serum albumin in T-PBS [10 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.5% Tween 20] for 2 h at 37°C under gentle shaking. After washing 3 times with TBST, each for 15 min, nitrocellulose strips were incubated with a primary antibody in T-PBS. After the membranes had been washed with T-TBS, they were incubated with goat anti-rabbit IgG conjugated to horseradish peroxidase in T-TBS and visualized with an enhanced chemiluminescence detection system as directed by the manufacturer (Amersham Pharmacia Biotech. Inc.).
Antibodies
The primary antibody used for immunoblotting was an anti-CagA polyclonal antibody (Austral Biologicals, San Ramon, CA).
Nucleotide sequence accession numbers
The sequences obtained here were deposited in GenBank under accession numbers GQ396658-GQ396660.
Statistical analysis
Statistical analysis was done using Microsoft Excel 2003 (Microsoft Corporation, Redmont, USA) software.
Results
Among 73 individuals included in the study, 40 had clinical features of DU and 33 were NUD or asymptomatic subjects. In 30 out of 40 (75%) DU and 22 out of 33 (67%) NUD or asymptomatic subjects, evidence of H. pylori infection was confirmed from the isolation of this bacterium by culture method, and these were included in the further study.
Characterization of cag pathogenicity island (cag PAI) of H. pylori
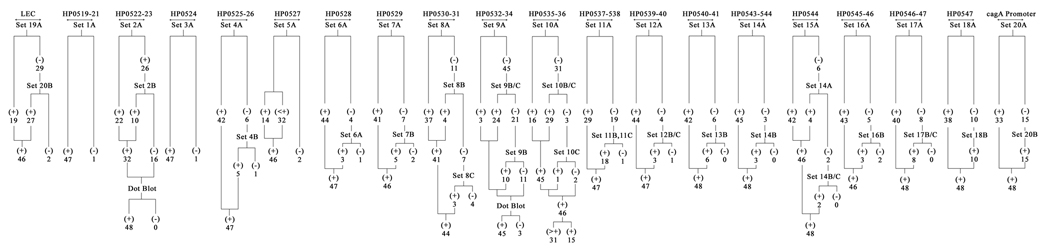
In this study, H. pylori strains from 52 clinical isolates [DU (n=30), NUD/AV (n=22)] were analyzed. Of the 52 strains, 92.3% (n=48) were cag PAI-positive and the rest 7.7% (n=4) lacked the entire cag PAI genes. The cag PAI positivity includes both intact cag PAI and partially deleted cag PAI. The cag PAI was defined as intact if all the gene sets of the cag PAI were present, whilst negative cag PAI was defined if none of the gene sets were present and gave a 550-bp amplicon with empty-site PCR. Partially deleted cag PAI was defined where a few, but not the whole set of cag PAI genes were present (Ikenoue et al., 2001). In our study using both PCR and dot-blot hybridization (DBH) methods, we found that 43 (82.6%) of the 52 strains had intact cag PAI, and 5 (9.6%) strains had partially deleted cag PAI (Table 2 and Figs. 1 and 2). In the cag PAI empty-site PCR where primers were designed from the flanking region of the cag PAI, 4 strains gave an amplicon of 550 bp indicating a complete lack of the cag PAI. Of the 5 partially deleted strains, 3 – I-110 (HP0535-HP0536), I-338 (HP0530-HP0531), and OSC36B (HP0532-HP0534) – had deletion in 2 genes, confirmed by both DBH and PCR assays, while the remaining 2 strains had deletions in multiple genes, and one further strain was devoid of the entire left end region of the cag PAI (data not shown).
Fig. 1.
Schematic representation of the distribution of the genes in cag PAI by PCR using 83 primers in various combinations and dot-blot hybridization among the H. pylori strains in Kolkata. Here, ‘+’ means positive by PCR and amplicon size is the same as in 26695, whereas ‘>+’ indicates positive but amplicon size is more than that of 26695 which is used as a reference positive control.
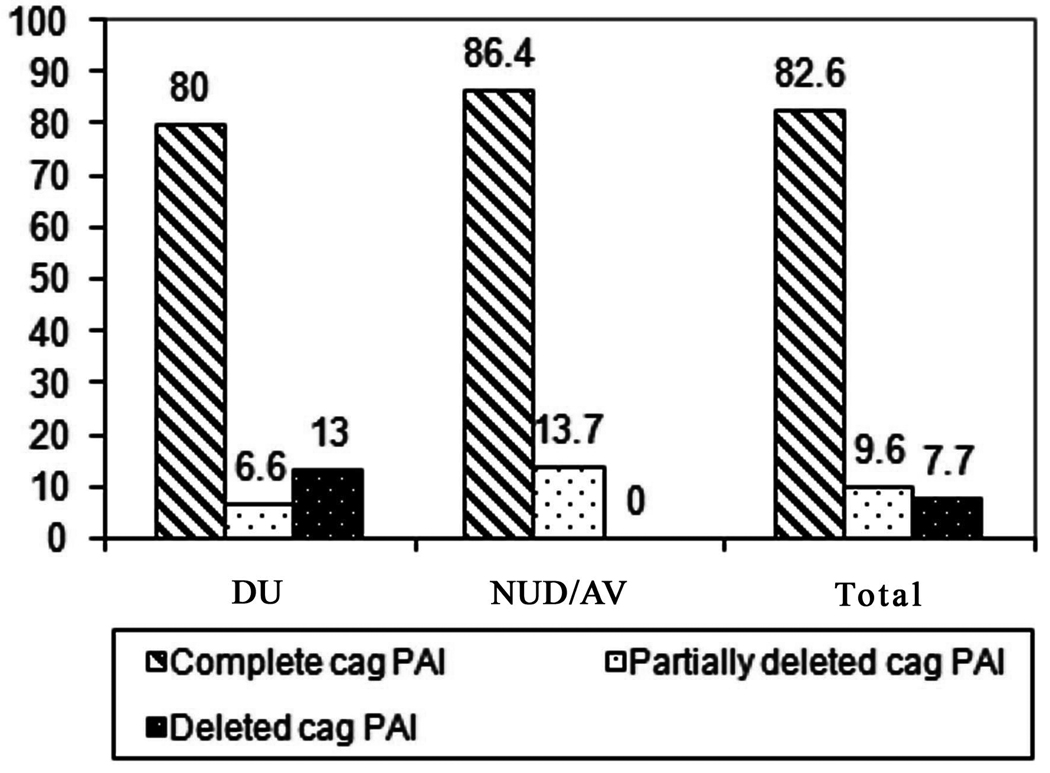
Fig. 2.
