Abstract
Cytochrome (cyt) P450s hydroxylate a variety of substrates that can differ widely in their chemical structure. The importance of these enzymes in drug metabolism and other biological processes has motivated the study of the factors that enable their activity on diverse classes of molecules. Protein dynamics have been implicated in cyt P450 substrate specificity. Here, 2D IR vibrational echo spectroscopy is employed to measure the dynamics of cyt P450cam from Pseudomonas putida on fast timescales using CO bound at the active site as a vibrational probe. The substrate-free enzyme and the enzyme bound to both its natural substrate, camphor, and a series of related substrates are investigated to explicate the role of dynamics in molecular recognition in cyt P450cam and to delineate how the motions may contribute to hydroxylation specificity. In substrate-free cyt P450cam three conformational states are populated, and the structural fluctuations within a conformational state are relatively slow. Substrate binding selectively stabilizes one conformational state, and the dynamics become faster. Correlations in the observed dynamics with the specificity of hydroxylation of the substrates, the binding affinity, and the substrates’ molecular volume suggest that motions on the hundreds of ps timescale contribute to the variation in activity of cyt P450cam towards different substrates.
I. Introduction
Protein dynamics are thought to play an important role in the activity of the cytochrome (cyt) P450s,1–3 a family of monooxygenases of substantial biological and medicinal importance.4,5 Cyt P450 enzymes display a remarkable ability to act on diverse classes of compounds that can differ vastly in their size, structure, and chemical nature.6 Also, the specificity of their activity for different substrates varies widely for individual enzymes of the family. Structural studies suggest the promiscuity of cyt P450s results from the high plasticity of their active sites.7–11 While large structural changes are evident from comparison of some cyt P450 crystal structures, often substrate binding leads to only localized changes in protein structure, such as small loop rearrangement, helix distortion and displacement, side chain rotation, and solvent rearrangement.12–15 The motions involved in these smaller structural changes occur on fast timescales that have been historically difficult to investigate.
Among cyt P450s, the paradigmatic cyt P450cam from Pseudomonas putida shows relatively high specificity for small, hydrophobic compounds that are similar to its physiological substrate, camphor.16,17 The high specificity is thought to result from relatively constrained active site dynamics.2,12 While cyt P450cam does act on a number of compounds, lower substrate binding affinity, hydroxylation efficiency, and stereo/regiospecificity are observed compared to camphor, the natural substrate.18–20 For example, camphor hydroxylation occurs with 100% specificity at the 5′-exo carbon, but hydroxylation of a similar compound, norcamphor, results in only 60% of the corresponding product.20 Calculations suggest that the variations in activity cannot be explained by differences in the chemical reactivity of the substrates, and that the fluctuating protein environment contributes substantially to the differences.21
Only small differences are observed among the numerous crystal structures solved for cyt P450cam bound to different substrates.14,16,22–25 However, the temperature factors from these structures16,22–25 and a recent structure of the substrate-free enzyme in a new conformation26 suggest localized motions in cyt P450cam vary among the substrate-free and different substrate-enzyme complexes. The possibility that dynamics play an important role in the relative specificity of cyt P450cam is supported also by molecular dynamics simulations2 and numerous other experimental studies; for example, experiments involving hydrostatic and osmotic pressure variations,27–30 CO rebinding kinetics,31,32 linear infrared spectroscopy,30,33,34 and both linear and time-resolved visible spectroscopy.35,36 While these studies suggest that differences in fast timescale motions exist among cyt P450cam-substrate complexes, a direct measurement of dynamics is important to verify the role of dynamics and also define how dynamics might contribute to biological function. The technique of two-dimensional infrared (2D IR) vibrational echo spectroscopy enables the direct measurement of the fast structural dynamics of cyt P450cam for various bound substrate complexes.
2D IR spectroscopy has been applied previously to characterize the dynamics in a number of proteins and investigate their involvement in ligand binding,37–40 protein folding,41–43 and other biological processes.44–49 In studies of heme proteins, heme-bound CO has provided a particularly useful vibrational probe of dynamics.38,42,44,45,48 The stretch of CO occurs in a spectral region free of other protein vibrations, its large transition dipole leads to strong signals, and the local-mode nature of its vibration simplifies spectral interpretation. Perhaps most advantageous is the sensitivity of the CO vibrational frequency to the electric field resulting from the protein environment. The vibrational spectrum of CO bound to cyt P450cam iron-heme is highly dependent on the presence and nature of ligands bound in the active site.30,33,34 As the CO environment changes due to protein structural motions, the CO vibrational frequency undergoes corresponding fluctuations. 2D IR vibrational echo spectroscopy can be used to measure the timescales and amplitudes of these frequency fluctuations, and thus characterize the dynamics of the CO’s protein environment.
In this study, 2D IR vibrational echo spectroscopy was used to assess the contribution of protein dynamics to substrate specificity and binding affinity in cyt P450cam. The influence of substrate binding on cyt P450cam motion was investigated by measuring the dynamics of substrate-free and substrate-bound proteins. In addition, the involvement of protein motions in controlling the selectivity of hydroxylation of different substrates was examined by characterizing the dynamics of cyt P450cam-CO bound to substrates with mild variations from the camphor structure (see Figure 1); that is, camphane, adamantane, norcamphor, and norbornane. The study finds that substrate binding stabilizes a single dominant conformation of the substrate-free enzyme and increases the rate of structural fluctuations that lead the protein to sample its conformational substates within this dominant conformation. Further, the 2D IR data demonstrate that motions occurring on the hundred picosecond timescale are correlated to the selectivity of the activity and to the binding affinity of cyt P450cam for different substrates, supporting the involvement of fast protein structural dynamics.
Figure 1.

Structures of cyt P450cam and substrates. A) Structure of the cyt P450cam-CO-camphor complex in the immediate vicinity of the CO ligand (PDB entry 1T87). Shown are the heme, the CO distal ligand, the Cys357 proximal ligand, the bound camphor substrate, and several residues of helix I, which forms one wall of the active site. B) Structures of substrates whose complexes with cyt P450cam were investigated with 2D IR vibrational echo spectroscopy.
II. Experimental Procedures
A. Sample preparation
Cyt P450cam was produced by recombinant expression with plasmid pDNC-334A in E. coli strain NCM53350 (kindly provided by the Pochapsky group at Brandeis University). Experiments were performed with cyt P450cam C334A, which has been shown to display activity identical to wild-type but decreased propensity for aggregation.51 Protein expression and purification were performed as previously reported.52,53 Cyt P450cam used in these experiments showed an absorbance ratio 392 nm/280 nm greater than 1.6 and less than 5% of the inactive P420 form by UV/Vis spectroscopy.
To prepare the complexes with the unnatural substrates, camphor was removed by passage over Sephadex G25 in 50 mM Tris-MOPS, pH 7.4, 20% w/v glycerol. The protein was exchanged by concentration and dilution into 50 mM sodium phosphate, pH 7.0, 100 mM KCl in D2O containing 20% w/v glycerol. The glycerol was found to prevent accumulation of cyt P420 in the substrate-free enzyme during 2D IR data acquisition. Experiments were also performed with the camphor complex in a sample containing no glycerol, and showed no significant differences. To prepare the norcamphor complex, the enzyme was exchanged into buffer containing 85 mM norcamphor. All other substrates were first dissolved in ethanol to 100–700 mM before addition to the concentrated protein solution (2–3 mM) to final concentrations of 4–23 mM, such that the final protein solution contained no greater than 5% substrate-free protein35 or ethanol. The samples were clarified through 0.45 μm filters before being reduced with ten-fold molar excess sodium dithionite. The cyt P450cam-CO complexes were formed by passage of a gentle stream of CO gas over the reduced protein sample.
