Centrosomal localization of kinase-active CK1δ is required for neurite outgrowth in response to Wnt-3a.
Abstract
Previously we determined that Dishevelled-2/3 (Dvl) mediate Wnt-3a–dependent neurite outgrowth in Ewing sarcoma family tumor cells. Here we report that neurite extension was associated with Dvl phosphorylation and that both were inhibited by the casein kinase 1 (CK1) δ/ε inhibitor IC261. Small interfering RNAs targeting either CK1δ or CK1ε decreased Dvl phosphorylation, but only knockdown of CK1δ blocked neurite outgrowth. CK1δ but not CK1ε was detected at the centrosome, an organelle associated with neurite formation. Deletion analysis mapped the centrosomal localization signal (CLS) of CK1δ to its C-terminal domain. A fusion protein containing the CLS and EGFP displaced full-length CK1δ from the centrosome and inhibited Wnt-3a–dependent neurite outgrowth. In contrast to wild-type CK1ε, a chimera comprised of the kinase domain of CK1ε and the CLS of CK1δ localized to the centrosome and rescued Wnt-3a–dependent neurite outgrowth suppressed by CK1δ knockdown. These results provide strong evidence that the centrosomal localization of CK1δ is required for Wnt-3a–dependent neuritogenesis.
Introduction
The Wnts comprise a large family of secreted lipid-modified glycoproteins that have a variety of activities during embryonic development and promote tissue homeostasis in the adult (Klaus and Birchmeier, 2008). They are particularly important in the development of the nervous system where they participate in several morphogenetic events including neural tube closure, formation of specific brain structures, as well as the induction and migration of neural crest cells (Ciani and Salinas, 2005; Malaterre et al., 2007). Wnts also stimulate axonal remodeling, pathfinding, dendritic arborization, and synaptogenesis (Salinas and Zou, 2008).
Several Wnt signaling mechanisms have been implicated in neurite outgrowth (Ciani and Salinas, 2005; Endo and Rubin, 2007; Sánchez-Camacho and Bovolenta, 2009). They are mediated by various Wnts and the Frizzled seven-pass transmembrane Wnt receptors or the atypical tyrosine kinase Wnt receptor, Ryk/Derailed (Yoshikawa et al., 2003; Lu et al., 2004; Liu et al., 2005). Wnt-7a promoted axonal remodeling of mossy fibers in mouse cerebellum by stabilizing microtubules via a mechanism that involved Dishevelled 1 (Dvl-1) and inhibition of glycogen synthase kinase 3β (GSK-3β; Krylova et al., 2000; Ciani et al., 2004). Activation of Dvl-1, Rac1, and c-Jun N-terminal kinase (JNK) by Wnt-7b stimulated dendritic arborization in hippocampal neurons (Rosso et al., 2005). Axon specification in hippocampal neurons was induced by Wnt-5a through a process that relied on interaction of Dvl-2 with atypical PKC-ζ (Zhang et al., 2007), an enzyme that also mediated Wnt-4–dependent extension of commissural axons (Lyuksyutova et al., 2003; Wolf et al., 2008).
As noted in the previous paragraph, Dvl isoforms contribute to Wnt-dependent neurite outgrowth in a variety of ways. Dvls function as positive effectors in the canonical Wnt/β-catenin pathway as well as in the noncanonical planar cell polarity (PCP) and calcium-dependent pathways (Gao and Chen, 2010). Many of their activities have been associated with specific molecular domains and presumed to be regulated by phosphorylation. Dvls have dozens of potential phosphorylation sites and are substrates for several kinases, including casein kinase 1 (CK1), CK2, and protein kinase C (Wallingford and Habas, 2005). In particular, several articles suggest a functional connection between CK1 and Dvls (Bryja et al., 2007a,b; 2008).
The CK1 family of evolutionarily conserved serine-threonine kinases consists of seven isoforms in mammals (α, β, γ1, γ2, γ3, δ, and ε). These enzymes share a highly related kinase domain but differ considerably in the length and sequence of their N- and C-terminal regions. The C-terminal domains have a role in the contrasting activities and regulation of the various isoforms (Graves and Roach, 1995; Gross and Anderson, 1998; Dahlberg et al., 2009). CK1 enzymes participate in multiple processes including DNA repair, cell cycle progression, and circadian rhythm (Gross et al., 1997; Lowrey et al., 2000). All the isoforms except CK1γs phosphorylate Dvl in vivo (McKay et al., 2001). However, accounts of the functional consequences associated with Dvl phosphorylation vary widely. CK1ε was initially identified as a positive regulator of the β-catenin pathway in Xenopus via a Dvl-dependent mechanism (Peters et al., 1999; Sakanaka et al., 1999). Another report claimed that CK1ε-dependent Dvl phosphorylation caused a shift from JNK to β-catenin signaling in Drosophila (Cong et al., 2004). However, others observed that CK1ε stimulated PCP signaling (Strutt et al., 2006), or both PCP and β-catenin signaling in Drosophila after Dvl phosphorylation (Klein et al., 2006). Alternatively, inhibition of CK1δ/ε blocked Wnt-3a–dependent Dvl phosphorylation in a rat dopaminergic cell line, but did not prevent activation of the β-catenin pathway (Bryja et al., 2007a). The same group also documented Wnt-5a–dependent Dvl phosphorylation by CK1δ/ε and linked it to dopaminergic differentiation (Schulte et al., 2005; Bryja et al., 2007b). Subsequently, they suggested that Dvl phosphorylation by CK1δ/ε triggered a switch from Rac1 activation to stimulation of another noncanonical signaling mechanism (Bryja et al., 2008).
Previously, we reported that Wnt-3a induced neurite outgrowth in Ewing sarcoma family of tumor (ESFT) cells via a noncanonical mechanism that required Frizzled-3, Dvl-2/3, and JNK activation (Endo et al., 2008). Now we describe a connection between Dvl phosphorylation and neurite outgrowth. Although CK1δ and CK1ε both contributed to Dvl phosphorylation, only CK1δ was required for Wnt-3a–dependent neurite extension. CK1δ, but not CK1ε was strongly localized to the centrosome, an organelle that functions in neurite formation (de Anda et al., 2005; Higginbotham and Gleeson, 2007), and displacement of CK1δ from the centrosome was associated with inhibition of neurite outgrowth. Moreover, a chimera comprised of the kinase domain of CK1ε and the centrosomal localization signal (CLS) of CK1δ rescued neurite outgrowth when expression of endogenous CK1δ was inhibited by siRNA. These findings demonstrated a surprising difference in function of CK1δ and CK1ε and established the importance of CK1δ centrosomal localization for Wnt-3a–dependent neurite outgrowth.
Results
Dvl-2/3 phosphorylation is associated with Wnt-3a–dependent neurite outgrowth
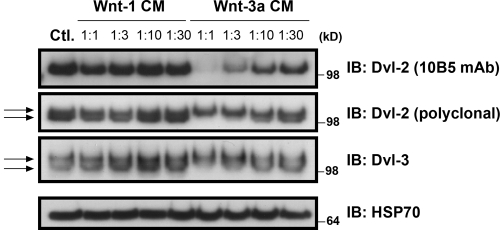
Wnt-3a stimulates neurite outgrowth in a variety of ESFT cell lines including TC-32 cells. In contrast, Wnt-1 conditioned medium (CM) failed to elicit neurite outgrowth (Endo et al., 2008). To investigate differences in the signaling downstream of Wnt-1 and Wnt-3a, we examined dose-dependent changes in Dvl phosphorylation after treatment of TC-32 cells. Wnt-3a CM induced a mobility shift of Dvl-2 and Dvl-3 in SDS-PAGE and decreased the recognition of Dvl-2 by mAb 10B5 (Fig. 1), all signs of Dvl phosphorylation (González-Sancho et al., 2004). No changes in Dvl mobility or immunoreactivity were seen with Wnt-1 CM. However, TC-32 cells were able to respond to Wnt-1 in other ways, as indicated by the stabilization of β-catenin (Endo et al., 2008). The differential effects of Wnt-3a and Wnt-1 on Dvl phosphorylation suggested a potential connection between these post-translational modifications and Wnt-3a–dependent neurite outgrowth.
