Abstract
Summary: Microbial evolution and subsequent species diversification enable bacterial organisms to perform common biological processes by a variety of means. The epsilonproteobacteria are a diverse class of prokaryotes that thrive in diverse habitats. Many of these environmental niches are labeled as extreme, whereas other niches include various sites within human, animal, and insect hosts. Some epsilonproteobacteria, such as Campylobacter jejuni and Helicobacter pylori, are common pathogens of humans that inhabit specific regions of the gastrointestinal tract. As such, the biological processes of pathogenic Campylobacter and Helicobacter spp. are often modeled after those of common enteric pathogens such as Salmonella spp. and Escherichia coli. While many exquisite biological mechanisms involving biochemical processes, genetic regulatory pathways, and pathogenesis of disease have been elucidated from studies of Salmonella spp. and E. coli, these paradigms often do not apply to the same processes in the epsilonproteobacteria. Instead, these bacteria often display extensive variation in common biological mechanisms relative to those of other prototypical bacteria. In this review, five biological processes of commonly studied model bacterial species are compared to those of the epsilonproteobacteria C. jejuni and H. pylori. Distinct differences in the processes of flagellar biosynthesis, DNA uptake and recombination, iron homeostasis, interaction with epithelial cells, and protein glycosylation are highlighted. Collectively, these studies support a broader view of the vast repertoire of biological mechanisms employed by bacteria and suggest that future studies of the epsilonproteobacteria will continue to provide novel and interesting information regarding prokaryotic cellular biology.
INTRODUCTION
Among the currently described eubacterial phyla, the Proteobacteria represent the largest and one of the most diverse groups of organisms (237). This phylum consists of over 200 genera and contains the majority of Gram-negative species (237). The group consists of five subdivisions: Alpha-, Beta-, Gamma-, Delta-, and Epsilonproteobacteria. Although the subdivisions evolved from a common ancestor, the estimated divergence dates indicate that individual proteobacterial subdivisions branched separately and in a particular order, with the delta subdivision having branched first, approximately 2.38 billion years ago, followed by the epsilon, alpha, beta, and gamma subdivisions (237, 536).
Although the gammaproteobacteria appear to have branched most recently, this subdivision contains many of the model species used to study biological processes of microorganisms, especially those of bacterial pathogens. Among these gammaproteobacteria, Escherichia coli and Salmonella are often used to model genetic and cellular biology mechanisms, whereas Haemophilus influenzae serves as a prototype for natural transformation of DNA. Other proteobacterial subdivisions also contain model species such as Neisseria spp., betaproteobacteria that are often studied to understand mechanisms of DNA uptake, recombination, and antigenic variation. While studies of these organisms provide useful paradigms for important biological processes, these paradigms often do not accurately represent mechanisms employed by delta- and epsilonproteobacteria.
Members of the epsilonproteobacteria represent a physiologically and ecologically diverse group of organisms that are quite evolutionarily distinct from the more commonly studied proteobacteria such as E. coli and Salmonella spp. (45, 536). One common factor shared by these organisms is that the niches they inhabit are often found in extreme, geographically distinct environments. From the high temperature and pressure of deep-sea hydrothermal vents to the highly acidic gastric mucosa or to microbial mats found in sulfidic caves and springs, these environments clearly require a unique repertoire of cellular survival processes that makes the epsilonproteobacteria unique (156, 440, 490, 556). Despite the nearly ubiquitous nature of this class of bacteria, the epsilonproteobacteria remain one of the most understudied groups of bacterial species. Taxonomically, the epsilonproteobacteria can be grouped into two orders: the Nautiliales and the Campylobacterales. Phylogenetic analysis of 16S rRNA gene sequences suggests that individual strains or species cluster together based on ecotype and metabolic capability (74). The most commonly studied genera within the epsilonproteobacteria are Helicobacter spp. and Campylobacter spp., which are members of the order Campylobacterales.
Campylobacter spp. are Gram-negative, microaerophilic, spiral-shaped, motile bacteria. These bacteria are normal intestinal inhabitants of a wide variety of animals and avian species but often are pathogens of humans. Globally, Campylobacter spp. are a major cause of acute gastroenteritis, with Campylobacter jejuni and Campylobacter coli responsible for the majority of cases of disease (11, 282, 544). Transmission to humans usually occurs by food- or waterborne routes. In humans, C. jejuni colonizes the intestinal epithelium and often causes a mild, watery diarrhea to a severe, bloody diarrheal illness, which is estimated to affect over 2 million individuals annually in the United States alone (11, 190, 545, 592). C. jejuni infection is also associated with the development of serious immune-mediated neurological sequelae known as Guillain-Barré syndrome (GBS) (reviewed in reference 282).
The Helicobacter spp. can be divided into two major categories, gastric and nongastric Helicobacter spp., with the latter group including enterohepatic Helicobacter spp. Although both groups are Gram-negative, spiral-shaped microaerophiles, these bacteria employ biological systems that are specific for their respective niches. Helicobacter pylori is a gastric species that is the major pathogen among all Helicobacter spp. Arguably one of the most successful human pathogens, H. pylori chronically colonizes approximately half of the world's population (62, 149). In developing countries, the prevalence of H. pylori infection can be >80%, whereas the number of people colonized in developed countries is typically under 40% (491). Because no environmental reservoirs have been discovered for H. pylori, transmission to humans is thought to occur via person-to-person contact, primarily at a young age (173, 174, 302, 365, 476). Persistent infection is commonly associated with outcomes that range from asymptomatic carriage and mild gastritis to more severe diseases such as peptic ulcer disease and gastric cancer (62, 63, 162, 474, 475, 583, 590). Given the length of colonization and its burden of disease, H. pylori is clearly a significant human pathogen worldwide.
Because Helicobacter and Campylobacter spp. are medically important human pathogens, they are the best studied of the epsilonproteobacteria. As such, the genes and biological processes that these bacteria employ are often compared with those found in model pathogens such as E. coli and Salmonella spp. While studies performed using these model organisms have contributed greatly to our knowledge of basic biology and pathogenesis, these model species represent only a small fraction of known bacterial diversity. As such, the biological strategies employed by these organisms do not always accurately represent those of other bacterial species. This fact has become increasingly evident as we discover more about the basic biological systems used by pathogenic Helicobacter and Campylobacter spp. The overarching goal of this review is to present five major biological processes employed by the pathogenic epsilonproteobacterial species H. pylori and C. jejuni and to compare how the mechanisms involved in these processes differ from those observed in the model systems. Specifically, we discuss flagellar gene expression and biosynthesis, DNA transformation and recombination, iron homeostasis and iron-responsive regulation by Fur, adherence and invasion mechanisms, and protein glycosylation.
FLAGELLAR GENE REGULATION AND BIOSYNTHESIS
Swimming motility in bacteria is promoted by a rotary nanomachine that consists of a flagellum to provide the physical force of movement and a chemosensory system to control the proper direction of movement. Chemotactic motility, the specific migration of a bacterium toward or away from components in the microenvironment, is required for initiating and maintaining interactions of C. jejuni and H. pylori with hosts. Aflagellated or motile but chemotaxis-defective mutants of C. jejuni are attenuated for persistent commensal colonization of poultry and infection of human volunteers (60, 254, 438, 633, 651). Similarly, motility and chemotaxis of H. pylori are required to initiate and maintain infection in murine and gnotobiotic piglet models of colonization (17, 152, 186, 465, 596). H. pylori chemotactic responses are also required to promote association with gastric cells in the stomachs of gerbils and to elicit inflammatory responses, a noted consequence of H. pylori infection and pathogenesis (407, 642).
Flagellar gene expression and biosynthesis have been investigated extensively through genetic and biochemical analyses of Salmonella spp. and E. coli. Cumulative results from these studies were combined to form a model to understand the regulation of flagellar gene expression and biosynthesis, which has been used to compare these processes in other bacteria. However, studies of C. jejuni and H. pylori have revealed that the epsilonproteobacteria use a number of factors that are either absent or not required for motility in Salmonella spp. and E. coli. As subsequent studies revealed, these proteins in C. jejuni and H. pylori impart alternative means of controlling flagellar gene expression or spatial and numerical parameters of flagellar biosynthesis.
Transcriptional Regulatory Cascades for Flagellar Gene Expression
The paradigm of E. coli and Salmonella.
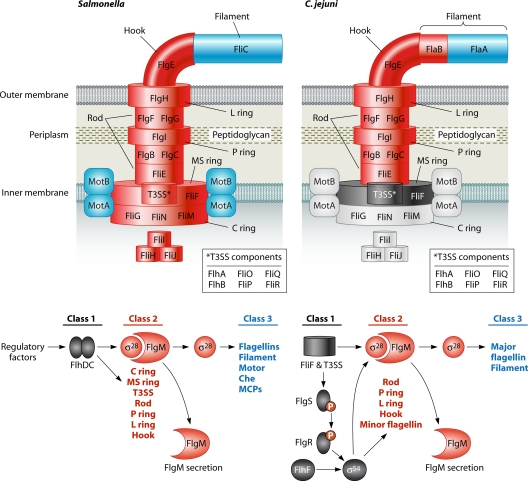
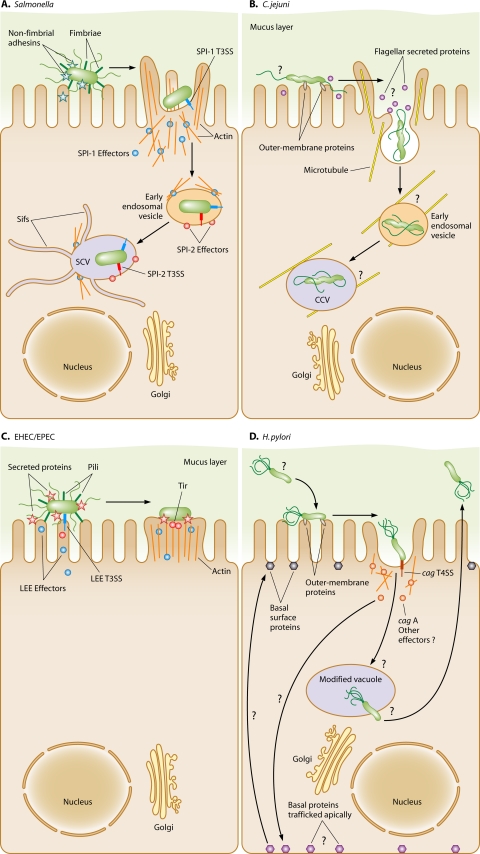
Despite the diversity of bacterial species, the structure of the flagellar organelle is relatively conserved from one bacterium to another. Depending on the organism, more than 25 different proteins form the flagellum, with another 10 to 60 proteins forming the chemosensory system (389). The flagellum consists of a hook-basal body (HBB) complex and an extracellular filament. The HBB complex is divided into a few substructures (described below) that originate from the cytoplasmic face of the inner membrane and extend to the hook on the extracellular surface of the outer membrane (Fig. 1). The filament is composed largely of hundreds of flagellin subunits and a few other minor proteins that assist filament biogenesis. The chemosensory system consists of chemoreceptors that usually sense a specific nutritional component, a phosphorelay signaling system that transmits signals from the chemoreceptors to the flagellar motor, and a methylation system to fine-tune chemoreceptor-induced signaling (see reference 616 for a general review). Depending on whether the sensed environmental factor is an attractant or repellant, the chemosensory system influences the clockwise or counterclockwise rotation of the flagellum, which ultimately directs the bacterium toward more favorable microenvironments for growth.
FIG. 1.
Comparison of the flagellar structures and regulatory cascades of Salmonella spp. and E. coli and those of C. jejuni and H. pylori. Proteins of the flagellar structures are color coded based on which class of the respective genes are found in the flagellar transcriptional hierarchies, using black (class 1), red (class 2), blue (class 3), and gray (outside the flagellar transcriptional hierarchy). Some genes are not exclusive to one class, but these are not included in this figure for simplicity. fliA (encoding σ28) and flgM are included as class 2 genes in C. jejuni, but these genes are not entirely dependent on σ54 for expression. Note that not all proteins required for flagellar biosynthesis are shown for simplicity in comparing structures and the transcriptional regulatory cascades. The most current proposed flagellar regulatory cascade for C. jejuni is shown as a representative for H. pylori, but more detailed analysis of the H. pylori system is required to verify this transcriptional hierarchy.
A bacterium must coordinate the expression of each gene encoding the structural or chemosensory components as well as promote the orderly secretion and/or interaction of each protein to build a properly functioning flagellar organelle. As elucidated for Salmonella spp. and E. coli, flagellar gene expression usually follows a tightly regulated, temporal, and hierarchal transcriptional pattern (312, 356). This pattern of expression allows bacteria to synthesize the flagellar structure from the inside out, beginning with the base of the flagellum at the cytoplasmic face of the inner membrane and extending outward to the extracellular tip of the filament (92, 390). Flagellar genes in Salmonella spp. and E. coli are divided into three classes, depending on their temporal order of expression (Fig. 1) (312, 356). Class 1 genes, which are transcribed first, include flhDC. These genes encode the heteromeric FlhDC master transcriptional regulator that directly binds to and activates the transcription of promoters of multiple operons containing the class 2 genes (374, 627). Class 2 genes encode multiple proteins that form the HBB complex, σ28, and FlgM, which is the anti-σ factor for σ28. The HBB consists of components that stretch from the cytoplasm to the extracellular surface (Fig. 1). These parts include the cytoplasmic ring (C ring) at the base of the flagellum and the inner membrane MS ring (consisting of a multimer of FliF). The components of a type III secretion system (T3SS) are embedded in the center of the MS ring to form the flagellar secretory apparatus. Motor proteins (MotA and MotB) associate with the C and MS rings to form the stator element of the flagellar motor. Bacterial envelope components of the HBB complex include multiple proteins that form the rod and the structural rings located in the peptidoglycan (P ring) and outer membrane (L ring). The hook is the most distal part of the HBB complex and is located on the surface of the outer membrane.
After FlhDC-mediated transcription of class 2 genes, the first components of the flagellum thought to be assembled are the MS ring and the T3SS (consisting of the FlhA, FlhB, FliO, FliP, FliQ, and FliR proteins) within the MS ring, although the order of assembly of the MS ring and the T3SS is unclear (for detailed reviews, see references 92 and 390). Attached to the cytoplasmic side of the MS ring is the C ring, which has dual functions in flagellar motility (188, 323): (i) it is the rotor and switch component of the flagellar motor (276, 393, 555, 657, 658), and (ii) it serves as an inverted cup to efficiently deliver proteins to the flagellar secretory apparatus in the center of the MS ring (159, 213). The flagellar secretory apparatus facilitates the ordered secretion of most flagellar proteins located beyond the inner membrane, beginning with the rod proteins. Flagellar proteins, in general, are secreted into the hollow center of the flagellum at the T3SS-rod juncture, transit to the tip of the sprouting flagellar structure, and polymerize on the growing tip. Thus, the flagellum is a conduit for transport of its own secretion substrates. As the rod is polymerized, it breaches the structural barrier of the peptidoglycan and outer membrane by passing through the center of the flagellar P and L rings, respectively. Note that FlgI and FlgH, which form the P and L rings, respectively, are likely secreted by the Sec type 2 secretory system (T2SS) instead of the flagellar T3SS (266, 294). After completion of rod assembly, the hook proteins are secreted to form a fulcrum on the bacterial surface.
A critical checkpoint in the flagellar transcriptional hierarchy occurs upon completion of the HBB complex. σ28, encoded by fliA, is required for expression of class 3 genes (312, 461). These genes encode the flagellins and other minor proteins of the filament, the stator components of the flagellar motor (MotA and MotB), and the chemosensory system. Even though fliA is a class 2 gene and is expressed with the genes encoding the HBB complex, σ28 is inhibited from interacting with RNA polymerase (RNAP) by the product of another class 2 gene, FlgM (84, 85, 356, 462). Once the HBB complex is complete, FlgM is secreted from the bacterium by the flagellar T3SS (272, 312, 355). As a result, σ28 is relieved from repression, allowing it to interact with RNAP to promote transcription of class 3 genes. Thus, expression of the flagellin, filament, motor, and chemosensory genes is coupled to a structural step in flagellar biosynthesis.
Transcriptional hierarchy for flagellar genes in C. jejuni and H. pylori.
The flagellar filaments of many Campylobacter and Helicobacter spp. are composed of two different flagellins, FlaA and FlaB (368, 378). FlaA is abundant in the filament and is required for full motility (229, 232, 297, 571, 581, 631, 632). FlaB is a minor flagellin, thought to be localized primarily to the basal portion of the filament, and its requirement for wild-type levels of motility varies with the species. The flaA and flaB alleles of these organisms are expressed from different promoters, with characteristic binding sequences for σ28 and σ54, respectively (229, 232, 368, 455, 571, 581, 631). Thus, two different alternative σ factors are required for flagellar gene expression in Campylobacter and Helicobacter spp., unlike the case in Salmonella spp. and E. coli. Along with flaB, many genes for the flagellar rod and hook proteins in C. jejuni and Helicobacter spp. are dependent on σ54 for expression and contain conserved σ54-binding sites in the respective promoters (9, 80, 255, 328, 447, 464, 473, 534, 564, 651).
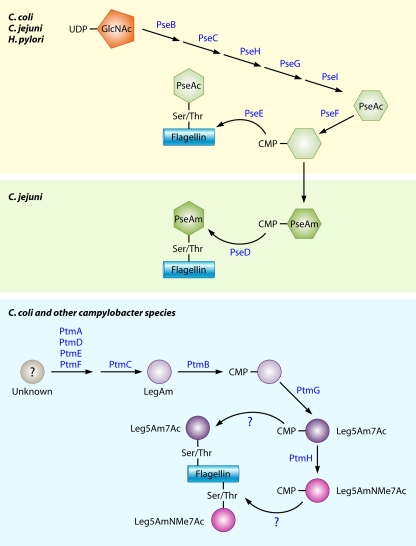
An interesting observation on C. jejuni and H. pylori is the relative conservatism of the use of σ54 for gene expression. In many bacteria, σ54 is required for expression of multiple operons, which often encode proteins for diverse biological functions (318, 351). Specificity for transcription of operons for one biological process in a single bacterium is often maintained by activation of σ54 through a characteristic trans-activator protein (often called an “enhancer-binding protein”) to promote transcriptional initiation (reviewed in references 351 and 451). However, almost all σ54-dependent genes in C. jejuni and H. pylori encode proteins that are structural components of the flagellum (80, 447). Therefore, σ54 is limited almost exclusively to flagellar gene expression in these bacteria. pseB is one σ54-dependent gene of C. jejuni that does not encode a structural component of the flagellum. However, PseB is an enzyme required for pseudaminic acid (PseAc) biosynthesis, a carbohydrate modification of the flagellin necessary for filament biogenesis (218; see below).
As mentioned above, σ54 requires a characteristic enhancer-binding protein to activate transcription of target genes. The prototypical member of this class of transcriptional activators is NtrC, which is found in many gammaproteobacteria (143). Phosphorelay from a cognate sensor histidine kinase to a specific aspartic acid of an NtrC-like response regulator usually activates the protein so that it can interact with target promoters and σ54. ATP hydrolysis by the regulator assists in transitioning DNA to an open complex formation.
Genetic screens in C. jejuni have identified multiple genes required for expression of the σ54 regulon (253, 255). Two genes revealed from these studies are flgS and flgR, which encode a sensor histidine kinase and an NtrC-like response regulator, respectively, that together form a σ54-activating two-component system in both C. jejuni and H. pylori (68, 255, 300, 301, 447, 564, 651). Other genes identified in C. jejuni that are required to stimulate σ54 activity are flhA, flhB, fliP, and fliR, which encode components of the T3SS, and flhF, which encodes a GTPase found mainly in polarly flagellated bacteria (255). Deletion of any of these genes in C. jejuni causes large reductions in expression of many σ54-dependent genes (80, 255, 300, 651). Similarly, mutation of flhA, flhB, and flhF homologs in H. pylori also reduces expression of the σ54 regulon (7, 447, 525).
These findings suggest that activation of σ54 is multifactorial, likely requiring the integrated activities of three separate systems, namely, the flagellar T3SS, the FlgSR two-component system, and FlhF (Fig. 1). Strong evidence from C. jejuni supports a model in which the T3SS influences activation of the FlgSR system, with the FlhF GTPase functioning possibly outside and downstream of this pathway for full expression of the σ54 regulon (40, 255, 300). As with most two-component regulatory systems, phosphorelay from the FlgS sensor kinase to the FlgR response regulator is required for expression of the σ54 regulon (300, 301, 651). Furthermore, in mutants lacking a component of the secretory apparatus, FlgS and FlgR are produced but are evidently defective in phosphorelay and do not activate σ54 (300). These results suggest that phosphorelay through FlgSR is dependent on the T3SS. Genetic studies have revealed that specific FlgR mutant proteins which do not require FlgS for activation and stimulation of σ54 can partially suppress the phenotype of mutants lacking components of the T3SS. These results suggest that FlgSR and the flagellar T3SS likely function within the same signaling pathway, with FlgSR acting downstream of the apparatus to stimulate σ54-dependent expression of flagellar genes (300, 301).
In contrast to C. jejuni flgS or T3SS mutants, these partially FlgS- and T3SS-independent FlgR mutant proteins do not suppress the phenotype of an flhF deletion mutant for expression of σ54-dependent flagellar genes (40). In addition, a double mutant lacking flhF and a secretory apparatus is more defective for expression of σ54-dependent flagellar genes than either single mutant. These results suggest that the FlhF GTPase functions outside the T3SS-FlgSR pathway for activation of expression of σ54-dependent flagellar genes. Current hypotheses propose that FlhF may function downstream of or converge with the T3SS-FlgSR pathway to activate σ54. Further analysis of FlhF demonstrated that full GTPase activity of FlhF is not required for expression of σ54-dependent flagellar genes (40). Mechanistic details regarding how FlhF influences activation of expression of σ54-dependent genes in a GTPase-independent manner remain to be elucidated.
For C. jejuni, the flagellar T3SS has been characterized further to determine its requirements for activating the FlgSR system and expression of the σ54 regulon. Specific point mutations or domain deletions in FlhB (a component of the T3SS) or deletion of fliI (encoding an ATPase that likely increases the efficiency of secretion of flagellar proteins through the apparatus [422, 479]) results in mutants that assemble secretion machineries but are severely hindered for secretion of the major flagellin FlaA (300). However, these mutants are not defective for expression of σ54-dependent flagellar genes, perhaps indicating that the flagellar T3SS does not have to be fully secretion competent to activate FlgSR (Fig. 1). If secretion is not required by the apparatus for FlgS to detect a signal, then formation of the T3SS may be a signal detected by FlgS to initiate phosphorelay to FlgR. Since FlgS is a cytoplasmic sensor kinase (300), it is conceivable that FlgS may detect a specific epitope on the cytoplasmic face of an assembled secretory apparatus to determine that the T3SS has formed. This hypothesis suggests that FlgS may physically interact with an epitope of a single protein or a multiprotein epitope of the T3SS once it has assembled. Detection of this type of signal would then lead to autophosphorylation of FlgS that culminates in activation of σ54.
Inclusion of σ54 in the flagellar transcriptional hierarchies of Campylobacter and Helicobacter spp. is a significant departure from the cases for Salmonella spp. and E. coli. The reason for the use of this alternative σ factor in the flagellar regulatory cascades of these epsilonproteobacteria is not entirely clear. The vast majority of σ54-dependent genes of both C. jejuni and H. pylori include those that encode the flagellar rod and hook proteins, which are dependent on the flagellar T3SS for transport out of the cytoplasm (Fig. 1). By making expression of these genes dependent on the secretory system, FlgSR, and σ54, a checkpoint within the bacterium is created: the T3SS must be formed before it initiates a regulatory cascade necessary to promote expression of its secretion substrates. Expression of σ54-dependent genes is tightly controlled through the requirements of (i) a highly specific binding site for σ54 in the promoters of target genes and (ii) an enhancer-binding protein that is often activated by a signal transduction system involving a cognate sensor kinase. These requirements likely prevent transcription of the rod and hook genes when their expression is not needed. Therefore, tight control of σ54 activity by a regulatory cascade consisting of the flagellar T3SS and the FlgSR system, along with FlhF in C. jejuni and H. pylori, seems to ensure that production of the rod and hook proteins occurs only in bacteria that have formed a system to secrete flagellar substrates. On the other hand, use of σ54 in flagellar transcriptional cascades is, curiously, a common theme in polarly flagellated bacteria (121, 330, 331, 494, 604). The biosynthetic processes required to generate a flagellum at a bacterial pole may require an ordered production of proteins that can be mediated only by the tight control of expression of the respective genes through mechanisms to activate σ54.
The requirement of σ28 for expression of flagellar genes and the control of σ28 activity are similar between gamma- and epsilonproteobacteria. In C. jejuni and Helicobacter spp., σ28 is required for expression of class 3 genes, which include the major flagellin gene, flaA, along with those encoding minor filament proteins (80, 255, 298, 368, 571). Like the case in gammaproteobacteria, FlgM associates with σ28 to inhibit its activity and releases σ28 from inhibition when FlgM is secreted out of the cytoplasm by the HBB (106, 298, 511, 650). Unlike the case in Salmonella spp. and E. coli, genes encoding the motor proteins and various components of the chemosensory system in C. jejuni and H. pylori are not a part of the σ28 regulon (80, 447). Expression of these genes is likely not controlled by the flagellar transcriptional hierarchy and may be constitutive. A curious finding noted in C. jejuni is that FlgM associates with σ28 to inhibit its activity at 37°C but not at 42°C, which is the body temperature of this organism's natural avian hosts (650). This finding possibly suggests that σ28 activity may be unregulated during commensal colonization of these hosts.
Using the Salmonella sp. and E. coli transcriptional cascades as a model, a regulatory hierarchy for flagellar gene expression can be constructed for C. jejuni and H. pylori (Fig. 1). This model is based mainly on the more detailed analyses of the molecular mechanisms in C. jejuni, but studies of H. pylori suggest that the transcriptional hierarchies of these two bacteria are likely similar. Class 1 genes include rpoN (encoding σ54) and genes that encode the flagellar T3SS, the FlgSR system, and the FlhF GTPase, which are all required to activate σ54. Stimulation of σ54 activity results in expression of the class 2 genes, which encode primarily the flagellar rod, ring, and hook proteins. Also potentially included as class 2 genes are fliA and flgM, which encode σ28 and its anti-σ factor, since some evidence exists that expression of these genes is partially σ54 dependent (80, 650). Class 3 genes include those dependent on σ28 for expression, which include flaA, genes encoding minor filament proteins, and a few nonflagellar genes that may encode proteins involved in virulence mechanisms (217, 489). This transcriptional hierarchy accounts for the requirement of two alternative σ factors for tight and ordered control of flagellar genes for proper biosynthesis of the flagellar organelle.