For the IR experiments the samples were placed between two CaF2 windows with a 50 μm Teflon spacer. The UV/Vis and linear FT IR spectra of the samples were recorded before and after 2D IR spectral acquisition to ensure sample integrity. The linear IR spectra of samples studied with 2D IR spectroscopy showed CO bands of 10–20 mOD on a ~0.35 OD background.
B. Vibrational Spectroscopy
Linear FT IR spectra were acquired at 1 cm−1 resolution on a Bruker Vertex 70 spectrometer. The time-resolved infrared experiments were performed as previously described54,55 with 120 fs, ~5 μJ pulses at 1950 cm−1 generated with an ultrafast mid-IR laser system consisting of a Ti:Sapphire oscillator/regenerative amplifier pumped optical parametric amplifier. Briefly, the 2D IR echo experiments involved application of three mid-IR light pulses to the sample (~0.8 μJ per pulse at the sample) with the times between the first and second pulse and the second and third pulse referred to as τ and Tw, respectively.55 At a time ≤ τ after the third pulse, a vibrational echo is emitted by the sample in a unique direction. The vibrational echo pulse is overlapped with another IR pulse, called the local oscillator, for heterodyne detection and to provide phase information for the vibrational echo signal. The combined vibrational echo/local oscillator pulse is passed through a monochromator onto an IR array detector, which records a spectrum that yields the ωm frequency axis (vertical axis), the axis of vibrational echo emission. Scanning τ produces an interferogram at each ωm. These interferograms are then Fourier transformed to produce the second, ωτ axis (horizontal axis) of the 2D IR spectrum. In the experiments, τ is scanned for fixed Tw to produce a 2D IR spectrum. Tw is then changed, and τ is again scanned to produce another 2D IR spectrum. The change in the spectra with Tw provides the dynamical information about the system.
Pump probe experiments are also performed to determine the vibrational lifetimes and for use in processing the 2D IR data. An ~3.5 μJ pump pulse is followed by a variably delayed,~0.4 μJ probe pulse. The pump-induced changes in the probe beam spectrum at each time delay were measured by dispersing the transmitted probe beam through the monochromator onto the array detector.
C. Data Analysis
The linear FT IR spectra were background-corrected by subtracting the solvent FT IR spectrum, followed by fitting and subtracting a polynomial function to the spectral regions away from the CO band. This function was used to make small residual base line corrections. The resulting spectra were then fit to a Gaussian function, or a sum of Gaussian functions for the substrate-free protein and norbornane complex. The time-resolved pump probe spectra were used to determine the vibrational lifetimes (T1) of the CO in each cyt P450cam sample. At each frequency of a spectrum, the pump-induced difference in the probe beam intensity as function of the probe delay time was fit to an exponential decay.
Protein structural fluctuations cause the CO stretch frequency to evolve in time. The time dependent change in frequency is called spectral diffusion. The frequency-frequency correlation function (FFCF) connects the waiting time dependent changes in the 2D band shapes caused by spectral diffusion to the time dependence of the structural changes of the proteins. The center line slope (CLS) method is used to determine the FFCF from 2D and linear spectra. 56,57 This method provides an accurate way to extract the FFCF and also provides a useful quantity to plot.56,57 At a particular ωτ, a slice through the 2D spectrum, projected onto the ωm axis, is a spectrum with a peak at a particular ωm value. Many such slices taken over a range of ωτ values produce a set of points. Connecting the resulting points yields the center line. In the absence of a homogeneous contribution, at Tw = 0 the slope of the center line would be 1. At sufficiently long time, when spectral diffusion has sampled all frequencies within the absorption spectrum, the 2D IR line shape would be circular, and the center line would be horizontal with a slope of zero. It has been shown theoretically that the Tw-dependent part of the normalized FFCF is directly related to the Tw dependence of the slope of the center line.56,57 Thus the slope of the center line, the CLS, will vary between a maximum of 1 at Tw = 0 and 0 in the limit of sufficiently long waiting time. The presence of a homogeneous contribution to the spectrum causes the initial value of the slope to be less than 1 at Tw = 0 (see below).
The multiple time scale dynamics were modeled by a multiexponential form of the FFCF, C(t).
| (1) |
For the ith dynamical process, Δi is the range of CO frequencies sampled due to protein structural fluctuations, and τi is the time constant of these fluctuations. This form of the FFCF has been widely used and in particular found applicable in studies of the structural dynamics of heme-CO proteins.38,42,44–46,54,58–60 The experimental time window is limited by the vibrational lifetime decay to several times T1, the vibrational lifetime, which reduces the signal to zero. Occurrence of very slow structural fluctuations on timescales longer than the experimental time window, if present, will appear as the static term Δs2 in the FFCF.
If Δτ < 1 for one component of the FFCF, then Δ and τ cannot be determined separately, but rather gives rise to a motionally narrowed homogeneous contribution to the absorption spectrum with pure dephasing width given by , where is the pure dephasing time, and Γ is the pure dephasing line width. The total homogeneous dephasing time, T2, also has contributions from the vibrational lifetime and orientational relaxation, Tor. T2 is given by
| (2) |
In these experiments, orientational relaxation of the entire protein is so slow that its contribution can be neglected. Detailed procedures for converting the CLS measurement into the FFCF have been described previously.56,57 By combining the CLS with the linear absorption spectrum, the full FFCF is obtained including the homogeneous component. Then using the vibrational lifetime, and Γ are obtained from Equation 2.
Due to the overlap of the three bands in the substrate-free FT IR spectrum (see Figure 2A below), the range used to fit the center line was restricted as follows. A cut along the diagonal was taken from each 2D spectrum. The diagonal spectrum was fit to a sum of three Gaussian bands, which were then used to calculate the relative amplitude of the bands at each frequency. Regions containing greater than 85% amplitude of a band were used to determine the CLS for that band.
Figure 2.
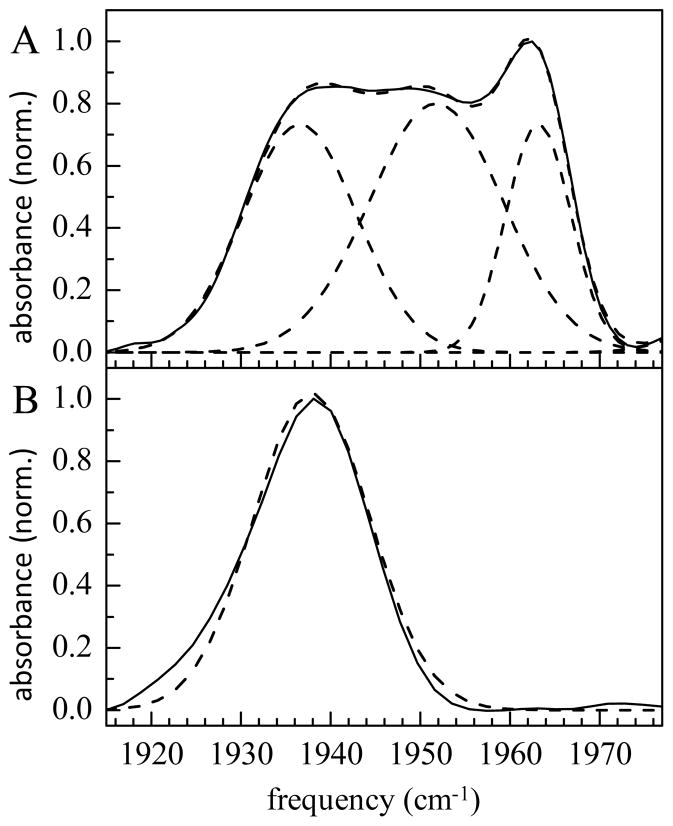
Linear FT IR spectra (solid lines), along with the Gaussian fits (dashed lines) to the spectra, of A) substrate-free cyt P450cam-CO and B) its complex with camphor.