Figure 1.
Differential response of TC-32 cells to dilutions of Wnt-1 and Wnt-3a CM. After 3 h incubation, whole-cell lysates were immunoblotted with antibodies to Dvl-2, Dvl-3, and HSP70, the last serving as a loading control. Control cells (Ctl.) were incubated with serum-free culture medium for 3 h before processing. Arrows highlight doublet bands indicative of phosphorylation.
CK1δ and CK1ε both phosphorylate Dvl-2/3, but only CK1δ is required for neurite outgrowth induced by Wnt-3a
A screen of small molecule inhibitors targeting kinases known to phosphorylate Dvls revealed that IC261, a preferential inhibitor of CK1δ/ε (Mashhoon et al., 2000), blocked Wnt-3a–dependent Dvl-2 phosphorylation and neurite extension. To confirm that CK1δ and CK1ε were responsible for the Dvl-2 mobility shift, we pretreated TC-32 cells with siRNA reagents directed against CK1δ or CK1ε alone or in combination before incubation with Wnt-3a. Western blot analysis verified that the siRNA reagents specifically inhibited the expression of the corresponding CK1 isoforms (Fig. 2 A). Immunoblotting with the Dvl-2 mAb 10B5 showed that knockdown of each CK1 isoform decreased the Wnt-3a–dependent mobility shift and loss of epitope recognition, whereas the simultaneous knockdown of CK1δ/ε had a stronger effect. The CK1δ/ε siRNA reagents also decreased the Dvl-2 mobility shift and enhanced 10B5 cross-reactivity in the absence of added Wnt-3a, implying that in the basal state Dvl-2 was partially phosphorylated by CK1δ and CK1ε (Fig. 2 A).
Figure 2.
CK1δ and CK1ε both contribute to Dvl phosphorylation but have contrasting roles in neurite outgrowth. (A) TC-32 cells were treated with siRNA reagents targeting expression of luciferase (negative control), CK1δ, and/or CK1ε, and subsequently incubated for 3 h with serum-free culture fluid or 1:10 dilution of Wnt-3a CM. Cell lysates were immunoblotted for Dvl-2, CK1δ, CK1ε, and HSP70. (B) Neurite outgrowth analysis in TC-32 cells treated with siRNA reagents directed against luciferase, CK1δ, or CK1ε, followed by 3 h incubation in the presence or absence of Wnt-3a. The percentage of cells with long neurites was determined and normalized to the percentage observed in cells treated with luciferase siRNA in the absence of Wnt-3a. Results are the means ± SD of three independent experiments. **, P < 0.01; *, P < 0.05. (C) Representative image of phalloidin 488–stained TC-32 cell treated with CK1ε siRNA and no Wnt-3a. Bar, 20 µm. (D) Representative images of phalloidin 488–stained TC-32 cells treated with CK1ε siRNA vs. CK1ε + CK1δ siRNA. Bar, 20 µm. (E) Neurite outgrowth analysis in TC-32 cells treated with siRNA reagents directed against luciferase, CK1δ, and/or CK1ε. Results are presented as described in B. ***, P < 0.001. (F) Immunoblot analysis of CK1δ, CK1ε, and HSP70 in TC-32 cell lysates after siRNA treatment described in E.
When cells were treated with CK1δ/ε siRNA reagents in the neurite outgrowth assay, we obtained a surprising result. Although CK1δ knockdown markedly inhibited Wnt-3a–induced neurite formation (Fig. 2 B), CK1ε knockdown increased neurite formation in the absence of exogenous Wnt-3a, and there was no additional stimulation by Wnt-3a (Fig. 2, B and C). The CK1δ requirement was confirmed when neurite outgrowth was rescued by expression of an siRNA-resistant CK1δ construct. Simultaneous knockdown of CK1δ and CK1ε prevented the neurite outgrowth observed when only CK1ε expression had been suppressed (Fig. 2, D–F), further emphasizing the importance of CK1δ for neurite formation.
CK1δ but not CK1ε is strongly localized to the centrosome
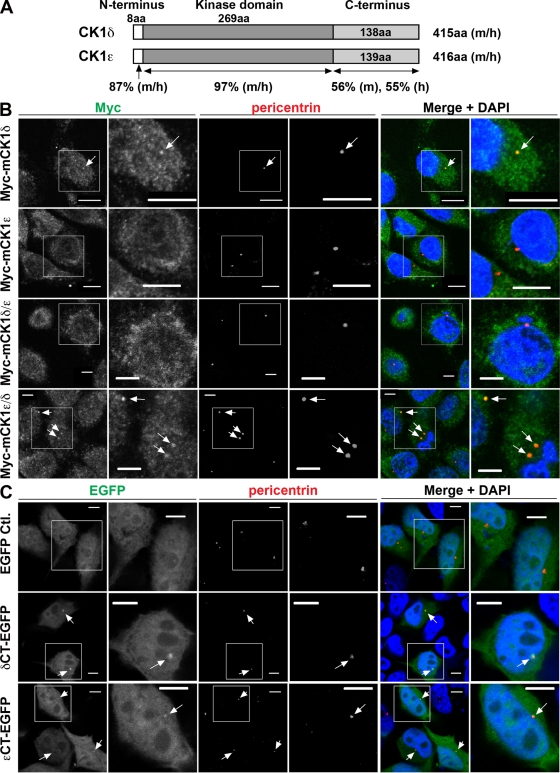
Because neurite outgrowth and axonal specification are dependent on the activity of the centrosome (de Anda et al., 2005; Higginbotham and Gleeson, 2007), we examined the centrosomal distribution of CK1δ and CK1ε in TC-32 cells. Confocal microscopy of methanol-fixed cells showed an intense signal for CK1δ that colocalized with the centrosomal marker γ-tubulin (Fig. 3 A). A weaker signal was detected in the cytoplasm and in neurites (Fig. 3 A). Analysis of CK1ε distribution revealed a diffuse pattern and little colocalization with pericentrin, another centrosomal marker (Fig. 3 B). To ensure that the contrast in centrosomal localization of CK1δ and CK1ε was not due to differences in detection conditions, experiments were performed with HeLa cells expressing Myc-tagged CK1δ or CK1ε and co-stained with Myc and pericentrin antibodies. As in TC-32 cells, only CK1δ colocalized with the centrosomal marker (Fig. 4).
Figure 3.
Contrasting centrosomal localization of endogenous CK1δ and CK1ε. (A) TC-32 cells were cultured in RPMI medium (Control) in the absence or presence of Wnt-3a, fixed in methanol, and stained for CK1δ, the centrosomal marker γ-tubulin, and DNA (DAPI). Arrows point to colocalized signals. Bars, 10 µm. (B) TC-32 cells were cultured as in A, fixed in formaldehyde, and stained for CK1ε, the centrosomal marker pericentrin, and DNA (DAPI). Bars: (top panels) 20 µm; (bottom panels) 5 µm.
Figure 4.