Percentage of intact cag PAI and partially deleted cag PAI and deleted cag PAI among the Kolkata strains with different clinical outcome. In total, 82.6% of the strains have intact cag PAI while only 9.6% of the strains have partially deleted cag PAI. Only 7.7% have cag PAI with a complete deletion. The frequency of complete cag PAI, partially deleted cag PAI, and deleted cag PAI among the duodenal ulcer patients and asymptomatic volunteers/non-ulcer dyspepsia were 80% and 86.4%, 6.6% and 13.7%, and 13% and 0%, respectively.
PCR using multiple primer sets and dot-blot hybridization analysis
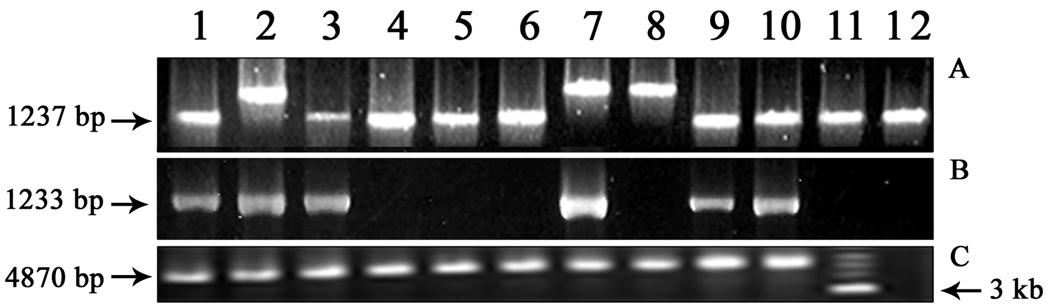
Analysis of the HP0519-521 regions showed that 10% of the strains produced around 150-bp higher amplicon (1414 bp) than the standard strain 26695 (1264 bp) using primers PAI-1S and PAI-1AS (Fig. 3A). After first screening by PCR, it was found that few primer pairs of the initial sets of primers (primer set A) failed to provide any amplicon in most of the Kolkata strains analyzed in this study (Fig. 1). These strains were subjected to DBH to evaluate whether the ORFs were truly absent or were falsely negative by PCR. It was found that most strains that were detected negative for a gene by PCR using primers from primer set A for a segment of a gene, hybridized with the corresponding probe in dot blot. For example, during analysis of HP0522-523 regions, 16 out of 48 cag PAI-containing strains were negative by PCR using primer sets A and B (Fig. 3B), but all (100%) PCR-negative strains showed positive signals after DBH with the specific probe generated by the same primers from 26695. Later on, primers designed from HP0519 (PAI-1S) and HP0524 (PAI-6AS) yielded around 4.8 amplicon size which is same as that of 26695 from the earlier PCR-negative strains indicating that these strains possessed the HP0522-523 regions (Fig. 3C). Similarly, for HP0532-HP0534 gene, 45 strains were negative by PCR with primer set PAI new2F-PAI 25AS (Fig. 4A) but after DBH with the specific probe generated by the same primer set from 26695, 45 out of 48 cag PAI-positive strains yielded positive signals (Fig. 4B). Those genes, which were not amplified by PCR with the primer set A, were successfully amplified with the second or the third sets of primers or by both sets (primer sets B and C) as given in Table 2. It is evident that using only primer set A, a mean of 29% (range 6.4–93.8%) of the genes could not be detected. Of these 29% undetected genes, primer sets B and C could detect cag PAI genes in a mean of 84% (range 38.5–100%) cases. Thus, the sensitivity of all the the primer sets A, B, and C in combination was 95% (Table 2).
Fig. 3.
(A) PCR assay of representative strains from Kolkata using primer set PAI-1S and PAI-1AS amplifying part of HP0519, HP0520, and HP0521. Lane 1 represents 26695 while lanes 2–12 represent different clinical isolates from Kolkata.6
(B) PCR assay of the same set of strains as in (3A) using primer set PAI-3S-PAI-3AS amplifying part HP0522 and HP0523.
(C) PCR assay of representative strains using primers PAI-1S and PAI-6AS designed from HP0519 and HP0524 gene of 26695, respectively. Lane 1 represents 26695, lanes 2–10 represent the strains that were negative for HP0522-523 by PCR, lane 11 is a 1-kb marker, and lane 12 indicates a cag PAI-negative strain as a negative control.
Fig. 4.
(A) Representative PCR result of HP0532–HP0534 gene using primer set PAInew2F-PAI25AS among the Kolkata strains. Lane 1, 100-bp marker (New England Biolab); lane 2, positive control; lane 5, 9, and 15 gave positive amplicons; lane 16, negative control. PCR assay yielded amplicons in 3 strains among 48 cag PAI-positive strains tested with the primer pair PAInew2F-PAI25AS.