III. Results and Discussion
Figure 2A displays the FT IR absorption spectrum of substrate-free cyt P450cam-CO along with a fit to three Gaussians functions. The fit provides the center frequency and line width of each band. The spectrum of unbound cyt P450cam-CO contains three distinct overlapping bands ranging in frequency from 1937 to 1963 cm−1. The relative amplitudes of these bands depend on glycerol concentration, as previously observed.30 At the glycerol concentration used in the 2D IR experiments (20% w/v), the amplitudes of the three bands are roughly equal. Upon addition of the natural substrate camphor, the spectrum of cyt P450cam-CO is dominated by a single band at 1938 cm−1 with a line width of 15 cm−1. The spectrum is shown in Figure 2B with a Gaussian fit. As observed previously,30,33,34 the frequencies and line widths of the CO vibrational band vary with the nature of a bound substrate (see Table 1). Single absorption peaks are found in the spectra of all substrate complexes investigated here except for the norbornane complex, which shows a very small low frequency shoulder. The FT IR spectra are in accord with previous studies,34 with the exception of the spectrum of the camphane complex. While previously three absorption peaks at 1941, 1955, and 1962 cm−1 were observed, in our study the cyt P450cam-CO-camphane FT IR spectrum shows one peak with a frequency of 1955 cm−1 when care was taken to assure that binding with the substrate was complete.
Table 1.
Absorption Spectrum Parameters and Lifetimes
As shown below by the 2D IR vibrational echo experiments, all of the absorption lines for the unbound protein and the protein complexes are inhomogeneously broadened. Thus, the spectral line width reflects the distribution of protein structures that occurs in the ensemble of proteins in the sample. In all cases, binding unnatural substrates shifts the CO band to higher frequency relative to binding camphor. While the line width depends on the structure throughout the protein,61 the center CO frequency in heme proteins is particularly sensitive to the local environment. In myoglobin-CO (MbCO), with the imidazole side group of the distal histidine His64 in the pocket, there are two peaks, denoted A1 and A3, that have been attributed to different configurations of the imidazole relative to the CO.54,59,62 Changes in the peak frequency of the CO in heme proteins have been attributed to changes in the electric field at the CO. Two mechanisms have been put forward. One mechanism uses direct Stark effect coupling of the proteins’ electric field at the CO. Simulations of MbCO and mutant MbCO using this direct Stark coupling mechanism obtained the FFCF from the fluctuating electric field along the CO bond with the Stark coupling constant as a single adjustable parameter.54,59,62 The FFCF was used to calculate the IR absorption spectrum and 2D IR vibrational echo data. The results yield the A1 and A3 bands with virtually the correct line shapes, but with a small error in the frequency separation between the two bands. The simulated FFCFs for the two bands show decay times within factors of ~2 of the experimentally observed dynamics within each substate. In the second mechanism, the electric field results in changes in the extent of back bonding from the Fe-heme π electron system to the CO antibonding π molecular orbital resulting in shifts in frequencies for different proteins and different protein substates.63–65
The structures of the cyt P450cam substrate complexes show possible contacts between the CO and the substrate and possibly one or more water molecules in the active site pocket. In the structure of cyt P450cam-CO-camphor complex,25 the camphor sits above the CO, well packed within the active site by protein side chains. Cyt P450cam structures with smaller unnatural substrates show the substrates bound further from the heme plane than camphor.16,24 This may permit the introduction of water molecules into the active site that either direct electron density toward the CO or act as a dielectric shield of the electric fields generated from more distant regions of the protein, providing a possible structural interpretation for the higher CO frequencies of these complexes. In contrast, the FT IR spectrum of substrate-free cyt P450cam indicates population of at least three, distinct conformational substates (see Figure 2A). The lowest frequency peak is similar in frequency and line width to the camphor complex (see Figure 2 and Table 1). As with the unnatural substrate complexes, the two high frequency lines observed in the substrate-free protein have been postulated to result from variation in water content and orientation,30 but could also result from variation in the positions of side groups in the pocket.
IR pump-probe experiments were performed on each cyt P450cam-CO complex to determine the CO stretching mode’s vibrational lifetime. The lifetimes for CO bound in the different complexes and the substrate-free protein range between 18–27 ps (see Table 1) and increase linearly with increasing frequency, as previously observed in studies of vibrational relaxation in MbCO and metal-carbonyl complexes.66–68 The vibrational lifetimes make a small contribution to the homogenous line width, ~0.25 cm−1.
2D IR spectra of cyt P450cam-CO (see examples in Figure 3) were obtained for varying waiting times Tw to examine the dynamics in the substrate-free protein and the camphor, camphane, norcamphor, adamantane, and norbornane complexes. All spectra show peaks due to the 0–1 transitions along the diagonal (red) as well as negative peaks (blue), which arise from vibrational echo emission at the 1–2 transition frequencies, shifted to lower frequency along the ωm axis by the vibrational anharmonicity (25 cm−1).45 In Figure 3, the negative going 1–2 peaks can be seen in the panels for the substrate-free complex. The other spectra in Figure 3 have been cropped so that the 0–1 peak can be shown more clearly. All of the 2D IR spectra display small additional peaks that arise from fifth order processes.69 These are discussed in the Supporting Information and are shown plainly in Figure SI-1 of the Supporting Information. For example, the slight positive peak at ωτ of 1940 cm−1 and ωm of 1963 cm−1 in the 2D IR spectrum of the substrate-free enzyme at Tw = 24 ps (lower left panel of Figure 3) is a 5th order signal, not an exchange peak.48,62,70 The 5th order contributions to the signals do not affect the data analysis that determines the dynamics using the 0–1 diagonal peaks69 as discussed in the Supporting Information. A complete description of the 5th signals will be presented in a separate publication.
Figure 3.