C-terminal domain of CK1δ is necessary and sufficient for centrosomal localization. (A) Schematic diagram of CK1δ and CK1ε protein sequences. Numbers of amino acid (aa) residues in domains from mouse (m) and human (h) proteins are indicated along with the percent sequence identity of CK1δ and CK1ε domains in each species. Domain sequences were obtained at http://www.uniprot.org and amino acid sequence analysis was performed with resources at http://www.ebi.ac.uk/Tools/clustalw2/index.html. (B) Immunofluorescent staining of HeLa cells stably transfected with lentiviral vector encoding Myc-tagged full-length mouse CK1δ or CK1ε, or chimeras in which their C-terminal domains were interchanged (mCK1δ/ε contains the C-terminal domain of CK1ε; mCK1ε/δ contains the C-terminal domain of CK1δ). Cells were fixed in methanol, stained for Myc, pericentrin, and DNA (DAPI). Bars: (top two rows) 10 µm; (bottom two rows) 5 µm. Arrows point to colocalized signals. Magnified area corresponds to the box in adjacent panel to the left. (C) Immunofluorescent signal in HeLa cells transiently expressing EGFP, δCT-EGFP, or εCT-EGFP. Cells were fixed in formaldehyde and co-stained with pericentrin antibody and DAPI. Bars, 5 µm. Arrows point to colocalized signals. Magnified area corresponds to the box in adjacent panel to the left.
To further evaluate the centrosomal distribution of CK1δ and CK1ε, Pearson’s correlation coefficient was calculated for each of the Myc-tagged CK1 proteins and centrosomal pericentrin in transiently transfected TC-32 cells (Zinchuk et al., 2007). The correlation coefficient for CK1δ (0.466 ± 0.147, n = 13) was significantly greater (P < 0.001) than that for CK1ε (0.195 ± 0.073, n = 12), reinforcing the conclusion that CK1δ exhibited a much stronger association with the centrosome.
Centrosomal localization signal of CK1δ is in the C-terminal domain
CK1δ and CK1ε are 87% and 97% identical in their N-terminal and kinase domains, respectively, but only 55–56% identical in their C-terminal domains (Fig. 4 A). We hypothesized that the C-terminal sequences accounted for the differences in their centrosomal distribution. To test this idea, colocalization experiments were performed in HeLa cells that expressed Myc-tagged wild-type CK1 isoforms or chimeras in which the C-terminal domains had been interchanged. Co-staining with antibodies to Myc and pericentrin demonstrated that only wild-type CK1δ and the chimera containing its C-terminal domain (δCT) showed a clear association (Fig. 4 B). This suggested that δCT was required for centrosomal distribution, a conclusion that was confirmed when a CK1δ derivative lacking δCT failed to localize to the centrosome (Fig. S1). To determine whether δCT was sufficient for centrosomal localization, δCT sequence was linked to cDNA encoding enhanced GFP (EGFP). Transient expression in HeLa cells followed by confocal microscopy revealed that δCT-EGFP bound to the centrosome, whereas EGFP did not (Fig. 4 C). A similar construct containing the C-terminal domain of CK1ε exhibited weaker centrosomal localization (Fig. 4 C). Taken together, these findings established that δCT was both necessary and sufficient for CK1δ binding to the centrosome.
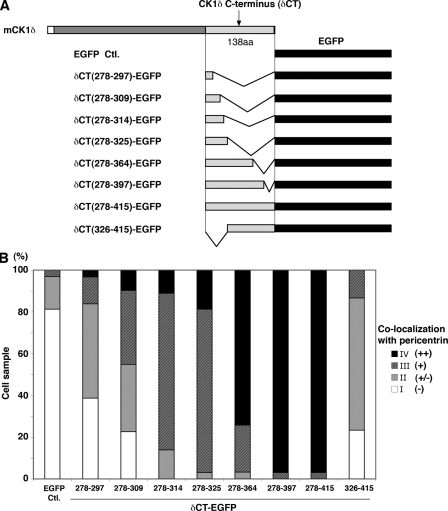
To map the CLS within δCT, a series of δCT-EGFP truncation mutants were generated and their colocalization with pericentrin was examined in transiently transfected HeLa cells (Fig. 5 and Fig. S2). A strong centrosomal signal was seen in >70% of cells expressing the mutant lacking residues 365–415 (δCT(278–364)-EGFP). The derivative lacking residues 326–415 (δCT(278–325)-EGFP) also localized to the centrosome in a large majority of cells, although the signal intensity was diminished. Further deletion of C-terminal sequences resulted in a progressively decreasing proportion of cells in which co-staining with pericentrin was observed. A complementary construct lacking residues 278–325 exhibited little association with the centrosome. These results demonstrated that residues 278–325 were necessary but not sufficient for a strong association with the centrosome. We concluded that the CLS was comprised of residues 278–364. Interestingly, this region contains most of the evolutionarily conserved differences between CK1δ and CK1ε (Fig. S3).
Figure 5.
Centrosomal localization signal of CK1δ was delineated by deletion mutant analysis. (A) Schematic diagram of EGFP fusion proteins containing varying segments from the C-terminal domain of mouse CK1δ (boundaries of segments are indicated by amino acid residue numbers). (B) Centrosomal localization of δCT-EGFP fusion proteins. For each of the indicated δCT-EGFP fusion proteins, colocalization with pericentrin was ascertained in ∼30 cells after transient transfection of HeLa cells. Semi-quantitative analysis was based on the intensity of EGFP signal that colocalized with pericentrin relative to EGFP signal elsewhere in the cell. Intense signal that colocalized with pericentrin was scored as ++ (black), colocalizing signal intensity comparable to that seen elsewhere in the cell was + (dark gray), weak signal was +/− (light gray), and no signal was − (white). The bar graph displays the percentage of cells in each category for all the fusion proteins. See also Fig. S2.
EGFP fusion proteins containing an intact CLS displaced full-length CK1δ from the centrosome and blocked neurite outgrowth
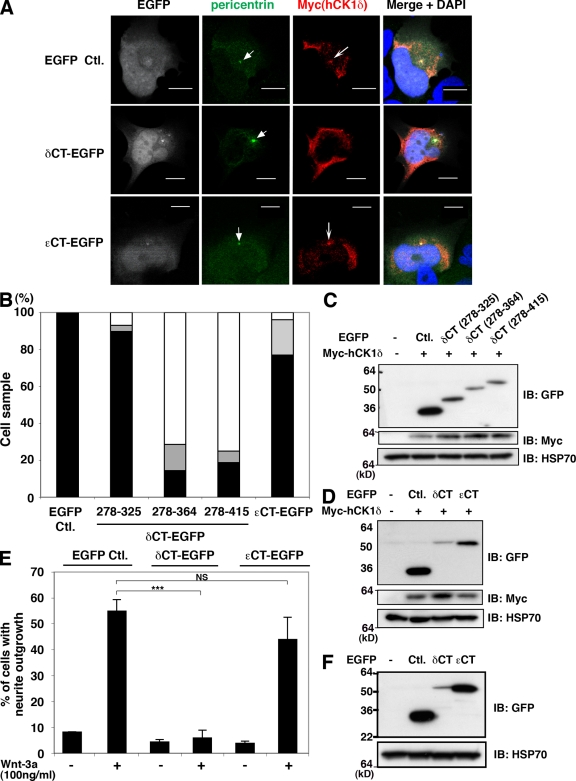
To test the functional relevance of centrosomal CK1δ, we first determined that EGFP derivatives containing the CLS could prevent the accumulation of CK1δ at the centrosome. Myc-tagged CK1δ was required in these experiments because the antibody used to detect endogenous CK1δ cross-reacts with the C-terminal domain. δCT/EGFP and δCT(278–364)-EGFP each inhibited the centrosomal localization of Myc-tagged CK1δ when the constructs were coexpressed in TC-32 cells (Fig. 6, A and B). Semi-quantitative analysis indicated that the centrosomal staining pattern was absent from at least 70% of the ∼30 cells examined (Fig. 6 B). In contrast, when cells coexpressed a truncation mutant lacking approximately half of the CLS, δCT(278–325)-EGFP, only ∼10% lacked the centrosomal staining pattern (Fig. 6 B). EGFP also did not impede the centrosomal distribution of Myc-CK1δ (Fig. 6, A and B). Moreover, εCT/EGFP had little effect on CK1δ centrosomal localization, even when εCT/EGFP was detected at the centrosome (Fig. 6, A and B). Immunoblotting confirmed that the failure to block centrosomal localization was not due to low levels of fusion protein expression (Fig. 6, C and D).