(B) Dot-blot hydbridization result of representative strains showed positive signals of the PCR-negative strains using the PAInew2F-PAI25AS-amplified 1-kb fragment from 26695 as probe. ‘1’ in figure denotes AM1, cag PAI-negative strain (as a negative control), and ‘2’ is 26695 (as a positive control), and ‘3–12’ denotes the same strains as in lanes 2–11 of Fig. 4A.
cagY gene (HP0527) of H. pylori
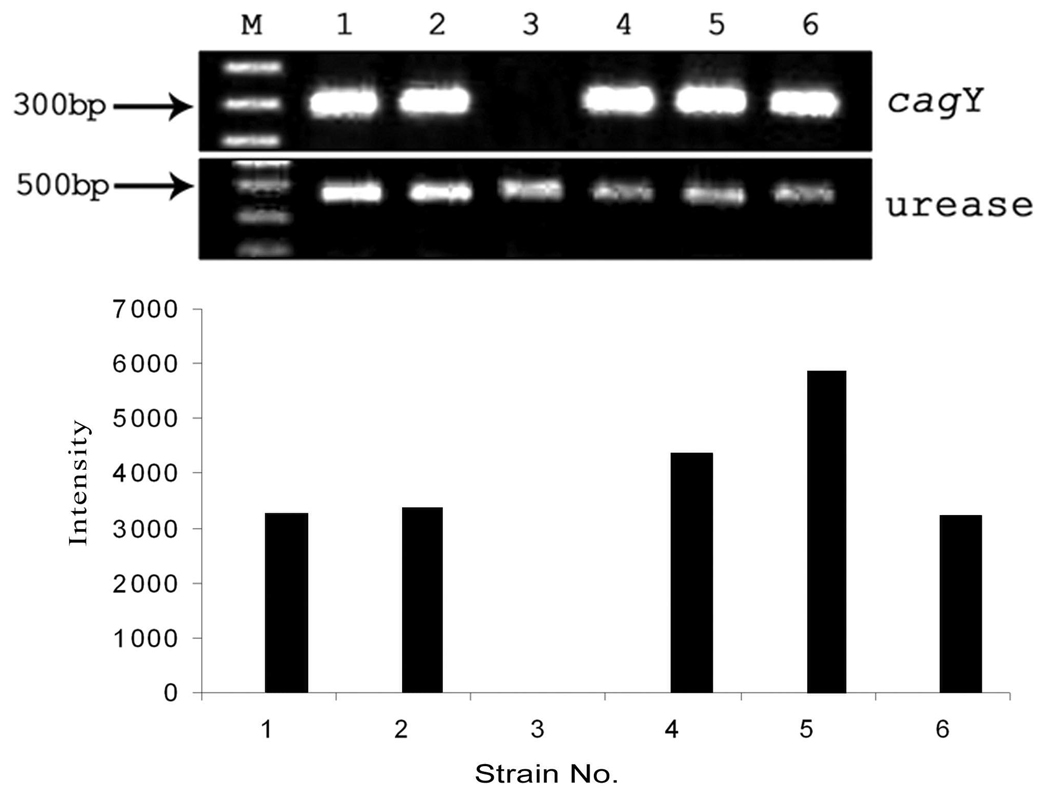
CagY protein, homologue of virB10, is an outer membrane protein and plays an important role in the cag PAI as it encodes for type IV export machinery. To understand the distribution of the gene encoding this CagY protein among the cag PAI-positive strains, primers (PAI14S and PAI15AS) were designed from HP0527 and HP0528 genes of 26695. PCR assay using these 2 primers was able to amplify 46 strains (95.8%) among the 48 cag PAI-containing strains. Interestingly, 32 of 48 (66.7%) strains produced around 400-bp shorter (1.1 kb) amplicon size than the standard 26695 strain while 14 of 48 strains (29.1%) gave an amplicon same as that of the strain 26695 (1.5 kb) (Fig. 5A and 5B). Sequence analysis of the shorter sequences revealed that the strains having a shorter amplicon had a 390-bp deletion, although this deletion did not alter the reading frame (Fig. 5C). RT-PCR analysis revealed that the number of repeats at the 5’ end of HP0527 does not have any effect on the level of mRNA transcription (Fig. 6). Interestingly, one strain showed no mRNA transcript for cagY while others showed various levels of transcription (lane 3, Fig. 6).
Fig. 5.
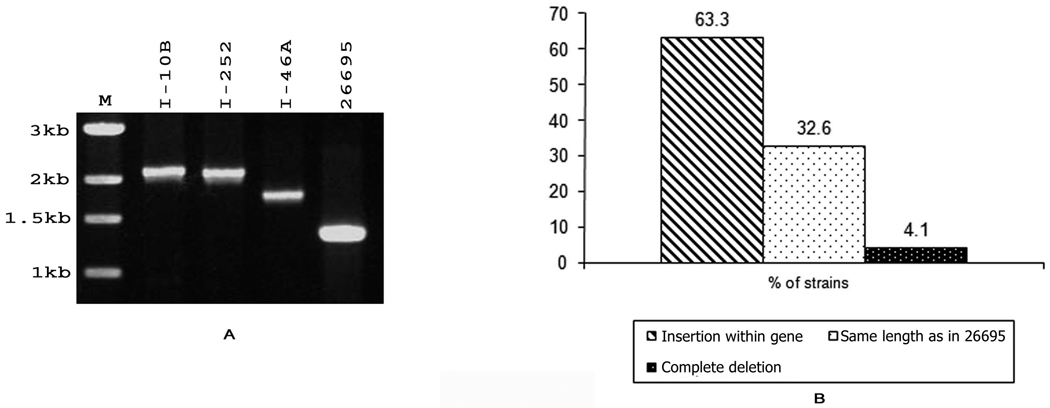
(A) PCR assay of the representative strains of H. pylori from Kolkata with primers PAI-14S and PAI15AS amplifying a part of HP0527 (VirB10) showed shorter amplicon in most of the strains as compared to 26695. Lane 1 represents 26695 while lanes 2–10 represent different strains isolated from Kolkata. (B) Percentage of H. pylori strains having partially deleted, complete, and totally deleted HP0527 gene of cag PAI among Kolkata strain. 66.7% of the strains gave a shorter amplicon, 29.1% of the strains gave the same amplicon as that of 26695 while only 4.1% of the strains showed complete deletion of the gene.
(C) Nucleotide sequence alignment of 26695 and one representative strain having shorter amplicon of the HP0527 gene of cag PAI from Kolkata. The first repeat of 390 bp is absent in those strains that have a shorter gene length in the HP0527 gene.
Fig. 6.