Mechanisms of expression of class 1 genes.
Class 1 genes in the flagellar transcriptional hierarchy of E. coli and Salmonella spp. are composed of flhDC (312, 356). In these bacteria, a diverse set of transcriptional regulators incorporate different environmental and intracellular signals to control flhDC expression. By having flhDC expression influenced by various factors, these bacteria are able to optimize the production of flagella under a variety of conditions, physiological states, or niches the bacteria encounter during infection of a host or in natural environments. Examples of these regulators and the various cues that influence their activity include CAP (utilization of carbon sources) (561, 659), DksA and ppGpp (nutrient limitation-induced stringent response) (3), the QseBC two-component regulatory system (quorum-sensing molecules) (100, 271, 563), the EnvZ-OmpR two-component system (osmolarity) (540), and the nucleoid-associated protein Fis (whose levels fluctuate under various conditions, such as the stringent response) (319).
flhDC expression is also repressed by transcriptional regulators that activate expression of other virulence genes. For example, FimZ activates transcription of fimA, encoding the major fimbrial subunit of the type I fimbriae, but represses expression of flhDC when overexpressed in Salmonella spp. (104). By governing expression of flhDC and fimA differentially through FimZ, a switch is created that allows bacteria either to adhere to host eukaryotic cells by the production of fimbriae or to swim through formation of flagella (104). Similarly, the RcsCSB system activates expression of genes required for capsular polysaccharide biosynthesis in E. coli and for intracellular growth in Salmonella spp. but represses flhDC expression (159, 187, 626, 652). Thus, motility is opposingly regulated with production of capsule or virulence genes required for growth within macrophages. The RtsAB system of Salmonella spp. may also opposingly control expression of invasion genes and those for motility (154). RtsA positively influences expression of invasion genes, but RtsB represses flhDC expression when overexpressed.
Other relatively less well-characterized transcriptional regulators have been shown to influence expression of flhDC in Salmonella spp. and E. coli. For example, two LysR-type regulators, LrhA of both Salmonella spp. and E. coli and HdfR of E. coli, repress flhDC expression (159, 334, 363). Furthermore, overexpression of the transcriptional regulators EcnR, SlyA, and PefI-SrgD also reduces expression of flhDC in Salmonella spp.(159, 652). Posttranscriptional regulation also influences flhDC expression in E. coli, as the stability of flhDC mRNA is dependent on the RNA-binding protein CsrA (637).
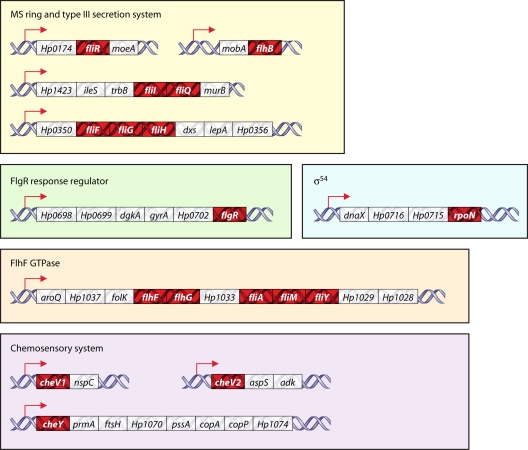
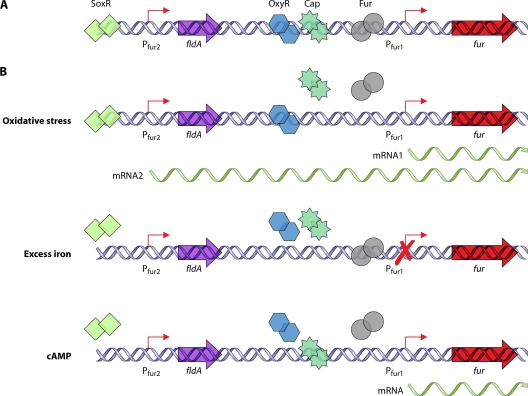
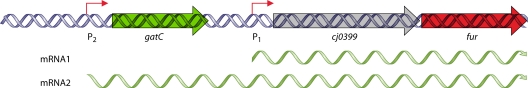
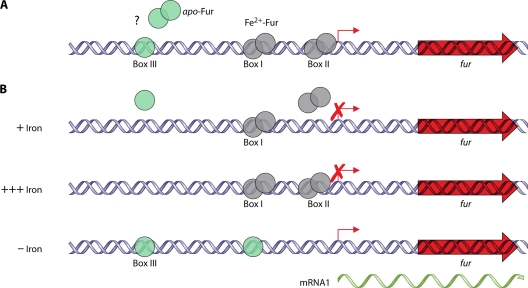
Studies have yet to identify any regulators or growth conditions that globally affect expression of class 1 genes of C. jejuni and H. pylori. One potential hypothesis is that expression of class 1 genes may be constitutive. Analysis of the global transcriptome of H. pylori recently provided a genome-wide map of numerous transcriptional start sites and potentially cotranscribed operons (534). From this analysis, it was found that many H. pylori class 1 genes are cotranscribed with housekeeping genes or genes that are thought to be essential for growth (Fig. 2). For instance, flhF is found in an operon with aroQ and folK, which encode proteins required for generation of some amino acids and folate, respectively. H. pylori rpoN is located within an operon with dnaX, which encodes a component of RNAP. Also, various che genes, which encode components of the chemosensory system, are found in operons with genes such as ftsH, aspC, and nspC that are likely required for optimal growth under certain conditions. Grouping of class 1 genes into these operons would ensure that the flagellar T3SS, FlhF, FlgSR, and σ54 are produced constitutively and available to promote expression of the class 2 genes when necessary. This type of global transcriptome analysis has not yet been performed for C. jejuni. However, considering that the genetic organization of many of these operons is similar in C. jejuni and H. pylori, it is likely that expression of many class 1 genes in C. jejuni is also constitutive (185, 473).
FIG. 2.
Operonic organization of many class 1 genes and che genes of H. pylori with essential or housekeeping genes. Many of these operons have been confirmed to be cotranscribed, as described by Sharma et al. (534). Flagellar genes are indicated in red. Gene designations are based on the genomic sequence of H. pylori strain 26695.
If class 1 genes are constitutively expressed, then the implication for C. jejuni and H. pylori is that as long as the proteins encoded by class 1 genes can initiate the necessary signaling steps to activate class 2 gene expression, these bacteria have the potential to always produce flagella, regardless of environmental or growth conditions. Since these bacteria are most often found associated with a host and motility is necessary for colonization of hosts, constitutive expression of flagella by these organisms may be a strategy to maintain fitness as infectious organisms. However, C. jejuni is transferred to new hosts through contact with fecal matter or handling and consumption of contaminated foods or water, suggesting that environmental reservoirs likely exist. It is unclear if flagellar biosynthesis occurs outside the hosts of C. jejuni and provides a survival advantage under these conditions.
One mechanism to turn off flagellar biosynthesis in Campylobacter species is phase variation (73, 251, 252, 315, 456, 471). In C. coli, flhA contains a homopolymeric tract of thymine residues in the 5′ end of the coding sequence that undergoes reversible and random alteration to effect production of the encoded flagellar T3SS protein (471). In C. jejuni, genetic targets of phase variation are flgS and flgR, encoding the two-component regulatory system necessary for σ54 activation (251, 252). Random and reversible alteration of homopolymeric and heteropolymeric nucleotide tracts within flgS and flgR, along with second-site intragenic and extragenic suppressor mutations, affect production of the FlgSR system to allow variable production of the flagella in vitro and in vivo to influence the colonization capacity of the bacterium. C. jejuni is potentially unique in that the FlgSR system is the only known two-component system where both regulatory factors undergo phase-variable production.
Spatial and Numerical Control of Flagellar Biosynthesis
Salmonella spp. and E. coli are peritrichous organisms, producing multiple flagellar organelles over the surfaces of the bacteria. Other bacteria, such as H. pylori and C. jejuni, in addition to Vibrio and Pseudomonas spp., exhibit a polar pattern of flagellar biosynthesis and produce only a limited number of flagella. H. pylori commonly produces up to six flagella, but only at one pole, whereas C. jejuni produces a single flagellum at one or both poles (200, 216, 480). Thus, rigorous spatial and numerical restrictions on flagellar biosynthesis that exist in C. jejuni and H. pylori are likely absent from Salmonella spp. and E. coli.
The only factor in Salmonella enterica serovar Typhimurium that has been noted to influence flagellar number is the level of expression of flhDC. In wild-type strains, individual bacteria normally produce four to six flagella (159). The level of flhDC expression appears to correlate directly with the number of flagella. Promoter mutations resulting in a 2-fold increase in expression of flhDC cause a doubling of flagellar number, presumably due to an increase in production of flagellar proteins available to build flagella (159).
Specific regulatory factors that govern the spatial and numerical aspects of flagellar biosynthesis have been identified and analyzed in C. jejuni and other polarly flagellated bacteria. Two of these factors are the FlhF GTPase and FlhG (annotated as FleN in Pseudomonas spp.), which appear unique to flagellar loci of polarly flagellated species of Vibrio, Pseudomonas, Campylobacter, and Helicobacter and are absent from peritrichous organisms such as Salmonella spp. and E. coli. Mutation of flhF and flhG in Vibrio and Pseudomonas spp. impacts flagellar biosynthesis in a variety of ways, by increasing or decreasing flagellar gene expression, altering flagellar number, or causing formation of lateral rather than polar flagella (109, 121, 122, 226, 352-354, 437, 468).
The FlhF proteins of polarly flagellated bacteria have a conserved GTPase domain, and GTP hydrolysis by C. jejuni has been demonstrated in vitro (40). Whereas deletion of flhF in C. jejuni results in significant reduction in expression of the σ54 regulon and motility, mutation of the GTPase domain of FlhF does not cause significant decreases in expression of σ54-dependent flagellar genes, but motility is reduced (40). Thus, in C. jejuni, FlhF is involved in flagellar biosynthesis in two different manners: an FlhF GTPase-independent process required for expression of σ54-dependent flagellar genes and an FlhF GTPase-dependent process involved in another aspect of flagellar motility.
The reduced motility phenotype of mutants producing FlhF proteins with lower GTPase activity appears to be due to improper placement or increased number of flagella, indicating that GTP hydrolysis by FlhF is a determinant for spatial and numerical control of flagellar biosynthesis (40). A significant proportion of a population of C. jejuni cells producing these FlhF mutant proteins either construct lateral flagella (rather than polar flagella) or construct more than one flagellum at a pole. Since flagellar biosynthesis begins at the inner membrane, with the assembly of the MS ring and the flagellar T3SS, one possible hypothesis is that GTP hydrolysis by FlhF may be required at an early step in flagellar biosynthesis by influencing the spatial placement of the MS ring, C ring, and T3SS at the bacterial poles and ensuring that only one apparatus is constructed per pole. Indeed, initial observations of polar localization of FlhF in C. jejuni may support the hypothesis that FlhF determines polar sites for flagellar biosynthesis (167). Because some of the GTPase-hindered FlhF mutants also produce truncated flagella or a flagellum only at one pole (40), GTP hydrolysis may also assist in forming a secretion-competent flagellar T3SS. The molecular mechanisms of how GTPase activity may influence these various aspects of flagellar biosynthesis remain to be characterized.
C. jejuni and H. pylori both encode FlhG, which has been shown in Vibrio cholerae and Pseudomonas spp. to regulate flagellar number (109, 121, 122, 352). V. cholera and Pseudomonas aeruginosa FlhG homologs repress the activity or transcription of a master transcriptional regulator (109, 122). Derepressed activation or expression of the regulator in flhG mutants artificially leads to an increase in flagellar gene expression relative to that in wild-type bacteria that likely directly results in the observed increased in polar flagellar numbers. If FlhG exerts numerical control of flagellar biosynthesis in C. jejuni and H. pylori, it likely does so by a different mechanism than that in V. cholerae and Pseudomonas spp., since C. jejuni and H. pylori lack an apparent master regulator atop the flagellar transcriptional hierarchy to control gene expression.
While the detailed molecular mechanisms that govern the spatial and numerical control of flagellar biosynthesis wait to be unraveled, the molecular mechanisms that influence flagellar number and placement are likely to differ between C. jejuni and H. pylori. In H. pylori, up to six flagella are naturally produced at a single pole, suggesting a more lenient control of flagellar numbers. In addition, the two poles of an H. pylori cell are apparently different, with only one supporting the biosynthesis of flagella in an individual bacterium, unlike in C. jejuni, where a flagellum is placed at both poles.
Differences in Filament Structure and Factors Involved in Flagellar Biosynthesis
Whereas flagellins of many bacterial species stimulate production of inflammatory mediators through activation of Toll-like receptor 5 (TLR5), the flagellins of Helicobacter spp. and C. jejuni do not (18, 248, 552). TLR5 recognizes a stretch of eight amino acids within the highly conserved D1 domain, which mediates interactions between adjacent flagellins to form the flagellar filament in Salmonella spp. (515, 663). This sequence is highly divergent in the flagellins of many epsilonproteobacteria, including Campylobacter and Helicobacter spp. When Salmonella enterica serovar Typhimurium produces a FliC flagellin harboring a change in TLR5 recognition pattern to resemble that of H. pylori FlaA, TLR5 is not stimulated (18). In addition, the bacterial strain is nonmotile, which highlights a major difference between the flagellins of the epsilonproteobacteria in building a filament (18). This difference in a small region of the D1 domain in the C. jejuni flagellin is believed to contribute to a looser packing of the flagellins within the flagellar filament (196).
A distinguishing feature of the flagellum of H. pylori is that the filament is encased in a sheath (200, 216). Most motile bacteria do not have a sheath; however, V. cholerae also produces a sheathed flagellum (184). The sheath extends the length of each filament and terminates in a bulb-like structure. The sheath appears to be composed of a bilayer membrane with lipooligosaccharide and an altered profile of fatty acids (201). However, no one specific fatty acid appears to be localized solely to the sheath relative to the cell body. Proteins are also found associated with the flagellar sheath, with the H. pylori adhesin A protein (HpaA) potentially enriched in the sheath compared to the bacterial surface (292, 385, 387). An understanding of how the sheath forms and its role in motility or the biology of H. pylori has remained elusive. One possible hypothesis is that the sheath prevents recognition of the flagellins by the immune system during infection. This idea is supported by the observation that only unsheathed flagella are recognized by flagellin-specific antibodies (344). Some mutations in flagellar genes have been associated with morphological changes in the sheath. For instance, mutation of flaA in Helicobacter spp. results in shortened flagella and shortened sheaths equal to the lengths of the flagella (297). Mutants lacking both flaA and flaB do not produce a filament, but occasionally a structure resembling an empty sheath without a filament is observed (297). Mutation of fliD, encoding the capping protein at the tip of the filament, causes variation in the lengths of filaments (325). The flagella of a fliD mutant often have empty sheathed extensions at the tip. In most cases, the terminal bulb does not form in fliD mutants. Additional analysis is required to understand how the sheath forms in relation to flagellar biosynthesis and whether the sheath plays a significant role in the biology of H. pylori.
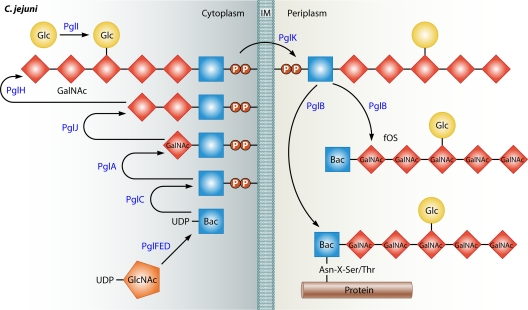
A few proteins have been identified in C. jejuni that are required for flagellar motility and are not present in Salmonella spp. and E. coli. These proteins may form additional structural components or modification of the flagellar basal body required for motility. FlgP and FlgQ are two proteins required for flagellar motility but not for flagellar biosynthesis (558). Initial analysis suggested that FlgP associates with the outer membrane and requires FlgQ for this localization, suggesting that FlgQ may have a chaperone-like activity for FlgP. Additionally, PflA is required for full motility in C. jejuni (660). Mutants lacking pflA produce flagella but are nonmotile, thus causing a paralyzed flagellum phenotype. Because of a predicted signal sequence in PflA, this protein may be secreted out of the cytoplasm and form part of the flagellar structure or motor. In addition to a requirement of the flagellins to be glycosylated by an O-linked protein glycsoylation system for flagellar biosynthesis (described below), modification of the FlgG rod protein with phosphoethanolamine is required for efficient flagellar biosynthesis and motility (116). How this modification to a rod protein or how FlgP, FlgQ, or PflA influences motility remains to be determined.
Summary
Due to a concerted effort by many laboratories over the past couple of decades, the importance of flagellar motility to Helicobacter and Campylobacter spp. for infection of hosts and the molecular mechanisms governing flagellar gene regulation and biosynthesis have contributed important knowledge concerning the biology of these epsilonproteobacteria. As such, these species have been useful systems for studying previously unrecognized alternatives to biosynthesis of bacterial organelles. So far, it is apparent that utilization of two alternative σ factors creates a different, highly ordered mechanism from those of gammaproteobacteria to regulate expression of flagellar genes and control proper biosynthesis of the flagella. Furthermore, Helicobacter and Campylobacter spp. also have additional controls to restrict the placement and number of flagella at the poles that are apparently absent in Salmonella spp. and E. coli. Considering the current knowledge, it is likely that C. jejuni and H. pylori will become model systems for understanding polar biosynthesis of organelles.
NATURAL TRANSFORMATION AND HOMOLOGOUS RECOMBINATION
The ability of bacteria to acquire exogenous DNA from the environment is important for increasing genetic variation and driving bacterial evolution. In bacteria, three distinct processes that mediate acquisition of exogenous DNA are conjugation, transduction, and transformation. Conjugation is the unidirectional transfer of DNA from one bacterium to another and requires a specialized pilus (360). Transduction is bacteriophage-mediated transfer of DNA between bacteria (364). Transformation is the process by which bacteria directly acquire naked DNA from the environment (144); bacteria that are able to obtain DNA in this manner are referred to as naturally transformable or naturally competent. The benefits of natural transformation for the recipient bacteria include the ability to acquire an exogenous source of nucleotides, a means to repair damaged chromosomal DNA, and a mechanism to acquire new genes (144, 177, 557). Thus, acquisition of exogenous DNA results in genetic diversity within a population of bacteria and provides the necessary framework for genomic evolution. As a result, this diversity often provides an in vivo fitness advantage for pathogenic bacteria (43, 125, 307, 580). Natural transformation is a multistep process that can be divided into the following steps: binding of DNA to the bacterial surface, transport of DNA across the bacterial membrane(s) into the cytoplasm, and integration of nonplasmid donor DNA into the recipient chromosome by homologous recombination. Here we present strategies that C. jejuni and H. pylori employ to complete this process compared to the model organisms Neisseria gonorrhoeae, H. influenzae (for both DNA uptake and processing), and E. coli (for homologous recombination).
DNA Uptake and Processing
Naturally competent bacteria encode highly specific systems that facilitate binding, uptake, transport, and recombination of exogenous DNA into the chromosome (90, 91, 144). These systems are composed of many proteins that may be specific for some species and are often expressed under particular physiological conditions (102, 147, 240, 388, 403). In Gram-negative bacteria, DNA uptake systems exhibit a high degree of similarity to two related macromolecular machines, the type IV pilus (T4P) and the T2SS (Fig. 3). Natural transformation in Gram-negative bacteria is best characterized for N. gonorrhoeae and H. influenzae; as such, we focus our discussion on DNA uptake and transport systems of these prototypical bacteria and compare how these systems differ from those employed by C. jejuni and H. pylori.
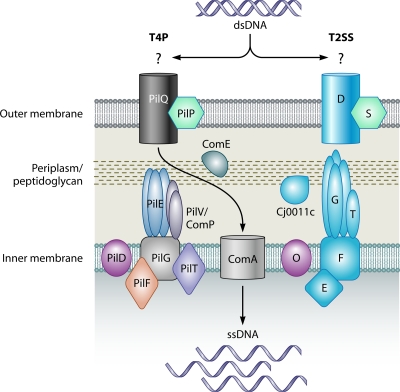
FIG. 3.
Components of the T4P and T2SS involved in transformation. (Left) Model of the T4P system, based on those previously reported (90, 91). Indicated components are named after the N. gonorrhoeae T4P. The outer membrane-spanning pore complex consists of PilQ and is assisted by the PilP pilot protein. The pilus/pseudopilus structure is composed of major (PilE) and minor (PilV or ComP) pilin proteins, which are processed by the prepilin peptidase, PilD. The T4P adhesin, PilC, associates with the distal end of the pilus structure (not shown). Pilus/pseudopilus biogenesis is also facilitated by the traffic NTPase PilF and the inner membrane protein PilG. Another traffic NTPase, PilT, is involved in disassembly of the pilus/pseudopilus. During twitching motility, the pilus complex polymerizes to facilitate attachment to a nearby surface. Once attached, the pilus complex depolymerizes to result in bacterial locomotion. In a similar manner, donor DNA may be taken into the cell by binding to an unidentified DNA receptor followed by retraction of the competence pseudopilus structure. Once inside the periplasmic space, the donor DNA may then be processed and translocated into the cytoplasm through ComA. DNA translocation may involve the periplasmic protein ComE. (Right) The T2SS contains several components that are homologous to the T4P system. The components shown are referred to by their respective letter designations, and the model is based on those reported previously (91, 516). Note that not all known T2SS components are shown and that some components may be species specific. Assisted by the S pilot protein, protein D forms the outer membrane-spanning pore complex. The pseudopilus-like structure is formed by G, as well as other minor pseudopilins, collectively represented by T, which are processed by the prepilin peptidase O. Other components involved in pseudopilus assembly are the inner membrane protein F and the traffic NTPase, E. The secretion-activating signal may be transmitted from E to the outer membrane by proteins C, M, and L (not shown). Known components of the C. jejuni Cts T2SS-like transformation system are depicted in blue. These components include homologs of the pore protein D (CtsD), the major and minor pseudopilins G (CtsG) and T (CtsT), the inner membrane protein F (CtsF), and the traffic NTPase E (CtsE) (641). Additionally, the periplasmic protein Cj0011c may be involved in transport of DNA in the periplasm. It is proposed that the T2SS pseudopilus acts as a piston, forcing secreted effectors from the periplasm into the extracellular environment. Although the mechanism for T2SS-mediated DNA uptake is currently unknown, it is possible that once donor DNA is bound, the T2SS works in reverse to transport DNA across the outer membrane and into the recipient cell.
DNA transport and the type IV pilus.
In N. gonorrhoeae, DNA uptake and transformation are mediated by T4P components (reviewed in references 91 and 239), which are also known as pilus/secretion/twitching motility/competence (PSTC) proteins (144). T4P are hair-like appendages found on the bacterial surface that mediate cellular adhesion and twitching motility, which is a form of bacterial translocation due to polymerization or extension of the pilus fiber, attachment to a nearby surface, and then depolymerization or retraction of the pilus (112, 404, 417, 543, 621). T4P are composed of a major pilin (PilE), a minor pilin (PilV), a traffic NTPase (PilT), an ATPase (PilF), a transmembrane secretin (PilQ), a prepilin processing peptidase (PilD), a pilot protein (PilP), and an adhesin (PilC) (1, 27, 57, 59, 66, 91, 141, 189, 295, 342, 509, 602, 644) (Fig. 3). Interestingly, while the presence of pili on the bacterial surface per se may not be required for DNA uptake, production of the major pilin is necessary (91, 206, 382, 509, 562). These findings suggest that some (but not all) components of T4P are involved in DNA uptake (141, 142, 189, 509, 602, 644).
Chen and Dubnau proposed a model for DNA uptake in N. gonorrhoeae where the core structural components of the T4P (such as PilQ, PilC, PilP, PilG, PilF, PilD, PilE, and a minor pilin) are assembled into a T4P-like structure termed a competence pseudopilus (90, 91). A key functional difference between the T4P and the competence pseudopilus is the differential production of two minor pilins, PilV and ComP, which facilitate T4P function and competence, respectively. Consistent with this model, production of the ComP pilin enhances natural transformation, while production of PilV hinders DNA uptake (1, 645). Additionally, DNA uptake requires two other accessory proteins, ComE and ComA (90, 91). Once exogenous DNA is taken up through the outer membrane channel (formed by PilQ), the periplasmic protein ComE binds the incoming DNA and transports it to the inner membrane protein ComA (169) (Fig. 3). This aspect of the proposed model is supported by the finding that comA mutants of N. gonorrhoeae are able to take up DNA from the environment but cannot transport the molecule into the cytoplasm (168). Upon association with ComA, the double-stranded DNA (dsDNA) molecule is processed so that only a single-stranded DNA (ssDNA) molecule is released into the cytoplasm to serve as a substrate for homologous recombination.
A key step in natural transformation is the binding of DNA at the bacterial surface. In N. gonorrhoeae, DNA uptake is facilitated by a specific 10-nucleotide DNA sequence (5′-GCCGTCTGAA-3′) (153, 215). This DNA uptake sequence (DUS) or uptake signal sequence (USS) is found in the N. gonorrhoeae and Neisseria meningitidis genomes, approximately once every 1 kb (123, 239, 550, 551), suggesting that the sequence is conserved to increase the likelihood and specificity of DNA acquisition between Neisseria spp. While the species specificity and nucleotide sequence requirements of DNA binding and uptake are clear, the DNA receptor in Neisseria spp. has not yet been identified.
Another prototype for natural transformation is the Gram-negative pathogen H. influenzae (214). H. influenzae is efficiently transformable and competence is induced under certain physiological conditions (214, 257, 406, 531). Natural transformation in H. influenzae is also comparable to that in Neisseria spp. in that DNA uptake is species specific and the efficiency of uptake varies between strains (406, 531). Transformation in H. influenzae is mediated by a specific DUS that is 11 nucleotides in length (5′-AAGTGCGGTCA-3′), though the first 9 nucleotides of this DUS appear to be sufficient for uptake (119, 182, 542). Similar to the case for Neisseria spp., while the H. influenzae DNA uptake sequence is known, the bacterial receptor responsible for this process has not been identified.
Natural transformation in C. jejuni.
Unlike N. gonorrhoeae, whose competence does not appear to be regulated (58), C. jejuni is most naturally transformable during early logarithmic growth (629). Compared to the well-studied models of DNA uptake and transport systems of N. gonorrhoeae and H. influenzae, many of the details of natural transformation in C. jejuni remain unknown. However, some important components that have been identified include a T2SS.