Figure 6.
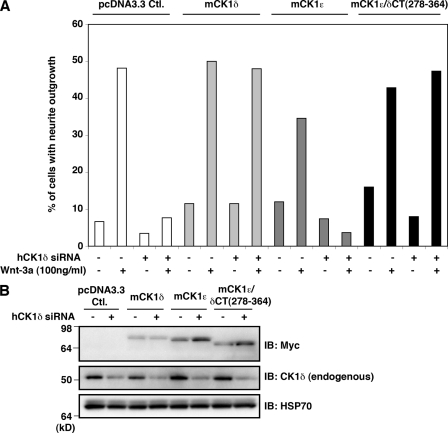
δCT-EGFP, but not εCT-EGFP, displaced CK1δ from the centrosome and inhibited Wnt-3a–dependent neurite outgrowth. (A) Representative confocal micrographs of TC-32 cells that were cotransfected with Myc-hCK1δ and the indicated EGFP constructs, and subsequently probed for Myc and EGFP distribution along with pericentrin and DNA (DAPI). Arrowheads highlight pericentrin signals, arrows indicate centrosomal localization of Myc-hCK1δ. Bars, 10 µm. (B) Semi-quantitative analysis of Myc-hCK1δ colocalization with pericentrin when coexpressed with the indicated EGFP constructs. Percentage of cells with clear colocalization is shown in black, questionable colocalization in gray, and no colocalization in white. Approximately 30 cells were analyzed in each treatment group. (C and D) Immunoblot analysis of the various EGFP derivatives transiently coexpressed in the centrosomal displacement experiments. (E) Wnt-3a–dependent neurite outgrowth in TC-32 cells transiently expressing EGFP, δCT-EGFP, or εCT-EGFP. The presence of neurites was quantified in ∼30 cells expressing the indicated EGFP proteins and incubated in the absence or presence of 100 ng/ml Wnt-3a for 3 h. Results are expressed as the mean ± SD of three independent experiments. ***, P < 0.001. (F) Immunoblot analysis of EGFP derivatives expressed in the neurite outgrowth experiments.
Expression of δCT-EGFP in TC-32 cells dramatically inhibited the neurite outgrowth normally elicited by Wnt-3a (Fig. 6 E). Alternatively, εCT-EGFP and EGFP did not block the response to Wnt-3a even though the proteins were expressed at higher levels than δCT-EGFP (Fig. 6, E and F). These results support the idea that the centrosomal localization of CK1δ is important for Wnt-3a–dependent neurite outgrowth.
Contrasting activity of CK1δ and CK1ε in neurite outgrowth is directly linked to centrosomal localization
To further address the potential relevance of centrosomal CK1δ for neurite outgrowth, we investigated the activity of a CK1ε derivative that contained the CLS of CK1δ in place of its own C-terminal domain. As with the CK1ε/δCT chimera (Fig. 4 B), this protein showed a strong centrosomal staining pattern (Fig. S4). Neurite extension after Wnt-3a administration was abrogated when TC-32 cells were pretreated with CK1δ siRNA targeting a 3′UTR sequence and rescued by the expression of full-length CK1δ (Fig. 7, A and B). Although neurite outgrowth was not maintained by the expression of full-length CK1ε, it was rescued by mCK1ε/δCT(278–364) (Fig. 7, A and B). Thus, the addition of sequence to CK1ε that anchored it like CK1δ to the centrosome was sufficient to enable Wnt-3a–dependent neurite outgrowth.
Figure 7.
mCK1ε/δCT(278–364) rescued Wnt-3a–dependent neurite outgrowth otherwise inhibited by CK1δ siRNA. (A) Wnt-3a–dependent neurite outgrowth in TC-32 cells treated with hCK1δ siRNA targeting 3′-UTR sequence and transiently transfected with empty vector (pcDNA3.3 Ctl.) or construct expressing mouse full-length CK1δ, full-length CK1ε, or chimera consisting of CK1ε N-terminal and kinase domains plus residues 278–364 from mouse CK1δ C-terminal domain. The presence of neurites was quantified in ∼30 cells expressing Myc-tagged CK1 protein for each treatment group. Data are from one of two experiments with similar results. (B) Immunoblot analysis of ectopically expressed Myc-tagged CK1 proteins, endogenous CK1δ, and HSP70 in the neurite outgrowth experiment. Results illustrate the efficacy and specificity of knockdown with siRNA directed against human CK1δ 3′-UTR sequence.
Discussion
The current study is the first to demonstrate a critical role for CK1δ in neurite formation. Knockdown of CK1δ expression by siRNA blocked Wnt-3a–dependent neurite outgrowth in TC-32 cells, and the CK1δ requirement was confirmed when transfection with an siRNA-resistant CK1δ cDNA restored neuritogenesis. Moreover, we determined that the centrosomal localization of CK1δ is important for neurite extension. Using CK1δ/ε chimeras and a series of CK1δ/EGFP fusion proteins we identified the CLS within the CK1δ C-terminal domain. Only EGFP fusion proteins containing this CLS displaced full-length CK1δ from the centrosome, mimicking results obtained with corresponding CLS/EGFP fusion proteins that displaced cyclins A and E from the centrosome (Matsumoto and Maller, 2004; Pascreau et al., 2010). Displacement of CK1δ from the centrosome was associated with inhibition of Wnt-3a–dependent neurite outgrowth, whereas expression of a similar fusion protein lacking the CLS or EGFP alone did not block neurite extension. Prior work demonstrated the importance of cell membrane or nuclear localization for the function of specific CK1 isoforms and implicated the C-terminal domain in this spatial regulation (Gross and Anderson, 1998; Robinson et al., 1999; Babu et al., 2002). In the present study, the C-terminal domain again was shown to specify a functionally important subcellular distribution, as the centrosomal localization of CK1δ was pivotal for Wnt-3a–dependent neurite outgrowth.
Our study revealed a surprising functional difference between CK1δ and CK1ε. Typically, they have been described as having similar or redundant activities, reflecting the 97% homology of their kinase domains (Knippschild et al., 2005). Both CK1δ and CK1ε contribute to the control of circadian rhythm (Lowrey et al., 2000; Lee et al., 2009), although recent studies with gene knockout mouse models and selective inhibitors for each kinase suggest that CK1δ has a stronger effect (Etchegaray et al., 2009; Meng et al., 2010). In our experimental system not only is the requirement of CK1δ for Wnt-3a–dependent neurite outgrowth significant, the stimulation of neurite extension after CK1ε siRNA treatment also is noteworthy. Apparently CK1ε has an inhibitory effect on neurite outgrowth, perhaps by competing with CK1δ for interaction with critical substrates. In this regard, we propose that their differential localization to the centrosome is critical: CK1ε elsewhere in the cell could prevent substrate access to CK1δ at the centrosome where phosphorylation presumably is crucial for neuritogenesis. Consistent with this view, CK1δ and Dvl-2/3 were required for neurite outgrowth induced by CK1ε siRNA knockdown (Fig. 2, D–F and Fig. S5).
The lack of substantial centrosomal localization by CK1ε reported in this paper differs from an earlier finding in which both CK1δ and CK1ε were identified at the centrosome (Milne et al., 2001). Moreover, in that article the kinase domain was alleged to be required for the centrosomal distribution. We believe our divergent results are attributable to differences in technique: the former study relied on conventional immunofluorescent microscopy rather than confocal microscopy and therefore may not have had sufficient resolution to draw definitive conclusions about the centrosomal distribution. Subsequently, the same investigators demonstrated that CK1δ and CK1ε associated with AKAP450 via the kinase domain (Sillibourne et al., 2002). Although AKAP450 localizes to the centrosome, it also is found at other sites such as the Golgi (Schmidt et al., 1999; Takahashi et al., 1999), where CK1δ and CK1ε have been detected (Milne et al., 2001). A recent report about CDK5RAP2 showed that it also localizes to the centrosome and Golgi, and that binding to AKAP450 was responsible for its Golgi but not centrosomal distribution (Wang et al., 2010). We suggest that AKAP450 binding to CK1δ and CK1ε may contribute to their Golgi distribution, whereas other binding partners are necessary for the centrosomal localization of CK1δ.
