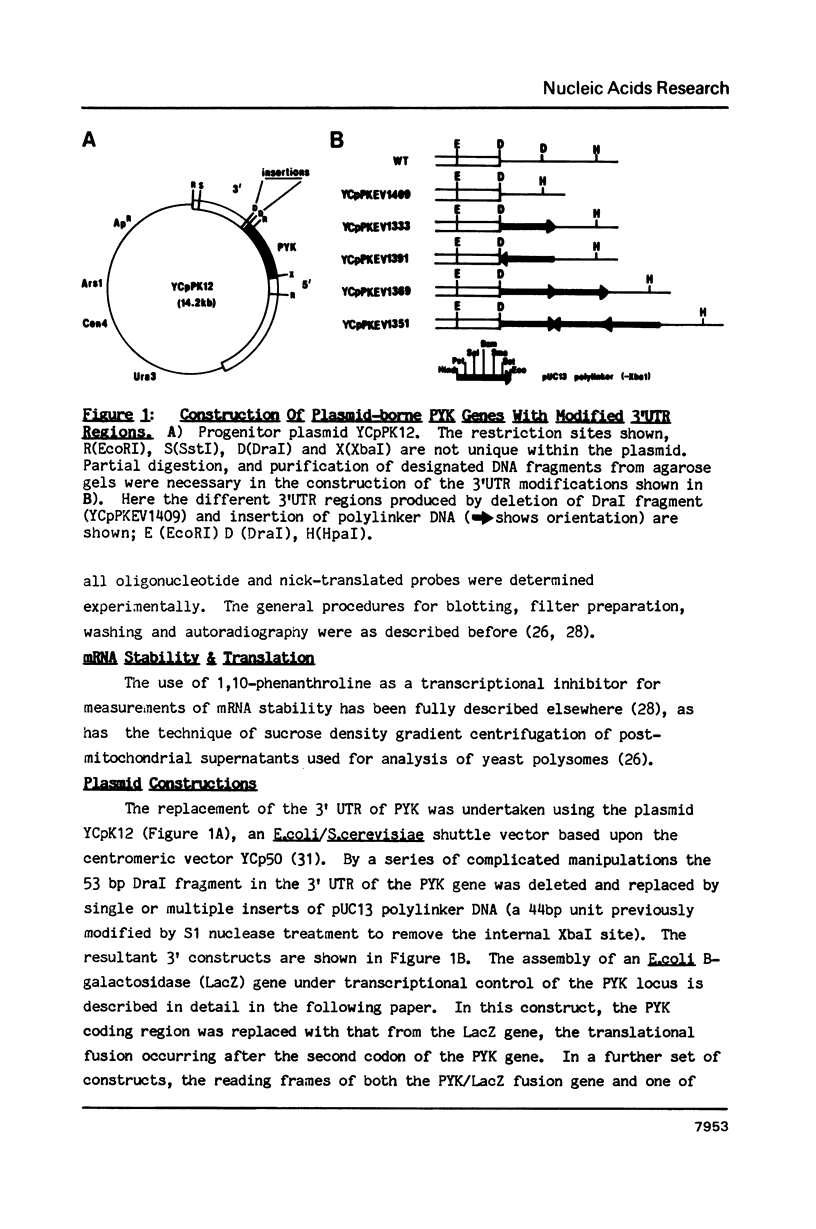
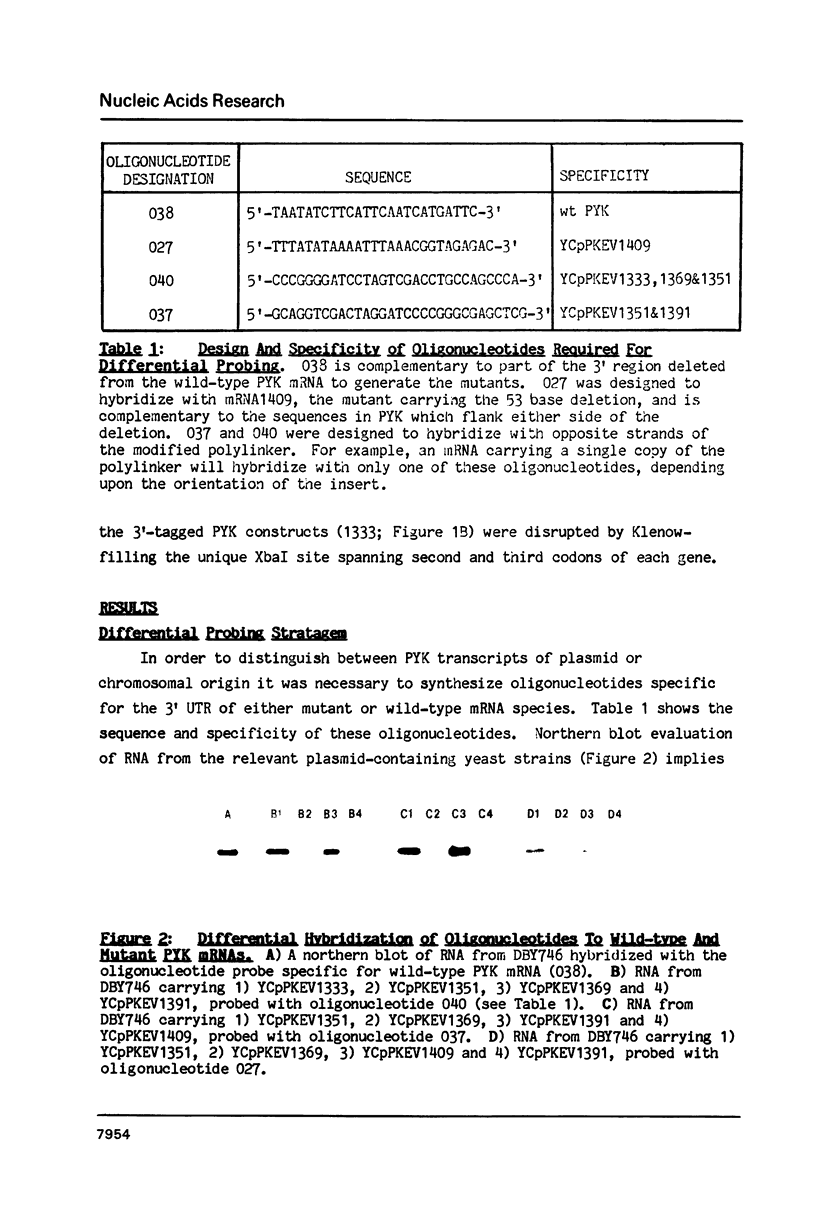
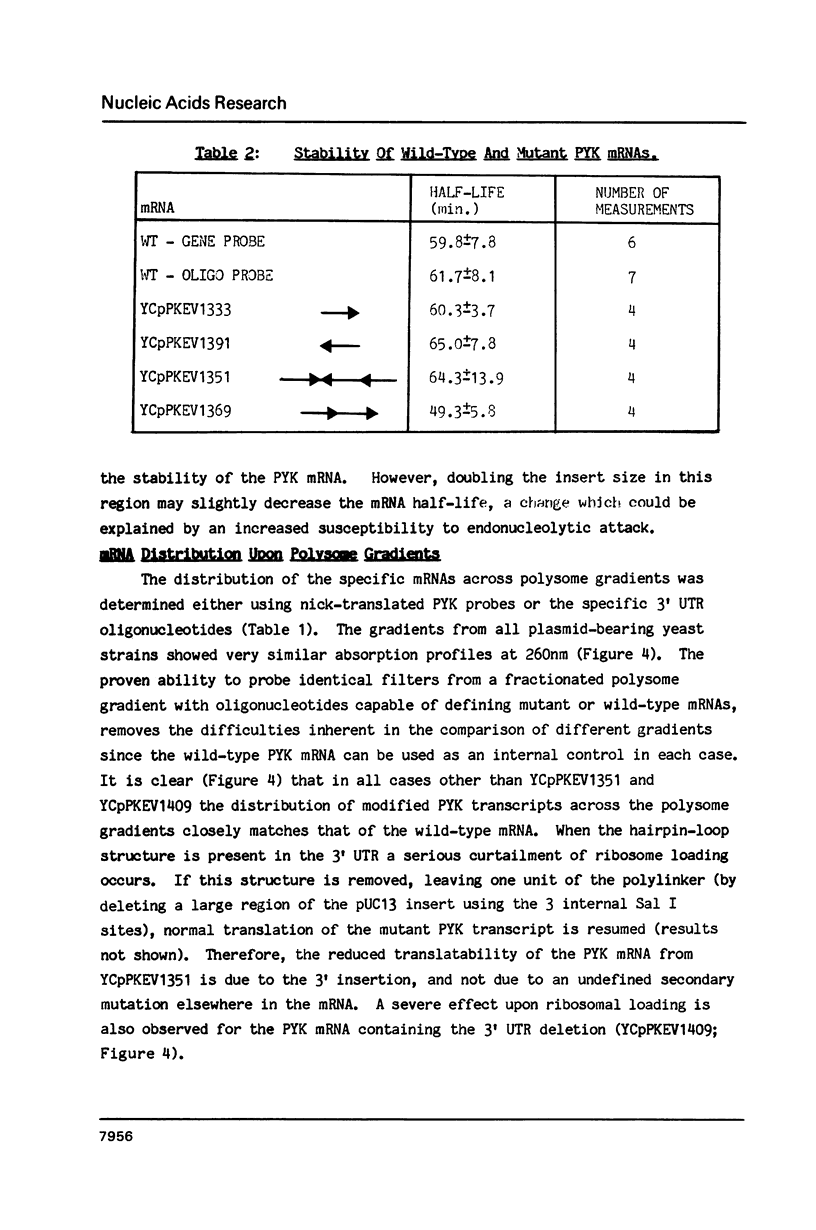
Abstract
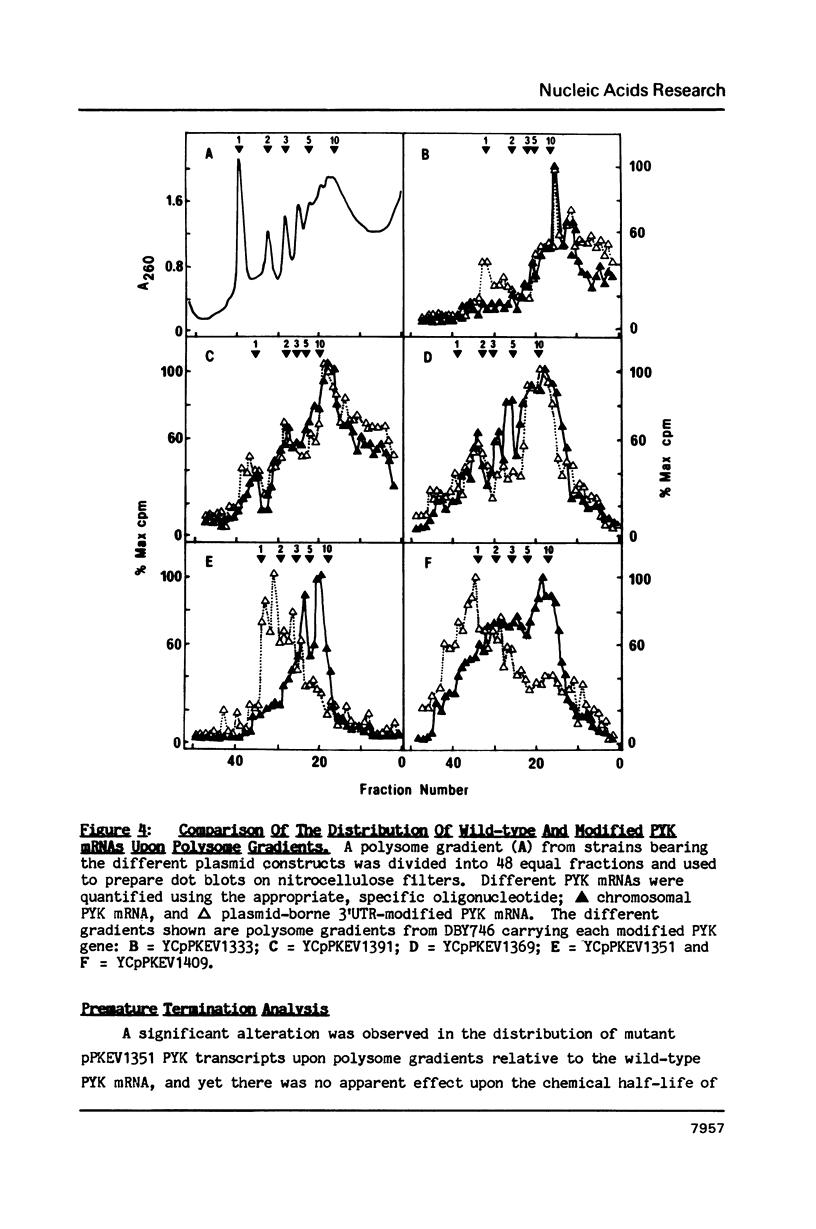
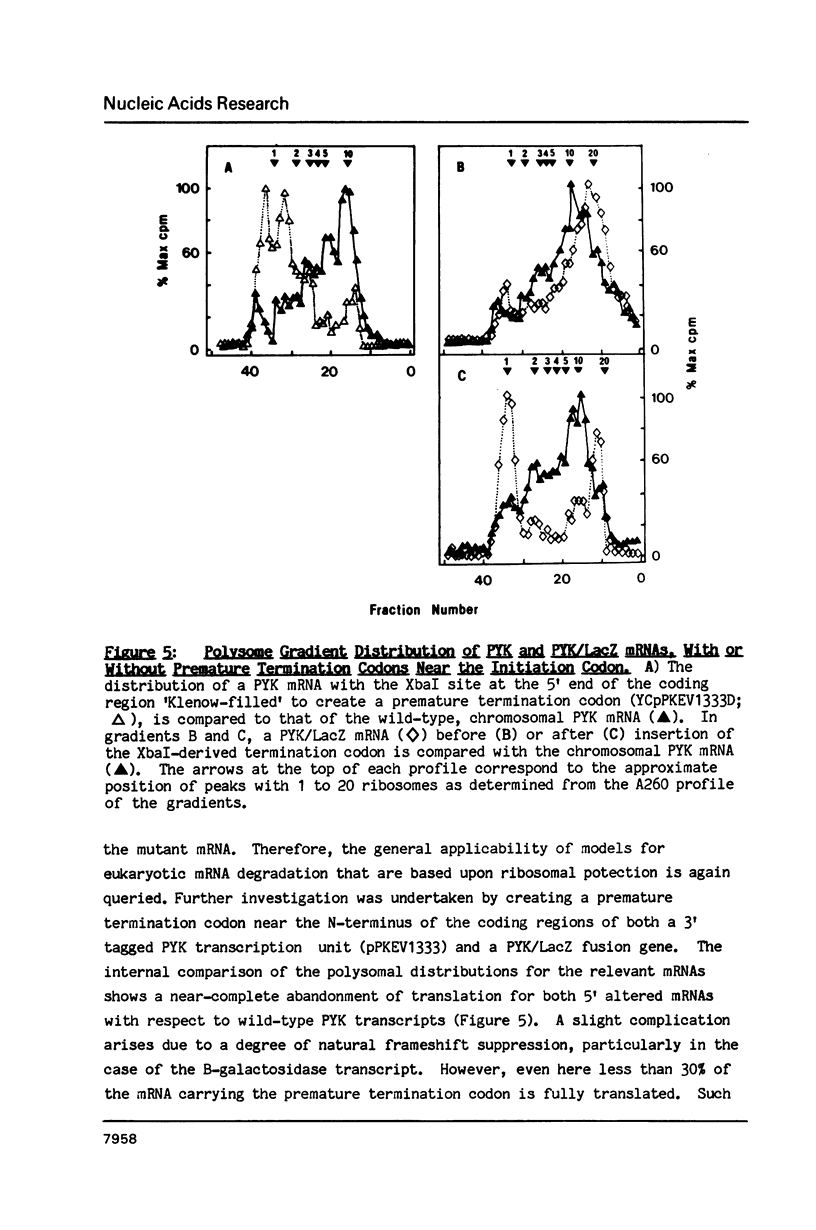
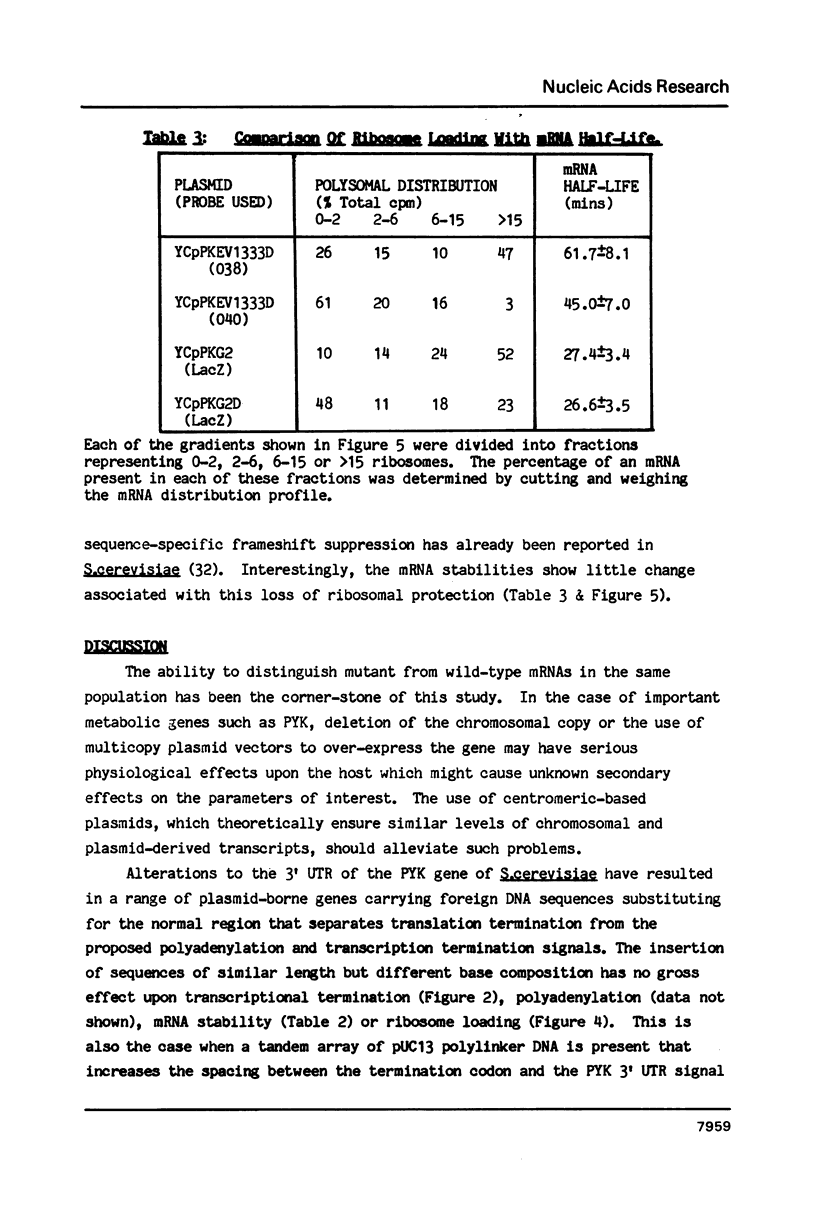
A 53 basepair deletion was constructed within the 3' untranslated region (3' UTR) of the yeast pyruvate kinase (PYK) gene borne upon a centromeric plasmid. Various modular assemblies of the pUC13 polylinker DNA (single unit = 44 bp) were used to replace the deleted region, and the effects of these modifications upon both transcript stability and translation ascertained in yeast. The use of a differential probing stratagem, based on the hybridisation of specific oligonucleotides to either pUC13 polylinker or unaltered PYK 3' UTR sequences, allowed for discrimination between mutant (plasmid borne) and wild-type (chromosomal) PYK transcripts. In no construct was there any significant alteration in mRNA stability, but translation of the PYK mRNA was severely curtailed by truncation of the 3' UTR or the presence of a strong hairpin-loop structure in the 3' UTR. A specific mutation in the