T2SSs are macromolecular machines that consist of at least 12 proteins (516). T2SSs are found frequently in Gram-negative bacteria, including many proteobacteria (99). Commonly studied T2SSs include those of Vibrio and Klebsiella spp., which are depicted in Fig. 3 (99, 516). Many components of the T2SSs are structurally and functionally similar to those of the T4P. For instance, the pilus-like portion of the T2SS is comprised of proteins that are analogous to the major and minor pilins of the T4P. Moreover, during assembly, these proteins are also processed by a prepilin peptidase (41, 150, 495). Additional T2SS components that have parallel functions in T4P are the outer membrane protein (analogous to PilQ in the T4P system) and the ATPases that provide the energy necessary for assembly of the T2SS. Another similarity between the systems is the requirement for pilus polymerization and depolymerization for function. Although the precise mechanism of secretion is not clear, it is hypothesized that secreted proteins are forced out through the outer membrane channel by polymerization of the T2SS pilus-like structure (176, 405, 516, 539); the extended pilus would then have to retract or depolymerize to secrete another protein. This type of action is analogous to the polymerization and depolymerization of the T4P during twitching motility (404, 417). Given this model of protein secretion, it is possible that T2SS-mediated DNA uptake mechanisms could operate in a similar manner, albeit in reverse. In this case, depolymerization of the T2SS pilus would facilitate donor DNA transport into the bacterial cell.
Using a genetic screen to analyze a C. jejuni transposon mutant library, Wiesner et al. identified components of a T2SS that are essential for natural transformation (641) (Fig. 3). The genes identified in this screen, termed cts genes (for Campylobacter transformation system), include ctsD, a pilQ homologue; ctsE, encoding an inner membrane-bound DNA receptor that is homologous to ComEA of Bacillus subtilis and PilT of N. gonorrhoeae; and ctsF, which encodes an inner membrane protein similar to PilG of N. gonorrhoeae. In addition to these core components of the DNA uptake T2SS, a gene that encodes a pseudopilin-like protein was also identified and termed ctsG (641). Based on sequence similarity at the N terminus, CtsG may be cleaved by a prepilin peptidase similar to the pseudopilin in N. gonorrhoeae (90, 91). Another gene identified in this screen, ctsT, also encodes a protein that contains a predicted prepilin peptidase cleavage site. Taken together, these data suggest that C. jejuni may employ a DNA uptake mechanism that is similar to the one proposed for N. gonorrhoeae (90, 91). Other identified genes that are important for natural transformation in C. jejuni include ceuB, proC, ctsW, ctsR, ctsP, and dprA (589, 641). The role of CeuB in DNA uptake is not readily apparent; however, CeuB is involved in the transport of iron across the cytoplasmic membrane and may possibly transport other substrates (472). The role in natural transformation of ProC, an enzyme that is involved in proline biosynthesis, is unclear. CtsW has limited homology to ComFA of B. subtilis, which is required for transport of DNA across the cytoplasmic membrane (145). Since a C. jejuni ctsW mutant is able to take up DNA similarly to the wild-type strain, CtsW likely functions in natural transformation downstream of DNA uptake (641). ctsR and ctsP encode proteins with no known homologs, and the roles of these proteins in natural transformation remain to be determined (641). Finally, C. jejuni also encodes a homolog of the DNA processing A protein (DprA) that is functionally similar to that of H. pylori (589). C. jejuni DprA has been shown to be required for uptake of plasmid DNA but, interestingly, is expendable during uptake of chromosomal DNA (589). This function is in direct contrast to DprA of H. influenzae, which is implicated in natural transformation of chromosomal but not plasmid DNA (317). The fact that C. jejuni carries proteins with similarity to those of known DNA uptake systems, as well as less conserved proteins that are also involved in transformation, perhaps suggests that this bacterium has evolved alternative strategies other than the prototypical T4P/T2SS to facilitate DNA uptake from the environment.
One unique aspect of natural transformation in some strains of C. jejuni is the presence of a plasmid-based DNA uptake system that shows similarity to a type IV secretion system (T4SS) rather than the traditional T4P/T2SS (31). Partial sequencing of the pVir plasmid in C. jejuni strain 81-176 identified three genes with similarity to the comB8 to -10 (also referred to as virB) genes of H. pylori (discussed below), as well as one additional gene whose sequence is similar to that of a gene found in the H. pylori cag pathogenicity island (31). Mutation of the pVir comB10 allele results in an 80% decrease in the frequency of transformation (31), confirming the involvement of at least one of these genes in natural transformation. Interestingly, the comB10 mutant is also attenuated for both adherence and invasion (31), which perhaps suggests that the natural transformation process provides a fitness advantage during infection.
Finally, C. jejuni also encodes two components that are likely involved in DNA transport into the cytoplasm. Cj1211 is a predicted inner membrane protein with homology to ComEC of H. pylori (284). Based on this similarity, Cj1211 may be involved in the transport of DNA into the cytoplasm, much like ComA in N. gonorrhoeae (91); mutation of this gene abolishes natural transformation of C. jejuni (284). Cj0011c is a periplasmic protein that shares partial homology to the ComEA DNA binding protein of B. subtilis (285). This protein binds both ssDNA and dsDNA, but binding is not sequence specific. While the loss of Cj0011c does not eliminate natural transformation, cj0011c mutant strains are transformed up to 50-fold less frequently than wild-type strains (285).
Unlike the case for N. gonorrhoeae and H. influenzae, DNA uptake in C. jejuni is not dependent on a specific nucleotide sequence. Nevertheless, DNA from Campylobacter spp. is transformed more efficiently than DNA from other bacterial species (629). By obtaining DNA from other strains, C. jejuni may acquire a new gene or a variant of a preexisting gene that provides a fitness advantage. Currently, there is a lack of information regarding whether the mechanisms employed for plasmid DNA transformation differ from those used during transformation of nonplasmid DNA. However, it is possible that there are at least some shared components for both types of DNA uptake and transformation. Although there is still much to be learned about natural transformation in this species, it is clear that C. jejuni employs unique mechanisms of DNA uptake and transformation relative to the prototypical systems found in N. gonorrhoeae and H. influenzae.
Natural transformation in H. pylori.
The majority of H. pylori strains are naturally transformable (260, 261, 278, 367, 444). However, unlike the case for other bacteria, natural transformation in H. pylori is mediated by a T4SS (Fig. 4) (262, 316). Typically, T4SSs transport macromolecules (protein or DNA) from the bacterial cytoplasm into the extracellular environment or a host cell (82, 97, 459). This translocation process is facilitated by a pilus-like structure that spans both the inner and outer bacterial membranes. The secretion substrates are recruited and subsequently loaded into the T4SS. In the case of T4SS-mediated injection into host cells, translocation of the substrates occurs after the tip of the secretion system comes in contact with a target cell.
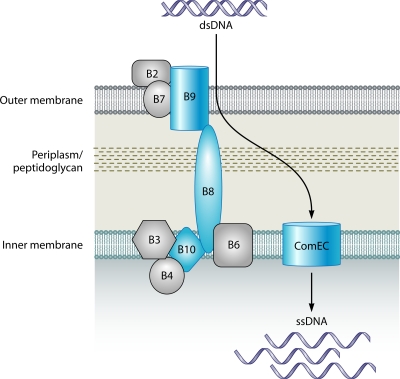
FIG. 4.
T4SS-related DNA uptake system of H. pylori and C. jejuni. The model of T4SS-mediated DNA uptake in H. pylori and C. jejuni is based on those proposed previously (261, 263, 548, 573). All components shown have been identified in H. pylori. The components also identified in C. jejuni are shown in blue (31). The transmembrane pore complex is composed of the Com proteins ComB6, ComB7, ComB8, ComB9, and ComB10. ComB3 is thought to be located in the inner membrane, but its function is unclear. Likewise, the precise function of ComB2 is not known; however, it is proposed that ComB2 forms the surface-exposed region of the T4SS. The ComB4 ATPase is thought to exist in a complex with ComB3 and may provide the energy necessary for T4SS biogenesis and/or substrate translocation. Once bound, through an unknown mechanism, donor DNA is taken up through the T4SS transmembrane complex into the periplasm. The periplasmic DNA is then processed and is likely translocated into the cytoplasm by the inner membrane-spanning channel ComEC. The processed single-stranded DNA can then be used as a substrate for homologous recombination.
The genes for the components of the H. pylori T4SS are located within the comB locus on the chromosome and are functionally separate from the Cag T4SS, which translocates the CagA protein into gastric epithelial cells (261, 262, 278, 459). comB consists of two separate operons, comB2 to -4 and comB6 to -10 (261, 316). The proteins of the ComB system are named after analogous proteins in the prototypical Vir T4SS of Agrobacterium tumefaciens (96). Genes in the first comB operon (comB2 to -4) encode proteins that are predicted to localize to the outer membrane (ComB2) or the inner membrane (ComB3) (Fig. 4) (316). The other gene in this operon, comB4, encodes an ATPase (261); however, whether ComB4 provides the energy for assembly of the ComB complex or directly facilitates DNA uptake remains to be elucidated. It has been hypothesized that ComB3 forms a multisubunit complex with the ComB4 ATPase at the inner membrane (95, 96, 316) (Fig. 4). However, the function of this protein complex is unknown. ComB2 has been proposed to form the surface-exposed structure of the T4SS, but the precise function of ComB2 remains unclear (316).
The second comB operon (comB6 to -10) contains genes that encode the inner membrane components of the T4SS. For instance, comB6 encodes a membrane-spanning protein essential for natural transformation (Fig. 4) (316). While not naturally transformable, an H. pylori comB6 mutant can be transformed by electroporation (316). Thus, ComB6 is a component of the DNA uptake apparatus and is not involved in later steps of natural transformation, such as homologous recombination. The amino acid sequence of ComB7 suggests that this protein is a lipoprotein that is localized to the periplasmic face of either the inner or outer membrane (261, 548). ComB7 and ComB9 both contain a conserved amino acid residue necessary to facilitate a disulfide interaction between the two proteins and provide stability for the T4SS. Furthermore, the amino acid sequence of ComB9 contains a transmembrane domain (261), suggesting inner membrane localization. Taken together, these data suggest that ComB7 and ComB9 interact to form a portion of the periplasm-spanning component of the T4SS. ComB8 and ComB10 are both inner membrane proteins and, along with ComB6, are thought to interact with the ComB7-ComB9 protein complex to complete the periplasmic and inner membrane-spanning portion of the DNA translocation channel (261). Furthermore, since overproduction of ComB9 and ComB10 (in the absence of the ComB7 and ComB8 proteins) results in specific degradation of these proteins, ComB7 and ComB8 likely play a role in stabilizing the other proteins in the ComB DNA translocation complex (261).
Another component of the H. pylori natural transformation system is a homolog of ComEC of B. subtilis, which is thought to be involved in the transport of DNA from the periplasm into the cytoplasm (662). Mutation of comEC in H. pylori results in a reduction of DNA binding and uptake, suggesting that the encoded protein is involved in both steps during natural transformation (662).
Recent studies on H. pylori DNA uptake have provided insight into the mechanism(s) involved in natural transformation (573) (Fig. 4). In H. pylori, ComB-mediated DNA uptake occurs preferentially at the poles of the cell, in a two-step process (573). dsDNA is taken up through the ComB T4SS, and subsequent processing and translocation into the cytoplasm are dependent on the ComEC membrane channel. It is currently unclear whether the dsDNA is processed by ComEC prior to translocation or the incoming DNA passes directly through this channel.
Additional H. pylori proteins serve as possible components of the natural transformation system. Despite being designated putative Com components, these proteins are not homologous to components of the T4SS or the T4P/T2SS and their genes are not located in close proximity to the comB2 to -4 and comB6 to -10 operons. For instance, HP1378 is a homolog of N. gonorrhoeae ComL, which is a peptidoglycan-linked lipoprotein in this species (192). Attempts to mutate the respective gene in H. pylori have been unsuccessful, which suggests that HP1378 may have an essential function, such as maintaining cell wall integrity (662).
ComH, a protein with no homology to other proteins, is also essential for natural transformation in H. pylori (546). comH is conserved across all tested H. pylori strains, suggesting the importance of ComH in H. pylori biology (546). Furthermore, ComH is required for natural transformation of both plasmid and genomic DNAs, which indicates that ComH functions during DNA uptake, transport, or processing rather than recombination (546). However, the mechanistic role of ComH in natural transformation remains to be determined.
Yet another constituent in the natural transformation process is DprA (19, 547). DprA in H. pylori is homologous to DprA of H. influenzae, which is essential for natural transformation of chromosomal DNA (317). In H. pylori, the function of DprA is unclear; however, the fact that an H. pylori dprA mutant does not completely lose the ability to be naturally transformed suggests that DprA may function to enhance the transformation process (19). Unlike DprA in H. influenzae, H. pylori DprA is involved in transformation of both plasmid and chromosomal DNAs, which perhaps suggests that this protein functions in DNA uptake or translocation rather than in integration (547).
Similar to that in C. jejuni, competence in H. pylori peaks during specific times during growth (42, 278). It has been demonstrated that induction of competence occurs in H. pylori in response to DNA damage (139). Upon DNA damage, H. pylori upregulates the expression of 41 genes, many of which are involved in DNA uptake and recombination. This upregulation is dependent on both RecA and the ComB system. Interestingly, one of the genes that is upregulated encodes a lysozyme homolog that may target and lyse neighboring cells to increase the amount of free DNA available for uptake. This free DNA would then be available for the remaining viable bacteria for DNA repair via homologous recombination. Furthermore, it is proposed that RecA may act as a sensor, recognizing the increased DNA uptake and further propagating the DNA damage response signal to create a positive-feedback loop (139). The precise mechanism by which RecA is able to sense these changes and propagate the response signal requires further study. Clearly, these findings highlight a unique DNA damage response that is different from the prototypical DNA damage responses employed by other bacteria. Similar to the case for C. jejuni, little is known about the differences in transformation of plasmid and nonplasmid DNAs. Futher studies are necessary to pinpoint the precise mechanistic differences in these two processes.
Homologous Recombination
The process of homologous recombination is essential to all organisms. Genetic exchange between two homologous DNA molecules helps to maintain chromosomal integrity and generate genetic diversity (345). In E. coli, there are at least 25 factors involved in homologous recombination. These factors include DNA helicases and topoisomerases, DNA binding proteins, DNases, ATPases, and nucleotide binding proteins (345). Furthermore, there are multiple recombination pathways that facilitate DNA recombination and repair, some of which share common components. The components and biochemical mechanisms of these various pathways have been reviewed previously (135, 345, 535, 549); thus, we focus our discussion on the major components involved in E. coli recombination as well as on how these components differ from those employed by C. jejuni and H. pylori.
The RecBCD recombination pathway of E. coli.
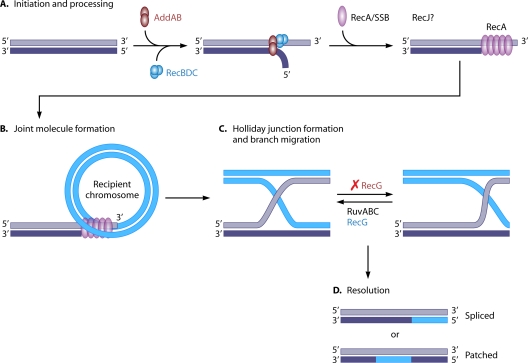
The RecBCD pathway is responsible for the majority of genetic recombination events that occur in E. coli and consists of four main steps (Fig. 5): (i) initiation, (ii) pairing and exchange of homologous DNA, (iii) heteroduplex (Holliday junction) formation and branch migration, and (iv) resolution of the DNA heteroduplex. Like many recombination pathways, the RecBCD pathway is dependent on the multifaceted RecA protein. In addition to RecA, RecBCD-mediated recombination is facilitated by the RecBCD enzyme complex (also known as exonuclease V [211]), the single-stranded DNA-binding protein (SSB), DNA polymerase I, DNA gyrase, DNA ligase, and the Holliday junction processing enzymes RuvA, RuvB, RuvC, and/or RecG (reviewed in references 135, 345, and 535).
FIG. 5.
dsDNA processing in homologous recombination. A simplified model of homologous recombination is shown. Component names in black are common to E. coli, H. pylori, and C. jejuni, those in red are found only in H. pylori and C. jejuni, and those in blue are found only in E. coli. (A) dsDNA is recognized and processed by either the RecBCD (in E. coli) or AddAB (in H. pylori and C. jejuni) complex. As the two strands become separated, the 5′ end is degraded preferentially, allowing RecA and SSB to bind to the exposed 3′ tail, forming the presynaptic complex. RecJ may also be involved in the conversion of dsDNA to a ssDNA substrate for RecA processing. (B) RecA then mediates the strand invasion reaction, which forms the joint molecule that scans the recipient DNA until a region of sufficient homology is found. (C) Holliday junction formation and branch migration are catalyzed by RuvAB. In E. coli, RecG also binds the Holliday junction structure and facilitates branch migration; however, H. pylori RecG (and possibly that of C. jejuni) binds the junction and inhibits branch migration and resolution. (D) The migrating DNA heteroduplex is recognized by the RuvC resolvase and is resolved to form either spliced or patched recombinant DNA molecules.
Initiation of homologous recombination begins when the donor dsDNA is processed by the dual helicase/nuclease function of the RecBCD enzyme complex. While RecC does not appear to have any conserved catalytic or enzymatic domains, RecB and RecD contain DNA helicase domains and RecB contains an additional nuclease domain (24, 135, 219). dsDNA processing by the RecBCD complex generates a 3′-ssDNA substrate that is bound by SSB and RecA (593).
RecA is widely conserved across bacterial species and mediates exchange of homologous DNA strands (504). Once bound to the ssDNA substrate, RecA polymerizes along the 3′ end of the molecule, forming a presynaptic complex (Fig. 5). This complex then scans the recipient chromosome for a region of homology. Once a region suitable for recombination is found, RecA initiates a DNA strand exchange reaction that forms a recombination intermediate or joint molecule (Fig. 5) (83, 335, 345, 346, 505, 638, 639). Formation of the joint molecule results in the development of a mobile DNA heteroduplex known as a Holliday junction (146, 264, 345). Once formed, the Holliday junction extends laterally along the recipient DNA molecule during branch migration, which is enhanced by the RuvAB protein complex (280, 435, 436, 477, 594, 605).
RuvA not only binds the Holliday junction but also assists in DNA strand separation and/or selective binding to the junction (496, 535). The lateral migration of the Holliday junction along the recipient DNA molecule is facilitated by the helicase activity of RuvB in complex with RuvA (Fig. 5) (280, 435, 436, 477). Finally, the migrating Holliday junction is resolved by the endonuclease activity of RuvC, which associates with the RuvAB complex (148, 281, 357, 434). Nucleolytic cleavage and subsequent resolution of the Holliday junction result in the formation of either patched or spliced DNA molecules (345, 535) (Fig. 5).
Additional recombination factors in E. coli.
In addition to the RecBCD pathway, E. coli produces several other factors involved in homologous recombination, including RecF, RecO, and RecR, which are part of the RecF pathway. This pathway is involved mainly in DNA damage and postreplication recombinational repair but also in the induction of the SOS response (61, 267, 412, 508, 628). Similar to the RecBCD pathway, RecFOR-mediated recombination is RecA dependent (392, 431). The RecFOR complex acts during the initiation stage of recombination by enhancing RecA loading onto SSB-containing ssDNA to accelerate DNA strand exchange independent of the RecBCD enzyme (345, 431). Two additional components in the E. coli RecF pathway are RecJ and RecN. RecJ contains 5′-ssDNA-specific exonuclease activity and is likely involved in processing dsDNA into a ssDNA substrate that is suitable for RecA activity (384). RecN contains a nucleoside triphosphate (NTP)-binding domain and is believed to stabilize 3′-ssDNA substrates used in DNA strand exchange (345, 375, 507). Finally, another component of the E. coli homologous recombination system is RecG, an ATP-dependent DNA helicase whose activity closely resembles that of RuvAB (376). Also like RuvAB, RecG specifically binds to Holliday junction structures and promotes branch migration (376, 640). Similar to that of the RuvAB proteins, the activity of RecG is not specific to one recombinational pathway and is involved in multiple kinds of DNA recombination and repair (345).
Homologous recombination in C. jejuni and H. pylori.
Compared to the well-studied recombination mechanisms of E. coli and H. pylori, there is currently a substantial gap in knowledge of homologous recombination in C. jejuni. Due to this void, the components and mechanisms of homologous recombination discussed here focus on what is known about the H. pylori system, with references to the known components of the C. jejuni recombination system provided when possible.
Despite differences in enzymatic components, the homologous recombination pathway in H. pylori consists of the same basic steps as that in E. coli. However, unlike the case in E. coli, presynaptic complex formation on dsDNA breaks in H. pylori is catalyzed primarily by the AddAB protein complex (Fig. 5) (15). Although the AddAB complex is structurally different from the RecBCD complex, both enzymes perform similar functions. Like the RecBCD complex in E. coli, AddAB exhibits ATP-dependent helicase and nuclease activities. Moreover, AddA is analogous to RecB, having both helicase and nuclease functions (15). However, unlike RecBCD, AddAB has dual-nuclease activity rather than dual-helicase activity. The second nuclease domain in AddAB is found within AddB (15). Both addA and addB mutants are deficient in DNA repair and homologous recombination and display a reduced capacity for colonization in a murine model of infection (15, 16). C. jejuni also appears to carry the AddAB enzyme complex (15). The high degree of homology of AddA and AddB of C. jejuni suggests that these proteins function in a similar manner to that of AddAB in H. pylori. However, the enzymatic activity of the putative C. jejuni AddAB complex has not been demonstrated.
Recent studies of H. pylori have implicated another enzyme complex in the processing of DNA during repair and RecA-mediated recombination initiation. A distant RecO ortholog was identified using bioinformatic analysis of the H. pylori genome (399). Although H. pylori RecO has relatively little homology to most other RecO orthologs, the overall structure of the protein suggests a conserved function. Mutational analysis in H. pylori indicates that RecO and RecR function together during intrachromosomal recombination (399). In addition, inactivation of either gene confers an increased sensitivity to DNA damage, suggesting that RecOR also participates in recombinational repair. Interestingly, disruption of the RecOR pathway does not appear to impair recombination after natural transformation, which suggests that RecOR-mediated recombination initiation may be subject to substrate specificity and may function primarily in types of DNA repair other than that of non-dsDNA breaks (399). This hypothesis is supported by subsequent findings indicating that the RecOR and AddAB recombination initiation pathways have little overlap in function and that disruption of the RecOR pathway has a greater impact than inactivation of AddAB on survival after UV irradiation (398). At this time, it is not clear whether C. jejuni also employs the RecOR pathway in a similar manner; however, a putative recO gene has been annotated in at least one C. jejuni genome (J. J. Gilbreath and D. S. Merrell, unpublished observation).
Once a DNA substrate has been processed by one of the presynaptic initiation enzyme complexes, RecA binds to the exposed 3′-ssDNA and forms the presynaptic nucleoprotein filament necessary to catalyze the homologous DNA strand exchange reaction. In H. pylori and C. jejuni, this process is likely quite similar to strand invasion in E. coli, although there are some differences in the H. pylori RecA protein itself. Overall, the H. pylori RecA sequence is very similar to those of other bacterial RecA proteins and shares 75% identity with C. jejuni RecA (524, 598). Like that of E. coli, RecA proteins in both H. pylori and C. jejuni are required for homologous recombination and DNA repair, and RecA activity may play a role in pH resistance in H. pylori (193, 234, 524, 598). H. pylori recA mutants are deficient in colonization in a murine model, which highlights the importance of H. pylori RecA in vivo (15). In contrast, C. jejuni recA mutants are able to colonize rabbits as well as the wild-type strain (234). One unique feature of H. pylori RecA is the requirement for posttranslational modification for full activity (181). Purified RecA from H. pylori displays altered electrophoretic mobility compared to the same protein purified from E. coli (524). Subsequent analysis of H. pylori RecA identified a putative N-linked glycosylation motif that is also conserved in C. jejuni RecA (181). Although the requirement for posttranslational modification has not been evaluated for C. jejuni, site-specific mutagenesis of this motif abrogates RecA modification in H. pylori, despite the fact that no N-linked protein glycosylation system has been found in H. pylori. The modification of H. pylori RecA is dependent on at least two other genes, galE and pmi (181). These findings are consistent with the hypothesis that RecA is modified by glycosylation, since galE and pmi both encode proteins involved in lipopolysaccharide (LPS) biosynthesis and perhaps in the generation of glycosylation substrates (181).
Another component involved in homologous recombination in H. pylori is RecN, which plays a role in both DNA repair and recombination: a recN mutant is more sensitive to DNA-damaging agents and oxidative stress and displays a 50-fold defect in homologous recombination (625). Given these sensitivities, it is perhaps not surprising that H. pylori recN mutants are unable to colonize as well as the wild-type strain in a mouse model of infection (625). While these results indicate that RecN plays an important role in H. pylori biology, the specifics of RecN function are not clear. One possibility is that RecN interacts with dsDNA breaks and recruits RecA to enhance the recombination process (625). This hypothesis is not unlike the proposed role of RecN in E. coli and is supported by the presence of a putative ATP/GTP binding site within a conserved SMC (structural maintenance of chromosome) domain (625). Genome sequence analysis of several C. jejuni strains indicated that RecN or a RecN homolog is present in the majority of strains (Gilbreath and Merrell, unpublished observations). Although the function of RecN in C. jejuni has not been determined, the overall similarity in recombination systems in C. jejuni and H. pylori may suggest that this protein behaves similarly to H. pylori RecN.
Similar to E. coli, both H. pylori and C. jejuni encode RecJ and RecR homologs (180; Gilbreath and Merrell, unpublished observations). While the genes for these proteins are annotated in some C. jejuni genomic sequences, the role that these putative homologs play in homologous recombination in H. pylori and C. jejuni will require future study. H. pylori also contains a RecG homolog, although the function of this enzyme is quite different from that in E. coli. As described above, E. coli RecG facilitates migration of Holliday junctions and can convert stalled replication forks into Holliday junctions, thus enhancing the recombination process (136, 345, 535, 541). In contrast, while still facilitating branch migration, H. pylori RecG actually inhibits recombination and competes with RuvB for binding to Holliday junction structures (Fig. 5) (308). Again, unlike the RecG pathway in E. coli, H. pylori Holliday junctions that are bound by RecG are not resolved. This phenomenon is likely due to the absence of the resolvase enzyme, RusA, that is usually associated with the RecG recombination pathway (308). Interestingly, H. pylori RecG is able to complement an E. coli recG mutant to the same extent as E. coli RecG (310). Thus, the species-specific antirepair function of the H. pylori RecG helicase may provide a means to maintain genetic diversity within a population of cells while still preserving critical genomic elements (308). A RecG homolog is also found in C. jejuni, but whether or not this protein also exhibits antirepair activity has not been determined. The fact that C. jejuni lacks the RecG pathway resolvase (RusA) highlights the possibility that the RecG helicase may have a similar function to that of H. pylori RecG.