The centrosomal distribution of CK1δ has potential significance that goes beyond neurite outgrowth. Detection of CK1δ at the mitotic spindle and induction of cytokinesis defects, mitotic arrest, centrosomal amplification, and the formation of multipolar spindle structures by IC261 suggest a broader role for CK1δ in centrosomal structure and function (Behrend et al., 2000; Stöter et al., 2005). Although a functional connection between CK1δ and Dvl has not been established in this context, a recent article demonstrated the presence of Dvl-2 at the mitotic spindle and a role in cell cycle regulation (Kikuchi et al., 2010). Dvl also contributes to the formation of motile cilia by facilitating the docking and planar polarization of the centrosomally derived basal bodies (Park et al., 2008). Furthermore, other Wnt pathway components, Axin2/conductin and β-catenin, have been identified at the centrosome and participate in centrosomal separation (Bahmanyar et al., 2008; Hadjihannas et al., 2010), a process that is disrupted by IC261 (Stöter et al., 2005). Future investigation will address the potential activity of CK1δ in such centrosomal processes. In this paper, we established a key role for CK1δ in Wnt-3a–dependent neurite outgrowth that underscores the importance of its localization at the centrosome.
Materials and methods
Recombinant protein and chemicals
Recombinant Wnt-3a was purchased from R&D Systems. Wnt-1 and Wnt-3a conditioned media (CM) were prepared as described previously (Endo et al., 2008). In brief, Wnt-1 CM was collected from a Wnt-1 stably transfected Rat-2 fibroblast line (kindly provided by Anthony Brown, Cornell Medical Center, New York, NY) after 72 h incubation in serum-free RPMI 1640 medium. Wnt-3a CM was obtained from a Wnt-3a stably transfected L929 clonal line after 72 h incubation in serum-free EMEM supplemented with nonessential amino acids, 2 mM l-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, and 100 µg/ml streptomycin. IC261 was purchased from EMD. Geneticin was obtained from Invitrogen.
Antibodies and reagents used for immunostaining
Alexa Fluor 488 phalloidin, Alexa Fluor 568 phalloidin, Alexa Fluor 488 goat anti–mouse IgG, Alexa Fluor 488 goat anti–rabbit IgG, Alexa Fluor 568 goat anti–rabbit IgG, and Alexa Fluor 660 goat anti–mouse IgG were from Invitrogen. Mouse anti-CK1δ antibody 128A was kindly provided by Eli Lilly. Mouse anti-CK1ε was from BD. Rabbit anti-pericentrin antibody and mouse anti-pericentrin antibody were from Abcam. 4′,6-diamidino-2-phenylindole (dihydrochloride; DAPI) and rabbit anti–γ-tubulin antibody were from Sigma-Aldrich. Rabbit anti-Myc antibody was from Cell Signaling Technology.
Antibodies used for Western blotting
Mouse anti-CK1δ (cat. no. sc-55553), mouse anti-Dvl-2 (10B5), rabbit anti-Dvl2 (H-75), mouse anti-Dvl-3 (4D3), and mouse anti-HSP70 antibodies were from Santa Cruz Biotechnology, Inc. Mouse anti-Myc antibody was from Invitrogen. Mouse anti-CK1ε antibody was from BD. Mouse anti-GFP was from Covance.
Recombinant DNA
pCS2+ myc-tagged hCK1δ and hCK1ε.
pCS2+ 6x myc-hCK1δ and pCS2+ hCK1ε were gifts from Dr. David Virshup (Institute of Medical Biology, Singapore). 6x myc tag sequence excised from pCS2+ 6x myc-hCK1δ with ClaI and StuI was inserted upstream of hCK1ε to obtain myc-tagged CK1ε construct.
Lentiviral expression constructs.
Four lentiviral constructs, pCMV12 6x myc-tagged mouse CK1δ, CK1ε, CK1δ/ε (CK1δ 1–277/CK1ε 278–416), and CK1ε/δ (CK1ε 1–277/CK1δ 278–415) were generated. First, entry clones were constructed by overlap extension PCR using cDNA clones purchased from Thermo Fisher Scientific. Lentiviral expression clones were constructed using Multisite Gateway recombinational cloning to link a promoter to the gene of interest. The backbone vector is a second-generation lentiviral vector based on the pFUFW backbone.
pcDNA3.3 constructs.
pcDNA3.3 6x myc-mCK1δ and 6x myc-mCK1ε were generated by TOPO cloning of PCR products amplified from the lentiviral constructs pCMV12 6x myc mCK1δ and 6x myc mCK1ε, respectively. PCR products (amplified with Expand High FidelityPLUS PCR system; Roche) were cloned into pcDNA3.3 TOPO vector (Invitrogen). Similarly, pcDNA3.3 6x myc-mCK1ε/δ was generated from lentiviral vector pCMV12 6x myc mCK1ε/δ, and used as a PCR template to generate deletion mutant 6x myc-mCK1ε/δCT (278–364). The PCR product (6x myc-mCKε/δCT 278–364) was cloned into pcDNA3.3 TOPO vector. pcDNA3.3 6x myc-mCK1δ-ΔCT and pcDNA3.3 6x myc-mCK1ε-ΔCT were obtained by TOPO cloning of PCR products amplified from the corresponding pcDNA3.3 full-length CK1 constructs, in each case amplifying codons 1–277 of the CK1 isoform coding sequence.
EGFP fusion constructs.
To generate δCT-EGFP, the construct containing the C-terminal domain of mouse CK1δ linked to EGFP, the δCT region (278–415) was PCR amplified from mouse CK1δ (entry clone used for lentiviral expression construct) and subcloned into pEGFP-N1 (Takara Bio Inc.). A forward primer containing XhoI site at the flanking region, and a reverse primer containing EcoRI site at the flanking region were used to amplify δCT. Both PCR product and pEGFP-N1 were digested with XhoI and EcoRI, ligated, and transformed. Deletion mutants (δCT-EGFP 278–297, 278–309, 278–314, 278–325, 278–364, 278–397, 326–415) were generated by PCR amplification (Expand Long Template PCR system; Roche). Using EcoRI-digested δCT-EGFP as a PCR template, PCR products were amplified with corresponding reverse primers, and one common forward primer, all of them containing EcoRI site at the flanking region. PCR products were digested with EcoRI, ligated, and transformed. A similar approach was used to generate εCT-EGFP, as εCT (278–416) was amplified from the mouse CK1ε entry clone and subcloned into pEGFP-N1. The fidelity of all the constructs generated in this study was verified by DNA sequence analysis in the DNA Sequencing MiniCore Facility at the National Cancer Instituseccte (Bethesda, MD).
Cell culture
The ESFT cell line TC-32 was maintained and plated on cell culture dishes, cluster plates, or glass coverslips that had been precoated with type I collagen solution (Sigma-Aldrich) as described previously (Endo et al., 2008). HeLa cells were maintained in DME (Invitrogen) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin in a 5% CO2 humidified 37°C cell culture incubator.
siRNA transfection
Double-stranded siRNA reagents directed against CK1δ and CK1ε were purchased from Thermo Fisher Scientific. CK1δ siRNA specifically targeting 3′UTR sequence was purchased from QIAGEN. Luc siRNA (target sequence: 5′-CGUACGCGGAAUACUUCGA-3′) was synthesized by Thermo Fisher Scientific.
siRNA transfection experiments in TC-32 cells were performed with the Amaxa system according to the manufacturer’s protocol, using 200 pmol of siRNA/106 cells. The effects of siRNA treatment were analyzed 48 h after transfection.