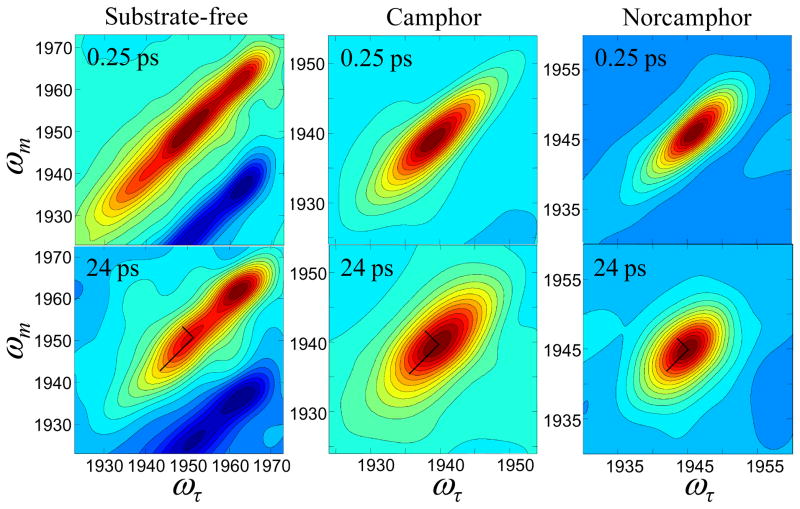
2D IR spectra of substrate-free cyt P450cam-CO (left), the complex with camphor (middle), and the complex with norcamphor (right), at short (upper) and long (lower) Tw delay times. The spectral region around the 0–1 transition is shown, with the contour lines representing a 5 % change in amplitude. To illustrate the differences in lineshapes, lines are drawn over five contours along the diagonal and antidiagonal in the 2D IR spectra with Tw of 24 ps.
The lowest frequency positive peak in the substrate-free spectrum appears to decrease anomalously quickly. This is because it has a somewhat shorter lifetime than the other two substates, and it has faster spectral diffusion than the 1952 cm−1 peak. The faster spectral diffusion reduces the 0–1 peak amplitude more, and it also causes more overlap with the corresponding negative going 1–2 peak, which is also broadening quickly.
The important feature in this experiment is the change in the shape of the 0–1 bands in the 2D IR spectra as Tw is increased, as illustrated in Figure 3. As the structure of the protein evolves in time, the CO frequency changes (spectral diffusion) because its frequency is sensitive to the evolving protein structure. Experiments and simulations on MbCO and experiments on neuroglobin-CO have shown that the CO frequency fluctuations are sensitive to global protein structural dynamics.54,60,62,71 At short waiting times, shown in the upper panels of Figure 3, the peaks in the spectra are substantially elongated along the diagonal, reflecting the incomplete sampling of the inhomogeneous distribution of states contained in the absorption line shape. The peak appears elongated along the diagonal because most of the CO molecules initially at frequencies ωτ have the same final frequencies ωm following the short Tw delay. The width perpendicular to the diagonal at very short time is caused by the motionally narrowed pure dephasing contribution with a small additional contribution from the vibrational lifetime. At longer waiting times, shown in the bottom panels of Figure 3, the protein has had time to sample more of its range of structures. The CO stretching mode no longer has the same initial (ωτ) and final (ωm) frequencies due to evolution of the protein structure during the time period Tw, and the bands in the spectra become less elongated along the diagonal (the major and minor axis are more similar). As can be seen in Figure 3, clear differences are apparent among the line shapes of the substrate-free, camphor bound, and norcamphor bound cyt P450cam in the 2D IR spectra at Tw = 24 ps. The differences are apparent from comparison of the ratios of the widths of the antidiagonal (width perpendicular to the principal diagonal) to the diagonal width. The ratio of the antidiagonal to the diagonal widths is a useful qualitative measure of the shapes of the bands. Using the largest five contours in the bottom panels of Figure 3 (see perpendicular lines on spectra), the ratios are 0.7, 0.5, and 0.3 for the norcamphor complex, the camphor complex, and the 1952 cm−1 substrate-free band, respectively.
To quantitatively compare the dynamics in the different cyt P450cam-CO complexes, the CLS was determined from the 2D IR spectra for each Tw. Figure 4A shows the CLS vs. Tw for P450cam with its natural substrate camphor and a fit to the data. Figure 4B displays the CLS time dependence for the three substrate-free bands. Because of the overlap of these bands and the short lifetimes of the lowest frequency bands, the time range for two of the bands is limited. The fit to the data for these two bands has been extended as an aid to the eye for comparison with the other curves. Figure 4C shows the CLS decays for all of the different substrate complexes, including the camphor substrate. As discussed in Section II, Experimental Procedures, the CLS decays were analyzed in combination with the linear FT IR spectra and vibrational lifetimes to obtain the FFCFs, which quantify the dynamics associated with the fluctuating CO frequency. The parameters describing the FFCFs of the different cyt P450cam samples are listed in Table 2.
Figure 4.

CLS decay curves and corresponding exponential fits for A) cyt P450cam-CO bound with its natural substrate camphor. The two timescales seen in the decays are evident. B) the 1939 cm−1 (red), 1952 cm−1 (green), and 1963 cm−1 (blue) bands of substrate-free cyt P450cam-CO. The fits have been extended for two of the bands as an aid to the eye for comparing to the other curves. C) The different substrate complexes: camphor (black), camphane (blue), adamantane (green), norcamphor (purple), and norbornane (red).
Table 2.
Dynamic Parameters from 2D IR Spectra
| Substrate | T2* (ps) | Γ (cm−1) | Δ1 (cm−1) | ν1 (ps) | Δ2 (cm−1) | τ2 (ps) |
|---|---|---|---|---|---|---|
| Camphor | 5.6 ± 1.1 | 1.9 ± 0.4 | 2.8 ± 0.3 | 6.8 ± 1.4 | 5.4 ± 0.2 | 370 ± 65 |
| Camphane | 9.8 ± 3.7 | 1.1 ± 0.4 | 1.8 ± 0.3 | 5.5 ± 1.8 | 3.8 ± 0.1 | 300 ± 60 |
| Adamantane | 8.5 ± 1.1 | 1.3 ± 0.2 | 2.3 ± 0.3 | 1.6 ± 0.6 | 4.1 ± 0.07 | 260 ± 40 |
| Norcamphor | 6.0 ± 2.2 | 1.8 ± 0.7 | 1.8 ± 0.1 | 5.8 ± 1.0 | 3.2 ± 0.1 | 110 ± 6 |
| Norbornane | 12 ± 0.4 | 0.86 ± 0.1 | 2.4 ± 0.5 | 2.2 ± 1.2 | 4.7 ± 0.06 | 230 ± 30 |
| Unbound 1937 cm−1 | 14 ± 4.6 | 0.78 ± 0.3 | 3.3 ± 0.5 | 17 ± 6.0 | 5.3 ± 0.4 | ∞ |
| 1952 cm−1 | 38 ± 7.3 | 0.28 ± 0.1 | 1.9 ± 0.2 | 9.8 ± 3.2 | 6.3 ± 0.05 | ∞ |
| 1963 cm−1 | 7.5 ± 2.7 | 1.4 ± 0.5 | 1.7 ± 0.1 | 11 ± 2.1 | 2.9 ± 0.04 | ∞ |
The ultrafast protein dynamics plus contributions from the solvent fluctuations produce a motionally narrowed Lorentzian component of the absorption spectrum,54 which is characterized by the pure dephasing time, , and the pure dephasing line width, Γ. Both of these parameters are given in Table 2. The pure dephasing times of ~6–12 ps obtained for the cyt P450cam-CO substrate complexes (see Table 2) are within the range of the 3–12 ps times observed in other heme protein-CO complexes such as Mb,72 neuroglobin,46,60 horseradish peroxidase,38 and a thermophilic cyt c 552.61,73 In previous studies, comparisons of wild-type and mutant proteins suggested contributions to the homogeneous dephasing might arise from fast fluctuations of polar groups positioned near the CO molecule. For example, introduction of a glutamine side chain in cyt c 55273 and removal of a histidine side chain in Mb72 led to decreased and increased dephasing times, respectively, presumably due to the manipulation of a rapidly fluctuating polar group in the heme pocket. In comparing the pure dephasing times for different bound substrates, it would be expected that contributions from fluctuations of the solvent would be very similar. Of the cyt P450cam-CO complexes studied here, the camphor and norcamphor complexes displayed the shortest pure dephasing times. These substrates are the only ones with carbonyl groups (see Figure 1), suggesting that this polar moiety may influence the pure dephasing of the proximal CO. Consistent with this possibility, the vibrational bands of the substrate-free protein showed slower homogeneous dephasing times. While fluctuation of the carbonyl group itself, or its hydrogen bond with residue Tyr67, may be directly responsible for the faster homogenous dephasing observed in the camphor and norcamphor complexes, the presence of the carbonyl may also indirectly influence the CO vibration by promoting a structure that directs another fluctuating polar group, such as a water molecule or the hydroxyl group of Thr252, toward the CO molecule.