RT-PCR analysis of mRNA levels for the detection of the cagY gene using primers HPO527F and HPO527R and urease gene using primers UreBF and UreBR. M, 100-bp marker strains. Lane 1, 26695; lane 2, I-34; lane 3, S(New England Biolab). Lanes 1–6, signifies amplification cagY and urease gene of H. pylori an61; lane 4, San74; lane 5, San10; lane 6, San54. Lanes 1and 4 denote the presence of 2 complete units (390 bp each) and one incomplete unit; lanes 2, 5, and 6 denote the presence of one complete and one incomplete unit. Results under the panels A and B estimate the amount of cagY gene transcripts by semiquantitative RT-PCR. Data for cagY mRNA were normalized to data for urease. The results were analyzed with Quantity One software (Bio-Rad).
HP0535 gene of cag PAI
With primer pair PAI27S and PAI27AS designed from HP0535 and HP0536 gene of 26695, only 16 out of 48 cag PAI-positive strains gave a positive amplicon. All the strains, which did not amplify with PAI27S and PAI27AS, were evaluated with 2 more internal primers. These 2 primer sets, PAI27S-HP0535AS2 and HP0535S2-PAI27AS (Table 1a and 1b), were able to amplify all except 2 strains (93.5%, 29/31). However, among 46 strains that amplified, 31 gave a higher amplicon as shown in Fig. 7A and 7B.
Fig. 7.
(A) PCR amplification of representative H. pylori strains from Kolkata showed variations in the gene length of HP0535-HP0536 using primer set PAI27S-PAI27AS. Lane 1, 1-kb marker (New England Biolab); lanes 2–5, I-110, I-252, I-46A, and 26695. I-252 and I-110 gave a 2-kb amplicon while I-46A gave about 1.8-kb amplicon. Among 47 strains that amplified with the primer pair, 31 gave a higher amplicon than that of 26695.
(B) Percentage of H. pylori strains showing variation in the gene length of HP0535–HP0536 gene using primer PAI27S-PAI27AS isolated from patients in Kolkata. About 63.3% of the Kolkata strains have an insertion within the gene.
The cagA gene and cagA promoter region and LEC of the cag PAI
The primers cag5cf and cag3cr were designed from conserved regions to amplify 350 bp of the cagA gene (Chattopadhyay et al., 2004). Fourty-eight strains gave a 350-bp amplicon with cag5cf and cag3cr primers showing that all the cag PAI-positive strains were cagA-positive. All the cagA-positive strains were positive for the ORF located in the extreme left of the cag PAI, namely HP0520, -521, and -522 (annotated in the 26695 genome). A pair of oligonucleotide primers was used to detect the presence of the cagA promoter region and the LEC (left-end region of cag PAI) containing both inside and outside genes of cag PAI (Ikenoue et al., 2001). The promoter region of the cagA gene was found in all the cag-positive strains (Fig. 1). With internal primers (LECF1-LECR1) for the left-end region, only 19 of 48 cag-positive (39.6%) strains were amplified. With primer pair LECF2-LECR2, 95.8% cagA-positive strains (46/48) gave a positive amplicon.
CagA expression and translocation
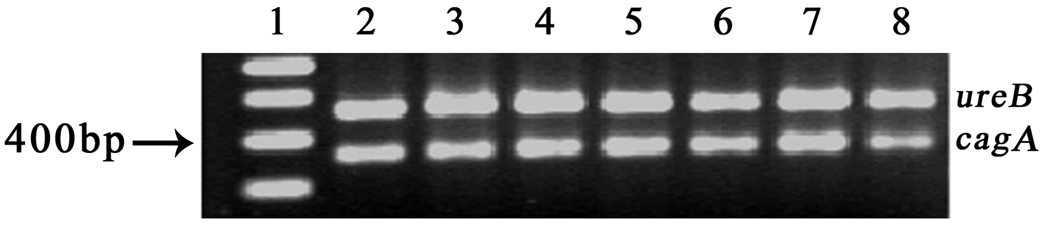
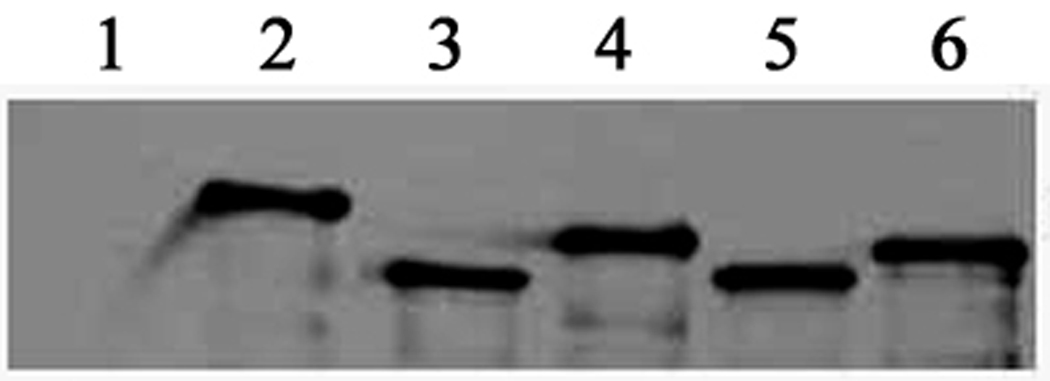
Then we addressed the question whether the cagA gene of H. pylori strains isolated from DU and NUD/AV can uniformly express the mRNA and the protein. First, we performed the RT-PCR analysis of the cagA gene of representative strains from both groups. All the tested strains isolated from both DU and NUD/AV subjects expressed the cagA transcripts at the transcriptional level indicating the intact cagA promoter regions of these strains (Fig. 8). We then went on to perform the CagA expression by Western blot analysis after infecting the AGS cells for 5 h with H. pylori strains isolated from both groups (Fig. 9). Expression of CagA indicated that the type IV secretion system (T4SS) is functional in the NUD/AV group also as CagA is translocated from bacteria to the host cell through this machinery.
Fig. 8.
RT-PCR analysis of mRNA levels for the detection of the cagA gene of H. pylori strains isolated from NUD/AV (lanes 2, 6, 7, and 8) and DU (lanes 3–5) using primers Cag 5cf and Cag 3cr and ureB using primers UreBF and UreBR. Lane 1 is a 100-bp marker.
Fig. 9.