DNA transfection
For transient transfection of HeLa cells, Lipofectamine 2000 (Invitrogen) was used. 1 d before transfection, HeLa cells were seeded on glass coverslips and placed in 24-well cell culture plates. Transfection was performed as described in the manufacturer’s protocol with cells 80–90% confluent. For instance, 2 µg DNA was used with 5 µl of Lipofectamine for each transfection with 24-well cell culture plates. For transient transfection of TC-32 cells, Amaxa transfection or PolyJet (SignaGen) was used. The effects of DNA transfection were analyzed 72 h (Amaxa) or 48 h (PolyJet) after transfection.
Combined transfection of DNA and siRNA (rescue experiment)
Cotransfection of pcDNA3.3 6x myc-mCK1δ, 6x myc-mCK1ε, or 6x myc mCK1ε/δCT (278–364) constructs and siRNA targeting hCK1δ 3′UTR sequence were performed with Amaxa system or GenMute (SignaGen). For Amaxa transfection, 106 cells were resuspended with 2 µg DNA and 200 pmol of siRNA, and placed in 6-well and 24-well plates. 72 h later, cells in 6-well plate were harvested for Western blotting, and cells placed in 24-well plates were treated with 100 ng/ml recombinant Wnt-3a for 3 h and fixed with formaldehyde to analyze neurite outgrowth. For GenMute, TC-32 cells were plated on the day before transfection on 6-well and 24-well plates. Cells were 50–60% confluent on the day of transfection. 0.5 µg DNA and 5 pmol of siRNA were used for 6-well plate, 0.25 µg DNA and 2.5 pmol siRNA were used for 24-well plate. 48 h later, cells in 6-well plates were harvested for immunoblotting, cells in 24-well plates were treated with 100 ng/ml recombinant Wnt-3a for 3 h, and fixed with formaldehyde to analyze neurite outgrowth.
Lentiviral expression
Lentiviral particles were produced by transient transfection of HEK293T cells. 2 d after transfection, the cell culture medium was harvested and concentrated ∼10-fold with Amicon Ultra-15 (Millipore) and stored at −80°C. On the day before transfection, HeLa cells were plated in a 6-well plate at a density that reached 80–90% confluency the next day. On the day of transfection, 0.2 ml of concentrated lentiviral particle was added to each well filled with 1 ml of complete growth medium and 8 µg/ml of polybrene (Millipore). 24 h later the medium was replaced with fresh complete medium without polybrene. After another 24 h geneticin was added to cell culture medium (400 µg/ml) to obtain stable transfectants. Fresh complete medium supplemented with geneticin was provided every 2 d. 1 wk later cells were subjected to Western blotting to verify recombinant protein expression.
Immunofluorescent analysis
TC-32 or HeLa cells were seeded on 12-mm-diam glass coverslips (Thermo Fisher Scientific) in complete growth medium. For TC-32 cells, collagen-coated coverslips were used. Depending on the combination of antibodies and reagents, different fixatives were used (Table S1). For methanol (MeOH) fixation, cells were first washed once with PBS, then with PHEM (60 mM Na-Pipes, 25 mM Na-Hepes, 10 mM Na-EGTA, and 2 mM MgCl2, pH 6.9), followed by treatment with PHEM containing 0.19 M NaCl, 1% Saponin, 10 µM Taxol, and 0.1% DMSO for 5 min at room temperature (RT) to extract and stabilize tubulin. Extracted cultures were immersed in MeOH at −30°C for 10 min, rehydrated by rinsing in PBS three times, and treated with blocking solution (5% BSA in PBS) for 30 min at 37°C. Primary antibody/antibodies was/were added with 2.5% BSA in PBS, and cell samples were incubated for 60 min at 37°C or overnight at 4°C. After washing three times with PBS, cell samples were incubated with the secondary antibody reagent(s) and DAPI with 2.5% BSA in PBS for 45 min at RT. After washing three times with PBS, coverslips were mounted on glass slides (VWR Scientific) using ProLong Gold Antifade reagent (Invitrogen). Formaldehyde fixation was performed as described previously (Endo et al., 2008). In brief, cells were fixed with freshly prepared 3.7% formaldehyde for 15 min at RT and permeabilized with 0.1% Triton X-100 in PBS for 5 min. After blocking with 5% BSA in PBS for 1 h at RT, primary antibody/antibodies was/were added with 2.5% BSA in PBS, and cell samples were incubated for 60 min at 37°C or overnight at 4°C, followed by the same procedure used for the MeOH fixation method described above.
Cell imaging
Fluorescent images were collected with a laser-scanning confocal microscope (510 LSCM; Carl Zeiss, Inc.), using a 63x objective (Carl Zeiss, Inc.). Zeiss LSM Image Browser version 4.0.0.157 was used for image processing, and composite figures were prepared with Adobe Photoshop CS2 v9.0.2 (Adobe Systems, Inc.).
Quantitative colocalization analysis
To further examine the colocalization of Myc-tagged CK1δ and CK1ε with pericentrin at the centrosome in TC-32 cells, Pearson’s correlation coefficient was calculated with Imaris x64 (v7.0.0) image visualization software (Bitplane, Inc.).
Quantitative analysis of neurite outgrowth
Stimulation of neurite outgrowth was monitored as described previously (Endo et al., 2008).
Immunoblotting
To detect CK1δ, CK1ε, and Dvl, 80–90% confluent monolayers of TC-32 cells that had been seeded in 6- or 12-well cell culture plates were serum starved overnight. For immunoblot analysis to verify siRNA knockdown of endogenous proteins, TC-32 cells transfected with siRNA were seeded in 6- or 12-well cell culture plates and harvested 48 h after transfection. After incubation for the indicated time, cells were rinsed twice with PBS, lysed with buffer (50 mM Hepes, pH 7.5, 50 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM sodium pyrophosphate, 50 mM NaF, 1 mM sodium vanadate, 10 µg/ml aprotinin, 10 µg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride), and processed for SDS-PAGE and Western blot analysis as described previously (Endo et al., 2008). In brief, cell lysates were clarified by centrifugation and protein concentration was determined with Protein Assay reagent (Bio-Rad Laboratories). For all immunoblotting, 30 µg of protein was loaded per lane in 10% or 4–20% polyacrylamide Tris-glycine gels. After SDS-PAGE, the proteins were transferred to Immobilon P membrane (Millipore), which was blocked with 5% milk, incubated with primary antibody overnight at 4°C, and subsequently incubated with horseradish peroxidase–labeled secondary antibody. The proteins were visualized with SuperSignal Femto Chemiluminescent reagents (Thermo Fisher Scientific) and BioMax film (Kodak).
Statistical analysis
The significance of differences in data obtained from neurite outgrowth assays was determined with Student’s t test. The differences were considered to be significant when the P value was less than 0.05.
Online supplemental material
Fig. S1 shows immunofluorescent staining of TC-32 and HeLa cells transiently expressing Myc-tagged mouse CK1δ or CK1ε lacking their respective C-terminal domains. Fig. S2 illustrates the rating system used in Fig. 5 B to analyze the centrosomal distribution of δCT-EGFP. Fig. S3 shows the centrosomal localization signal of CK1δ and the C-terminal ends of various deletion constructs. Fig. S4 shows the centrosomal localization of mCK1ε/δCT(278–364). Fig. S5 shows that Dvl-2/3 siRNA blocked neurite outgrowth induced by CK1ε siRNA. Table S1 provides a summary of conditions (antibodies, other reagents, fixation procedure) used for cell staining in various experiments. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201011111/DC1.