The genomes of H. pylori and C. jejuni also encode another recombination-limiting factor, MutS2. As a whole, the MutS family of proteins is often involved in the recognition and repair of mismatched DNA base pairs (220, 442). However, in H. pylori, MutS2 is not involved in mismatch repair and actually inhibits intergenomic recombination (309, 487). This suppression of homologous recombination may provide H. pylori with another means to control genetic diversity or to aid in the recognition of illegitimate recombination events (307, 487). The function of MutS2 in C. jejuni recombination or mismatch repair remains to be determined.
The final steps of homologous recombination in H. pylori and C. jejuni are thought to be catalyzed much like they are in E. coli. ruvA, ruvB, and ruvC are found in both H. pylori and C. jejuni (308; Gilbreath and Merrell, unpublished observations). The RuvABC pathway has been shown to be the major recombination pathway in H. pylori (308). Although few detailed analyses of RuvABC function have been reported for either of these species, the necessity of RuvB for recombination and repair has been demonstrated for H. pylori (308). These data, along with the highly conserved nature of the RuvABC pathway in bacteria, suggest that this pathway is the primary way that H. pylori (and likely C. jejuni) promotes branch migration and subsequent resolution of Holliday junctions during homologous recombination (Fig. 5).
Summary
The mechanisms of natural transformation and homologous recombination employed by C. jejuni and H. pylori are quite distinct from those of commonly studied Gram-negative bacteria. Indeed, H. pylori and C. jejuni encode DNA uptake and processing systems that are mostly unique to these epsilonproteobacteria and that contain components more akin to those of Gram-positive species such as B. subtilis than to those of model Gram-negative species. In addition, the mechanism of DNA uptake and processing used by H. pylori also closely resembles that of B. subtilis (573). Consistent with this line of comparisons, the use of the AddAB enzyme complex rather than the RecBCD system in recombination substrate processing more closely resembles the mechanisms used by Gram-positive bacteria than the prototypical E. coli pathways. Taken together, these data further highlight the ways in which C. jejuni and H. pylori have evolved unique processes to thrive in their respective niches.
IRON HOMEOSTASIS AND IRON-RESPONSIVE GENE REGULATION
Iron is one of the most abundant elements on the planet and is a basic requirement for life. In bacteria, iron plays a critical role in processes such as metabolism, electron transport, oxidative stress responses, and regulation of gene expression (65, 164, 245, 291, 400, 408). Although iron is essential for most bacteria, it also catalyzes free radical formation through Fenton chemistry (Fe2+ + H2O2 → Fe3+ + OH + OH−), which results in cellular damage. As such, homeostasis between iron acquisition and storage is crucial for bacterial survival. The ability to achieve the proper balance of iron homeostasis is complicated by the environment in which bacteria live. For instance, in anaerobic or acidic environments, iron is found in its soluble ferrous form (Fe2+) and can be acquired readily by most microbes that thrive in these situations. However, iron acquisition by aerobic microorganisms is more challenging: in the presence of oxygen, iron is quickly oxidized to an insoluble, biologically unavailable ferric form (Fe3+). As such, bacteria have evolved many strategies to obtain and sequester ferric iron. Common strategies include solubilization of free iron by metal-chelating siderophores (115, 445, 499, 575) and confiscation of host iron-bound molecules such as heme, transferrin, hemoglobin, and lactoferrin (67, 108, 225, 329, 623).
Expression of many iron uptake and storage systems is iron dependent. A key component in the regulation of these systems is the ferric uptake regulator (Fur), which typically behaves as a transcriptional repressor by binding specific DNA sequences found within cognate promoters (34, 164, 242, 244, 245). Fur binding obstructs association of RNAP within these promoters, thus preventing transcription. The nature and number of iron acquisition systems that bacteria produce usually reflect the type of environment in which the bacteria live. The goal of this section is to compare and contrast iron acquisition, storage, and iron-responsive regulatory systems found within the prototypical bacterium E. coli to those found within C. jejuni and H. pylori.
Iron Uptake and Transport
Ferrous iron uptake and transport.
Under anaerobic conditions, iron is found in the more physiologically accessible ferrous form (Fe2+), which can be acquired by bacteria through the utilization of dedicated uptake systems (Table 1). For example, E. coli acquires ferrous iron through the FeoABC uptake system (222, 306). FeoB is a cytoplasmic membrane protein with an N-terminal GTP binding/hydrolysis domain that is required for ferrous iron transport (396). The specific role of FeoA in iron transport remains unclear, although both FeoA and FeoB are required for full function (222). FeoC is a putative transcriptional regulator and is also required for full FeoB function (222). This system has been shown to be important for growth in vivo, as E. coli feoB mutants are defective in host colonization (577).
TABLE 1.
Iron sources and relevant uptake systems in E. coli, C. jejuni, and H. pylori
| Organism and iron source | Uptake system | Reference(s) |
|---|---|---|
| E. coli | ||
| Ferri-enterochelin | Fep, Cir, Fiu | 386, 449 |
| Ferri-hydroxamatesa | FhuA | 172, 303, 343 |
| Rhodotorulic acid | FhuE | 519 |
| Heme/hemoglobin | ChuA, Hbp | 466, 603 |
| Ferrous iron | Feo | 243, 306 |
| Ferric citrate | Fec | 493 |
| C. jejuni | ||
| Ferri-enterochelin | CfrA, CfrB, Ceu | 37, 233, 467, 502, 655 |
| Ferrichrome | CfuA | 195 |
| Rhodotorulic acid | P19, Cj1588-Cj1663 | 283, 574 |
| Heme | Chu | 503 |
| Transferrin | Cj0173c-Cj0178 (Cfbp/Ctu) | 419, 611 |
| Lactoferrin | Cj0173c-Cj0178 (Cfbp/Ctu) | 419, 611 |
| Ovotransferrin | Cj0173c-Cj0178 (Cfbp/Ctu) | 419, 611 |
| Ferrous iron | Feo | 439 |
| H. pylori | ||
| Ferrous iron | Feo | 599, 614 |
| Hemoglobin | FrpB2 | 212, 533 |
| Transferrin | Unknown | 533 |
| Lactoferrin | Unknown | 274, 533 |
| Heme | Unknown | 134, 649 |
| Ferric citrate | Fecb | 117, 599, 614 |
Hydroxamates include both aerobactin and ferrichrome.
The function of the Fec system in H. pylori has not been fully verified.
In H. pylori, FeoB is the major ferrous iron transport protein and is required for colonization in a mouse model of infection (614). Unlike the ferrous iron transport system in E. coli, the genome of H. pylori does not contain feoA or feoC homologs (599). As seen with E. coli FeoB, the N-terminal region of H. pylori FeoB contains two regions that show similarity to nucleotide-binding domains, thus suggesting possible ATPase activity (614). However, the mechanism of ferrous iron transport through FeoB in H. pylori remains unclear.
Similar to H. pylori, C. jejuni strains appear to encode FeoB but not FeoC homologs (498). However, many feoB alleles contain significant mutations that are predicted to eliminate FeoB function (185, 263). Furthermore, targeted mutation of feoB does not hinder ferrous iron uptake, which suggests that C. jejuni contains unidentified genes for an alternative ferrous iron uptake system (498).
Siderophores.
Siderophores are small organic molecules that specifically bind, solubilize, and aid in the delivery of ferric iron into bacteria. Under oxygen-rich conditions, ferrous iron (Fe2+) is oxidized to the highly insoluble ferric (Fe3+) form, which makes iron acquisition in bacteria more challenging. One mechanism for acquiring ferric iron is through the secretion of siderophores that bind the metal; the ferri-siderophores are subsequently taken up by an energy-dependent, receptor-mediated mechanism. Examples of siderophores include enterochelin and aerobactin of E. coli, with enterochelin perhaps being the best characterized of the siderophores (132, 199, 359, 622). Siderophore biosynthesis and processing have been the subjects of many investigations and comprehensive overviews (445, 499, 500, 622).
In addition to encoding their own iron acquisition systems, some bacteria also possess the ability to scavenge siderophores produced by other microbes (described below). Whether synthesized endogenously or scavenged from other bacterial species, ferri-siderophore complexes are too large to diffuse freely across the bacterial outer membrane. Instead, they require specific outer membrane protein receptors to mediate uptake and internalization. Not including the ferric citrate outer membrane transport protein (discussed below), E. coli produces at least five of these iron transport proteins (Table 1): Cir, Fiu, and FepA, for ferri-enterochelin transport (386, 449); FhuA, for transport of ferri-hydroxamates (172, 303, 343); and FhuE, for ferri-rhodotorulic acid transport (519). Once the iron-bound ligand has been translocated into the periplasm, it is escorted to its cognate cytoplasmic membrane transporter protein by a periplasmic binding protein (PBP). In E. coli, the PBPs FepB, FecB, and FhuD are responsible for trafficking of the ferri-siderophore ligands from FepA, FecA, and FhuA/FhuE, respectively (391, 493, 565, 566). Once bound to its cognate ligand, the PBP shuttles the ferri-siderophore to its respective ABC transporter, which hydrolyzes ATP to undergo a conformational change for transport of the ferri-siderophore into the cytoplasm.
Unlike E. coli, C. jejuni does not appear to produce endogenous siderophores. An early study by Field et al. reported siderophore production in C. jejuni (175), but subsequent genomic sequence analyses revealed that most C. jejuni strains do not possess identifiable siderophore biosynthesis genes (185, 263, 473). Similar to E. coli, C. jejuni possesses dedicated uptake systems for a variety of exogenously produced ferri-siderophores, including those for ferri-enterochelin (37, 233, 502), ferrichrome (37, 195), and ferri-rhodotorulic acid (574) (Table 1). Like the Fep system of E. coli, the ferri-enterochelin uptake system in C. jejuni consists of an outer membrane receptor protein (CfrA), a periplasmic binding protein (CeuE), and the CeuBCD transporter complex (502). The membrane-spanning domains are composed of CeuB and CeuC, while CeuD serves as the ATPase. The overall importance of this system in C. jejuni is suggested by the fact that the outer membrane receptor protein, CfrA, is conserved among a large subset of C. jejuni isolates and is essential for ferri-enterochelin uptake as well as for in vivo fitness in a chick model of colonization (467). Structural data suggest that the lack of conservation of charged and aromatic residues seen in CfrA could affect substrate binding affinity, thus providing C. jejuni with a broader range of siderophore binding than that provided by FepA of E. coli (81). Given the biological implications of utilizing multiple exogenous iron sources and the inability of C. jejuni to synthesize siderophores endogenously, the promiscuity of CfrA for multiple siderophores is an attractive hypothesis. A second ferri-enterochelin receptor was recently identified (655). This receptor, designated CfrB, shares ∼34% similarity with CfrA, and expression of cfrB is regulated by iron. CfrB is highly conserved among C. coli primary isolates as well as several isolates of C. jejuni. Mutation of cfrB greatly reduces colonization in a chick model of infection, indicating that this iron uptake receptor is important in vivo (655).
Other components of the ferri-enterochelin iron uptake system appear to be more conserved. For instance, the subunit organization of the C. jejuni ferri-enterochelin uptake (Ceu) system is analogous to that in E. coli (Fep). The Ceu permease consists of two distinct polypeptides, CeuB and CeuC, and a single protein, CeuD, which hydrolyzes ATP (502). CeuE is proposed to be the PBP of this ferri-enterochelin uptake system. Unlike mutation of cfrA, mutation of ceuE does not abrogate growth when bacteria are supplemented with ferri-enterochelin, suggesting that other ABC transporters in C. jejuni may share some degree of functional redundancy in iron uptake (502).
Another undercharacterized iron uptake system in C. jejuni is encoded by the cj1658 to cj1663 genes (420). This locus encodes the periplasmic P19 protein (Cj1659), the putative membrane protein Cj1658 (also called Cftr1), and the putative ABC transporter proteins Cj1661 to -1663 (86, 283, 420, 473). Expression of p19 is iron regulated, and the protein binds both ferric iron and copper (86); however, the role of P19 in copper uptake/homeostasis is not fully understood. Mutational analysis of these genes further suggests that this locus encodes a functional iron uptake system, as neither p19 nor cj1658/cftr1 mutants are able to utilize ferri-rhodotorulic acid and the loss of P19 hinders growth under iron-limited conditions (86, 574). Furthermore, p19 and cj1658 homologues are involved in iron uptake in Yersinia pestis (77). Taken together, these data support the hypothesis that the p19 and cj1658 genes encode proteins involved in iron uptake. In addition, C. jejuni may encode proteins homologous to the Fhu ferrichrome uptake system, but functional demonstration of this uptake system requires further experimentation (195).
The diverse iron uptake systems found in Helicobacter spp. reflect the respective site of colonization for each species. Nongastric Helicobacter spp., such as Helicobacter cinaedi, Helicobacter fennelliae, Helicobacter bilis, and Helicobacter hepaticus, inhabit nutritionally competitive niches. Thus, similar to E. coli and C. jejuni, this subset of Helicobacter spp. produce siderophores and can utilize iron from human and bovine transferrin and lactoferrin, as well as heme, hemoglobin, and ferric citrate (134). In contrast, since they are the only characterized bacterial residents within the stomach, the gastric Helicobacter spp., i.e., H. pylori, Helicobacter felis, Helicobacter acinonyx, and Helicobacter mustelae, face little competition for nutrients from other bacterial species and are not forced to compete with other microbes for ferri-siderophores. Instead, gastric Helicobacter spp. have evolved the ability to acquire iron from their only major competitor, the mammalian host (Table 1).
Iron acquisition from host sources such as heme, hemoglobin, lactoferrin, and transferrin.
In addition to competing with other microbial species for iron acquisition, commensal and pathogenic bacteria are in fierce competition with their hosts. In mammalian hosts, extracellular iron is stored temporarily by high-affinity chelating molecules such as heme, transferrin, and lactoferrin. Other than iron bound to hemoglobin (within erythrocytes), the primary mammalian iron carrier protein is transferrin (for a review on mammalian iron homeostasis, see reference 20). At mucosal surfaces, however, lactoferrin is the major iron-chelating molecule. During infection, these iron-containing molecules represent a readily available iron source for bacteria.
To facilitate growth in a host, some pathogenic E. coli strains benefit from the ability to acquire iron from one of the most common iron sources within the host, hemoglobin. Iron acquisition from hemoglobin is mediated by the secretion from E. coli of a hemoglobin protease, Hbp (466). Hbp degrades hemoglobin, binds the iron-containing heme molecule, and is important for full virulence. However, in contrast to C. jejuni and H. pylori, pathogenic strains of E. coli do not utilize iron from lactoferrin or transferrin.
Iron acquisition is important for C. jejuni in the context of both human and avian hosts. Differences in iron sources within these hosts mean that C. jejuni must be able to utilize a variety of host iron-containing molecules. Within the human host, iron is available in the form of heme/hemoglobin (described above), as well as transferrin and lactoferrin. Within the avian host, iron is found in serum and egg white, in an ovotransferrin-bound form. Of the possible iron sources available, C. jejuni can utilize the following: hemin and hemin-hemopexin complexes, hemoglobin and hemoglobin-haptoglobin complexes, and transferrin proteins (419, 486). Two heme uptake proteins (encoded by cj0177 and cj0178) have been shown to bind heme in vitro (87), but the roles of these proteins in heme uptake have not been determined conclusively. A hemin uptake system is produced by many C. jejuni strains (420). In this system, ChuA is the outer membrane protein receptor, ChuD is a putative PBP, and ChuB and ChuC are predicted to be the membrane-spanning protein and the ATPase component of the ABC transporter, respectively (473). Furthermore, some strains contain chuZ, which encodes a heme oxygenase (503). The chu system is present in all currently sequenced C. jejuni strains, and chuZ appears to be highly conserved among clinical isolates (420, 503). As with other C. jejuni iron uptake systems, the ChuA outer membrane receptor is required for heme utilization, but the PBP and ABC transporter components are not essential (503). This finding further suggests that a functional redundancy in iron transport exists in C. jejuni, similar to earlier reports on E. coli O157:H7 (603).
Recent studies have shown that C. jejuni utilizes human transferrin, lactoferrin, and avian ovotransferrin in a receptor-specific, contact-dependent manner (419). The genes encoding this system are grouped into two transcriptional units. The ABC transporter, ATPase, and putative PBP are encoded by cfbpC, cfbpB, and cfbpA, respectively. These genes are grouped in an operon upstream of ctuA, which encodes the outer membrane receptor (265). Previous studies have shown that CtuA is required for colonization in the chick model of infection (467). CtuA appears to be a multispecific receptor that is able to transport ferri-transferrin, ferri-lactoferrin, and ferri-ovotransferrin, which is a unique function among iron uptake systems (419). Mutation of ctuA results in decreases in growth and the ability to utilize human ferri-lactoferrin; however, uptake of this molecule is not abrogated completely (419). Thus, C. jejuni may possess alternative ferri-lactoferrin uptake systems.
During colonization of the gastric mucosa, Helicobacter spp. have the opportunity to scavenge host iron-containing molecules. Among the gastric Helicobacter spp., H. pylori appears unique in its ability to utilize host lactoferrin (274). In addition, a recent study using defined minimal media showed that H. pylori is also able to utilize both human and bovine forms of lactoferrin and transferrin (533). Interestingly, H. pylori preferentially binds the apo forms of lactoferrin and transferrin, which may suggest that preferential binding to the apo forms limits H. pylori pathogenicity to allow for a persistent infection without excessive damage to the host (533).
The high turnover rate of epithelial cells from mucosal surfaces results in bioavailability of iron in the form of heme. As such, H. pylori has evolved mechanisms to use iron bound to heme while inhabiting the gastric mucosa. Worst et al. previously described several iron-repressible outer membrane proteins that specifically bind to heme, although these proteins were not subsequently identified or characterized further (649). However, the importance of heme utilization is further substantiated by the ability of most Helicobacter spp. to utilize heme for growth and by the identification of a heme oxygenase homolog, HugZ, in H. pylori (134, 236).
Given that common hallmarks of chronic H. pylori infection include gastritis and peptic ulcers and that ulceration may lead to bleeding, hemoglobin may provide the bacterium with yet another iron source. Indeed, many Helicobacter spp. are capable of utilizing hemoglobin. For H. pylori, the hemoglobin receptor was identified as FrpB2 and specific hemoglobin utilization was clearly confirmed using a chemically defined growth medium (212, 533). Analysis of the predicted FrpB2 structure revealed an overall similarity to the hemoglobin-binding protein, ChuA, of E. coli O157:H7 strains (212). Whether they scavenge iron from ferri-siderophores produced by other bacterial species or from iron-bound host proteins, the iron uptake systems of C. jejuni and H. pylori suggest that these pathogens have evolved iron acquisition strategies that are well suited to life in their respective niches.
Iron uptake from host sources via ferric citrate (Fec) uptake systems.
The ferric citrate uptake system is another well-studied bacterial iron uptake mechanism. In E. coli, this system includes five transport proteins (FecABCDE), the FecR signal transducer, and the FecI σ factor (69, 70, 324, 566). Under conditions of low iron, expression of this uptake system is induced by ferric citrate, which is bound by the outer membrane receptor FecA and subsequently internalized into the periplasm (493, 617). FecA not only mediates the uptake of ferric citrate but also transmits a signal through the cytoplasmic membrane protein FecR, which subsequently activates the FecI σ factor (69, 157, 158, 247, 324). FecI then associates with RNAP and activates transcription of the fecABCDE operon (23, 395). FecB serves as the PBP that shuttles ferric citrate to the cytoplasmic ABC transporter, which is composed of FecCDE (69, 566).
A bona fide ferric citrate uptake system has not been identified in C. jejuni; however, H. pylori does encode such a system (599, 614). The H. pylori genome carries three putative fecA homologues (fecA1, fecA2, and fecA3) but does not appear to carry fecI and fecR. Transcriptional analysis of the H. pylori fecA genes indicates that expression of these genes is partially growth phase dependent and is mediated by the metal-binding regulatory protein Fur or NikR (117). The fecA1 gene is expressed throughout the growth cycle, whereas fecA2 and fecA3 are expressed preferentially during the late and early exponential phases, respectively (117). Furthermore, expression of both fecA1 and fecA2 is regulated by iron and Fur, whereas fecA3 expression is Fur independent and not iron responsive. Instead, expression from the fecA3 promoter is nickel responsive and is regulated by the nickel-responsive regulatory protein NikR (117). Given the regulatory interplay among Fur and NikR regulons, it is possible that Fur-dependent derepression is an indirect effect resulting from changes in NikR expression. Although it is clear that expression of these genes is controlled by metal, the specific function of the fecA genes in H. pylori remains to be elucidated.
Iron Storage
Depending on the environmental conditions, bacteria may be exposed to toxic amounts of heavy metals such as zinc, cobalt, copper, nickel, or iron. In bacteria, heavy metals are exported through transporters of the cation diffusion facilitator (CDF) and resistance-nodulation-cell division (RND) families (448). Given the abundance and requisite nature of iron, one could imagine that iron efflux systems would be advantageous in bacterial life. Nevertheless, evidence for iron export systems in bacteria remains scarce. One such system is the FieF (previously named YiiP) iron/zinc efflux system found in E. coli (223). Similar iron export systems have not been described for either C. jejuni or H. pylori.
In the absence of dedicated export systems, bacteria maintain intracellular iron stores by using molecules known as ferritins and bacterioferritins (22). E. coli possesses both ferritins (FtnA and FtnB) and a bacterioferritin (Bfr) (21). Both types of proteins form spherical structures composed of 24 subunits and a hollow iron-containing core region. FtnA is the major iron storage protein of E. coli, responsible for storage of more than half of the total cellular iron (2). Bacterioferritins contain 12 heme molecules per subunit (22), a feature not associated with ferritins. Bacterioferritins typically store less iron than ferritins and, like the closely related Dps proteins, may play a role in oxidative stress responses (2).
A single ferritin-like iron storage protein has been described for C. jejuni (619). The Campylobacter ferritin, Cft, displays a high degree of similarity to the H. pylori ferritin, Pfr, and a lesser degree of similarity to the ferritin found in E. coli. Similar to other bacterial ferritins, Cft binds iron and does not contain heme, and expression of cft is regulated by iron (467). Mutation of cft in C. jejuni reduces growth under iron-limiting conditions and increases susceptibility to oxidative damage, suggesting that Cft plays a role in iron storage and the oxidative stress response (618). In addition to Cft, C. jejuni also produces a single putative bacterioferritin (Dps) that is homologous to the neutrophil-activating protein (NapA) of H. pylori (611). Dps binds ∼40 iron atoms per monomer and protects against hydrogen peroxide stress (277). Despite the presence of a Fur binding site upstream of dps, studies have shown that it is expressed constitutively under both excess iron and iron-depleted conditions (277). Thus, while the major role of Dps in Campylobacter spp. is likely to provide protection from oxidative stress, it may also function in iron storage. Recent studies have also implicated C. jejuni Dps in modulation of the host immune system in GBS, although this effect may be secondary to the primary role(s) of Dps in C. jejuni physiology (484, 485). A third putative iron-binding protein, Cj0241c, is well conserved in currently sequenced C. jejuni strains (611; Gilbreath and Merrell, unpublished observations). Bioinformatic evidence suggests that this protein is a member of the bacteriohemerythrin family and contains conserved oxygen and metal binding sites, as well as an oligomerization interface that suggests a multimeric higher-order structure capable of binding iron and possibly oxygen.
In H. pylori, Pfr serves as the major iron storage molecule. Like the ferritins of other bacteria, Pfr is a nonheme iron storage protein whose production is regulated in response to iron (54). The importance of Pfr is illustrated by the fact that this iron storage protein is necessary for survival during iron starvation and is required for efficient colonization in a gerbil model of infection (620). pfr expression is regulated in a manner unique to H. pylori (described further below), in which transcription is repressed by apo-Fur (54). Like C. jejuni, H. pylori also contains a bacterioferritin-like protein of the Dps family, NapA, which binds iron and DNA (but only in the iron-bound form) (601, 624). NapA also rapidly sequesters free iron, which suggests a role for NapA in protection of DNA from iron-catalyzed free radical damage rather than simple iron storage (624). Unlike Dps proteins of E. coli and C. jejuni, NapA is a major immunogen in vivo and activates the formation of reactive oxygen species in human mononuclear leukocytes and neutrophils (166, 518, 624). Similar to C. jejuni Dps, the importance of NapA in H. pylori is highlighted by the fact that napA mutants are deficient in colonization in a murine model of infection (624). The fact that all C. jejuni and H. pylori strains encode iron storage and additional iron-scavenging proteins highlights the importance of maintaining proper intracellular iron levels in these bacteria.
Fur Regulation and Fur-Regulated Genes
Maintaining the balance between sufficient iron and levels that lead to iron toxicity is crucial, and therefore this balance must be tightly regulated. In bacteria, the primary regulatory checkpoint in iron homeostasis is iron uptake, and expression of genes encoding factors involved in this process is regulated at the transcriptional level. The major transcription factor responsible for this regulation is Fur, which functions through the utilization of ferrous iron as a cofactor (34, 245). Identified in E. coli over 25 years ago (245), Fur is a histidine-rich, ∼17-kDa homodimeric metalloprotein that contains two distinct functional domains: an N-terminal domain responsible for DNA binding and a C-terminal domain that mediates oligomerization of the protein (578). Fur functions by iron-dependent binding to promoter regions of iron-regulated genes at a conserved binding sequence known as the Fur box (132, 163, 164). These DNA sequences typically overlap the −10 and −35 regions of Fur-regulated promoters so that Fur binding prevents the association of RNAP.
Initial reports indicated that the consensus Fur binding sequence was a 19-bp palindromic DNA sequence, GATAATGATAATCATTATC (132). Subsequent analysis of Fur-DNA interactions revealed that the functional E. coli Fur binding sequence actually consists of three or more hexameric repeats of a GATAAT sequence (163). The specific model of Fur-DNA interaction has since been refined further by Baichoo and Helmann, who suggested that Fur dimers interact with a specific 15-bp inverted heptamer repeat (7-1-7) (36). The 7-1-7 inverted repeat model of Fur-DNA interaction essentially includes the basic components of both the 19-bp and GATAAT models.
Regulation of fur expression in E. coli differs significantly from that of C. jejuni and H. pylori (Fig. 6). Analysis of the E. coli fur promoter reveals two closely spaced transcriptional start sites (131). Within the start sites is a Fur binding site that overlaps the core promoter elements. Directly upstream of the −35 region is a binding site for the catabolite-activating protein (Cap) and an OxyR binding site (131, 667). The positioning of these regulatory sites indicates that expression of fur is autoregulatory and is responsive to both cyclic AMP (cAMP) and oxidative stress. Depending on environmental conditions, E. coli fur can also be expressed as part of a polycistronic mRNA from the SoxRS-regulated fldA promoter, which is upstream of fur (667).