AGS cells were cocultured with H. pylori strains from DU (lanes 3 and 4) and NUD/AV (lanes 2, 5, and 6) subjects for 5 h at 37°C before the cells were lysed and the samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting with polyclonal anti-CagA antibody. Lane 1 indicates the result of AM1 (one cag PAI-negative strain) infected AGS cell.
Correlation with clinical outcome
The frequency of complete cag PAI, partially-deleted cag PAI, and deleted cag PAI among the duodenal ulcer patients and NUD or AV were 80% (24/30) and 86.4% (19/22), 6.6% (2/30) and 13.7% (3/22), and 13% (4/30) and 0%, respectively (Table 3). These differences were, however, not significant (p>0.05). All the 48 cagA-positive strains isolated from DU patients and asymptomatic individuals carried the gene cagE. cagT gene was present in 100% of the DU patients and in 86.4% (19/22) of the strains from NUD or AV. All the 4 cag PAI-negative strains belonged to the DU group.
Table 3.
Distribution pattern of intact cag PAI and partially deleted cag PAI/cag PAI− in H. pylori strains isolated from DU and NUD/AV subjects in Kolkata, India.
Among 6 strains, 4 strains produced total cag PAI−, and the remaining 2 amplified partially deleted cag PAI.
All 3 strains yielded partially deleted cag PAI.
Discussion
The severity of H. pylori-related disease correlates with the presence of a cag pathogenicity island (cag PAI) in western countries. Genetic diversity within the cag PAI may have a profound effect on the pathogenic potential of the infecting strain, and the cag PAI has been studied in different H. pylori populations by various methods including PCR, Southern blotting, dot blot, and by long-distance PCR (Slater et al., 1999; Audibert et al., 2001; Ikenoue et al., 2001). In this study, we comprehensively analyzed 52 H. pylori strains isolated from DU and NUD/AV from Kolkata, India, by PCR using 83 primers sets. Our results showed that 92.3% of the strains were positive for cagA. These strains were also positive for the promoter region of cagA. Strains that were cagA-negative (7.7%) were also negative for all the genes of cag PAI. We found a partially deleted cag PAI in only 9.6% of the strains, while a complete cag PAI was present in the 82.6% of the strains. These data are consistent with our previous observation (Mukhopadhyay et al., 2000) and are different from the reports from East Asia (Japan and China) (Ito et al., 1997; Pan et al., 1997) and western countries (Nilsson et al., 2003). An analysis from East Asia (Ito et al., 1997; Pan et al., 1997) and South Africa (Kidd et al., 2001) revealed that 100% of the H. pylori strains are cag+. Studies in Swedish (Nilsson et al., 2003) and French populations (Jenks et al., 1998) indicated the frequency of the cagA gene in 83% and 87.7%, respectively. Our results on partially deleted cag PAI are similar to the observations from the East Asia (Ikenoue et al., 2001) and also from the Western world (Censini et al., 1996; Jenks et al., 1998; Maeda et al., 1999; Nilsson et al., 2003). Indian populations belong to a diverse set of culture and language groups that are largely endogamous. As a result of evolutionary antiquity and endogamy, populations of India show a high genetic differentiation and extensive structuring (Majumder, 2010). The geographical distribution of the language groups in India is largely non-overlapping. Linguistic differences of populations provide the best explanation of genetic differences observed in this region of the world (Majumder, 2010). Interestingly, a recent report from the southern part of India (Ali et al., 2005) showed the presence of intact cag PAI in 39.7% and partially deleted cag PAI in 58% of the strains. These strains were collected from the population of South India of the Telugu linguistic group who are mainly Dravidian and married consanguineously for millennia and are different from other Indian populations (Ahmed et al., 2003). The genetic separation of this population from other Indian communities has already been reported (Bamshed et al., 1996). A low prevalence of intact cag PAI was also reported from Karachi, Pakistan, with a missing cagE and cagA promoter region (Yakoob et al., 2009).
Therefore, our results showed the diversity of the prevalence of cag PAI among the different regions of India. Another study (Kauser et al., 2004) reported the analysis of cag PAI of H. pylori strains from various regions of the world including Japan, India, England, and Latin American countries using the same primer set with the prevalence of intact cag PAI in 57.1%, 12%, 3%, and 18.6% of the cases, respectively. This observation is markedly different from other reports on intact cag PAI positivity from the same regions (Jenks et al., 1998; Ikenoue et al., 2001). Some studies might have used smaller numbers of primer sets to analyze the cag PAI region, and this may lead to a lower percentage of intact cag PAI. Moreover, the same primer sets may not work in different regions as region-specific variations in the cag PAI have been reported from different parts of the world. In fact, 29% of the cag PAI genes of Kolkata strains were also undetected in our initial PCR assay using primer set A. However, our subsequent PCR analyses using primer set B and primer set C (Table 2) increased the detection level to 95%.
The relationship between the presences of complete, partially deleted, and absent cag PAI with duodenal ulceration is controversial. In our study, cag PAI was present in 86.6% and 100% of DU and NUD/AV, respectively. The complete cag PAI and partially deleted cag PAI were present in 80% and 6.7% of DU and 86.4% and 13.6% of NUD/AV, respectively, while 13.9% of DU and 0% of NUD/AV were cag−. These results are in accord with our previous reports, which showed a high prevalence of cag+ strains in both peptic ulcer patients and benign-disease populations (Mukhopadhyay et al., 2000). Many studies in European and Asian populations (Jenks et al., 1998; Maeda et al., 1999; Kidd et al., 2001; Nilsson et al., 2003) have reported the presence of increased intact cag PAI in duodenal ulcer patients compared to gastritis controls. However, several reports indicated a lack of correlation between the presence of cag PAI and peptic ulcer disease (Peters et al., 2001; Zhang et al., 2005). Moreover, cag PAI-negative strains, like the present study, have been detected among duodenal ulcer patients (Nilsson et al., 2003). Interestingly, in the present study, all the cag− strains were isolated from DU patients. This may also be due to the fact that the cag− strains that we have isolated do not represent the entire H. pylori population of the whole stomach since different H. pylori strains may have colonized different parts of the stomach. It is possible that cag+ H. pylori strains were also present in a different area of the stomach of the same DU patient. Moreover, our unpublished data strongly suggest that the coexistence of multiple H. pylori strains in a single host, unlike the situation in Europe and North America (Taylor et al., 1995; Marshall et al., 1996; Shortridge et al., 1997), are quite common in India. Moreover, Yakoob et al. (2000) showed that 2 different H. pylori strains may coexist in an infected individual and may not be uniformly distributed among biopsy sites. It has been shown that if both forward and reverse primers are selected from same ORF, it produces better results than taking 2 primers from 2 different ORFs especially in cases of detection of false-negative PCR. Therefore, we used pairs of oligonucleotide primers to detect the presence of the cagA promoter region and the LEC containing both inside and outside genes of cag PAI (Ikenoue et al., 2001). In our study, LEC1 was found to be present in only 19 of the 48 (39.6%) cag-positive strains amplified, and LEC2 was present in 46 of the 48 cag-positive strains (95.8%). These observations are similar to those of other studies (Ikenoue et al., 2001).