Acknowledgments
We thank Eli Lilly for providing the CK1δ mAb 128A, David Virshup for pCS2+ 6x myc-hCK1δ and pCS2+ hCK1ε constructs, Anthony Brown for the Rat2 fibroblast/Wnt-1 line that was the source of Wnt-1 conditioned medium, and Dom Esposito (Advanced Technology Program, SAIC-Frederick) for preparation of Gateway entry clones and lentiviral expression constructs.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Abbreviations used in this paper:
- CK
- casein kinase
- CLS
- centrosomal localization signal
- CM
- conditioned medium
- Dvl
- Dishevelled
- EGFP
- enhanced GFP
- ESFT
- Ewing sarcoma family tumor
References
- Babu P., Bryan J.D., Panek H.R., Jordan S.L., Forbrich B.M., Kelley S.C., Colvin R.T., Robinson L.C. 2002. Plasma membrane localization of the Yck2p yeast casein kinase 1 isoform requires the C-terminal extension and secretory pathway function. J. Cell Sci. 115:4957–4968 10.1242/jcs.00203 [DOI] [PubMed] [Google Scholar]
- Bahmanyar S., Kaplan D.D., Deluca J.G., Giddings T.H., Jr, O’Toole E.T., Winey M., Salmon E.D., Casey P.J., Nelson W.J., Barth A.I. 2008. beta-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 22:91–105 10.1101/gad.1596308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrend L., Milne D.M., Stöter M., Deppert W., Campbell L.E., Meek D.W., Knippschild U. 2000. IC261, a specific inhibitor of the protein kinases casein kinase 1-delta and -epsilon, triggers the mitotic checkpoint and induces p53-dependent postmitotic effects. Oncogene. 19:5303–5313 10.1038/sj.onc.1203939 [DOI] [PubMed] [Google Scholar]
- Bryja V., Schulte G., Arenas E. 2007a. Wnt-3a utilizes a novel low dose and rapid pathway that does not require casein kinase 1-mediated phosphorylation of Dvl to activate beta-catenin. Cell. Signal. 19:610–616 10.1016/j.cellsig.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Bryja V., Schulte G., Rawal N., Grahn A., Arenas E. 2007b. Wnt-5a induces Dishevelled phosphorylation and dopaminergic differentiation via a CK1-dependent mechanism. J. Cell Sci. 120:586–595 10.1242/jcs.03368 [DOI] [PubMed] [Google Scholar]
- Bryja V., Schambony A., Cajánek L., Dominguez I., Arenas E., Schulte G. 2008. Beta-arrestin and casein kinase 1/2 define distinct branches of non-canonical WNT signalling pathways. EMBO Rep. 9:1244–1250 10.1038/embor.2008.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciani L., Salinas P.C. 2005. WNTs in the vertebrate nervous system: from patterning to neuronal connectivity. Nat. Rev. Neurosci. 6:351–362 10.1038/nrn1665 [DOI] [PubMed] [Google Scholar]
- Ciani L., Krylova O., Smalley M.J., Dale T.C., Salinas P.C. 2004. A divergent canonical WNT-signaling pathway regulates microtubule dynamics: dishevelled signals locally to stabilize microtubules. J. Cell Biol. 164:243–253 10.1083/jcb.200309096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong F., Schweizer L., Varmus H. 2004. Casein kinase Iepsilon modulates the signaling specificities of dishevelled. Mol. Cell. Biol. 24:2000–2011 10.1128/MCB.24.5.2000-2011.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlberg C.L., Nguyen E.Z., Goodlett D., Kimelman D. 2009. Interactions between Casein kinase Iepsilon (CKIepsilon) and two substrates from disparate signaling pathways reveal mechanisms for substrate-kinase specificity. PLoS ONE. 4:e4766 10.1371/journal.pone.0004766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Anda F.C., Pollarolo G., Da Silva J.S., Camoletto P.G., Feiguin F., Dotti C.G. 2005. Centrosome localization determines neuronal polarity. Nature. 436:704–708 10.1038/nature03811 [DOI] [PubMed] [Google Scholar]
- Endo Y., Rubin J.S. 2007. Wnt signaling and neurite outgrowth: insights and questions. Cancer Sci. 98:1311–1317 10.1111/j.1349-7006.2007.00536.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y., Beauchamp E., Woods D., Taylor W.G., Toretsky J.A., Uren A., Rubin J.S. 2008. Wnt-3a and Dickkopf-1 stimulate neurite outgrowth in Ewing tumor cells via a Frizzled3- and c-Jun N-terminal kinase-dependent mechanism. Mol. Cell. Biol. 28:2368–2379 10.1128/MCB.01780-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray J.P., Machida K.K., Noton E., Constance C.M., Dallmann R., Di Napoli M.N., DeBruyne J.P., Lambert C.M., Yu E.A., Reppert S.M., Weaver D.R. 2009. Casein kinase 1 delta regulates the pace of the mammalian circadian clock. Mol. Cell. Biol. 29:3853–3866 10.1128/MCB.00338-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C., Chen Y.G. 2010. Dishevelled: The hub of Wnt signaling. Cell. Signal. 22:717–727 10.1016/j.cellsig.2009.11.021 [DOI] [PubMed] [Google Scholar]
- González-Sancho J.M., Brennan K.R., Castelo-Soccio L.A., Brown A.M. 2004. Wnt proteins induce dishevelled phosphorylation via an LRP5/6- independent mechanism, irrespective of their ability to stabilize beta-catenin. Mol. Cell. Biol. 24:4757–4768 10.1128/MCB.24.11.4757-4768.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves P.R., Roach P.J. 1995. Role of COOH-terminal phosphorylation in the regulation of casein kinase I delta. J. Biol. Chem. 270:21689–21694 10.1074/jbc.270.21.12717 [DOI] [PubMed] [Google Scholar]
- Gross S.D., Anderson R.A. 1998. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 10:699–711 10.1016/S0898-6568(98)00042-4 [DOI] [PubMed] [Google Scholar]
- Gross S.D., Simerly C., Schatten G., Anderson R.A. 1997. A casein kinase I isoform is required for proper cell cycle progression in the fertilized mouse oocyte. J. Cell Sci. 110:3083–3090 [DOI] [PubMed] [Google Scholar]
- Hadjihannas M.V., Brückner M., Behrens J. 2010. Conductin/axin2 and Wnt signalling regulates centrosome cohesion. EMBO Rep. 11:317–324 10.1038/embor.2010.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H.R., Gleeson J.G. 2007. The centrosome in neuronal development. Trends Neurosci. 30:276–283 10.1016/j.tins.2007.04.001 [DOI] [PubMed] [Google Scholar]
- Kikuchi K., Niikura Y., Kitagawa K., Kikuchi A. 2010. Dishevelled, a Wnt signalling component, is involved in mitotic progression in cooperation with Plk1. EMBO J. 29:3470–3483 10.1038/emboj.2010.221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaus A., Birchmeier W. 2008. Wnt signalling and its impact on development and cancer. Nat. Rev. Cancer. 8:387–398 10.1038/nrc2389 [DOI] [PubMed] [Google Scholar]
- Klein T.J., Jenny A., Djiane A., Mlodzik M. 2006. CKIepsilon/discs overgrown promotes both Wnt-Fz/beta-catenin and Fz/PCP signaling in Drosophila. Curr. Biol. 16:1337–1343 10.1016/j.cub.2006.06.030 [DOI] [PubMed] [Google Scholar]
- Knippschild U., Gocht A., Wolff S., Huber N., Löhler J., Stöter M. 2005. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell. Signal. 17:675–689 10.1016/j.cellsig.2004.12.011 [DOI] [PubMed] [Google Scholar]