While the homogeneous dephasing is responsible reduction of the Tw = 0 value of the CLS from 1, the slower dynamics give rise to spectral diffusion and the Tw dependent decay of the CLS curve. The amplitudes and time scales associated with the spectral diffusion are caused by protein structural fluctuations. As discussed above, the FFCF quantifies the pure dephasing and spectral diffusion dynamics. The FFCFs determined by 2D IR spectroscopy show protein dynamics induced spectral diffusion of all cyt P450cam-CO samples occurring on roughly two timescales (see Table 2). A shorter timescale component of ~6 ps is observed for camphor, camphane, and norcamphor complexes, while faster corresponding dynamics with a correlation time of ~2 ps are found for the adamantane and norbornane complexes. The short timescale of these dynamics suggests that they arise from motions involving relatively small groups, such as side chain fluctuations or motions of active site water molecules. The dynamics on this timescale are significantly slower in the substrate-free cyt P450cam-CO. The FFCFs determined from all three bands of the substrate-free enzyme display relatively slow correlation times of ~10–17 ps. As the substrate is replaced by water molecules in the substrate-free enzyme,22,26 active site water motions are a likely contributor to the observed differences in dynamics. It is interesting to note that comparable timescales of 13 ps74 and 18 ps75 have recently been measured for constrained water at water-surfactant interfaces, depending on the nature of the interface, with IR polarization selective pump-probe spectroscopy.
The slowest dynamics measured in this study of cyt P450cam with bound substrates occur on the hundreds of ps timescale. Dynamics occurring on time scales out to several times the longest Tw contribute to the observed evolution of the 2D spectra.76 In fitting the substrate bound data, allowing a variable offset (infinite time component) in addition to the bi-exponential parameters led to convergence to an offset of zero; that is, no dynamics slower than τ2, the slowest component of the FFCF reported in Table 2. However, it is not possible to completely rule out the existence of even slower dynamics that occur outside of the experimental time window of ~100 ps. The approximately two-fold larger value of Δ2 (root mean frequency fluctuation amplitude) compared to Δ1 for all of the substrate bound complexes show that these slower motions make the greatest contribution to the vibrational linewidth. The FFCFs of the substrate complexes have dynamics with correlation times of 100–370 ps (Table 2). The slow timescale suggests that this component arises from motions that involve larger groups than give rise to the faster dynamics. Molecular dynamics simulations of MbCO find that dynamics throughout the protein contribute to the FFCF,61 and this is likely also the case for the FFCFs of the cyt P450cam complexes. Previous studies employing 2D IR chemical exchange spectroscopy detected 50–100 ps A1-A3 substate switching dynamics in MbCO.48,62,70 X-ray crystallography studies77 and molecular dynamics simulations70 suggest that the observed A1-A3 substate switching involves the motion of the F-helix, although the proximate cause is the change in orientation of the imidazole side group of the distal histidine, H64.54,62,71 In cyt P450cam, motions of helix I are possibly involved in the 100–370 ps dynamics.
While the CLS data for the substrate bound complexes show bi-exponential decays, the substrate-free enzyme show different behavior, with no decay after a Tw time of ~20 ps (see Figure 4). (The timescale of the slowest dynamics for the lowest frequency state could not be reliably determined.) The slowest timescale component in the CLS data for the higher frequency bands of the substrate-free enzyme are well fit by a single exponential decay and a constant offset. The offset arises from a component of the dynamics detected by the 2D IR experiments that is so slow that it falls outside the experimental time window of ~100 ps. There are structures of the substrate-free enzyme that are not sampled on the timescale of the experiment, indicating that the energy landscape contains a range of populated substates separated by relatively high barriers. In comparison, the 100–370 ps dynamics observed for the substrate bound complexes demonstrates that substrate binding produces energy landscapes with relatively lower barriers and therefore faster interconversion among structures. With a substrate bound, the enzyme accesses all of the structural configurations that give rise to the inhomogeneous spectral line on a time scale of <1 ns within experimental error.
Considered together, the 2D IR spectra of the substrate-free enzyme and the substrate bound complexes illuminate the nature of substrate binding in cyt P450cam. The earliest models of protein-ligand binding invoked a lock-in-key mechanism wherein a protein has been tailored through evolution to possess a perfectly complementary binding pocket for its specific ligand.78 This simple model involves no protein motion, and is not supported by the data presented here for cyt P450cam. The models of conformational selection79 and induced-fit80,81 ligand binding include a contribution from protein dynamics, and fundamentally differ in whether protein motions occur before (conformational selection) or in response to (induced-fit) ligand binding. Although such distinctions are instructive, the actual binding mechanisms of enzymes may not be described completely by a single model. Different experimental techniques may reveal specific aspects of ligand binding.
The linear IR spectra support a conformational selection mechanism of binding in cyt P450cam. The multi-component spectrum of the substrate-free IR spectrum reduces to a single dominate band upon binding in any of the substrate complexes. This change in the absorption spectrum implies population of and interconversion among at least three distinct conformational states in the absence of a substrate and the selective stabilization and population of one state when a substrate binds. It is important to note that the single peak in a substrate bound enzyme spectrum is inhomogeneously broadened, demonstrating that there are still a vast number of structural conformations associated with the state stabilized by substrate binding. While the linear data support a conformational selection binding mechanism, the 2D IR data also show strong evidence of an induced-fit mechanism, as the dynamics within the conformational states are altered as a result of substrate binding. The differences in dynamics are seen clearly from the values reported in Table 2. Both the shorter and longer timescale dynamics are faster in the substrate bound complexes compared to the substrate-free enzyme states. Therefore, overall the 2D IR data demonstrate that substrate binding not only selectively stabilizes a particular substate of a fluctuating substrate-free protein, but that the presence of the substrate also leads to more rapid sampling of the structures that exist within the selected substate that give rise to the inhomogeneously broadened absorption spectrum.
Examination of the crystal structure of the cyt P450cam-CO-camphor tertiary complex reveals several aspects of the active site whose motions can contribute to the dynamics observed by 2D IR spectroscopy.25 The CO ligand is directed toward Gly249 within a helical bulge of distal helix I formed by an unusual hydrogen bond between the backbone carbonyl of Gly248 and the hydroxyl of the side chain of Thr252 (Figure 1).25 It is likely that the CO vibrational frequency is highly sensitive to motions of helix I, and any changes in the enzyme that influence helix I’s motion will be manifested in the time dependence of the 2D IR spectra. Possible motions are apparent from comparison with the structure of the dioxygen complex, which shows a reduction in helix I distortion and rotation of the hydroxyl group of Thr252 toward the dioxygen ligand.