N-terminal coding sequences, which created a premature termination codon in both a 3' 'tagged' PYK plasmid and a PYK/LacZ fusion gene, aborted the translation of a majority of their transcripts but left their chemical half-lives unaltered. This observation is at variance with some previously published data (Losson & Lacroute (1979) Proc Natl Acad Sci USA 76, 5134; Pelsey & Lacroute (1984) Curr Genet 8, 277), but is consistent with our own earlier observation that there is no obvious link between ribosome loading and mRNA stability in yeast (Santiago et al. (1986) Nucleic Acids Res 14, 8347). Possible reasons for this disparity are discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albrecht G., Krowczynska A., Brawerman G. Configuration of beta-globin messenger RNA in rabbit reticulocytes. Identification of sites exposed to endogenous and exogenous nucleases. J Mol Biol. 1984 Oct 5;178(4):881–896. doi: 10.1016/0022-2836(84)90317-6. [DOI] [PubMed] [Google Scholar]
- Baim S. B., Pietras D. F., Eustice D. C., Sherman F. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol. 1985 Aug;5(8):1839–1846. doi: 10.1128/mcb.5.8.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Brawerman G. Determinants of messenger RNA stability. Cell. 1987 Jan 16;48(1):5–6. doi: 10.1016/0092-8674(87)90346-1. [DOI] [PubMed] [Google Scholar]
- Brock M. L., Shapiro D. J. Estrogen stabilizes vitellogenin mRNA against cytoplasmic degradation. Cell. 1983 Aug;34(1):207–214. doi: 10.1016/0092-8674(83)90151-4. [DOI] [PubMed] [Google Scholar]
- Busslinger M., Portmann R., Birnsteil M. L. A regulatory sequence near the 3' end of sea urchin histone genes. Nucleic Acids Res. 1979 Jul 11;6(9):2997–3008. doi: 10.1093/nar/6.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capasso O., Bleecker G. C., Heintz N. Sequences controlling histone H4 mRNA abundance. EMBO J. 1987 Jun;6(6):1825–1831. doi: 10.1002/j.1460-2075.1987.tb02437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber R. F., Culbertson M. R. Codon recognition during frameshift suppression in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Oct;4(10):2052–2061. doi: 10.1128/mcb.4.10.