FIG. 6.
Genetic organization and simplified regulation of the E. coli fur locus. (A) The primary fur promoter lies directly upstream of the fur coding region. Transcription typically initiates from one of two proximally located transcriptional start sites (TSSs), as indicated by Pfur1, and is regulated by Fur, Cap, and/or OxyR (131, 667). A secondary SoxR-regulated promoter upstream of the fldA coding sequence may also drive expression of fur under oxidative stress conditions (667). (B) (Top) Under conditions of oxidative stress, fur expression is activated from the proximal promoter TSSs (Pfur1) and/or from the fldA promoter (Pfur2) by SoxR (667), resulting in the production of either a monocistronic mRNA (mRNA1) or a polycistronic mRNA that also contains fldA (mRNA2). (Middle) When excess iron is present, fur expression is repressed in an autoregulatory manner. (Bottom) fur expression from the proximal promoter is also activated in response to increases in cAMP, which results in the production of a monocistronic mRNA.
Whether Fur is acting as a repressor or an activator, the regulatory role of Fur is global. Perhaps not surprisingly, the majority of Fur-regulated promoters are associated with genes involved in iron homeostasis or genes encoding iron-containing proteins. For example, E. coli genes that are regulated by classical Fe2+-Fur repression include those involved in enterochelin biosynthesis (entABCDEFG) (72, 408); ferri-enterochelin uptake (fepA) and transport (fepBCDEG) (72, 245); aerobactin biosynthesis and uptake (iucABCD-iutA) (132); ferrichrome uptake (fhuABCD) (245); ferric dicitrate uptake (fecA), transport (fecBCDE) (158), and regulation (fecI and fecR) (70, 158, 245); ferri-rhodotorulic acid uptake (fhuE) (408); energy coupling mechanisms (tonB, exbB, and exbD) (408); and ferrous iron uptake (feoABC) (306). Experiments using comparative genomics and the Fur titration assay (FURTA) also demonstrated Fur regulation of additional iron-related genes (469, 576), which adds to the line of evidence suggesting that Fur is a global regulator.
Prior to the discovery of the regulatory RNA RyhB (discussed below), available data suggested that Fur behaved as more than just a repressor. In E. coli, several genes appeared to be activated by Fe2+-Fur. These genes included ftnA (ferritin), bfr (bacterioferritin), sdhCDAB (succinate dehydrogenase), acnA (aconitase), fumA (fumarase), and sodB (superoxide dismutase). However, in 2002, Massé and Gottesman described RyhB, a small RNA (sRNA) that is negatively regulated by Fe2+-Fur (402). RyhB (also identified as sraI by Argaman et al. [25]) is a regulatory RNA transcribed from the intergenic region between yhhX and yhhY. The regulatory effect of this sRNA requires a direct interaction with the RNA chaperone protein Hfq and results in degradation of target mRNAs (402). Because expression of this sRNA is repressed by Fe2+-Fur, the downstream derepression of RyhB target genes under conditions of high iron appeared to be due to Fur-mediated activation. A subsequent microarray analysis of RyhB-dependent regulation in E. coli identified at least 56 genes whose expression is affected by RyhB. These data suggest an integral role for sRNA in regulation of iron homeostasis (401). Interestingly, the list of genes regulated by RyhB is nearly identical to those once thought to be activated by Fe2+-Fur. Thus, it was assumed that these genes, which appeared to be activated by Fe2+-Fur, were all negatively regulated by RyhB. However, subsequent analysis by Nandal et al. indicated that ftnA (encoding a ferritin previously identified to be under the control of RyhB) is actually directly activated by Fe2+-Fur through competition with the histone-like nucleoid protein H-NS (441). Furthermore, a study by McHugh et al. (after the discovery of RyhB) reported Fe2+-Fur activation of 42 genes in E. coli (408). The vast majority of these genes were identified by Massé as RyhB regulated. Taken together, these data indicate the need for further experiments to show whether the effect of RyhB on gene expression is direct or indirect and to determine possible alternative regulatory mechanisms involved in iron metabolism.
C. jejuni encodes two Fur homologs (Fur and PerR) which are involved in iron-responsive transcriptional regulation (613). C. jejuni Fur is able to partially complement an E. coli fur mutant but is not recognized by E. coli Fur antisera (647). These data suggest that C. jejuni Fur differs significantly from E. coli Fur, despite the fact that the Fur proteins share ∼40% identity (611). PerR, the peroxide stress regulator, constitutes the other Fur homologue and has been shown to regulate expression of katA (encoding catalase) and ahpC (encoding alkyl hydroperoxidase), which are involved in the C. jejuni oxidative stress response (613). Collectively, these data suggest that unlike Fur, PerR is not a global regulator in C. jejuni and regulates expression of only a defined collection of genes.
As discussed above, Fur represses gene expression by binding to specific target sequences known as Fur boxes. Promoter sequence analysis of Fur-regulated genes indicates that the C. jejuni consensus Fur box sequence does not correspond to that found in E. coli (611). Instead, a modified E. coli Fur box consisting of six consecutive repeats of a 5′-NAT-3′ sequence was proposed and then used by van Vliet et al. to identify putative Fur boxes in C. jejuni (611). The C. jejuni Fur box was further expanded to a 19-bp putative consensus (5′-GATAATGATAATCATTATC-3′) (467). However, the validity of this proposed consensus requires further experimental confirmation. Since C. jejuni encodes two Fur family homologues, Fur and PerR, there is likely a high degree of similarity in the Fur and PerR binding sequences (611). Thus, this similarity could potentially confound the elucidation of Fur and PerR boxes.
In contrast to E. coli fur, C. jejuni fur is not expressed from its own promoter (612); instead, fur is likely transcribed as part of an operon with upstream and downstream genes (Fig. 7). Directly upstream of fur are two genes, cj0399 and gatC, which are expressed from iron-independent promoters that can drive fur expression (612). The absence of a promoter directly upstream of fur and the lack of iron-responsive regulation of fur are features unique to C. jejuni. These observations suggest that changes in fur expression may be dependent on environmental stimuli other than iron availability, although the nature of these stimuli has not been demonstrated.
FIG. 7.
Genetic organization of the C. jejuni fur locus. In the absence of its own promoter, fur expression in C. jejuni is driven by one of two distal promoters. Transcription initiated from the P1 promoter results in the production of a polycistronic mRNA containing both cj0399 and fur coding regions (mRNA1). Alternatively, expression from the P2 promoter produces an mRNA carrying gatC, cj0399, and fur (mRNA2). Neither of these promoters is iron responsive or Fur regulated, indicating that fur expression is not autoregulatory but instead responds to unknown environmental signals (612).
Global transcriptional analysis of C. jejuni in response to iron identified 208 genes whose expression is significantly altered after the addition of iron under steady-state conditions (467). Additionally, approximately twice as many genes display transient differential expression shortly after iron supplementation, which indicates a significant global response to increased iron availability in C. jejuni (467). Genes repressed after the addition of iron include those for iron uptake or storage systems, components of the oxidative stress response, and uncharacterized functions (265, 467). Moreover, several genes involved in energy metabolism are significantly upregulated in response to iron, and iron may also play at least some role in regulating members of the N-linked protein glycosylation pathway as well as glycoproteins (467).
Fur regulation in H. pylori is a complex story that has not yet been unraveled fully. While the sequence of H. pylori Fur is relatively similar to that of E. coli Fur, it is functionally quite different (56, 79). Similar to Fur proteins in E. coli and C. jejuni, H. pylori Fur can act as a classic transcriptional repressor, utilizing iron as a cofactor (55). However, unlike Fur in other organisms, H. pylori Fur has also been shown to act as an apo-repressor as well as an activator (in both the iron-bound and apo forms) (6, 118, 128, 130, 161, 197). Iron-bound Fur repression in H. pylori is similar to that seen in many other organisms: Fur is thought to bind to Fur boxes as a dimer and to repress transcription in an iron-dependent manner.
Similar to the case in E. coli and C. jejuni, H. pylori Fe2+-Fur also targets promoter regions that contain specific sequences (Fur boxes). Though the Fur binding sequence in H. pylori has not been characterized fully, a consensus sequence based on known Fur-regulated promoters (5′-NNNNNAATAATNNTNANN-3′) was proposed (416). Currently, a more complete consensus has yet to be determined. Furthermore, detailed analysis of apo-Fur target sequences has been performed with only two promoters, those of pfr and sodB (encoding superoxide dismutase) (54, 161). In both instances, apo-Fur binds specifically to promoters under metal-free conditions. In the case of sodB, a single nucleotide substitution in the Fur box results in altered apo-Fur regulation (78). Promoter sequence comparison of these two genes failed to highlight a putative binding sequence, and homology to the proposed Fe2+-Fur consensus is not evident (79, 161). This difference in sequence is perhaps not surprising given the functional difference of Fe2+-Fur- and apo-Fur-targeted promoters; furthermore, the presence or absence of metal cofactors may result in conformational changes in protein structure that affect DNA binding specificity (111, 482).
One highly complex example of iron-responsive regulation found among genes in the H. pylori Fur regulon is that of the fur promoter itself (Fig. 8). As seen in E. coli, expression of the fur promoter in H. pylori is autoregulated (131, 128). This iron-responsive autoregulation provides bacteria with a sensitive mechanism to control iron levels within the cell (128). In contrast to expression of fur in C. jejuni, where transcription is initiated from promoters distal to the fur gene itself (612), H. pylori fur is transcribed from its own promoter, which has three distinct Fur boxes (128). The Fur box with the highest binding affinity under iron-replete conditions (box I) overlaps the −35 region of the promoter, consistent with Fe2+-Fur repression due to inhibition of RNAP interaction at the promoter. An additional Fe2+-Fur binding site (box II) lies further downstream (positions +19 to −13) and may provide stronger or more complete repression under conditions where high concentrations of iron are present in the cell. The third Fur binding site (box III) is located between the −87 and −104 nucleotides and is bound by apo-Fur. In contrast to the repression seen by Fur binding to boxes I and II, apo-Fur binding to box III results in the activation of fur expression (129). The combination of both Fe2+-Fur repression and apo-Fur activation in the autoregulation of fur expression allows H. pylori to tightly control production of this multifunctional regulatory protein.
FIG. 8.
Autoregulation of H. pylori fur. The model shown was based on the work of Delany et al. (128, 129). (A) The relative positions of the two Fe2+-Fur binding sites and the single apo-Fur binding site are indicated. Fe2+-Fur binds DNA as a dimer (111, 128, 482); however, the oligomeric state of apo-Fur is unknown, as indicated by the question mark. For simplicity, apo-Fur is shown to function as a monomer. (B) (Top) Under iron-replete conditions, Fe2+-Fur binds the high-affinity site in box I and represses fur expression. (Middle) Under conditions of high/excess iron, Fe2+-Fur also binds the lower-affinity site in box II, resulting in a more complete repression of fur expression. (Bottom) In the absence of iron or under iron-restricted conditions, box II is not bound by Fur and fur expression is derepressed. In addition, apo-Fur binds the target sequences in box III and box I, activating fur transcription (129).
Similar to that in C. jejuni, the iron-responsive regulatory network in H. pylori is extensive and contains genes that serve diverse functions. Transcriptional profiling identified 97 genes that are regulated by iron and 43 genes whose expression is Fur dependent (160). A subsequent study examined the role of Fur in iron- and acid-responsive regulation (197). Collectively, these studies identify a variety of genes encompassing diverse functional categories and thus highlight the role of Fur as a global regulator. H. pylori genes repressed by Fe2+-Fur include those involved in iron uptake and transport (such as feoB, fecA1-3, frpB, exbB, and ceuE), energy metabolism, cellular processes, and protein synthesis, as well as many additional genes with unknown or hypothetical functions (160, 197, 416).
Adding yet another level of complexity to regulation by H. pylori Fur is the observation that several genes appear to be upregulated by Fe2+-Fur (197). Detailed analysis of Fur regulation of the nifS promoter suggests that Fur can in fact act as a transcriptional activator. Under iron-replete conditions, Fur was found to protect two distinct regions within the nifS promoter (6). Although there are no clear Fur boxes in the nifS promoter, the Fur binding regions were reported to be 150 and 200 nucleotides upstream from the transcriptional start, which could be consistent with an orientation of Fur needed to activate transcription. Another set of genes reported to be induced by Fe2+-Fur encode the 2-oxoglutarate oxidoreductase complex, OorDABC (197). These components were also reported to be induced by Fe2+-Fur in C. jejuni (265). In H. pylori, the upstream region of these genes appears to contain a putative Fur box (based on a consensus proposed by Merrell et al. [416]), but direct activation by Fur has not yet been demonstrated for H. pylori.
apo-Fur has been shown to regulate expression of pfr and sodB (54, 78, 160, 161). Other genes putatively repressed by apo-Fur include serB (encoding a phosphoserine phosphatase), genes involved in energy metabolism, such as hydA and hydB, and several genes with unknown or hypothetical function (160). Regulation by the iron-free form of Fur has not been demonstrated definitively for any other organism, although indirect evidence suggests that apo-Fur regulation may exist in C. jejuni and a subspecies of Desulfovibrio (50, 265). Overall, apo-Fur regulation is seen as somewhat controversial due to the previous discovery of Fur-regulated sRNAs such as RyhB. One hypothesis that could possibly explain the apparent regulatory role of apo-Fur in H. pylori is the existence of a Fur-regulated sRNA. As discussed above, Fur-sRNA interactions are well described for E. coli, where RyhB affects expression of several genes previously thought to be Fur regulated (402). However, data supporting the sRNA hypothesis are lacking for H. pylori, since only a few sRNAs have been studied to date and the published H. pylori genomes lack a RyhB homologue (653, 654). Despite this, recent publication of the H. pylori primary transcriptome has indicated that a large number of sRNAs exist (534). Based on sequence similarity, several of these putative sRNAs may be involved in regulation of Fur target genes. Candidate sRNAs (denoted by the prefix “HPnc”) and putative Fur-regulated targets include HPnc1680 (cheV), HPnc3280 (pfr and serB), and HPnc7370 (fecA). Further studies will be necessary to determine what, if any, role these putative sRNAs play in iron-responsive regulation in H. pylori.
Summary
In most living organisms, the essential but toxic nature of iron requires tightly regulated homeostatic mechanisms. The iron uptake systems found in pathogenic bacteria such as C. jejuni and H. pylori have evolved to ensure that these bacteria are able to acquire iron from the most prevalent sources within their respective environments. As inhabitants of the avian and human intestinal tracts, C. jejuni strains are in competition for iron, not only with other intestinal microbes but also with the host. To survive within this competitive environment, C. jejuni has evolved mechanisms for acquiring iron from other bacterial species as well as from human and avian hosts (452). In contrast, as the lone known bacterial resident of the gastric mucosa, H. pylori competes for iron solely with the human host. Thus, H. pylori has evolved specific iron acquisition mechanisms that primarily allow the bacterium to compete for host-derived sources of iron (134, 533). Regardless of the colonization site, both C. jejuni and H. pylori tightly control the expression of iron uptake and storage systems by using Fur. Furthermore, and possibly due to the relative paucity of regulatory proteins in the bacterium, H. pylori Fur has evolved the ability to function in an extended regulatory capacity. Collectively, the iron homeostatic mechanisms employed by C. jejuni and H. pylori highlight yet another example of how these epsilonproteobacteria function outside the classical systems.
BACTERIAL INTERACTIONS WITH GASTROINTESTINAL EPITHELIUM
For pathogenic bacteria to cause disease, they must first successfully colonize the host. The process of colonization is multifactorial, involving many components that assist bacteria in finding and surviving in their proper niche in the host. Thus, different pathogens have developed factors that aid in colonization. Notably, the repertoires of tools used by different pathogens of humans are affected by the site of bacterial colonization. Salmonella spp., E. coli, C. jejuni, and H. pylori each initiate disease in humans by first colonizing mucosal surfaces. Cytotoxin-producing, noninvasive E. coli strains, such as enteroaggregative E. coli (EAEC), enterohemorrhagic E. coli (EHEC), and enteropathogenic E. coli (EPEC), and invasive pathogens such as Salmonella spp., enteroinvasive E. coli (EIEC), and C. jejuni are major causes of diarrheal disease (443). H. pylori infects the gastric mucosa to elicit an inflammatory response, which can lead to gastritis (397, 432, 433). Chronic inflammation, along with the destruction of gastric epithelial cells, can result in more severe outcomes of H. pylori infection, including duodenal ulcers and gastric cancer.
To initiate infection, these pathogens first attach to the mucosal epithelium of different regions of the gastrointestinal tract. Each bacterium must then manipulate the epithelium to create a niche that supports replication and persistence in the human host. Salmonella spp., E. coli, C. jejuni, and H. pylori use different adhesins and colonization factors that target different receptors on epithelial cells. Salmonella spp. and C. jejuni are able to subsequently invade epithelial cells and persist in an intracellular niche (Fig. 9 A and B). However, the strategies by which these two bacterial species promote invasion and survive intracellularly occur via different virulence factors and mechanisms. In contrast, E. coli and H. pylori typically remain associated with the surfaces of epithelial cells. These bacteria have been observed to invade epithelial cells to a certain extent, but the significance of an intracellular lifestyle is unclear. Instead, E. coli and H. pylori appear to efficiently manipulate host cell biology to create an extracellular niche for replication (Fig. 9C and D). This section outlines the strategies used by each pathogen to promote adherence and subsequent survival strategies within or outside epithelial cells.
FIG. 9.
Comparison of the interactions of S. enterica serovar Typhimurium, C. jejuni, EHEC (or EPEC), and H. pylori with epithelial cells. (A) Adherence of Salmonella spp. to epithelial cells is mediated by both fimbrial and nonfimbrial adhesins. Subsequent translocation of effector proteins by the SPI-1 T3SS leads to actin rearrangements that result in membrane ruffling and invasion of the bacterium. Translocation of SPI-2 effectors results in maturation of the initial vesicle to the SCV. The SCV is modified, resulting in the formation of Sifs and migration of the SCV to a location near the Golgi apparatus to serve as an intracellular replicative niche. (B) C. jejuni adherence likely involves flagellar motility and a collection of outer membrane proteins. Subsequent invasion and intracellular survival mechanisms may be mediated by a variety of proteins, with some possibly secreted by the flagellar machinery. Invasion of host cells by C. jejuni is a microtubule-dependent process that results in membrane protrusions. C. jejuni initially enters the host cell in an endocytic vacuole that matures to a specialized CCV. The CCV is trafficked to a perinuclear location in a process involving microtubules. Question marks indicate factors and steps in CCV maturation that are unknown. (C) EHEC and EPEC use a variety of pili and other proteins to mediate a localized adherence pattern to intestinal epithelial cells. Subsequent translocation of T3SS effector proteins causes the polymerization of actin filaments and the formation of A/E lesions. A/E lesions are characterized by loss of microvilli and the formation of pedestal structures that promote tight adherence of the bacteria. (D) H. pylori subsists mainly in the mucous layer covering the gastric epithelium. Adherence of H. pylori is influenced by flagellar motility and a series of outer membrane proteins that use blood group antigens as specific receptors. Translocation of CagA and possibly other effectors by the Cag T4SS intoxicates the host cell to result in disruption of normal signaling pathways, loss of polarity, and dysregulation of cellular trafficking. These processes are thought to possibly redirect nutrients to the apical surface to promote growth of H. pylori. Evidence suggests that a small percentage of H. pylori organisms invade host cells and escape a modified vacuole before returning to the mucous layer. Question marks indicate unknown mechanisms of vacuole escape and release from the epithelial cell, as well as factors that may be redirected to the apical surface to promote H. pylori growth.
Adherence to Gastrointestinal Epithelium
Salmonella spp. and E. coli.
The virulence mechanisms employed by Salmonella spp. and E. coli have been studied extensively and form the prototypical model for pathogenesis of disease by enteric bacteria (for general reviews, see references 101, 120, 138, and 178). These organisms utilize both plasmid-encoded and chromosomally encoded virulence factors to facilitate host interactions. Several of these virulence factors are found within pathogenicity islands, which are distinct genomic regions that are often acquired through horizontal gene transfer (178, 523). Two of the best-characterized Salmonella pathogenicity islands (SPIs) are SPI-1 and SPI-2. SPI-1, SPI-2, and the locus of enterocyte effacement (LEE) pathogenicity island found in EPEC and EHEC strains each encode a T3SS and associated effector proteins that are injected into epithelial cells to mediate many of the changes in host cell biology required to initiate adherence or invasion or to maintain associations with host cells (178, 179, 194, 270, 523).
Adherence of Salmonella spp. and E. coli to intestinal epithelial cells is facilitated by cellular appendages, outer membrane proteins, and secreted proteins that interact with host cell receptors (Fig. 9A and C). Salmonella spp. adhere by a variety of fimbriae, each of which may exhibit a specific tissue tropism (609). These fimbriae include type 1 fimbriae (Fim), long polar fimbriae (Lpf), plasmid-encoded fimbriae (Pef), and aggregative fimbriae (Agf). These fimbriae have been shown to be involved in adhesion to various cell types and in colonization in animal models of infection (12, 46-49, 582). Additionally, insertional inactivation of genes in the putative bcf, stb, stc, std, and sth fimbrial operons of Salmonella spp. results in reduced cecum and fecal recovery in mice relative to that with wild-type bacteria 1 month after infection but not after 5 days, indicating that these operons may encode fimbriae required for persistent infection (273, 636).
In addition to fimbriae, the genome of Salmonella enterica serovar Typhimurium also encodes several nonfimbrial adhesins. SPI-4 encodes the secreted adhesion SiiE, which mediates adherence to the apical face of polarized epithelial cells (202, 203). In addition, MisL and ShdA are two outer membrane proteins that bind fibronectin, a component of the extracellular matrix (140, 326, 327, 430, 530). misL mutants are highly attenuated for colonization in a 2-week-old chick model of infection (430), while shdA of Salmonella spp. is expressed during infection of the murine cecum and is required for persistence (327).
Adherence of EHEC and EPEC strains is thought to be a two-step process that involves various adhesins that mediate initial interactions to bring the bacteria in contact with a host cell (termed localized adherence), followed by a LEE-dependent process that results in a tight, intimate adherence to the cells (Fig. 9C) (reviewed in reference 453). Like Salmonella spp., EHEC and EPEC both produce surface appendages that mediate adherence to host cells. The E. coli common pilus (ECP) and the bundle-forming pilus (BFP) are two prominent pili that promote adherence to various cell lines in vitro (103, 501, 514). However, adherence of E. coli strains is also mediated by nonpilus factors. The EHEC factor for adherence (Efa-1) is a nonpilus adhesin that has been implicated in both in vitro adherence and in vivo colonization of the bovine intestinal epithelium (33, 446, 572). In addition to being a conduit for transport of secreted effector proteins, the EspA filament of the LEE-encoded T3SS has also been implicated in adherence of EPEC strains (103). Additionally, the EHEC virulence plasmid pO157 encodes the Etp T2SS along with YodA and StcE, two Etp-secreted factors that influence adhesion processes (228, 259). yodA mutants of EHEC demonstrate reduced adherence to HeLa cells and are less competitive for colonization of the rabbit intestine (259).
After initial localized adherence, EHEC and EPEC induce a characteristic histopathological change in host epithelial cells known as attaching and effacing (A/E) lesions (333, 427). These lesions are characterized by loss of microvilli, intimate attachment of the bacterium to the host cell, and the production of pedestals, which are morphological projections of the eukaryotic cell with bacteria attached at the tips that mediate a close association between the host and pathogen (Fig. 9C) (427). Both EHEC and EPEC require the LEE-encoded T3SS to mediate A/E lesion formation (523). One effector protein encoded by the LEE that is translocated into the host cell by the T3SS is Tir (322). This effector serves as a receptor for the EHEC and EPEC outer membrane protein intimin (encoded by eae) (322, 506). Tir has additional functions (described below) that alter the host cell architecture to allow EHEC and EPEC to form pedestals to maintain association with the epithelial cells.
C. jejuni and H. pylori.
The genomes of C. jejuni and H. pylori lack genes encoding fimbrial or pilus systems to mediate adherence (9, 185, 473). Instead, these pathogens utilize a different repertoire of adherence determinants to facilitate interactions with host cells. Flagellar motility of C. jejuni is recognized as a major determinant that promotes interactions with human intestinal epithelial cells (221, 340, 413, 632, 660). However, the exact role of flagella in mediating adherence of C. jejuni to intestinal cells is unclear. Different groups have reported contradictory observations concerning the dependency of FlaA and the flagellar filament on adherence of C. jejuni. Some of these discrepancies are due to differences in strains and mutants studied or to differences in experimental procedures. Mutation of flaA or shearing of the filament was shown to reduce C. jejuni adherence to INT407 cells, regardless of centrifugation-mediated contact of the bacteria with the surfaces of epithelial cells (413, 660). These results suggest that the flagellar structure is directly involved in adherence, possibly serving as an adhesin. However, in another study, nonmotile mutants that lack flagella were reported to adhere to cells equally as well as wild-type bacteria when the bacteria were allowed to contact epithelial cells via centrifugation, suggesting that the role of the flagellum may be to steer the bacterium to the surface of an epithelial cell so that a nonflagellar adhesin can initiate adherence (210, 221). Even within the same study, two different strains demonstrated flagellum-dependent and -independent adherence, even with centrifugation (340). Thus, flagellar motility is likely involved in adherence, but in a strain-specific manner. Furthermore, chemotactic motility is also required for C. jejuni to mediate adherence, as a cheY mutant, which is defective in proper chemotaxis, exhibits decreased adherence to epithelial cells in vitro and is attenuated for colonization in a ferret model of disease (661). Even though the exact influence of motility in adherence has not been determined precisely, motility likely makes C. jejuni more fit for mediating adherence.
The C. jejuni flagellum has been implicated in secretion of proteins that influence host cell interactions (94, 339, 340). The flagellar secretion machinery demonstrates significant homology to the T3SSs of many bacterial pathogens (107, 270). As such, the flagellar systems of some pathogenic bacteria have been found to secrete virulence factors involved in pathogenesis (204, 362, 664). One C. jejuni protein that is secreted in a flagellum-specific manner is FlaC (559). FlaC shows significant homology to the FlaA and FlaB flagellins of the flagellar filament. However, flaC mutants are not defective for motility, as analyzed by in vitro methods (305, 559). Instead, purified FlaC has been shown to bind to HEp-2 human epidermoid carcinoma cells, and a flaC mutant is unable to invade HEp-2 cells as well as a wild-type strain does (559).