Differences in dynamics are observed not only between substrate-bound and substrate-free states of cyt P450cam, but also among different substrate complexes. The motions on the hundreds of picosecond timescale are found to be the slowest in cyt P450cam bound to its natural substrate, camphor, implying that the barriers to structural fluctuations on this timescale are greatest for this complex. In comparison, the τ2 correlation time is several times shorter for the norcamphor complex. From a simple Eyring model of kinetics,82 comparison of the longer timescales components suggest the barriers to the corresponding structural changes in the camphor complex are roughly 30% greater than in the norcamphor complex, and are consistent with optimal packing of the natural substrate in the cyt P450cam active site. The faster dynamics of the norcamphor complex indicate relatively low kinetic barriers among structural conformations and hence a smoother energy landscape. Conversely, the slower dynamics in the complex with the natural substrate indicate relatively high barriers to motion, leading to the picture of a more “rugged” energy landscape.
With the exception of the outlier norcamphor, the dynamics on this longer timescale in general become faster with decreasing molecular volume of the bound substrate. It is important to note that the error bars, which are based on sensitivity analysis of the fits, overlap for some the different substrate bound FFCFs. However, each fit was unique in the sense that different initial guesses for the parameters always converged to the identical fit values. The trend in the values suggests that imperfect packing of the smaller unnatural substrates leads to a reduction in the barriers to motions in the enzyme. However, the molecular volume cannot be the sole factor contributing to the variation in dynamics measured for the different substrate complexes, as norcamphor is slightly larger in volume than norbornane yet displays significantly faster dynamics (see Table 2). Comparison of norcamphor to the other substrate structures (Figure 1) indicates the dynamics result from a small volume combined with the presence of a polar carbonyl group. Perhaps the small size of the molecule permits increased motion of the active site generally, similar to the other small substrates, but that fluctuation of the polar carbonyl group of norcamphor leads to increased motion in the enzyme. While the carbonyl group forms a hydrogen bond to Tyr67 in the crystal structures of both camphor and norcamphor cyt P450cam complexes, molecular dynamics simulations of only the norcamphor complex observed transient breaking of this bond and formation of a new Try67-Thr100 hydrogen bond.17
The unique dynamics observed in the norcamphor complex are particularly notable given the low stereo/regiospecificity of the hydroxylation of the norcamphor substrate. While 100% and 90% 5′-exo hydroxylation product is obtained from camphor and camphane, respectively,19 the activity of cyt P450cam toward norcamphor results in only 60% of the 5′-exo hydroxylation product.20 (100% 1-hydroxyl-adamantane product is obtained,18 but due to symmetry this compound has only two hydroxylation sites. Data could not be found for norbornane, which also has high symmetry.) The correlation between the dynamics of the complexes and their stero/regiospecificity with different substrates is consistent with the involvement of protein dynamics in the selectivity of cyt P450cam activity. While the actual chemical reaction likely involves a low probability incursion to a high energy state, it seems plausible that the more rugged landscape would serve to restrict the particular sequences of structural fluctuations that can lead to the transition state, and thus enhance the regioselectivity of camphor hydroxylation. In contrast, the smoother energy landscape of the norcamphor complex may lead to a more permissive trajectory to the transition state and thus allow the reaction to proceed with the “incorrect” carbon centers of norcamphor.
In addition, the 2D IR results suggest that the dynamics of the complexes may influence the binding affinity, as the dissociation constants and τ2 are also generally correlated. Those substrates with smaller KD values for binding to cyt P450cam (increased binding affinity) show longer τ2 correlation times in the FFCFs of the substrate complexes (Tables 1 and 2). Thus the binding affinity increases with slower dynamics. This effect is not likely due to conformational entropy changes upon binding, as more constrained substrate complexes might be expected to lead to a greater entropic penalty for binding. It is more likely that the higher barriers that result in slower dynamics are due to more enthalpically favorable interactions within the tighter complexes. One may speculate that the restricted dynamics of the bound state can lead to greater retention of the substrate in the pocket. This would decrease the rate of substrate release out of the active site, thereby decreasing KD. Such a possibility is consistent with the common observation for other proteins that the dissociation constants are typically governed by the off rates to a greater extent than the on rates.83
A study that used 2D IR vibrational echo spectroscopy with azide bound at the active site of formate dehydrogenase also found a reduction of slower timescale dynamics when the enzyme bound a cofactor.84 The authors suggest enzyme rigidity to be a signature of the transition state structure and that the sub-picosecond dynamics might reflect motions that modulate the donor-acceptors distance in enzymatic hydrogen tunneling. The study of formate dehydrogenase did not investigate a range of substrates to address differences in chemical reactivity or binding. In contrast to the formate dyhdrogenase study and the results presented here, earlier studies that investigated the changes in dynamics upon binding of a variety of ligands to horseradish peroxidase (HRP) found that ligand binding leads to selective population of one conformational state, but found slower dynamics upon substrate binding.38 However, the physiological substrate for HRP is unknown. Therefore, the HRP complexes investigated may not be representative of naturally evolved enzyme-ligand binding. Here, the natural substrate, camphor, and closely related compounds were studied. The results of this study indicate stronger substrate binding and greater regioselectivity correlate with slower dynamics of the enzyme-substrate complexes.
IV. Concluding Remarks
Protein dynamics have been invoked to explain the large variation in specificity toward different substrates displayed by the cyt P450 family of enzymes.1,11,85 Similarly, protein dynamics are believed to be involved in the differential activity toward structurally similar substrates displayed by the relatively substrate-specific cyt P450cam.18,35,36 In this study, the dynamics in cyt P450cam were directly characterized using 2D IR vibrational echo spectroscopy. The data show that, in the absence of substrates, cyt P450cam-CO occupies at least three distinct conformational substates that give rise to three peaks in IR absorption spectrum of the CO stretching mode (see Figure 2). Interconversion among these substates occurs on a relatively slow timescale, a nanosecond or longer. Faster motions (picosecond timescales) lead the enzyme to sample a range of the structures within each substate that give rise to the inhomogeneously broadened absorption line. The binding of substrates stabilizes a particular substate of the substrate-free enzyme resulting in a single peak in the absorption spectrum. Within the selected substate, the rate of structural fluctuations that lead the enzyme to sample its structural conformations is increased relative to the substrate-free dynamics. These results indicate that substrate binding leads to lower barriers between different structures. Such rapid sampling may enhance the rate of obtaining particular catalytically productive structures.
2D IR vibrational echo spectroscopy found that dynamics occur in cyt P450cam substrate complexes on a wide range of timescales. The results suggest that dynamics on the hundreds of ps timescale show a correlation with the specificity of cyt P450cam on the different substrates. The 370 ps dynamics observed in the enzyme bound to the natural substrate camphor are replaced by faster, 110 ps motions in the complex with norcamphor. These changes are correlated with a reduction in stereo/regiospecificity of hydroxylation from 100% for camphor to only 60% for norcamphor. Furthermore, the correlation times of the hundreds of picoseconds component of the dynamics in the enzyme-substrate complexes generally increase with the molecular volumes of the bound substrates and with binding affinity, indicating that motions on this timescale contribute to differences in substrate binding. The complex with the natural substrate, camphor, shows slower motions compared to the unnatural substrate complexes, indicating higher barriers to protein structural changes. The enzyme has evolved to optimally bind camphor and likely to restrict the structural fluctuations that may lead to the transition state such that hydroxylation occurs only at a specific carbon center. Thus, overall the data support the involvement of dynamics in enzyme function, and in particular, the specificity of hydroxylation by cyt P450cam.