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil A., Proudfoot N. J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3' end formation. Cell. 1987 May 8;49(3):399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- Gorski K., Roch J. M., Prentki P., Krisch H. M. The stability of bacteriophage T4 gene 32 mRNA: a 5' leader sequence that can stabilize mRNA transcripts. Cell. 1985 Dec;43(2 Pt 1):461–469. doi: 10.1016/0092-8674(85)90176-x. [DOI] [PubMed] [Google Scholar]
- Graves R. A., Pandey N. B., Chodchoy N., Marzluff W. F. Translation is required for regulation of histone mRNA degradation. Cell. 1987 Feb 27;48(4):615–626. doi: 10.1016/0092-8674(87)90240-6. [DOI] [PubMed] [Google Scholar]
- Guyette W. A., Matusik R. J., Rosen J. M. Prolactin-mediated transcriptional and post-transcriptional control of casein gene expression. Cell. 1979 Aug;17(4):1013–1023. doi: 10.1016/0092-8674(79)90340-4. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Herskowitz I. Structure of a yeast pheromone gene (MF alpha): a putative alpha-factor precursor contains four tandem copies of mature alpha-factor. Cell. 1982 Oct;30(3):933–943. doi: 10.1016/0092-8674(82)90298-7. [DOI] [PubMed] [Google Scholar]
- Lillehaug J. R., Kleppe K. Kinetics and specificity of T4 polynucleotide kinase. Biochemistry. 1975 Mar 25;14(6):1221–1225. doi: 10.1021/bi00677a020. [DOI] [PubMed] [Google Scholar]
- Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981 Sep 24;293(5830):311–314. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- Losson R., Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., Kleid D. G. Nucleotide sequence of the rightward operator of phage lambda. Proc Natl Acad Sci U S A. 1975 Mar;72(3):1184–1188. doi: 10.1073/pnas.72.3.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse D. E., Yanofsky C. Polarity and the degradation of mRNA. Nature. 1969 Oct 25;224(5217):329–331. doi: 10.1038/224329a0. [DOI] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5' noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell. 1985 Mar;40(3):515–526. doi: 10.1016/0092-8674(85)90200-4. [DOI] [PubMed] [Google Scholar]
- Piechaczyk M., Yang J. Q., Blanchard J. M., Jeanteur P., Marcu K. B. Posttranscriptional mechanisms are responsible for accumulation of truncated c-myc RNAs in murine plasma cell tumors. Cell. 1985 Sep;42(2):589–597. doi: 10.1016/0092-8674(85)90116-3. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. The end of the message. Nature. 1982 Aug 5;298(5874):516–517. doi: 10.1038/298516a0. [DOI] [PubMed] [Google Scholar]
- Rabbitts P. H., Forster A., Stinson M. A., Rabbitts T. H. Truncation of exon 1 from the c-myc gene results in prolonged c-myc mRNa stability. EMBO J. 1985 Dec 30;4(13B):3727–3733. doi: 10.1002/j.1460-2075.1985.tb04141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago T. C., Bettany A. J., Purvis I. J., Brown A. J. Messenger RNA stability in Saccharomyces cerevisiae: the influence of translation and poly(A) tail length. Nucleic Acids Res. 1987 Mar 25;15(6):2417–2429. doi: 10.1093/nar/15.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago T. C., Purvis I. J., Bettany A. J., Brown A. J. The relationship between mRNA stability and length in Saccharomyces cerevisiae. Nucleic Acids Res. 1986 Nov 11;14(21):8347–8360. doi: 10.1093/nar/14.21.8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Blundell M., Kennell D. Translation and mRNA decay. Mol Gen Genet. 1978 Apr 6;160(2):121–129. doi: 10.1007/BF00267473. [DOI] [PubMed] [Google Scholar]
- Shaw G., Kamen R. A conserved AU sequence from the 3' untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986 Aug 29;46(5):659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- Simcox A. A., Cheney C. M., Hoffman E. P., Shearn A. A deletion of the 3' end of the Drosophila melanogaster hsp70 gene increases stability of mutant mRNA during recovery from heat shock. Mol Cell Biol. 1985 Dec;5(12):3397–3402. doi: 10.1128/mcb.5.12.3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanssens P., Remaut E., Fiers W. Inefficient translation initiation causes premature transcription termination in the lacZ gene. Cell. 1986 Mar 14;44(5):711–718. doi: 10.1016/0092-8674(86)90837-8. [DOI] [PubMed] [Google Scholar]
- Sutton A., Broach J. R. Signals for transcription initiation and termination in the Saccharomyces cerevisiae plasmid 2 micron circle. Mol Cell Biol. 1985 Oct;5(10):2770–2780. doi: 10.1128/mcb.5.10.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci U S A. 1986 May;83(10):3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. Mutationally altered 3' ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J Mol Biol. 1984 Jul 25;177(1):107–135. doi: 10.1016/0022-2836(84)90060-3. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]