For a few adhesins of C. jejuni, host cell receptors have been identified. CadF and FlpA are two outer membrane proteins that bind fibronectin and are required for adherence to human intestinal or chicken hepatocarcinoma cells (336, 337, 341, 426). cadF and flpA mutants are both attenuated for colonization of chicks, indicating that the encoded proteins are likely required for interactions with the avian gut mucosa to promote commensalism (183, 337, 341, 671). Another C. jejuni protein implicated in adherence is JlpA, a surface-associated lipoprotein that promotes binding to the host cell heat shock protein 90α (Hsp90α) (290). Mutation of jlpA results in reduced adherence to HEp-2 cells (289). Furthermore, purified recombinant JlpA is able to bind to HEp-2 cells, and exogenous addition of the protein inhibits adherence of C. jejuni to these cells (289). However, diminished adherence of a jlpA mutant was not observed with T84 cells (454), suggesting that this protein may mediate tropism for different cell types.
Other factors that have been implicated in adherence of C. jejuni to host cells include the CapA autotransporter protein, plasmid-encoded factors, and a capsular polysaccharide. CapA has also been demonstrated to have both in vitro and in vivo functions in adherence to Caco-2 cells and persistence in the chick intestinal tract, respectively (26). In addition, some strains of C. jejuni harbor pVir, a plasmid that has been implicated in virulence in a ferret model of disease (31). This plasmid encodes parts of a putative T4SS that appears to influence DNA transformation (as discussed above) (31). Mutation of two pVir genes, comB3 and virB11, causes a reduction in adherence to INT407 cells (31). Additionally, one nonproteinaceous factor of C. jejuni that has also been associated with adherence to human intestinal epithelial cells is a surface capsular polysaccharide (29, 32). Another saccharide structure involved in adherence of C. jejuni is the N-linked protein glycosylation system that modifies certain periplasmic and outer membrane proteins with specific glycans (described below) (585). However, it is currently unknown how this glycosylation system impacts adhesion mechanisms.
H. pylori infection is believed to result in a vast majority of bacteria residing in an extracellular niche. Sampling of gastric mucus from infected gerbils revealed that approximately one-third of the bacteria were tightly associated or freely swimming in the mucus within 5 μm atop the gastric epithelium, with the remaining bacteria swimming in distal regions of the mucous layers (529). Attachment to host cells has been observed to occur preferentially near cell-cell junctions (13). Various H. pylori proteins mediate adherence at different stages of infection, which may explain why multiple adhesins are expressed and influence host cell interactions (369). Like C. jejuni flagella, H. pylori flagella are required for colonization in vivo, but it is unclear if flagella are used solely for motility or function directly as an adhesin (151).
Two prominent adhesins of H. pylori are BabA and SabA (275, 394). These proteins are outer membrane proteins that recognize sialylated and nonsialylated host Lewis (Le) antigens present on the surfaces of epithelial cells (64, 170, 275, 394, 429). BabA binds primarily Leb and related fucosylated ABO blood group antigens (64, 275). Preference for BabA-mediated adherence to Leb antigens may correlate with the disproportionate increase in peptic ulcers seen in H. pylori-infected individuals with blood group O (64). Another adhesin, SabA, binds the sialylated Lex antigen with a lower affinity than that of BabA for its receptors (394). Production of Lex appears to increase on the apical gastric epithelium upon H. pylori-mediated inflammation (394, 538). SabA-mediated binding of Lex likely creates additional binding sites for persistence in individuals who have succumbed to inflammation and gastritis.
Several outer membrane proteins with unknown receptors are also involved in promoting H. pylori adherence and in vivo growth. Insertional inactivation of horB reduces adherence to the human gastric adenocarcinoma epithelial cell line AGS, as well as colonization of murine stomachs (554). The heat shock protein HopZ is also capable of mediating H. pylori adherence to AGS cells, but this adherence mechanism appears to be strain specific (481, 656). Additional adhesins include AlpA and AlpB, which are essential for binding human gastric biopsy sections (127, 458). H. pylori strains that lack these adhesins are also less competitive than wild-type strains in a guinea pig model of infection, indicating that they are potentially involved in important adherence mechanisms that contribute to pathogenesis (127).
Invasion of Intestinal Epithelium and Intracellular Replication
Creation of an intracellular niche for Salmonella spp.
T3SSs are protein delivery machines produced by several pathogens that are often required for infection and pathogenesis of disease. While the secretion machinery itself is well conserved between species, the secreted effector proteins vary greatly, which allows for distinctive modes of interaction with the host cell that often disrupt host cell signaling and architecture (105, 194, 205, 270). Salmonella enterica serovar Typhimurium produces two well-characterized T3SSs with associated effectors that are encoded on separate SPIs. These T3SSs and respective effectors mediate different steps in host cell invasion. Whereas the SPI-1 T3SS mediates cytoskeletal rearrangements that are important for host cell invasion, the SPI-2 T3SS is required for events after entry that support intracellular replication and survival (241, 607). Immunofluorescence microscopy studies revealed that translocation of SPI-1 effectors into the host cell results in rapid polymerization of the cytoskeleton microfilament actin, resulting in the formation of membrane protrusions that ultimately extend toward and engulf the bacteria in a process termed membrane ruffling (293). The complex, multifaceted cascade of effectors that are required for alteration of signaling pathways and cytoskeletal rearrangements are the subjects of many extensive reviews (120, 478, 522, 607). In brief, major bacterial effectors that induce membrane ruffling and phagocytosis of Salmonella are SopE2, which is the exchange factor for the host cell GTP-binding proteins Cdc42 and Rac-1, and SopB, an inositol polyphosphate phosphatase (246, 570, 595, 668). These bacterial and host proteins are required to induce actin filament polymerization through the Arp2/3 complex and phagocytosis, which mediate entry of the bacterium into the host cell (39, 646, 668). Additionally, SipC is another SPI-1-encoded effector that nucleates actin assembly, whereas SipA directly binds actin and the actin-bundling protein T-plastin (88, 249, 424, 669, 670). The actions of these effectors stabilize filaments and inhibit depolymerization to facilitate membrane ruffling (424, 669, 670). The SPI-1 effector SptP acts as a GTPase-activating protein that reverses alterations to the cytoskeleton by inducing depolymerization, returning the host cell membrane to its original state (191).
Invasion of intestinal epithelial cells results in localization of Salmonella spp. within a specialized vacuole that does not follow the normal lysosomal maturation pathway, which would typically result in destruction of the bacteria (10, 178). This specialized vacuole, termed the Salmonella-containing vacuole (SCV), supports replication of bacteria (Fig. 9A) (10, 178, 179, 606). It is clear that T3SS effectors of both the SPI-1 and SPI-2 systems of Salmonella spp. are critical for SCV development. However, due to some functional redundancy between effectors, it has been difficult to discern which SPI-1 or SPI-2 effectors are responsible for specific events.
Upon invasion, expression of SPI-1 genes is decreased, whereas that of SPI-2 genes is induced (517). SPI-1-encoded effectors such as SopB and SipA are involved in the latter stages of invasion to complete phagocytosis and formation of the initial SCV (71, 120, 256). SopB inhibits lysosomal fusion of the maturing SCV by reducing the negative surface charge of the vacuolar membrane (38). The SPI-2 effectors are secreted across the vacuolar membrane into the host cell for further maturation of the SCV and movement of the SCV to a juxtanuclear region near the microtubule-organizing center (MTOC) (418, 513, 634). The effectors SseF, SseG, and SifA aid in trafficking and maintenance of the SCV at this locale (4, 126, 238, 288, 350, 497). SifA also aids in the formation of Salmonella-induced filaments (Sifs) emanating from the SCV (Fig. 9A) (421, 569). Additionally, SseI, SspH2, and SteC are involved in a process known as vacuole-associated actin polymerization (VAP) that manipulates F-actin to induce formation of an actin meshwork around the SCV (415, 418, 488, 648). As a result of these alterations, the SCV becomes more stable and intercepts proteins from the Golgi body for optimization as a replicative niche (349).
Invasion and intracellular survival of C. jejuni.
Like Salmonella spp., C. jejuni has the ability to invade human intestinal epithelial cells and reside in an intracellular niche for replication. However, the virulence factors and molecular mechanisms delineating such events for C. jejuni have only begun to be identified. Unlike Salmonella spp., which use T3SSs and translocated effector proteins to manipulate host cells for invasion and intracellular survival, C. jejuni lacks genes encoding these virulence factors. Instead, C. jejuni is thought to take advantage of the flagellar T3SS to secrete potential effector proteins that may assist in invasion and intracellular survival processes (217, 339, 559).
A class of C. jejuni proteins known as the Campylobacter invasion antigens (Cia) have been proposed to be secreted by the flagellar T3SS when bacteria are cocultured with intestinal epithelial cells (94, 339). However, these proteins do not appear to be required for motility; instead, the Cia proteins may be required for invasion of C. jejuni into host cells. Two of the identified secreted proteins are CiaB, which shares only limited homology to SipB of Salmonella spp., and CiaC, which lacks homology to other proteins (94, 339). C. jejuni mutants lacking each protein are impaired for invasion of INT407 cells (94, 339). However, the attenuated invasion phenotype of the ciaB mutant is not observed for a different strain of C. jejuni with T84 cells (454). These results suggest that Cia-mediated invasion may be strain or cell type specific. The functions of CiaB and CiaC and the identifications of other potential Cia proteins remain to be determined. However, CiaB may play a role in secretion, since a ciaB mutant demonstrates a defect in secretion of other potential Cia proteins (339).
Even though σ28 is involved mainly in expression of flagellar genes in many bacteria, C. jejuni uses σ28 for expression of some genes not involved in motility (80, 217, 489). Instead, some of these σ28-dependent genes have been implicated in invasion or toxicity of intestinal epithelial cells (217, 489). One of these genes, fspA, appears as two alleles (fspA1 and fspA2) in C. jejuni strains (489). FspA1 and FspA2 are both secreted in a flagellum-dependent manner but are not required for invasion. Instead, FspA2 is able to induce apoptosis in INT407 cells (489). Another σ28-dependent gene involved in invasion of intestinal cells is cj0977 (217). Initial analysis indicated that Cj0977 is cytoplasmic and is not secreted by the flagellar T3SS. This protein was first shown to not be required for motility in semisolid motility agar (217). However, another study concluded that a cj0977 mutant demonstrated a motility defect in liquid broth, which may have contributed to an observed in vitro invasion defect (454).
Certain surface structures of C. jejuni also influence invasion processes. Campylobacter spp. produce sialylated lipooligosaccharide in the outer leaflet of the outer membrane, which has been shown to influence invasion (311, 383). Additionally, the surface capsular polysaccharide not only potentiates adherence but also is required for invasion of host cells (29, 32). How these factors affect invasion or possibly intracellular survival remains unknown.
Like Salmonella spp., C. jejuni alters host cell biology to trigger invasion, but the uptake mechanism diverges from that of Salmonella. Rather than being mediated by actin, C. jejuni invasion occurs through a novel microtubule-dependent process (Fig. 9B) (269, 460). Inhibitors of microtubule polymerization have been shown to prevent uptake of C. jejuni (460). In addition, immunofluorescence has shown that the bacterium interacts with host cell membrane protrusions containing microtubules and not actin filaments (269).
Little is known about the mechanisms of C. jejuni intracellular survival and replication after invasion. Within intestinal epithelial cells, C. jejuni survives within a modified membrane-bound compartment termed the Campylobacter-containing vesicle (CCV), where it is capable of replication (Fig. 9B) (338, 510, 635). Immunofluorescence microscopy indicates that the CCV avoids lysosomal fusion (635). However, the mechanism by which C. jejuni matures in the CCV is unknown. Immunofluorescence microscopy also indicates that C. jejuni colocalizes with microtubules after uptake and migrates in parallel with these polymers, to the perinuclear region in close proximity to the Golgi apparatus, by a dynein-dependent mechanism (269, 635). Details regarding the formation of the CCV and the role of this specialized vacuole in the intracellular lifestyle of C. jejuni await further characterization.
One intriguing finding regarding the intracellular lifestyle of C. jejuni is that the bacterium appears to undergo significant physiological changes, such as oxygen sensitivity and alterations in metabolism, that likely influence the ability of C. jejuni to replicate intracellularly (454, 635). In a transposon mutagenesis screen to identify mutants of C. jejuni attenuated for invasion, mutations in aspA, aspB, and sodB were found to not be required for invasion per se but were necessary for intracellular survival (454). AspA and AspB are involved in aspartate and glutamate catabolism, which produces fumarate, a metabolite that can serve as both a carbon source and an electron donor in C. jejuni (532, 553). Since the intracellular survival defect of aspA and aspB mutants could be restored by prior growth in the presence of fumarate, it was concluded that a fumarate-dependent process may prime the cells for intracellular growth (454). Mutation of sodB, which encodes a superoxide dismutase, results in decreased intestinal cell invasion and decreased colonization of MyD88-deficient mice (454, 483). The viability of the sodB mutant decreases over time after invasion, indicating that SodB assists in intracellular survival (454). SodB likely quenches reactive oxygen intermediates once C. jejuni is internalized by the host cell to protect against oxidative damage (76).
Alteration of Host Cells To Create an Extracellular Niche
Intimate adherence of E. coli strains.
Unlike Salmonella spp. and C. jejuni, the major strains of E. coli that promote diarrheal disease, EHEC and EPEC, are thought to be largely noninvasive and persist by associating with the surfaces of epithelial cells. EHEC and EPEC first promote a localized adherence and then a tight association with cells known as intimate adherence (reviewed in reference 453). Intimate adherence results in a gross alteration of host cell architecture and in formation of characteristic A/E lesions with the formation of pedestals that extrude from the surface of the epithelial cell with bacteria tightly associated at the tip (Fig. 9C) (322, 333, 427, 506). To induce these morphological changes in host cells, EHEC and EPEC manipulate intracellular actin polymerization from an extracellular location.
A T3SS effector with a major role in host cell actin rearrangement and pedestal formation is Tir, which inserts into the host cell membrane to serve as the receptor for the E. coli intimin protein (227, 322, 506). In addition, host cell kinases phosphorylate tyrosine, serine, and threonine residues of Tir, which results in blockage of inhibitory pathways for actin polymerization (198, 227, 630). As a result, EHEC and EPEC manipulate actin polymerization to form the pedestals necessary to initiate and maintain intimate adherence (133, 227). The Tir proteins of EPEC and EHEC are not interchangeable and use different pathways to alter actin polymerization (133, 321). Phosphorylation of EPEC Tir allows for the formation of a docking site for Nck adaptor proteins (227). This binding of Nck proteins results in the loss of autoinhibition of N-WASP, which culminates in activation of Arp2/3 and in actin polymerization (227). However, EHEC strains use an Nck-independent pathway by injecting a non-LEE-encoded effector, EspFu, into host cells (75, 133, 198). EspFu directly relieves N-WASP from autoinhibition, which begins a cascade of events leading to actin polymerization (75). Despite the impressive morphological changes mediated by EHEC and EPEC that result in the formation of A/E lesions, it is still unclear what biological benefit these changes impart for the bacterium during infection of the human host.
Manipulation of cell polarity by H. pylori to form a replicative niche.
Observations of H. pylori during infection of humans or rodents indicate that the bacterium is predominantly attached to gastric epithelium or freely swimming in the region of the mucous layer directly above the cells (250, 529, 567). However, there have been reports that this tight association with the gastric epithelium may contribute to invasion and formation of an intracellular niche for H. pylori (13, 98, 165, 358, 579). Some of these studies have also found that intracellular H. pylori resides in a vacuole or in autophagocytic vesicles that temporarily support replication (13, 98). At later times postinfection, H. pylori is either killed in these vesicles or released extracellularly, which may enable the bacterium to infect other cells (13, 98). Whereas the relevance of a potential intracellular lifestyle is currently unclear, invasion of H. pylori may allow temporary protection from antibiotics, possibly indicating a survival strategy that contributes to a notable failure of antibiotic therapy upon H. pylori infection in humans (414).
In contrast, more details are known about the extracellular lifestyle of H. pylori during infection. H. pylori can adhere to gastric epithelial cells and efficiently replicate on the surfaces of the cells to form microcolonies (14, 591). Major determinants of H. pylori that are required for efficient replication on the apical surface of gastric cells are the Cag T4SS and the CagA effector protein (591). CagA is injected into host cells and phosphorylated by host cell kinases to result in CagA-mediated changes in cellular signaling pathways (30, 286, 459, 568). CagA has been demonstrated to have multiple effects on host cell biology (14, 35, 258, 512, 666). CagA-mediated changes include disruption of host cell polarity and tight junctions, induction of migration of intoxicated cells, and alteration of cellular morphology to an elongated spindle shape known as the hummingbird phenotype. However, exactly how CagA-mediated effects benefit the biology of H. pylori during infection has been unclear.
Some clues have been provided regarding how CagA contributes to H. pylori infection. Prominent CagA-induced phenotypes include disruption of cellular polarity and mislocalization of proteins normally targeted to apical or basolateral surfaces or to tight junctions (Fig. 9D) (14, 512, 666). These observations formed the foundation for the proposal that H. pylori may manipulate cellular polarity to benefit in vivo growth. Evidence supporting this hypothesis includes the observation that H. pylori replicates and forms microcolonies on the apical surface of polarized epithelial monolayers close to the junction between cells (591). Whereas both wild-type and cagA mutant strains can replicate on the basolateral side of the polarized monolayers, a cagA mutant is unable to grow on the apical surface. However, treatment of the polarized monolayer with inhibitors that target the atypical protein kinase C (aPKC)/Par1b pathway, which maintains cellular polarity, allows a cagA mutant to replicate on the apical surface as well as wild-type bacteria do (591). These results suggest that CagA may disrupt cellular polarity to possibly redirect nutrient transport to the apical surface to support H. pylori replication.
Another major virulence factor that likely assists in H. pylori pathogenesis and enhances nutrient acquisition is the VacA vacuolating cytotoxin (425). VacA was first identified due to its ability to induce cytoplasmic vacuolation of eukaryotic cells (110, 366). Further investigations revealed that VacA is able to create ion-selective channels in the plasma membranes of cells (124, 470, 584, 600). VacA has been shown to increase the permeability of the plasma membranes of eukaryotic cells to low-molecular-weight molecules and ions such as urea, carbonate, iron, and nickel, which are necessary for H. pylori growth (124, 470, 584, 600). Thus, the combined activity of CagA and VacA may be optimal for nutrient acquisition during an extracellular lifestyle on the surface of the gastric epithelium.
Additional studies indicate that CagA specifically targets the Par1b kinase to prevent its phosphorylation by aPKC, likely causing mislocalization of Par1b to the apical surface to contribute to polarity defects (512). Interactions with and inhibition of Par1b also allow CagA to interact with SHP2, which is then able to induce cell spreading and the characteristic hummingbird phenotype (512). Thus, H. pylori may manipulate host cell polarity to turn the gastric epithelium into a replicative niche while also inducing changes in host cell architecture and motility that may lead to cellular turnover, damage, and inflammation.
Summary
By analysis of the virulence processes that contribute to pathogenesis of disease, it is clear that gastrointestinal pathogens are equipped with a variety of weapons that enable diverse mechanisms for interactions with host cells. Studies of the pathogenic mechanisms of gammaproteobacteria such as Salmonella spp. and E. coli have provided detailed models of molecular manipulation of host cell biology by injected bacterial effector proteins that behoove the pathogens. We are only now beginning to understand the virulence factors of C. jejuni and H. pylori and how these factors confer a fitness advantage in vivo. Whereas C. jejuni is similar to Salmonella spp. in terms of being able to invade and survive intracellularly in epithelial cells, this bacterium clearly manipulates the host cell differently. Notably, C. jejuni lacks a prototypical T3SS and alters microtubule organization instead of actin polymerization. H. pylori resides extracellularly, like EHEC and EPEC, but appears to manipulate polarity to extract nutrients from host cells to facilitate replication and persistence on the mucosal surface. Continued exploration will likely reveal novel virulence mechanisms of these epsilonproteobacteria.
PROTEIN GLYCOSYLATION SYSTEMS
Protein glycosylation was once thought to be an activity confined to eukaryotic and archaeal systems. However, with technological advancements in glycobiology, protein glycosylation has been observed in some eubacterial species as well. Studies of Campylobacter spp., in particular, have gone far to invalidate the dogma of exclusivity of protein glycosylation to eukaryotes, with the identification of two different protein glycosylation systems within these bacteria. In addition to producing surface sugar structures such as a capsular polysaccharide and sialylated lipooligosaccharide, Campylobacter spp. modify flagellins with an O-linked glycan and produce an abundant periplasmic saccharide that is also found as an N-linked glycan on at least 30 different periplasmic or outer membrane proteins (for reviews, see references 235, 314, 377, and 586). Helicobacter spp. also produce an O-linked glycosylation system to modify flagellins, and these systems are essential in Campylobacter spp. and H. pylori for biogenesis of the flagellar filament. O-linked glycosylation systems have previously been identified in beta- and gammaproteobacteria such as E. coli, Neisseria spp., and Pseudomonas spp. (reviewed in references 5, 53, and 587). However, N-linked glycosylation is rare in bacteria, with only Campylobacter spp. and H. influenzae demonstrating this activity to date (224, 371, 588, 665). The glycosylation systems in Campylobacter and Helicobacter spp. are required for various biological activities and consequently influence interactions with various hosts.
O-Linked Glycosylation Systems
Limited complexity of O-linked protein glycosylation in E. coli.
O-linked protein glycosylation results in attachment of a glycan to the hydroxyl oxygen of a serine or threonine residue of a target protein. Bacterial species such as E. coli, Pseudomonas aeruginosa, and Neisseria spp. contain functional O-linked glycosylation systems (reviewed in references 5, 44, and 53), but this activity has not been observed in Salmonella spp. In E. coli, O-linked protein glycosylation is limited to three different autotransporter proteins, AIDA-1, TibA, and Ag43 (51, 52, 89, 155, 370, 537). Unlike the complex O- and N-linked glycosylation systems of Campylobacter spp., which require multiple enzymes for biosynthesis and linkage of the respective glycans (described below), the E. coli glycosylation machinery is relatively simple. In E. coli, a single, specific glycosyltransferase promotes the addition of heptoses to multiple serine or threonine residues of a corresponding autotransporter protein (51, 52, 89, 155, 332, 370, 428). However, complementation studies have shown that these glycosyltransferases can mediate the characteristic glycosylation of noncognate autotransporters (332, 428, 537). The heptose sugars used as substrates for modification of E. coli proteins are derived from the LPS biosynthesis pathway rather than being synthesized by specific enzymes dedicated solely to forming the O-linked glycan (52). Glycosylation of the E. coli autotransporters appears to increase the stability of the proteins or assist the proteins in serving as adhesins (52, 89, 155, 332, 370, 537).
O-linked glycosylation of Campylobacter and Helicobacter flagellins.
Campylobacter and Helicobacter spp. use an O-linked glycosylation system for modification of flagellins, which is essential for filament formation and motility (137, 218, 231, 299, 373, 410, 521, 597). Evidence for posttranslational modification of the Campylobacter flagellins was first observed in C. coli, and the process was then characterized for both C. coli and C. jejuni (8, 381). Subsequently, other epsilonproteobacteria, including H. felis and H. pylori, were found to also glycosylate flagellins (296, 299, 521). Initial work suggested that the attached glycans were composed of sialic acid due to lectins specific for this saccharide binding to flagellins (137). However, the structures of the glycans were subsequently found to be composed of two structurally similar 9-carbon saccharides, pseudaminic acid (Pse5Ac7Ac; PseAc) and legionaminic acid (LegAm), depending on the bacterial species (380, 411, 521, 597). PseAc is the major glycan found on the Campylobacter and Helicobacter flagellins (380, 409, 410, 520, 521, 597). However, C. jejuni flagellins also contain modified versions of PseAc, including those with an acetamidino substitution (Pse5Am7Ac; PseAm), an O-acetyl addition (Pse5Ac7Ac8OAc), or an N-acetylglutamine attachment (Ps5Am7Ac8GlnAc) (520, 597). In contrast, the glycans of H. pylori flagellins are modified only with PseAc (521). Some C. jejuni and C. coli strains also modify flagellins with LegAm derivatives containing acetamidino and N-methylacetamidoyl additions (resulting in Leg5Am7Ac and Leg5AmNMe7Ac, respectively) (230, 380, 411, 597). These LegAm derivatives are structurally similar to the PseAm glycan of C. jejuni flagellins.
The glycosylation of Campylobacter flagellins contributes 10% of the mass of the proteins, making these flagellins the most heavily glycosylated bacterial proteins known to date (597). Glycosylation of H. pylori flagellins is also extensive and is responsible for 4% of the total protein mass (520, 521). The FlaA flagellins of C. jejuni and C. coli are glycosylated on 16 to 19 serine or threonine residues, whereas H. pylori FlaA is modified on 7 residues (521). Additionally, the H. pylori FlaB minor flagellin is modified on 10 serines or threonines (521). Glycosylation of FlaB of Campylobacter spp. has been reported to occur but currently remains uncharacterized (377). For the purposes of this section of the review, “flagellin” is used to refer mainly to FlaA. The glycosylated serines or threonines of the Campylobacter and Helicobacter flagellins are found within the proposed D2 and D3 domains of the protein (521, 597). For Salmonella FliC, these domains are the exposed regions of the flagellins once they are assembled into the filament (663). Assuming that the Campylobacter and Helicobacter flagellins are folded similarly once incorporated into the filament, modification of the serines or threonines within the D2 and D3 domains may result in coverage of the external surface of the filament with glycans. Analysis of the sequences surrounding the glycosylated residues has not revealed a strict consensus sequence for modification (597). Instead, serines or threonines immediately downstream of hydrophobic amino acids within the D2 and D3 regions may be favored for glycosylation. In addition, glycosylation of 3 of the 19 serines or threonines of the C. jejuni flagellin is required for complete filament biosynthesis and motility (167).
Glycosylation of the Campylobacter flagellins occurs independently of the FlgSR two-component system, the flagellar T3SS, and FlhF (167), which indicates that flagellin glycosylation likely occurs in the cytoplasm before secretion of the proteins. As described above, these three sets of proteins are required for expression of the C. jejuni σ54 regulon (40, 80, 253, 255, 300, 301, 651). One gene required for O-linked glycosylation of the flagellins is pseB, which is also part of the σ54 regulon (80, 218, 410). pseB encodes the enzyme that converts the starting compound UDP-α-d-GlcNac to the first intermediate of the PseAc biosynthesis pathway (Fig. 10) (114, 410, 527, 528). In a mutant that lacks σ54, pseB expression is reduced 4-fold, indicating that expression of this gene may also rely on σ70 (218). Thus, at least one component of the PseAc pathway is partly coexpressed with flagellar operons, which links these two systems for proper flagellar biosynthesis. However, glycosylation of flagellins still occurs in C. jejuni mutants defective for expression of σ54-dependent genes (167). Thus, the level of σ54-independent expression of pseB is sufficient for glycosylation of flagellins during in vitro growth. Determining whether full expression of pseB is required for glycosylation of the Campylobacter flagellins in vivo or in some other environmental setting remains to be done.