The cagY gene encoding a protein, homologue of virB10, is relatively large (5–6 kb) and includes 2 repetitive regions (Liu et al., 1999), and an amplified fragment produced by PAI14S and PAI15AS primers contained the first repetitive region, which exists in the 5’ end of the gene (from the 9th AA in 26695). In our study, for cagY gene, 66.7% of the strains gave a shorter amplicon than 26695. We also observed that the repetitive region consists of 2 complete units (390 bp each) and one incomplete unit (171 bp) or one complete and one incomplete unit. Further, analysis showed that the strains having shorter amplicons lacked one complete unit (390 bp) and therefore had a 390-bp deletion, although this deletion did not affect the reading frame and mRNA expression. Moreover, CagA expression of H. pylori-infected AGS cells by Western blot analysis indicated that T4SS is uniformly functional in DU and NUD/AV groups (even when HP0527 is shorter) as CagA is translocated from bacteria to the host cell through this machinery. The HP0535 gene was found to be highly diverse among the Kolkata strains. We found that insertion occurs within the HP0535-HP0536 gene. Among 46 strains that amplified, 31 (67.4%) gave a higher amplicon with respect to 26695 when PCR was done for HP0535 and HP0536 gene.
In conclusion, it was found that 82.6% of the H. pylori strains of Kolkata had a complete cag PAI, but 92.3% of the strains carried the cagA gene. Thus, the presence of the cagA gene does not necessarily indicate the presence of intact cag PAI and cannot be used invariably as a marker of intact cag PAI or as a marker for virulence, especially in the context of Indian H. pylori strains. Although the cag PAI is an important virulence component for infection, the determination of the direction of disease development (gastritis, ulcers, or cancer) is likely to involve a highly complex interplay of many bacterial and/or host factors. The differences in the development of H. pylori-induced diseases could also be due to the fact that the strains, upon colonization, may modify their abilities to induce epithelial cell responses (e.g. IL-8 secretion) as part of their adaptation to the changing conditions within the host milieu (Backert et al., 2004). In order to unravel the hidden features of H. pylori–host interactions, our results suggest that future work should be directed toward the identification of the host genetic background, other translocated bacterial virulence factors, and probable environmental factors, which may play a crucial role in the formation of H. pylori-related gastroduodenal disorders.
Acknowledgement
We would like to thank Mr. Manash Ray for his technical support during the preparation of the manuscript. The work was supported in part by the Indian Council of Medical Research, Government of India; the Japan Initiative for Global Research Network on Infectious Diseases, Ministry of Education, Culture, Sports, Science and Technology of Japan; and the Department of Biotechnology (No. BT/PR10407/BRB/10/604/2008).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed N, Khan AA, Alvi A, Tiwari S, Jyothirmayee CS, Kauser F, Ali M, Habibullah CM. Genomic analysis of Helicobacter pylori from Andhra Pradesh, South India, molecular evidence for three major genetic clusters. Current. Sci. 2003;85:1579–1586. [Google Scholar]
- Ali M, Aleem AK, Santosh TK, Niyaz A, Venkateswar RL, Habibullah CM. Association between cag-pathogenicity island in Helicobacter pylori isolates from peptic ulcer, gastric carcinoma, and non-ulcer dyspepsia subjects with histological changes. World J. Gastroenterol. 2005;11:6815–6822. doi: 10.3748/wjg.v11.i43.6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asahi M, Azuma T, Ito S, Ito Y, Suto H, Nagai Y, Tsubokawa M, Tohyama Y, Maeda S, Omata M, Suzuki T, Sasakawa C. Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J. Exp. Med. 2000;191:593–602. doi: 10.1084/jem.191.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton JC, Cao P, Peek RM, Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori, association of specific vacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- Atherton JC, Peek RM, Jr, Tham KT, Cover TL, Blaser MJ. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- Audibert C, Burucoa C, Janvier B, Fauchere JL. Implication of the structure of the Helicobacter pylori cag pathogenicity island in induction of interleukin-8 secretion. Infect. Immun. 2001;69:1625–1629. doi: 10.1128/IAI.69.3.1625-1629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, N.Y.: Greene Publishing and Wiley-Interscience; 1993. [Google Scholar]
- Azuma T, Yamakawa A, Yamazaki S, Ohtani M, Ito Y, Muramatsu A, Suto H, Yamazaki Y, Keida Y, Higashi H, Hatakeyama M. Distinct diversity of the cag pathogenicity island among Helicobacter pylori strains in Japan. J. Clin. Microbiol. 2004;42:2508–2517. doi: 10.1128/JCM.42.6.2508-2517.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Schwarz T, Miehlke S, Kirsch C, Sommer C, Kwok T, Gerhard M, Goebel UB, Lehn N, Koenig W, Meyer TF. Functional analysis of the cag pathogenicity island in Helicobacter pylori isolates from patients with gastritis, peptic ulcer, and gastric cancer. Infect. Immun. 2004;72:1043–1056. doi: 10.1128/IAI.72.2.1043-1056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Fraley AE, Crawford MH, Cann RL, Busi BR, Naidu JM, Jorde LB. mtDNA variation in caste populations of Andhra Pradesh, India. Hum. Biol. 1996;68:1–28. [PubMed] [Google Scholar]
- Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Dutta S, Chowdhury A, Chowdhury S, Mukhopadhyay AK, Rajendran K, Bhattacharya SK, Berg DE. Virulence genes in Helicobacter pylori strains from West Bengal residents with overt H. pylori-associated disease and healthy volunteers. J. Clin. Microbiol. 2002;40:2622–2625. doi: 10.1128/JCM.40.7.2622-2625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay S, Patra R, Ramamurthy T, Chowdhury A, Santra A, Dhali GK, Bhattacharya SK, Berg DE, Nair GB, Mukhopadhyay AK. Multiplex PCR assay for rapid detection and genotyping of Helicobacter pylori directly from biopsy specimens. J. Clin. Microbiol. 2004;42:2821–2824. doi: 10.1128/JCM.42.6.2821-2824.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa P. Human gastric carcinogenesis, a multistep and multifactorial process – first American Cancer Society Award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxity and duodenal ulcer. Proc. Natl. Acad. Sci. USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A, Telford JL, Giudice GD, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- Cover TL, Tummuru MLR, Cao P, Thompson SA, Blaser MJ. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J. Biol. Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- Datta S, Chattopadhyay S, Balakrish GB, Mukhopadhyay AK, Hembram J, Berg DE, Saha DR, Khan A, Santra A, Bhattacharya SK, Chowdhury A. Virulence genes and neutral DNA markers of Helicobacter pylori isolates from different ethnic communities of West Bengal, India. J. Clin. Microbiol. 2003;41:3737–3743. doi: 10.1128/JCM.41.8.3737-3743.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi H, Tsutsumi R, Muto S, Sugiyama T, Azuma T, Asake M, Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002a;295:683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. USA. 2002b;99:14428–14433. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PI, Hwang IR, Cittelly D, Lai KH, El-Zimaity HM, Gutierrez O, Kim JG, Osato MS, Graham DY, Yamaoka Y. Clinical presentation in relation to diversity within the Helicobacter pylori cag pathogenicity island. Am. J. Gastroenterol. 2002;97:2231–2238. doi: 10.1111/j.1572-0241.2002.05977.x. [DOI] [PubMed] [Google Scholar]
- Ikenoue T, Maeda AS, Gura KO, Akanuma M, Mitsuno Y, Imai Y, Yoshida H, Shiratori Y, Omata M. Determination of Helicobacter pylori virulence by simple gene analysis of the cag pathogenicity island. Clin. Diagn. Lab. Immunol. 2001;8:181–186. doi: 10.1128/CDLI.8.1.181-186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilver D, Arnqvist A, Ogren J, Frick IM, Kersulyte D, Incecik ET, Berg DE, Covacci A, Engstrand L, Boren T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigen revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- Ito Y, Azuma T, Ito S, Miyaji H, Hirai M, Yamazaki Y, Sato F, Kato T, Kohli Y, Kuriyama M. Analysis and typing of the vacA gene from cagA-positive strains of Helicobacter pylori isolated in Japan. J. Clin. Microbiol. 1997;35:1710–1714. doi: 10.1128/jcm.35.7.1710-1714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenks PJ, Megraud F, Labigne A. Clinical outcome after infection with Helicobacter pylori does not appear to be reliably predicted by the presence of any of the genes of the cag pathogenicity island. Gut. 1998;43:752–758. doi: 10.1136/gut.43.6.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauser F, Khan AA, Hussain MA, Carroll IM, Ahmad N, Tiwari S, Shouche Y, Das B, Alam M, Ali SM, Habibullah CM, Sierra R, Megraud F, Sechi LA, Ahmed N. The cag pathogenicity island of Helicobacter pylori is disrupted in the majority of patient isolates from different human populations. J. Clin. Microbiol. 2004;42:5302–5308. doi: 10.1128/JCM.42.11.5302-5308.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersulyte D, Mukhopadhyay AK, Velapatino B, Su W, Pan Z, Garcia C, Hernandez V, Valdez Y, Mistry RS, Gilman RH, Yuan Y, Gao H, Alarcon T, Lopez-Brea M, Nair GB, Chowdhury A, Datta S, Shirai M, Nakazawa T, Ally R, Segal I, Wong BCY, Lam SK, Olfat F, Boren T, Engstrand L, Torres O, Schneider R, Thomas JE, Czinn S, Berg DE. Differences in genotypes of Helicobacter pylori from different human populations. J. Bacteriol. 2000;182:3210–3218. doi: 10.1128/jb.182.11.3210-3218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd M, Lastovica AJ, Atherton JC, Louw JA. Conservation of the cag pathogenicity island is associated with vacA alleles and gastroduodenal disease in South African Helicobacter pylori isolates. Gut. 2001;49:11–17. doi: 10.1136/gut.49.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Mcdaniel TK, Falkow S, Karlin S. Sequence anomalies in the Cag7 gene of the Helicobacter pylori pathogenicity island. Proc. Natl. Acad. Sci. USA. 1999;96:7011–7016. doi: 10.1073/pnas.96.12.7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Yoshida H, Ikenoue T, Ogura K, Kanai F, Kato N, Shiratori Y, Omata M. Structure of cag pathogenicity island in Japanese Helicobacter pylori isolates. Gut. 1999;44:336–341. doi: 10.1136/gut.44.3.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder PP. The human genetic history of South Asia. Current Biol. 2010;20:R184–R187. doi: 10.1016/j.cub.2009.11.053. [DOI] [PubMed] [Google Scholar]
- Marshall DG, Coleman DC, Sullivan DJ, Xia H, O’Morain CA, Smyth CJ. Genomic DNA fngerprinting of clinical isolates of Helicobacter pylori using oligonucleotide probes containing repetitive sequences. J. Appl. Bacteriol. 1996;81:509–517. doi: 10.1111/j.1365-2672.1996.tb03540.x. [DOI] [PubMed] [Google Scholar]