Supplementary Material
Acknowledgments
We thank the National Institutes of Health (2-R01-GM061137-09) for support of this research. MCT also thanks the National Institutes of Health for a post doctoral fellowship (F32-GM090549).
Footnotes
Supporting Information Available: The supporting information contains figures of 2D IR spectra showing bands originating from fifth order interactions and the Tw dependence of the band amplitudes, as well as a brief theoretical explanation for the appearance of the fifth order signals. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Skopalík J, Anzenbacher P, Otyepka M. J Phys Chem B. 2008;112:8165. doi: 10.1021/jp800311c. [DOI] [PubMed] [Google Scholar]
- 2.Winn PJ, Lüdemann SK, Gauges R, Lounnas V, Wade RC. Proc Natl Acad Sci USA. 2002;99:5361. doi: 10.1073/pnas.082522999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wade RC, Motiejunas D, Schleinkofer K, Sudarko, Winn PJ, Banerjee A, Kariakin A, Jung C. Biochim Biophys Acta. 2005;1754:239. doi: 10.1016/j.bbapap.2005.07.044. [DOI] [PubMed] [Google Scholar]
- 4.Guengerich FP. Chem Res Toxicol. 2008;21:70. doi: 10.1021/tx700079z. [DOI] [PubMed] [Google Scholar]
- 5.Guengerich FP. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. de Montellano O, editor. Vol. 3. Plenum Press; New York: 2005. p. 377. [Google Scholar]
- 6.Schuler MA, Sligar SG. In: The Ubiquitous Roles of the Cytochromes: Metal Ions in Life Sciences. Sigel A, Sigel H, Sigel RKO, editors. Vol. 3. John Wiley & Sons, Ltd; West Sussex: 2007. p. 1. [Google Scholar]
- 7.Li HY, Poulos TL. Nature Struct Biol. 1997;4:140. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- 8.Yano JK, Koo LS, Schuller DJ, Li HY, de Montellano PRO, Poulos TL. J Biol Chem. 2000;275:31086. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 9.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. Proc Natl Acad Sci USA. 2003;100:13196. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poulos TL, Johnson EF. In: Cytochrome P450: Structure, Mechanism, and Biochemistry. de Montellano O, editor. Vol. 3. Plenum Press; New York: 2005. p. 87. [Google Scholar]
- 11.Ekroos M, Sjögren T. Proc Natl Acad Sci USA. 2006;103:13682. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet BM, Ringe D, Petsko GA, Sligar SG. Science. 2000;287:1615. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 13.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhotiw H. Science. 2004;305:683. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 14.Nagano S, Poulos TL. J Biol Chem. 2005;280:31659. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- 15.Sakurai K, Shimada H, Hayashi T, Tsukihara T. Acta Crystallogr F. 2009;65:80. doi: 10.1107/S1744309108044114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raag R, Poulos TL. Biochemistry. 1991;30:2674. doi: 10.1021/bi00224a016. [DOI] [PubMed] [Google Scholar]
- 17.Harris D, Loew G. J Am Chem Soc. 1995;117:2738. [Google Scholar]
- 18.White RE, Mccarthy MB, Egeberg KD, Sligar SG. Arch Biochem Biophys. 1984;228:493. doi: 10.1016/0003-9861(84)90015-8. [DOI] [PubMed] [Google Scholar]
- 19.Atkins WM, Sligar SG. J Biol Chem. 1988;263:18842. [PubMed] [Google Scholar]
- 20.Loida PJ, Sligar SG, Paulsen MD, Arnold GE, Ornstein RL. J Biol Chem. 1995;270:5326. doi: 10.1074/jbc.270.10.5326. [DOI] [PubMed] [Google Scholar]
- 21.Collins JR, Loew GH. J Biol Chem. 1988;263:3164. [PubMed] [Google Scholar]
- 22.Poulos TL, Finzel BC, Howard AJ. Biochemistry. 1986;25:5314. doi: 10.1021/bi00366a049. [DOI] [PubMed] [Google Scholar]
- 23.Poulos TL, Finzel BC, Howard AJ. J Mol Biol. 1987;195:687. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 24.Raag R, Poulos TL. Biochemistry. 1989;28:917. doi: 10.1021/bi00428a077. [DOI] [PubMed] [Google Scholar]
- 25.Raag R, Poulos TL. Biochemistry. 1989;28:7586. doi: 10.1021/bi00445a013. [DOI] [PubMed] [Google Scholar]
- 26.Lee YT, Wilson RF, Rupniewski I, Goodin DB. Biochemistry. 2010;49:3412. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diprimo C, Deprez E, Hoa GHB, Douzou P. Biophys J. 1995;68:2056. doi: 10.1016/S0006-3495(95)80384-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung C. BBA - Protein Struct M. 2002;1595:309. [Google Scholar]
- 29.Jung C, Bec N, Lange R. Eur J Biochem. 2002;269:2989. doi: 10.1046/j.1432-1033.2002.02980.x. [DOI] [PubMed] [Google Scholar]
- 30.Jung C, Ristau O, Schulze H, Sligar SG. Eur J Biochem. 1996;235:660. doi: 10.1111/j.1432-1033.1996.00660.x. [DOI] [PubMed] [Google Scholar]
- 31.Contzen J, Ristau O, Jung C. FEBS Lett. 1996;383:13. doi: 10.1016/0014-5793(96)00103-2. [DOI] [PubMed] [Google Scholar]
- 32.Tetreau C, Mouawad L, Murail S, Duchambon P, Blouquit Y, Lavalette D. Biophys J. 2005;88:1250. doi: 10.1529/biophysj.104.050104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung C, Hui Bon Hoa G, Schroeder K, Simon M, Doucet J. Biochemistry. 1992;31:12855. doi: 10.1021/bi00166a021. [DOI] [PubMed] [Google Scholar]
- 34.Jung C, Schulze H, Deprez E. Biochemistry. 1996;35:15088. doi: 10.1021/bi961507i. [DOI] [PubMed] [Google Scholar]
- 35.Schulze H, Hoa GHB, Jung C. BBA - Protein Struct M. 1997;1338:77. doi: 10.1016/s0167-4838(96)00192-6. [DOI] [PubMed] [Google Scholar]
- 36.Prasad S, Mitra S. Biochemistry. 2002;41:14499. doi: 10.1021/bi026379e. [DOI] [PubMed] [Google Scholar]
- 37.Bredenbeck J, Helbing J, Nienhaus K, Nienhaus GU, Hamm P. Proc Natl Acad Sci USA. 2007;104:14243. doi: 10.1073/pnas.0607758104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Proc Natl Acad Sci USA. 2007;104:2637. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandaria JN, Dutta S, Hill SE, Kohen A, Cheatum CM. J Am Chem Soc. 2008;130:22. doi: 10.1021/ja077599o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim M, Hamm P, Hochstrasser RM. Proc Natl Acad Sci USA. 1998;95:15315. doi: 10.1073/pnas.95.26.15315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung HS, Khalil M, Tokmakoff A. J Phys Chem B. 2004;108:15332. [Google Scholar]