FIG. 10.
O-linked protein glycosylation systems of Campylobacter and Helicobacter species. The O-linked protein glycosylation systems result in the production of different types of glycans linked to flagellin proteins. The PseAc pathway begins with UDP-GlcNAc and is relatively conserved in Campylobacter and Helicobacter spp. The PseAm pathway of C. jejuni converts CMP-PseAc to CMP-PseAm for addition of the glycan to flagellin. The LegAm pathway of C. coli begins with an unknown sugar to result in production of Leg5Am7Ac or Leg5AmNMe7Ac for addition to flagellins. The pathways of the O-linked glycosylation systems result in the production of nucleotide-linked glycans (CMP-glycans) for addition to flagellins. Due to the production of two different glycans within C. jejuni and C. coli, flagellin proteins are modified heterogeneously with different sugars. GlcNAc, N-acetylglucosamine; PseAc, pseudaminic acid; PseAm, pseudaminic acid with acetamidino addition; LegAm, legionaminic acid; Leg5Am7Ac, legionaminic acid with acetamidino modification; and Leg5AmNMe7Ac, legionaminic acid with N-methylacetamidoyl modification.
(i) Biosynthesis of Campylobacter and Helicobacter O-linked glycans.
In C. jejuni, C. coli, and H. pylori, the major flagellin glycan modification is PseAc, which is generated by multiple enzymes encoded by the pse genes (Fig. 10) (93, 113, 218, 231, 299, 373, 410, 457, 521, 526, 528, 560, 597). Much of the information regarding PseAc biosynthesis has been elucidated from studies of C. jejuni strain 81-176 (410). UDP-α-d-GlcNac is the precursor for PseAc and PseAm, which are formed through the actions of six enzymes (PseB, PseC, PseG, PseH, PseI, and PseF) (Fig. 10). Mutants lacking any of these enzymes fail to produce PseAc or PseAm and are nonmotile (410). Flagellins of a pseA mutant are modified only with PseAc, not with PseAm, indicating that PseA catalyzes production of PseAm from PseAc (Fig. 10) (410, 597). PseE and PseD do not appear to be involved in PseAc or PseAm biosynthesis, as mutants lacking the respective genes produce both glycans (410). Instead, PseE and PseD are thought to function as glycosyltransferases that add PseAc and PseAm, respectively, to the flagellins.
H. pylori has a simpler O-linked protein glycosylation system, primarily attaching only the PseAc glycan to flagellins (520, 521, 527). The H. pylori PseAc biosynthetic pathway uses enzymes that are homologs of the C. jejuni PseAc pathway (114, 299, 526-528). As with C. jejuni, mutation of any H. pylori pse gene results in unmodified flagellins (299, 521). The sequence of reactions in the H. pylori PseAc biosynthetic pathway has been verified in vitro, with PseAc being synthesized from the UDP-α-d-GalNac precursor by sequential addition of each purified enzyme (527).
The ptm genes in C. coli are required for generation of the LegAm derivatives Leg5Am7Ac and Leg5AmNMe7Ac (Fig. 10) (230, 411). These genes are absent from the well-studied C. jejuni strain 81-176, but some other C. jejuni strains appear to carry homologs of a few ptm genes (185, 473). A survey of C. jejuni isolates from animals and avian species has provided evidence for production of Leg5Am7Ac and Leg5AmNMe7Ac in these strains, indicating that some C. jejuni strains have the capacity to glycosylate flagellins with these saccharides (268). Mutation of the ptmA to -G genes in C. coli results in loss of all LegAm derivatives (230, 380, 409). In these mutants, flagellins are modified with PseAc and associated derivatives instead of a heterogeneous mixture of PseAc and LegAm glycans (380). The enzymes encoded by ptmA to -F are required for production of LegAm (Fig. 10) (409). A mutant lacking ptmG accumulates LegAm but is unable to synthesize the two LegAm derivatives Leg5Am7Ac and Leg5AmNMe7Ac, whereas a ptmH mutant accumulates Leg5Am7Ac but not Leg5AmNMe7Ac (409). These observations indicate that PtmG synthesizes the acetamidino addition to LegAm, which is then converted to Leg5AmNme7Ac by the action of PtmH (Fig. 10). Glycosyltransferases required for the addition of the LegAm derivatives to target serines or threonines of the flagellin have yet to be identified.
The O-linked protein glycosylation system has been studied extensively for only one strain of C. jejuni, which produces PseAc and PseAm. However, as mentioned above, other C. jejuni strains produde LegAm derivatives, and evidence has been obtained that some strains can produce additions of 2,3-di-O-methylglyceric acid or carboxyl groups to PseAc (268, 379, 608). This heterogeneity of glycosylation is reflected in the genomic loci for the O-linked protein glycosylation systems of Campylobacter spp., which are some of the most diverse genomic elements of these species. Continued exploration of the O-linked protein glycosylation systems of Campylobacter spp. will most likely continue to reveal much more diversity in these systems than is currently known.
(ii) Biological function of O-linked glycosylation systems.
Elucidation of the biological roles of the O-linked protein glycosylation systems of Campylobacter and Helicobacter spp. in promoting interactions with hosts is technically challenging because glycan synthesis is required for production of flagella. Thus, Campylobacter and Helicobacter mutants that eliminate glycan production altogether result in aflagellated bacteria, which complicates interpretation of experimental results because flagellar motility is required for many types of host-bacterium interactions. However, the role of specific glycans has been analyzed through the use of mutants that decorate flagellins with predominantly one glycan. These studies revealed that PseAm is required for optimal levels of adherence to and invasion of human intestinal epithelial cells as well as for virulence in a ferret model of pathogenesis (231). Furthermore, loss of production of LegAm derivatives from a strain of C. jejuni lowers the fitness of the bacterium for commensal colonization of chicks (268). These results suggest that glycan heterogeneity on C. jejuni flagellins is required for optimal interaction with various hosts. Because glycosylation occurs in the predicted D2 and D3 regions of the flagellins that likely form the exposed regions of the filament, the glycans may play a role in evading certain immune responses (597). Indeed, glycosylation of flagellin does contribute to antigenic variation and differential reactivity with sera, suggesting that this process could contribute to immune evasion in a host (8, 492).
The biological role of these modifications in filament biosynthesis is unclear. Hypotheses have been proposed that the glycans may (i) promote stability of the flagellins prior to secretion or (ii) initiate specific contacts between flagellin subunits during polymerization of the filament. Disruption of PseAc biosynthesis in C. jejuni or H. pylori correlates with reduced levels of flagellins in cell lysates relative to those for wild-type bacteria (218, 299, 521). In C. coli, which has two independent systems for the biosynthesis of PseAc and LegAm derivatives, elimination of either system does not affect the levels of flagellins in lysates (218). However, elimination of both glycan biosynthesis systems severely reduces levels of unsecreted flagellins (218). These observations indicate that flagellin glycosylation may be important for stability of the proteins. However, C. jejuni flagellar secretory apparatus mutants, which are hindered for flagellin secretion out of the cytoplasm, show similarly reduced levels of flagellins to those for a pseC mutant (167). Since flagellins are glycosylated in the absence of the flagellar secretory apparatus (167), these results indicate that reduced flagellin levels, regardless of glycosylation state, may be due to degradation in the absence of secretion. Interestingly, multiple studies have also reported that some mutants that fail to produce the glycans, and thus the filament, also appear to lack flagellar hooks (218, 231, 299). However, hook proteins do not appear to be glycosylated, at least as reported by one study (137). Thus, it is currently unclear if glycan production is directly involved in hook biosynthesis or if the lack of filament production in glycosylation-deficient mutants results in instability of the hooks.
It is possible that glycosylation of flagellins may be involved directly in subunit-subunit interactions that facilitate filament polymerization. As described above, the D1 domain of the Campylobacter and Helicobacter flagellins does not stimulate TLR5 (18). The D1 domain of the Salmonella FliC flagellin is involved in subunit-subunit contacts for filament polymerization (663). Considering that Campylobacter and Helicobacter spp. have altered D1 domains relative to that in Salmonella FliC, compensatory mutations in the flagellins may be necessary to promote stacking of the flagellins for proper synthesis of a functional filament. Alternatively, the glycan modifications may contribute to some confirmations that allow for filament biosynthesis with flagellins that have altered D1 domains in these bacteria.
As demonstrated with wild-type H. pylori and specific mutants of C. jejuni and C. coli, heterogeneity of glycan modifications is not required for filament biosynthesis (218, 380, 597). Thus, filament biosynthesis occurs as long as the flagellins are modified with one of the specific O-linked glycans. However, one study revealed that the properties of the filament appear to be different when it is modified by predominantly one glycan. In a C. coli pseB mutant that produces a filament modified solely by LegAm derivatives, the filament is easily dissociated by SDS, unlike the case for isogenic strains that modify flagellins with solely PseAc or a combination of PseAc and LegAm derivatives (218). These findings suggest that specific glycan modifications may influence filament stability under different environmental conditions.
Another altered phenotype observed in mutants that produce only homogenous glycan modifications is autoagglutination (AAG) (231, 268, 608). Campylobacter spp. demonstrate an AAG phenotype that likely impacts the ability to form microcolonies and biofilms, both of which may be important for interactions with intestinal epithelial cells and other aspects of colonization of hosts (60, 231, 268). Many factors have been linked to the AAG phenotype, including flagella (210, 423). More specifically, heterogeneity of O-linked glycosylation of flagella appears to be one factor that mediates AAG (231, 268, 608). Campylobacter mutants with limited O-linked glycan diversity are reduced for AAG, adherence to and invasion of INT407 cells, commensal colonization of chicks, and virulence in a ferret model of pathogenesis (231, 268, 608). In addition, the AAG phenotype in C. jejuni is dependent on the glycosylation of seven specific serine or threonine residues, including two serines whose glycosylation is required for wild-type filament biogenesis and motility (167). Because the glycans are thought to be on the exposed regions of the flagellins in the flagellar filament, it has been proposed that different glycans on the flagella of adjacent bacteria likely interact to promote AAG, which presumably provides an advantage during in vivo growth in a host.
N-Linked Protein Glycosylation System of C. jejuni
N-linked protein glycosylation results in attachment of a saccharide to the amide nitrogen of an asparagine residue. This type of glycosylation is rare in bacteria, with only C. jejuni and H. influenzae having demonstrated this activity (224, 371, 588, 665). The C. jejuni N-linked glycosylation system is complex, containing multiple biosynthetic enzymes for modification of many proteins, whereas the H. influenzae system is simpler, involving modification of a single protein with hexose subunits (224, 371, 372, 450, 665). Other epsilonproteobacteria, including a few Helicobacter species other than H. pylori, and a few deltaproteobacteria appear to contain genes that could function in N-linked protein glycosylation, but experimental confirmation of this activity is lacking (28, 287, 587). Because of the excellent progress in elucidating the N-linked protein glycosylation system of C. jejuni, this bacterium has become a model system for analysis of this type of glycobiology and glycoengineering.
Biosynthesis of N-linked glycan.
Evidence for a general protein glycosylation system in C. jejuni was first acquired when mutations in the 12-gene pgl locus diminished reactivity of proteins to typing antisera and GalNac-specific lectins such as soybean agglutinin (SBA) (371, 588). Using SBA affinity chromatography, more than 22 proteins were isolated and shown to have a glycan structure attached (371, 665). Nuclear magnetic resonance (NMR) analysis of one of these proteins revealed that the glycan structure is a heptasaccharide (665). This heptasaccharide consists of a linear chain of five GalNac residues (with one having a glucose attachment) connected to β-d-bacillosamine (2,4-diacetamido-2,4,6-trideoxy-β-d-glucopyranose) (Fig. 11), which is consistent with many studies identifying lectins specific for GalNac residues reacting with C. jejuni proteins in a pgl-dependent manner (371, 372, 588, 610, 665). Unlike the variability seen with the O-linked glycan attached to flagellins of Campylobacter spp., the N-linked glycan demonstrates very little variability (665).
FIG. 11.
N-linked protein glycosylation system of C. jejuni. The N-linked protein glycosylation system of C. jejuni begins with UDP-GlcNAc to result in a heptasaccharide that is linked to a lipid carrier on the cytoplasmic face of the inner membrane (IM). The heptasaccharide is flipped to the periplasm by PglK and is removed from the carrier by PglB to result in fOS or linkage to a protein. Glc, glucose; GlcNAc, N-acetylglucosamine; Bac, bacillosamine; GalNAc, N-acetylgalactosamine.
A conserved sequence for glycosylation in C. jejuni has been identified and consists of D/E-X-N-X-S/T (where N is the modified asparagine and X can be any amino acid except for proline) (348, 450, 665). This consensus sequence is nearly identical to the sequences of eukaryotic proteins modified by respective N-linked protein glycosylation systems. However, the requirement for aspartic acid or glutamic acid at the −2 position in the C. jejuni sequence is not favored in eukaryotic systems. Mutational analysis of C. jejuni proteins revealed that the consensus sequence is necessary but not always sufficient for glycosylation (347, 348). The lack of glycosylation at asparagine residues in some consensus sequence within C. jejuni proteins suggests that the local structure or flexibility of a protein may influence N-linked glycosylation of a specific site.
The pgl locus contains 12 genes that encode enzymes required for biosynthesis and linkage of the heptasaccharide to proteins in the periplasmic space (Fig. 11) (372, 588, 615). Formation of the glycan is a complex process whose chemistry has been detailed in various works and is summarized briefly below (207-209, 320, 372, 463). Biosynthesis of bacillosamine is a multistep process mediated by PglF, PglE, and PglD (Fig. 11). This saccharide is attached to undecaprenyl pyrophosphate (Und-PP) by PglC on the cytoplasmic face of the inner membrane. The glycosyltransferases PglA, PglJ, and PglH perform the sequential addition of five GalNac residues to the Und-PP-linked bacillosamine. PglA and PglJ transfer the first two GalNac residues, with PglH transferring the three terminal GalNac residues (Fig. 11). PglI transfers a glucose to one of the GalNac residues to complete the heptasaccharide. After biosynthesis of the glycan, the PglK flippase transports the lipid-linked heptasaccharide to the periplasm, where the PglB oligosaccharide transferase can attach the glycan to accepting asparagine residues of target proteins (Fig. 11).
Due to the typical simplicity of bacterial systems relative to those of eukaryotes, the C. jejuni Pgl system has become a useful model for studying aspects of glycobiology specific for N-linked protein glycosylation systems. Increasing the feasibility of analyzing the C. jejuni system is the demonstration that the entire pgl operon can be coexpressed with a natural target in E. coli to result in proper N-linked glycosylation of the protein (615). By using an E. coli-based system, future research may allow for greater advances in glycoengineering of proteins and tailoring of modifications for various biological or medical purposes.
Biological role of N-linked glycosylation in C. jejuni.
The biological function of the N-linked glycosylation system in C. jejuni has not been elucidated fully. As analyzed by in vitro and in vivo model systems, C. jejuni pgl mutants have demonstrated defects in host interactions. Mutation of pglE, pglF, pglH, pglB, pglD, or pglK causes significant reductions in commensal colonization of the chick intestinal tract (254, 313, 320). In addition, a complete Pgl system is required for wild-type levels of in vitro adherence to and invasion of human intestinal epithelial cells (313, 585). These studies suggest that N-linked protein glycosylation is required to mediate interactions with both human and avian hosts. However, it is not known specifically why this system is required for associations with hosts.
So far, over 30 C. jejuni N-linked periplasmic or outer membrane proteins have been identified (348, 665). It has been proposed that the glycan modifications may be required for the biological activity of the proteins. However, two studies have provided contradictory evidence for this hypothesis. Larsen et al. discovered that VirB10 (also known as ComB10), encoded by the pVir plasmid in some strains of C. jejuni, is glycosylated by the N-linked glycosylation system (361). virB10, along with other genes, encodes proteins with homology to components of T4SSs (31). VirB10 was found to be glycosylated on two different asparagine residues, and glycosylation of both sites is required for optimal stability of the protein (361). However, only one specific asparagine is required to be glycosylated for VirB10-dependent DNA transformation. Contrarily, Kakuda and DiRita discovered that the periplasmic protein Cj1496 is glycosylated at two asparagines by the Pgl system (304). Whereas Cj1496 is required for optimal levels of invasion of human intestinal epithelial cells and commensal colonization of the chick intestinal tract, mutation of the two glycosylation sites did not affect stability of the protein or cause a defect in invasion or colonization (304). Combined, these studies suggest that only a subset of glycosylated proteins may depend upon modification for stability or biological activity.
A recent study suggested that a major biological activity of the Pgl system may be to produce a free, unlinked periplasmic form of the heptasaccharide (452). By using semiquantitative mass spectrometry, Nothaft et al. discovered that approximately 90% of the glycan was found in a free oligosaccharide (fOS) form in the periplasm, and only 10% of the glycan was found to be N-linked to protein (452). Like that of the N-linked glycan, production of the fOS is dependent on many Pgl enzymes, including PglB, which suggests that PglB may promote fOS release from the lipid carrier in the absence of a protein acceptor (Fig. 11). Examination of various growth conditions revealed that high osmolarity due to increased NaCl concentrations reduced fOS production (452). Furthermore, mutation of pglB resulted in a significant decrease in the growth rate at high NaCl concentrations compared to that of the wild-type strain. Curiously, PglB-dependent fOS production was found to be reduced at high NaCl concentrations, but PglB-dependent N-linked glycosylation was not (452). These findings suggest that one function of the Pgl system is to produce periplasmic fOS to protect against fluctuations in different osmotic environments.
Initial evidence also suggests that the general protein glycosylation system may be important for influencing immune responses in a host (610). Macrophage galactose-type lectin (MGL), which is found on immature dendritic cells and subsets of macrophages, is able to bind N-linked proteins of C. jejuni by recognizing terminal GalNac residues (610). A C. jejuni pglA mutant that lacks N-linked glycan stimulates twice the amount of interleukin-6 (IL-6) production from human dendritic cells as that induced by wild-type bacteria, which suggests that the general protein glycosylation system may be important for dampening immune responses in the human host. Considering that the Pgl system can generate fOS (452), it is unknown if N-linked glycan or fOS may interact with MGL to alter immune responses.
Summary
The epsilonproteobacteria contain many fascinating biological systems. Among these systems are the O- and N-linked protein glycosylation systems, which function in the modification of proteins important for multiple physiological, colonization, and virulence properties. Not only are these systems important for host interactions, but the glycan structures also impact organelle development and, likely, bacterial cell structure. Furthermore, with the relative simplicity of bacterial systems compared to eukaryotic systems, along with great technological improvements in the analysis of carbohydrate synthesis, both Campylobacter and Helicobacter spp. have the opportunity to greatly impact the field of glycobiology by serving as model systems for understanding biosynthesis of these glycans.
CONCLUSIONS AND FUTURE DIRECTIONS
C. jejuni and H. pylori are medically important members of the physiologically diverse epsilonproteobacteria. Because this group of microorganisms is one of the most understudied, the types of biological systems employed by H. pylori and C. jejuni are often compared to those of enteric bacteria such as E. coli and Salmonella spp. While comparisons to these model pathogens can be useful and informative, the biological strategies employed by these prototypes do not always accurately represent those of more evolutionarily distant species such as H. pylori and C. jejuni. As this review illustrates, these two epsilonproteobacteria use highly distinct mechanisms to accomplish conserved processes compared to many model bacterial pathogens.
While flagellar systems in many motile bacterial species share common components and overall structure, a key aspect of flagellar motility that differs in C. jejuni and H. pylori is the use of two different σ factors (σ54 and σ28) to coordinate flagellar gene expression (9, 80, 229, 232, 255, 368, 447, 455, 464, 473, 534, 571, 581). Furthermore, unlike other bacteria that encode σ54, nearly all σ54-dependent genes in C. jejuni and H. pylori encode flagellar proteins (80, 217, 447). Activation of σ54-dependent gene expression relies on a complete T3SS to initiate a signal to activate the FlgSR two-component system (255, 300). However, how the T3SS may influence the FlgSR signaling system to activate σ54-dependent gene expression is unknown. The molecular mechanism and specific signals required for activation of this unique signaling pathway certainly merit further study.
Another common biological mechanism found in C. jejuni and H. pylori that differs from those in Salmonella spp. and E. coli is the spatial and numerical control of flagellar biosynthesis. While it is clear that flagellar components such as the FlhF GTPase influence these processes, the precise mechanism by which FlhF and possibly other factors regulate numerical and spatial organization of flagella is unknown. Additionally, H. pylori produces a sheathed flagellum in a process that not only diverges from that in the model enteric bacteria but also is different from that in C. jejuni (200, 216). How the sheath is formed and the role that it plays in H. pylori motility remain further interesting yet unanswered questions.
Both C. jejuni and H. pylori display enormous amounts of genetic diversity (171, 580, 643). Furthermore, maintaining inter- and intrastrain diversity has been shown to provide a fitness advantage in vivo (43, 125, 279, 307, 643). Because of this fact, the ability to acquire new genes or alleles is of the utmost importance. As naturally competent species, C. jejuni and H. pylori are able to acquire and maintain exogenous DNA from the environment (278, 444, 629). Generally speaking, this process can be broken into two distinct fundamental processes: DNA uptake/transport and, in the case of nonplasmid DNA, homologous recombination.
The natural transformation systems of C. jejuni and H. pylori are relatively similar to one another but are quite distinct from those of other bacteria. C. jejuni employs both T2SS- and T4SS-based DNA uptake systems, a quality so far demonstrated only for this species (31, 641). However, only a subset of the typically necessary components of these two systems has been identified. Additional studies are needed to complete our understanding of how these DNA transformation systems operate and to determine the extent to which these systems contribute to genetic diversity in C. jejuni. Likewise, T4SS-mediated DNA transformation in H. pylori is unique. While nearly all of the constituents of the classical T4SS have been identified, there are still many uncharacterized aspects of natural transformation in H. pylori. For instance, the precise role of non-ComB factors in the natural transformation process needs further study. Another paradigm-shifting facet of natural transformation in H. pylori is the upregulation of competence genes in response to DNA damage (139). In the proposed model, the DNA damage response signal is somehow detected and amplified by RecA and subsequently results in lysis of neighboring bacteria. This type of behavior in response to DNA damage has not been described for any other bacterial species to date. Certainly, a clearer picture of how this process is regulated will provide considerable insight into how H. pylori maintains a high level of genetic diversity.
Once inside the bacterial cell, exogenous DNA must be processed further and integrated into the recipient genome. Our knowledge about the mechanisms and components required for recombination in C. jejuni and H. pylori is somewhat lacking. What is clear is that these pathogens utilize a combination of systems found in both Gram-negative (RecOR and RecA) and Gram-positive (AddAB) bacteria. Furthermore, these systems do not show overlap in function like those of E. coli. These findings perhaps highlight the fact that these two epsilonproteobacteria are not as evolutionarily related to model microorganisms as once believed.
Iron is a critical factor for most organisms. As such, the iron uptake systems encoded by C. jejuni and H. pylori have evolved to ensure that these bacteria are able to acquire iron from the most prevalent sources within their respective environments. While much information concerning potential iron sources and iron uptake systems has been discovered for these pathogens, there are many intriguing questions remaining. For example, while both C. jejuni and H. pylori encode ferrous iron uptake systems (Feo), neither of these systems contains the accessory factors used by E. coli (498, 599). Furthermore, C. jejuni feoB mutants are still able to take up ferrous iron. Therefore, there are likely additional unidentified factors involved in ferrous iron uptake. Additionally, the C. jejuni membrane receptor CtuA shows multisubstrate specificity, which appears unique among iron uptake systems (419). Future studies are needed to identify the molecular determinants that facilitate this type of specificity.
Similar to most bacteria, C. jejuni and H. pylori control the expression of iron uptake and storage systems by using the ferric uptake regulator, Fur, However, there are several unique aspects of Fur regulation in these pathogens that remain poorly understood. H. pylori Fur has evolved the ability to regulate gene expression in both the iron-bound and apo forms (130, 161). Indirect evidence for apo-Fur regulation also exists for C. jejuni, although this type of regulation has yet to be demonstrated conclusively (265). Furthermore, although iron-bound Fur regulation is fairly well understood, a highly conserved consensus Fe2+-Fur recognition sequence has not yet been identified in either organism. In addition, the apo-Fur binding sequence in H. pylori has not been defined. The lack of a clear consensus apo-Fur binding site perhaps suggests that there are conformational differences in the DNA binding regions of Fe2+-Fur and apo-Fur. In addition, it is also currently unclear whether apo-Fur functions as an oligomer (similar to Fe2+-Fur) or whether an alternative form of the protein is required for this type of regulation. Moreover, direct Fe2+-Fur-mediated activation has been demonstrated definitively for only one gene (nifS) in H. pylori, and the possible mechanisms underlying activation have not been determined (6). Given the distal location of the Fur binding sites in H. pylori Fe2+-Fur-activated promoters (6, 197), it is not intuitively clear whether or not Fur would be able to interact directly with RNAP or whether another factor is involved in facilitating activation.
The recent publication of the H. pylori primary transcriptome highlights a possible role for regulatory RNAs in the iron-responsive regulatory networks of this organism. Sequence homology suggests that at least a few of these sRNAs may also influence expression of known or suspected Fur-regulated genes (534). However, further studies are needed to confirm these predictions as well as to answer many other questions. For instance, although expression of many of these putative sRNAs has been confirmed, a role for any of these sRNAs has not yet been determined. Additionally, many bacterial sRNAs require the RNA chaperone Hfq to regulate gene expression; however, an Hfq homologue has not been identified in H. pylori. While this type of analysis has not yet been performed with C. jejuni, the genomic similarities of these epsilonproteobacteria may allow researchers to determine whether similar sRNAs are present in C. jejuni. Certainly, many interesting questions remain to be answered concerning the unique mechanisms by which the epsilonproteobacteria acquire, store, and utilize iron.