- Krylova O., Messenger M.J., Salinas P.C. 2000. Dishevelled-1 regulates microtubule stability: a new function mediated by glycogen synthase kinase-3beta. J. Cell Biol. 151:83–94 10.1083/jcb.151.1.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Chen R., Lee Y., Yoo S., Lee C. 2009. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA. 106:21359–21364 10.1073/pnas.0906651106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Shi J., Lu C.C., Wang Z.B., Lyuksyutova A.I., Song X.J., Zou Y. 2005. Ryk-mediated Wnt repulsion regulates posterior-directed growth of corticospinal tract. Nat. Neurosci. 8:1151–1159 10.1038/nn1520 [DOI] [PubMed] [Google Scholar]
- Lowrey P.L., Shimomura K., Antoch M.P., Yamazaki S., Zemenides P.D., Ralph M.R., Menaker M., Takahashi J.S. 2000. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 288:483–492 10.1126/science.288.5465.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., Yamamoto V., Ortega B., Baltimore D. 2004. Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell. 119:97–108 10.1016/j.cell.2004.09.019 [DOI] [PubMed] [Google Scholar]
- Lyuksyutova A.I., Lu C.C., Milanesio N., King L.A., Guo N., Wang Y., Nathans J., Tessier-Lavigne M., Zou Y. 2003. Anterior-posterior guidance of commissural axons by Wnt-frizzled signaling. Science. 302:1984–1988 10.1126/science.1089610 [DOI] [PubMed] [Google Scholar]
- Malaterre J., Ramsay R.G., Mantamadiotis T. 2007. Wnt-Frizzled signalling and the many paths to neural development and adult brain homeostasis. Front. Biosci. 12:492–506 10.2741/2077 [DOI] [PubMed] [Google Scholar]
- Mashhoon N., DeMaggio A.J., Tereshko V., Bergmeier S.C., Egli M., Hoekstra M.F., Kuret J. 2000. Crystal structure of a conformation-selective casein kinase-1 inhibitor. J. Biol. Chem. 275:20052–20060 10.1074/jbc.M001713200 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Maller J.L. 2004. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 306:885–888 10.1126/science.1103544 [DOI] [PubMed] [Google Scholar]
- McKay R.M., Peters J.M., Graff J.M. 2001. The casein kinase I family in Wnt signaling. Dev. Biol. 235:388–396 10.1006/dbio.2001.0308 [DOI] [PubMed] [Google Scholar]
- Meng Q.J., Maywood E.S., Bechtold D.A., Lu W.Q., Li J., Gibbs J.E., Dupré S.M., Chesham J.E., Rajamohan F., Knafels J., et al. 2010. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc. Natl. Acad. Sci. USA. 107:15240–15245 10.1073/pnas.1005101107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne D.M., Looby P., Meek D.W. 2001. Catalytic activity of protein kinase CK1 delta (casein kinase 1delta) is essential for its normal subcellular localization. Exp. Cell Res. 263:43–54 10.1006/excr.2000.5100 [DOI] [PubMed] [Google Scholar]
- Park T.J., Mitchell B.J., Abitua P.B., Kintner C., Wallingford J.B. 2008. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat. Genet. 40:871–879 10.1038/ng.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascreau G., Eckerdt F., Churchill M.E., Maller J.L. 2010. Discovery of a distinct domain in cyclin A sufficient for centrosomal localization independently of Cdk binding. Proc. Natl. Acad. Sci. USA. 107:2932–2937 10.1073/pnas.0914874107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M., McKay R.M., McKay J.P., Graff J.M. 1999. Casein kinase I transduces Wnt signals. Nature. 401:345–350 10.1038/43830 [DOI] [PubMed] [Google Scholar]
- Robinson L.C., Bradley C., Bryan J.D., Jerome A., Kweon Y., Panek H.R. 1999. The Yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol. Biol. Cell. 10:1077–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso S.B., Sussman D., Wynshaw-Boris A., Salinas P.C. 2005. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat. Neurosci. 8:34–42 10.1038/nn1374 [DOI] [PubMed] [Google Scholar]
- Sakanaka C., Leong P., Xu L., Harrison S.D., Williams L.T. 1999. Casein kinase iepsilon in the wnt pathway: regulation of beta-catenin function. Proc. Natl. Acad. Sci. USA. 96:12548–12552 10.1073/pnas.96.22.12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas P.C., Zou Y. 2008. Wnt signaling in neural circuit assembly. Annu. Rev. Neurosci. 31:339–358 10.1146/annurev.neuro.31.060407.125649 [DOI] [PubMed] [Google Scholar]
- Sánchez-Camacho C., Bovolenta P. 2009. Emerging mechanisms in morphogen-mediated axon guidance. Bioessays. 31:1013–1025 10.1002/bies.200900063 [DOI] [PubMed] [Google Scholar]
- Schmidt P.H., Dransfield D.T., Claudio J.O., Hawley R.G., Trotter K.W., Milgram S.L., Goldenring J.R. 1999. AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J. Biol. Chem. 274:3055–3066 10.1074/jbc.274.5.3055 [DOI] [PubMed] [Google Scholar]
- Schulte G., Bryja V., Rawal N., Castelo-Branco G., Sousa K.M., Arenas E. 2005. Purified Wnt-5a increases differentiation of midbrain dopaminergic cells and dishevelled phosphorylation. J. Neurochem. 92:1550–1553 10.1111/j.1471-4159.2004.03022.x [DOI] [PubMed] [Google Scholar]
- Sillibourne J.E., Milne D.M., Takahashi M., Ono Y., Meek D.W. 2002. Centrosomal anchoring of the protein kinase CK1delta mediated by attachment to the large, coiled-coil scaffolding protein CG-NAP/AKAP450. J. Mol. Biol. 322:785–797 10.1016/S0022-2836(02)00857-4 [DOI] [PubMed] [Google Scholar]
- Stöter M., Bamberger A.M., Aslan B., Kurth M., Speidel D., Löning T., Frank H.G., Kaufmann P., Löhler J., Henne-Bruns D., et al. 2005. Inhibition of casein kinase I delta alters mitotic spindle formation and induces apoptosis in trophoblast cells. Oncogene. 24:7964–7975 10.1038/sj.onc.1208941 [DOI] [PubMed] [Google Scholar]
- Strutt H., Price M.A., Strutt D. 2006. Planar polarity is positively regulated by casein kinase Iepsilon in Drosophila. Curr. Biol. 16:1329–1336 10.1016/j.cub.2006.04.041 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Shibata H., Shimakawa M., Miyamoto M., Mukai H., Ono Y. 1999. Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the golgi apparatus. J. Biol. Chem. 274:17267–17274 10.1074/jbc.274.24.17267 [DOI] [PubMed] [Google Scholar]
- Wallingford J.B., Habas R. 2005. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development. 132:4421–4436 10.1242/dev.02068 [DOI] [PubMed] [Google Scholar]
- Wang Z., Wu T., Shi L., Zhang L., Zheng W., Qu J.Y., Niu R., Qi R.Z. 2010. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J. Biol. Chem. 285:22658–22665 10.1074/jbc.M110.105965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A.M., Lyuksyutova A.I., Fenstermaker A.G., Shafer B., Lo C.G., Zou Y. 2008. Phosphatidylinositol-3-kinase-atypical protein kinase C signaling is required for Wnt attraction and anterior-posterior axon guidance. J. Neurosci. 28:3456–3467 10.1523/JNEUROSCI.0029-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa S., McKinnon R.D., Kokel M., Thomas J.B. 2003. Wnt-mediated axon guidance via the Drosophila Derailed receptor. Nature. 422:583–588 10.1038/nature01522 [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhu J., Yang G.Y., Wang Q.J., Qian L., Chen Y.M., Chen F., Tao Y., Hu H.S., Wang T., Luo Z.G. 2007. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat. Cell Biol. 9:743–754 10.1038/ncb1603 [DOI] [PubMed] [Google Scholar]
- Zinchuk V., Zinchuk O., Okada T. 2007. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem. Cytochem. 40:101–111 10.1267/ahc.07002 [DOI] [PMC free article] [PubMed] [Google Scholar]