- Minohara Y, Boyd DK, Hawkins HK, Ernst PB, Patel J, Crowe SE. The effect of the cag pathogenicity island on binding of Helicobacter pylori to gastric epithelial cells and the subsequent induction of apoptosis. Helicobacter. 2007;12:583–590. doi: 10.1111/j.1523-5378.2007.00537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss SF, Sordillo EM, Abdalla AM, Makarov V, Hanzely Z, Perez-Perez GI, Blaser MJ, Holt PR. Increased gastric epithelial cell apoptosis associated with colonization with cagA+ Helicobacter pylori strains. Cancer Res. 2001;61:1406–1411. [PubMed] [Google Scholar]
- Mukhopadhyay AK, Kersulyte D, Jeong JY, Datta S, Ito Y, Chowdhury A, Chowdhury S, Santra A, Bhattacharya SK, Azuma T, Nair GB, Berg DE. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 2000;182:3219–3227. doi: 10.1128/jb.182.11.3219-3227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH Consensus Conference. Helicobacter pylori in peptic ulcer disease. NIH Consensus Development Panel on Helicobacter pylori in peptic ulcer disease. JAMA. 1994;272:65–69. [PubMed] [Google Scholar]
- Nilsson C, Sillén A, Eriksson L, Strand ML, Enroth H, Normark S, Falk P, Engstrand L. Correlation between cag pathogenicity island composition and Helicobacter pylori-associated gastroduodenal disease. Infect. Immun. 2003;71:6573–6581. doi: 10.1128/IAI.71.11.6573-6581.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science. 2000;287:1497–1500. doi: 10.1126/science.287.5457.1497. [DOI] [PubMed] [Google Scholar]
- Owen RJ, Peters TM, Varea R, Teare EL, Saverymuttu S. Molecular epidemiology of Helicobacter pylori in England, prevalence of cag pathogenicity island markers and IS605 presence in relation to patient age and severity of gastric disease. FEMS Immunol. Med. Microbiol. 2001;30:65–71. doi: 10.1111/j.1574-695X.2001.tb01551.x. [DOI] [PubMed] [Google Scholar]
- Pan ZJ, van der Hulst RW, Feller M, Xiao SD, Tytgat GN, Dankert J, van der Ende A. Equally high prevalences of infection with cagA-positive Helicobacter pylori in Chinese patients with peptic ulcer disease and those with chronic gastritis-associated dyspepsia. J. Clin. Microbiol. 1997;35:1344–1347. doi: 10.1128/jcm.35.6.1344-1347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsonnet J. Helicobacter and gastric adenocarcinoma. In: Parsonnet J, editor. Microbes and Malignancy, Infection as a Cause of Human Cancers. New York, N.Y.: Oxford University Press; 1999. pp. 372–408. [Google Scholar]
- Peek RM, Moss SF, Tham KT, Perez-Perez GI, Miller GG, Atherton JC, Holt PR, Blaser MJ. Helicobacter pylori cagA1 strains and dissociation of gastric epithelial proliferation from apoptosis. J. Natl. Cancer Inst. (Bethesda) 1997;89:863–868. doi: 10.1093/jnci/89.12.863. [DOI] [PubMed] [Google Scholar]
- Peters TM, Owen RJ, Slater E, Varea R, Teare EL, Saverymuttu S. Genetic diversity in the Helicobacter pylori cag pathogenicity island and effect on expression of anti-cagA serum antibody in UK patients with dyspepsia. J. Clin. Pathol. 2001;54:219–223. doi: 10.1136/jcp.54.3.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal ED, Cha J, Lo J, Falkow S, Tompkins LS. Altered states, involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. USA. 1999;96:14559–14564. doi: 10.1073/pnas.96.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge VD, Stone GG, Flamm RK, Beyer J, Versalovic J, Graham DY, Tanaka SK. Molecular typing of Helicobacter pylori isolates from a multicenter US clinical trial by ureC restriction fragment length polymorphism. J. Clin. Microbiol. 1997;35:471–473. doi: 10.1128/jcm.35.2.471-473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater E, Owen RJ, Williams M, Pounder RE. Conservation of the cag pathogenicity island of Helicobacter pylori, associations with vacuolating cytotoxin allele and IS605 diversity. Gastroenterology. 1999;117:1308–1315. doi: 10.1016/s0016-5085(99)70281-7. [DOI] [PubMed] [Google Scholar]
- Stein M, Bagnoli F, Halenbeck R, Rappuoli R, Fantl WJ, Covacci A. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 2002;43:971–980. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- Stein M, Rappuoli R, Covacci A. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. USA. 2000;97:1263–1268. doi: 10.1073/pnas.97.3.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor NS, Fox JG, Akopyants NS, Berg DE, Thompson N, Shames B, Yan L, Fontham E, Janney F, Hunter FM. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J. Clin. Microbiol. 1995;33:918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Tummuru MK, Cover TL, Blaser MJ. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori, evidence of linkage to cytotoxin production. Infect. Immun. 1993;61:1799–1809. doi: 10.1128/iai.61.5.1799-1809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn LJ, Figueiredo C, Sanna R, Blaser MJ, Quint WG. Distinct variants of Helicobacter pylori cagA are associated with vacA subtypes. J. Clin. Microbiol. 1999;37:2306–2311. doi: 10.1128/jcm.37.7.2306-2311.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakoob J, Fan XG, Hu GL, Yang HX, Liu L, Liu SH, Tan DM, Li TG, Zhang Z. Polycolonization of Helicobacter pylori among Chinese subjects. Clin. Microbiol. Infect. 2000;7:187–192. doi: 10.1046/j.1198-743x.2001.00226.x. [DOI] [PubMed] [Google Scholar]
- Yakoob J, Jafri W, Abbas Z, Abid S, Khan R, Jafri N, Ahmad Z. Low prevalence of the intact cag pathogenicity island in clinical isolates of Helicobacter pylori in Karachi, Pakistan. Br. J. Biomed. Sci. 2009;66:137–142. doi: 10.1080/09674845.2009.11730260. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Argent RH, Letley DP, Thomas RJ, Atherton JC. Tyrosine phosphorylation of CagA from Chinese Helicobacter pylori isolates in AGS gastric epithelial cells. J. Clin. Microbiol. 2005;46:786–790. doi: 10.1128/JCM.43.2.786-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]