- 42.Kim S, Chung JK, Kwak K, Bowman SEJ, Bren KL, Bagchi B, Fayer AA. J Phys Chem B. 2008;112:10054. doi: 10.1021/jp802246h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Backus EHG, Bloem R, Donaldson PM, Ihalainen JA, Pfister R, Paoli B, Caflisch A, Hamm P. J Phys Chem B. 2010;114:3735. doi: 10.1021/jp911849n. [DOI] [PubMed] [Google Scholar]
- 44.Finkelstein IJ, Goj A, McClain BL, Massari AM, Merchant KA, Loring RF, Fayer MD. J Phys Chem B. 2005;109:16959. doi: 10.1021/jp0517201. [DOI] [PubMed] [Google Scholar]
- 45.Finkelstein IJ, Zheng J, Ishikawa H, Kim S, Kwak K, Fayer MD. Phys Chem Chem Phys. 2007;9:1533. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa H, Finkelstein IJ, Kim S, Kwak K, Chung JK, Wakasugi K, Massari AM, Fayer MD. Proc Nat Acad Sci USA. 2007;104:16116. doi: 10.1073/pnas.0707718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolano C, Helbing J, Kozinski M, Sander W, Hamm P. Nature. 2006;444:469. doi: 10.1038/nature05352. [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa H, Kwak K, Chung JK, Kim S, Fayer MD. Proc Natl Acad Sci USA. 2008;105:8619. doi: 10.1073/pnas.0803764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim YS, Liu L, Axelsen PH, Hochstrasser RM. Proc Natl Acad Sci USA. 2009;106:17751. doi: 10.1073/pnas.0909888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamuro Y, Molnar K, Coales S, Ouyang B, Simorellis A, Pochapsky T. J Inorg Biochem. 2008;102:364. doi: 10.1016/j.jinorgbio.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nickerson DP, Wong LL. Protein Eng. 1997;10:1357. doi: 10.1093/protein/10.12.1357. [DOI] [PubMed] [Google Scholar]
- 52.Unger BP, Gunsalus IC, Sligar SG. J Biol Chem. 1986;261:1158. [PubMed] [Google Scholar]
- 53.O’Keefe DH, Ebel RE, Peterson JA. Methods Enzymol. 1978;103:151. doi: 10.1016/s0076-6879(78)52017-x. [DOI] [PubMed] [Google Scholar]
- 54.Merchant KA, Noid WG, Akiyama R, Finkelstein I, Goun A, McClain BL, Loring RF, Fayer MD. J Am Chem Soc. 2003;125:13804. doi: 10.1021/ja035654x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park S, Kwak K, Fayer MD. Laser Phys Lett. 2007;4:704. [Google Scholar]
- 56.Kwak K, Park S, Finkelstein IJ, Fayer MD. J Chem Phys. 2007;127:124503. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 57.Kwak K, Rosenfeld DE, Fayer M. J Chem Phys. 2008;128:204505. doi: 10.1063/1.2927906. [DOI] [PubMed] [Google Scholar]
- 58.Massari AM, Finkelstein IJ, Fayer MD. J Am Chem Soc. 2006;128:3990. doi: 10.1021/ja058745y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merchant KA, Thompson DE, Xu Q-H, Williams RB, Loring RF, Fayer MD. Biophys J. 2002;82:3277. doi: 10.1016/S0006-3495(02)75669-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ishikawa H, Kim S, Kwak K, Wakasugi K, Fayer MD. Proc Nat Acad Sci USA. 2007;104:19309. doi: 10.1073/pnas.0709760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massari AM, Finkelstein IJ, McClain BL, Goj A, Wen X, Bren KL, Loring RF, Fayer MD. J Am Chem Soc. 2005;127:14279. doi: 10.1021/ja053627w. [DOI] [PubMed] [Google Scholar]
- 62.Bagchi S, Nebgen BT, Loring RF, Fayer MD. J Am Chem Soc. 2010;132:18367. doi: 10.1021/ja108491t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spiro TG, Wasbotten IH. J Inorg Biochem. 2005;99:34. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 64.Park ES, Andrews SS, Hu RB, Boxer SG. J Phys Chem B. 1999;103:9813. [Google Scholar]
- 65.Phillips GN, Jr, Teodoro MN, Li T, Smith B, Olson JS. J Phys Chem B. 1999;103:8817. [Google Scholar]
- 66.Dlott DD, Fayer MD, Hill JR, Rella CW, Suslick KS, Ziegler CJ. J Am Chem Soc. 1996;118:7853. [Google Scholar]
- 67.Hill JR, Dlott DD, Rella CW, Peterson KA, Decatur SM, Boxer SG, Fayer MD. J Phys Chem. 1996;100:12100. [Google Scholar]
- 68.Hill JR, Dlott DD, Fayer MD, Rella CW, Rosenblatt MM, Suslick KS, Ziegler CJ. J Phys Chem. 1996;100:218. [Google Scholar]
- 69.Finkelstein IJ, McClain BL, Fayer MD. J Chem Phys. 2004;121:877. doi: 10.1063/1.1758940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bagchi S, Thorpe DG, Thorpe IF, Voth GA, Fayer MD. J Phys Chem B. 2010;114:17187. doi: 10.1021/jp109203b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Merchant KA, Noid WG, Thompson DE, Akiyama R, Loring RF, Fayer MD. J Phys Chem B. 2003;107:4. [Google Scholar]
- 72.Rector KD, Rella CW, Kwok AS, Hill JR, Sligar SG, Chien EYP, Dlott DD, Fayer MD. J Phys Chem B. 1997;101:1468. [Google Scholar]
- 73.Massari AM, McClain BL, Finkelstein IJ, Lee AP, Reynolds HL, Bren KL, Fayer MD. J Phys Chem B. 2006;110:18803. doi: 10.1021/jp054959q. [DOI] [PubMed] [Google Scholar]
- 74.Fenn EE, Wong DB, Fayer MD. Proc Nat Acad Sci USA. 2009;106:15243. doi: 10.1073/pnas.0907875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moilanen DE, Fenn EE, Wong D, Fayer MD. J Phys Chem B. 2009;113:8560. doi: 10.1021/jp902004r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bai YS, Fayer MD. Phys Rev B. 1989;39:11066. doi: 10.1103/physrevb.39.11066. [DOI] [PubMed] [Google Scholar]
- 77.Teeter MM. Protein Sci. 2004;13:313. doi: 10.1110/ps.03334304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis. Freeman; New York: 1999. [Google Scholar]
- 79.Mccammon JA, Northrup SH. Nature. 1981;293:316. doi: 10.1038/293316a0. [DOI] [PubMed] [Google Scholar]
- 80.Koshland J, Daniel E. Angew Chem, Int Ed. 1994;33:2375. [Google Scholar]
- 81.Koshland J, Daniel E. Adv Enzymol Relat Subj Biochem. 1960;22:45. doi: 10.1002/9780470122679.ch2. [DOI] [PubMed] [Google Scholar]
- 82.Eyring H. J Chem Phys. 1935;3:107. [Google Scholar]
- 83.Northrup S, Erickson H. Proc Natl Acad Sci. 1992;89:3338. doi: 10.1073/pnas.89.8.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bandaria JN, Dutta S, Nydegger MW, Rock W, Kohen A, Cheatum CM. Proc Natl Acad Sci USA. 2010;107:17974. doi: 10.1073/pnas.0912190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cojocaru V, Winn PJ, Wade RC. Biochim Biophys Acta. 2007;1770:390. doi: 10.1016/j.bbagen.2006.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.