A critical step in bacterial pathogenesis is the ability to efficiently colonize the host. Both C. jejuni and H. pylori have evolved a repertoire of virulence and colonization factors that facilitate host-pathogen interactions. A major factor in the ability of C. jejuni to initiate and maintain infection of humans is invasion of intestinal epithelial cells. Invasion is mediated partially by Cia effector proteins, which are secreted through the flagellar T3SS (94, 339). Curiously, the involvement of Cia proteins in invasion varies among C. jejuni strains and may be contingent upon the host cell type (454). Thus, there are likely other invasion factors that have not yet been identified. Another intriguing but poorly understood aspect of C. jejuni intracellular survival is precisely how the bacterium hijacks the microtubule network for invasion and exploits the host cell to induce CCV formation. In contrast to C. jejuni, H. pylori is not generally considered an invasive pathogen per se. However, some studies have reported invasion and intracellular survival during H. pylori infection (13, 98, 165, 358, 579). Since invasiveness does not seem to be the predominant lifestyle of H. pylori, future studies are required to better understand the biological significance of intracellular survival and replication during H. pylori infection, as well as the mechanisms used in these processes.
The protein glycosylation systems employed by C. jejuni and H. pylori are key elements in the biology of these microorganisms. The modification of flagellins with O-linked sugars is essential for flagellar biosynthesis. A leading hypothesis proposes that the O-linked glycans facilitate flagellar filament polymerization, but it is not understood how this might occur. In addition, the flagellin glycosylation loci and corresponding modifications are some of the most diverse elements of Campylobacter species. Analysis of a collection of diverse Campylobacter and Helicobacter species may illuminate additional levels of complexity in these systems.
N-linked protein glycosylation systems such as the one found in C. jejuni are rare in bacteria but common in eukaryotes. As such, C. jejuni is a good model system for understanding this biological process in both bacterial and eukaryotic systems. Because the C. jejuni N-linked glycosylation system is functional upon expression in E. coli, further analyses of the mechanisms and components involved in this pathway will likely pave the way for engineering recombinant glycoproteins; this type of glycoprotein engineering could have significant implications for biomedical sciences, such as for diagnostic or therapeutic purposes.
Although there are many understudied areas of C. jejuni and H. pylori biology, it is clear that these epsilonproteobacteria employ many unique and interesting biological systems. Further study of these systems will undoubtedly provide valuable insight into the biology of these fascinating microorganisms.
Acknowledgments
Research in the laboratory of David R. Hendrixson is supported by NIH grant R01 AI065539 and by National Research Initiative grants 2006-35201-17382 and 2009-35201-05039 from the USDA Cooperative State Research, Education, and Extension Service Food Safety Program. Research in the laboratory of D. Scott Merrell is made possible by grant AI065529 from the NIAID, grant R073PW from USUHS, and grant 300411-7.20-60393 from USMCI. Jeremy J. Gilbreath is supported by the Robert D. Watkins Graduate Research Fellowship from the American Society for Microbiology.
The contents of the manuscript are the sole responsibility of the authors and do not necessarily represent the official views of the NIH or the DOD.
Biography
 Jeremy J. Gilbreath was born in Kodiak, AK, and moved to rural Heber Springs, AR, where he was raised. Despite the close proximity of towns in rural Arkansas, he managed to live there for over 20 years without ever meeting his current Ph.D. mentor. After serving in the U.S. Navy, he obtained a B.S. in Clinical Laboratory Science from Idaho State University. True to his Alaskan heritage, while attending college in Idaho, Jeremy developed a passion for fly-fishing and spending time in the rugged mountain country. He is currently pursuing his Ph.D. in Emerging Infectious Diseases at the Uniformed Services University in Bethesda, MD, and was recently named an ASM Robert D. Watkins Graduate Research Fellow. Jeremy's interest in bacterial physiology and protein biochemistry is reflected in his research on iron-responsive gene regulation and the structure-function relationship between the iron-bound and apo forms of the Fur protein of Helicobacter pylori.
Jeremy J. Gilbreath was born in Kodiak, AK, and moved to rural Heber Springs, AR, where he was raised. Despite the close proximity of towns in rural Arkansas, he managed to live there for over 20 years without ever meeting his current Ph.D. mentor. After serving in the U.S. Navy, he obtained a B.S. in Clinical Laboratory Science from Idaho State University. True to his Alaskan heritage, while attending college in Idaho, Jeremy developed a passion for fly-fishing and spending time in the rugged mountain country. He is currently pursuing his Ph.D. in Emerging Infectious Diseases at the Uniformed Services University in Bethesda, MD, and was recently named an ASM Robert D. Watkins Graduate Research Fellow. Jeremy's interest in bacterial physiology and protein biochemistry is reflected in his research on iron-responsive gene regulation and the structure-function relationship between the iron-bound and apo forms of the Fur protein of Helicobacter pylori.
 William L. Cody is an adjunct faculty member at Richland College. His research interests include bacterial pathogenesis and the transcriptional activation of virulence factors. He received his B.S. at Xavier University of Louisiana and his Ph.D. in Microbiology from the University of Colorado, Denver. As a graduate student, he studied the AlgZR two-component signal transduction system and the regulation of hydrogen cyanide production in Pseudomonas aeruginosa. As a postdoctoral research scientist at the University of Texas Southwestern Medical Center at Dallas, he worked on research projects that included the characterization of small RNA-binding proteins and the analysis of capsule production in Campylobacter jejuni.
William L. Cody is an adjunct faculty member at Richland College. His research interests include bacterial pathogenesis and the transcriptional activation of virulence factors. He received his B.S. at Xavier University of Louisiana and his Ph.D. in Microbiology from the University of Colorado, Denver. As a graduate student, he studied the AlgZR two-component signal transduction system and the regulation of hydrogen cyanide production in Pseudomonas aeruginosa. As a postdoctoral research scientist at the University of Texas Southwestern Medical Center at Dallas, he worked on research projects that included the characterization of small RNA-binding proteins and the analysis of capsule production in Campylobacter jejuni.
 D. Scott Merrell was born and raised in rural Bald Knob, AR. Scotty left the south to complete his Ph.D. studies at Tufts Medical School and to conduct postdoctoral training at Stanford University. He is currently an Associate Professor in the Department of Microbiology and Immunology at the Uniformed Services University in Bethesda, MD. Intrigued by the stress response and how bacteria manage to colonize inhospitable environments, Scotty's research group studies colonization and virulence factors of the gastric pathogen Helicobacter pylori. Scotty is a former Applied Genomics of Infectious Diseases ID Training Fellow and Damon Runyon-Walter Winchell Cancer Fund Postdoctoral Fellow and received a Merck Irving S. Sigal Memorial Award for excellence in basic research in medical microbiology and infectious diseases. Despite his many years away from Arkansas, Scotty still confesses a secret passion for Southern-fried catfish, hush puppies, pickled green tomatoes, and the use of “y'all” in polite conversation.
D. Scott Merrell was born and raised in rural Bald Knob, AR. Scotty left the south to complete his Ph.D. studies at Tufts Medical School and to conduct postdoctoral training at Stanford University. He is currently an Associate Professor in the Department of Microbiology and Immunology at the Uniformed Services University in Bethesda, MD. Intrigued by the stress response and how bacteria manage to colonize inhospitable environments, Scotty's research group studies colonization and virulence factors of the gastric pathogen Helicobacter pylori. Scotty is a former Applied Genomics of Infectious Diseases ID Training Fellow and Damon Runyon-Walter Winchell Cancer Fund Postdoctoral Fellow and received a Merck Irving S. Sigal Memorial Award for excellence in basic research in medical microbiology and infectious diseases. Despite his many years away from Arkansas, Scotty still confesses a secret passion for Southern-fried catfish, hush puppies, pickled green tomatoes, and the use of “y'all” in polite conversation.
 David R. Hendrixson is an Associate Professor of Microbiology at the University of Texas Southwestern Medical Center at Dallas. After earning a B.S. in Microbiology at the University of Arkansas, he followed his interest in the biology of bacterial organisms by studying the adherence mechanisms of Haemophilus influenzae under the mentorship of Joseph W. St. Geme at Washington University in St. Louis, MO. After earning a Ph.D. in Molecular Microbiology and Microbial Pathogenesis, he received further training in bacterial genetics during his postdoctoral fellowship with Victor J. DiRita in the Department of Microbiology and Immunology at the University of Michigan Medical School. During this time, he developed many genetic tools for Campylobacter jejuni and employed various methodologies to uncover a variety of interesting genetic regulatory mechanisms and colonization factors of the bacterium. These pursuits continue in his own research program at the University of Texas Southwestern Medical Center.
David R. Hendrixson is an Associate Professor of Microbiology at the University of Texas Southwestern Medical Center at Dallas. After earning a B.S. in Microbiology at the University of Arkansas, he followed his interest in the biology of bacterial organisms by studying the adherence mechanisms of Haemophilus influenzae under the mentorship of Joseph W. St. Geme at Washington University in St. Louis, MO. After earning a Ph.D. in Molecular Microbiology and Microbial Pathogenesis, he received further training in bacterial genetics during his postdoctoral fellowship with Victor J. DiRita in the Department of Microbiology and Immunology at the University of Michigan Medical School. During this time, he developed many genetic tools for Campylobacter jejuni and employed various methodologies to uncover a variety of interesting genetic regulatory mechanisms and colonization factors of the bacterium. These pursuits continue in his own research program at the University of Texas Southwestern Medical Center.
REFERENCES
- 1.Aas, F. E., et al. 2002. Competence for natural transformation in Neisseria gonorrhoeae: components of DNA binding and uptake linked to type IV pilus expression. Mol. Microbiol. 46:749-760. [DOI] [PubMed] [Google Scholar]
- 2.Abdul-Tehrani, H., et al. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aberg, A., J. Fernandez-Vazquez, J. D. Cabrer-Panes, A. Sanchez, and C. Balsalobre. 2009. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J. Bacteriol. 191:3226-3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abrahams, G. L., P. Muller, and M. Hensel. 2006. Functional dissection of SseF, a type III effector protein involved in positioning the Salmonella-containing vacuole. Traffic 7:950-965. [DOI] [PubMed] [Google Scholar]
- 5.Abu-Qarn, M., J. Eichler, and N. Sharon. 2008. Not just for Eukarya anymore: protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 18:544-550. [DOI] [PubMed] [Google Scholar]
- 6.Alamuri, P., N. Mehta, A. Burk, and R. J. Maier. 2006. Regulation of the Helicobacter pylori Fe-S cluster synthesis protein NifS by iron, oxidative stress conditions, and fur. J. Bacteriol. 188:5325-5330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allan, E., N. Dorrell, S. Foynes, M. Anyim, and B. W. Wren. 2000. Mutational analysis of genes encoding the early flagellar components of Helicobacter pylori: evidence for transcriptional regulation of flagellin A biosynthesis. J. Bacteriol. 182:5274-5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alm, R. A., P. Guerry, M. E. Power, and T. J. Trust. 1992. Variation in antigenicity and molecular weight of Campylobacter coli VC167 flagellin in different genetic backgrounds. J. Bacteriol. 174:4230-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alm, R. A., et al. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 10.Alonso, A., and F. Garcia-del Portillo. 2004. Hijacking of eukaryotic functions by intracellular bacterial pathogens. Int. Microbiol. 7:181-191. [PubMed] [Google Scholar]
- 11.Altekruse, S. F., N. J. Stern, P. I. Fields, and D. L. Swerdlow. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Althouse, C., S. Patterson, P. Fedorka-Cray, and R. E. Isaacson. 2003. Type 1 fimbriae of Salmonella enterica serovar Typhimurium bind to enterocytes and contribute to colonization of swine in vivo. Infect. Immun. 71:6446-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Amieva, M. R., N. R. Salama, L. S. Tompkins, and S. Falkow. 2002. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell. Microbiol. 4:677-690. [DOI] [PubMed] [Google Scholar]
- 14.Amieva, M. R., et al. 2003. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300:1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amundsen, S. K., et al. 2008. Helicobacter pylori AddAB helicase-nuclease and RecA promote recombination-related DNA repair and survival during stomach colonization. Mol. Microbiol. 69:994-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amundsen, S. K., J. Fero, N. R. Salama, and G. R. Smith. 2009. Dual nuclease and helicase activities of Helicobacter pylori AddAB are required for DNA repair, recombination, and mouse infectivity. J. Biol. Chem. 284:16759-16766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andermann, T. M., Y. T. Chen, and K. M. Ottemann. 2002. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect. Immun. 70:5877-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen-Nissen, E., et al. 2005. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. U. S. A. 102:9247-9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the DprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews, N. C., and P. J. Schmidt. 2007. Iron homeostasis. Annu. Rev. Physiol. 69:69-85. [DOI] [PubMed] [Google Scholar]
- 21.Andrews, S. C., P. M. Harrison, and J. R. Guest. 1989. Cloning, sequencing, and mapping of the bacterioferritin gene (bfr) of Escherichia coli K-12. J. Bacteriol. 171:3940-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 23.Angerer, A., S. Enz, M. Ochs, and V. Braun. 1995. Transcriptional regulation of ferric citrate transport in Escherichia coli K-12. Fecl belongs to a new subfamily of sigma 70-type factors that respond to extracytoplasmic stimuli. Mol. Microbiol. 18:163-174. [DOI] [PubMed] [Google Scholar]
- 24.Aravind, L., K. S. Makarova, and E. V. Koonin. 2000. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 28:3417-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Argaman, L., et al. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 26.Ashgar, S. S., et al. 2007. CapA, an autotransporter protein of Campylobacter jejuni, mediates association with human epithelial cells and colonization of the chicken gut. J. Bacteriol. 189:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aukema, K. G., E. M. Kron, T. J. Herdendorf, and K. T. Forest. 2005. Functional dissection of a conserved motif within the pilus retraction protein PilT. J. Bacteriol. 187:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baar, C., et al. 2003. Complete genome sequence and analysis of Wolinella succinogenes. Proc. Natl. Acad. Sci. U. S. A. 100:11690-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachtiar, B. M., P. J. Coloe, and B. N. Fry. 2007. Knockout mutagenesis of the kpsE gene of Campylobacter jejuni 81116 and its involvement in bacterium-host interactions. FEMS Immunol. Med. Microbiol. 49:149-154. [DOI] [PubMed] [Google Scholar]
- 30.Backert, S., et al. 2000. Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell. Microbiol. 2:155-164. [DOI] [PubMed] [Google Scholar]
- 31.Bacon, D. J., et al. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bacon, D. J., et al. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-777. [DOI] [PubMed] [Google Scholar]
- 33.Badea, L., et al. 2003. Contribution of Efa1/LifA to the adherence of enteropathogenic Escherichia coli to epithelial cells. Microb. Pathog. 34:205-215. [DOI] [PubMed] [Google Scholar]
- 34.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 35.Bagnoli, F., L. Buti, L. Tompkins, A. Covacci, and M. R. Amieva. 2005. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc. Natl. Acad. Sci. U. S. A. 102:16339-16344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baichoo, N., and J. D. Helmann. 2002. Recognition of DNA by Fur: a reinterpretation of the Fur box consensus sequence. J. Bacteriol. 184:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baig, B. H., I. K. Wachsmuth, and G. K. Morris. 1986. Utilization of exogenous siderophores by Campylobacter species. J. Clin. Microbiol. 23:431-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakowski, M. A., et al. 2010. The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe 7:453-462. [DOI] [PubMed] [Google Scholar]
- 39.Bakshi, C. S., et al. 2000. Identification of SopE2, a Salmonella secreted protein which is highly homologous to SopE and involved in bacterial invasion of epithelial cells. J. Bacteriol. 182:2341-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balaban, M., S. N. Joslin, and D. R. Hendrixson. 2009. FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J. Bacteriol. 191:6602-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bally, M., et al. 1992. Protein secretion in Pseudomonas aeruginosa: characterization of seven xcp genes and processing of secretory apparatus components by prepilin peptidase. Mol. Microbiol. 6:1121-1131. [DOI] [PubMed] [Google Scholar]
- 42.Baltrus, D. A., and K. Guillemin. 2006. Multiple phases of competence occur during the Helicobacter pylori growth cycle. FEMS Microbiol. Lett. 255:148-155. [DOI] [PubMed] [Google Scholar]
- 43.Baltrus, D. A., K. Guillemin, and P. C. Phillips. 2008. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evolution 62:39-49. [DOI] [PubMed] [Google Scholar]
- 44.Banerjee, A., and S. K. Ghosh. 2003. The role of pilin glycan in neisserial pathogenesis. Mol. Cell. Biochem. 253:179-190. [DOI] [PubMed] [Google Scholar]
- 45.Battistuzzi, F. U., A. Feijao, and S. B. Hedges. 2004. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evol. Biol. 4:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baumler, A. J., and F. Heffron. 1995. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J. Bacteriol. 177:2087-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baumler, A. J., et al. 1996. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect. Immun. 64:61-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baumler, A. J., R. M. Tsolis, and F. Heffron. 1996. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect. Immun. 64:1862-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumler, A. J., R. M. Tsolis, and F. Heffron. 1996. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc. Natl. Acad. Sci. U. S. A. 93:279-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bender, K. S., et al. 2007. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 73:5389-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539-1546. [DOI] [PubMed] [Google Scholar]
- 52.Benz, I., and M. A. Schmidt. 2001. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol. Microbiol. 40:1403-1413. [DOI] [PubMed] [Google Scholar]
- 53.Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45:267-276. [DOI] [PubMed] [Google Scholar]
- 54.Bereswill, S., et al. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bereswill, S., et al. 1999. The ferric uptake regulator (Fur) homologue of Helicobacter pylori: functional analysis of the coding gene and controlled production of the recombinant protein in Escherichia coli. Med. Microbiol. Immunol. 188:31-40. [DOI] [PubMed] [Google Scholar]
- 56.Bereswill, S., F. Lichte, T. Vey, F. Fassbinder, and M. Kist. 1998. Cloning and characterization of the fur gene from Helicobacter pylori. FEMS Microbiol. Lett. 159:193-200. [DOI] [PubMed] [Google Scholar]
- 57.Biswas, G. D., S. A. Lacks, and P. F. Sparling. 1989. Transformation-deficient mutants of piliated Neisseria gonorrhoeae. J. Bacteriol. 171:657-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biswas, G. D., T. Sox, E. Blackman, and P. F. Sparling. 1977. Factors affecting genetic transformation of Neisseria gonorrhoeae. J. Bacteriol. 129:983-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biswas, G. D., S. A. Thompson, and P. F. Sparling. 1989. Gene transfer in Neisseria gonorrhoeae. Clin. Microbiol. Rev. 2(Suppl.):S24-S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Black, R. E., M. M. Levine, M. L. Clements, T. P. Hughes, and M. J. Blaser. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157:472-479. [DOI] [PubMed] [Google Scholar]
- 61.Blanar, M. A., S. J. Sandler, M. E. Armengod, L. W. Ream, and A. J. Clark. 1984. Molecular analysis of the recF gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 81:4622-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blaser, M. J. 1998. Helicobacter pylori and gastric diseases. BMJ 316:1507-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blaser, M. J., et al. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 64.Boren, T., P. Falk, K. A. Roth, G. Larson, and S. Normark. 1993. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262:1892-1895. [DOI] [PubMed] [Google Scholar]
- 65.Bou-Abdallah, F., A. C. Lewin, N. E. Le Brun, G. R. Moore, and N. D. Chasteen. 2002. Iron detoxification properties of Escherichia coli bacterioferritin. Attenuation of oxyradical chemistry. J. Biol. Chem. 277:37064-37069. [DOI] [PubMed] [Google Scholar]
- 66.Boyle-Vavra, S., and H. S. Seifert. 1996. Uptake-sequence-independent DNA transformation exists in Neisseria gonorrhoeae. Microbiology 142:2839-2845. [DOI] [PubMed] [Google Scholar]
- 67.Bracken, C. S., M. T. Baer, A. Abdur-Rashid, W. Helms, and I. Stojiljkovic. 1999. Use of heme-protein complexes by the Yersinia enterocolitica HemR receptor: histidine residues are essential for receptor function. J. Bacteriol. 181:6063-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brahmachary, P., M. G. Dashti, J. W. Olson, and T. R. Hoover. 2004. Helicobacter pylori FlgR is an enhancer-independent activator of σ54-RNA polymerase holoenzyme. J. Bacteriol. 186:4535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Braun, V., M. Braun, and H. Killmann. 2004. Ferrichrome- and citrate-mediated iron transport, p. 158-177. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 70.Braun, V., S. Mahren, and M. Oglerman. 2003. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr. Opin. Microbiol. 6:173-180. [DOI] [PubMed] [Google Scholar]
- 71.Brawn, L. C., R. D. Hayward, and V. Koronakis. 2007. Salmonella SPI1 effector SipA persists after entry and cooperates with a SPI2 effector to regulate phagosome maturation and intracellular replication. Cell Host Microbe 1:63-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brickman, T. J., B. A. Ozenberger, and M. A. McIntosh. 1990. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J. Mol. Biol. 212:669-682. [DOI] [PubMed] [Google Scholar]
- 73.Caldwell, M. B., P. Guerry, E. C. Lee, J. P. Burans, and R. I. Walker. 1985. Reversible expression of flagella in Campylobacter jejuni. Infect. Immun. 50:941-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Campbell, B. J., A. S. Engel, M. L. Porter, and K. Takai. 2006. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4:458-468. [DOI] [PubMed] [Google Scholar]
- 75.Campellone, K. G., and J. M. Leong. 2003. Tails of two Tirs: actin pedestal formation by enteropathogenic E. coli and enterohemorrhagic E. coli O157:H7. Curr. Opin. Microbiol. 6:82-90. [DOI] [PubMed] [Google Scholar]
- 76.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 78.Carpenter, B. M., et al. 2009. A single nucleotide change affects fur-dependent regulation of sodB in H. pylori. PLoS One 4:e5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carpenter, B. M., J. M. Whitmire, and D. S. Merrell. 2009. This is not your mother's repressor: the complex role of fur in pathogenesis. Infect. Immun. 77:2590-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carrillo, C. D., et al. 2004. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J. Biol. Chem. 279:20327-20338. [DOI] [PubMed] [Google Scholar]
- 81.Carswell, C. L., M. D. Rigden, and J. E. Baenziger. 2008. Expression, purification, and structural characterization of CfrA, a putative iron transporter from Campylobacter jejuni. J. Bacteriol. 190:5650-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cassuto, E., S. C. West, J. Podell, and P. Howard-Flanders. 1981. Genetic recombination: recA protein promotes homologous pairing between duplex DNA molecules without strand unwinding. Nucleic Acids Res. 9:4201-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chadsey, M. S., and K. T. Hughes. 2001. A multipartite interaction between Salmonella transcription factor sigma28 and its anti-sigma factor FlgM: implications for sigma28 holoenzyme destabilization through stepwise binding. J. Mol. Biol. 306:915-929. [DOI] [PubMed] [Google Scholar]
- 85.Chadsey, M. S., J. E. Karlinsey, and K. T. Hughes. 1998. The flagellar anti-σ factor FlgM actively dissociates Salmonella typhimurium σ28 RNA polymerase holoenzyme. Genes Dev. 12:3123-3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chan, A. C., et al. 2010. Structure and function of P19, a high-affinity iron transporter of the human pathogen Campylobacter jejuni. J. Mol. Biol. 401:590-604. [DOI] [PubMed] [Google Scholar]
- 87.Chan, A. C., B. I. R. F. Lelj-Garolla, K. A. Pedersen, A. G. Mauk, and M. E. Murphy. 2006. Cofacial heme binding is linked to dimerization by a bacterial heme transport protein. J. Mol. Biol. 362:1108-1119. [DOI] [PubMed] [Google Scholar]
- 88.Chang, J., et al. 2007. SipC multimerization promotes actin nucleation and contributes to Salmonella-induced inflammation. Mol. Microbiol. 66:1548-1556. [DOI] [PubMed] [Google Scholar]
- 89.Charbonneau, M. E., et al. 2007. O-linked glycosylation ensures the normal conformation of the autotransporter adhesin involved in diffuse adherence. J. Bacteriol. 189:8880-8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen, I., and D. Dubnau. 2003. DNA transport during transformation. Front. Biosci. 8:s544-s556. [DOI] [PubMed] [Google Scholar]
- 91.Chen, I., and D. Dubnau. 2004. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2:241-249. [DOI] [PubMed] [Google Scholar]
- 92.Chevance, F. F., and K. T. Hughes. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chou, W. K., S. Dick, W. W. Wakarchuk, and M. E. Tanner. 2005. Identification and characterization of NeuB3 from Campylobacter jejuni as a pseudaminic acid synthase. J. Biol. Chem. 280:35922-35928. [DOI] [PubMed] [Google Scholar]
- 94.Christensen, J. E., S. A. Pacheco, and M. E. Konkel. 2009. Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol. Microbiol. 73:650-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Christie, P. J., and E. Cascales. 2005. Structural and dynamic properties of bacterial type IV secretion systems. Mol. Membr. Biol. 22:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chu, Y.-T., Y.-H. Wang, J.-J. Wu, and H.-Y. Lei. 2010. Invasion and multiplication of Helicobacter pylori in gastric epithelial cells and implications for antibiotic resistance. Infect. Immun. 78:4157-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cianciotto, N. P. 2005. Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 13:581-588. [DOI] [PubMed] [Google Scholar]
- 100.Clarke, M. B., and V. Sperandio. 2005. Transcriptional regulation of flhDC by QseBC and sigma (FliA) in enterohaemorrhagic Escherichia coli. Mol. Microbiol. 57:1734-1749. [DOI] [PubMed] [Google Scholar]
- 101.Clarke, S. C., R. D. Haigh, P. P. Freestone, and P. H. Williams. 2003. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin. Microbiol. Rev. 16:365-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Claverys, J. P., and L. S. Havarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798-d1814. [DOI] [PubMed] [Google Scholar]
- 103.Cleary, J., et al. 2004. Enteropathogenic Escherichia coli (EPEC) adhesion to intestinal epithelial cells: role of bundle-forming pili (BFP), EspA filaments and intimin. Microbiology 150:527-538. [DOI] [PubMed] [Google Scholar]
- 104.Clegg, S., and K. T. Hughes. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Coburn, B., I. Sekirov, and B. B. Finlay. 2007. Type III secretion systems and disease. Clin. Microbiol. Rev. 20:535-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Colland, F., et al. 2001. Identification of the Helicobacter pylori anti-σ28 factor. Mol. Microbiol. 41:477-487. [DOI] [PubMed] [Google Scholar]
- 107.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 108.Cornelissen, C. N., and P. F. Sparling. 1994. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol. Microbiol. 14:843-850. [DOI] [PubMed] [Google Scholar]
- 109.Correa, N. E., F. Peng, and K. E. Klose. 2005. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 187:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cover, T. L., and M. J. Blaser. 1992. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J. Biol. Chem. 267:10570-10575. [PubMed] [Google Scholar]
- 111.Coy, M., and J. B. Neilands. 1991. Structural dynamics and functional domains of the fur protein. Biochemistry 30:8201-8210. [DOI] [PubMed] [Google Scholar]
- 112.Craig, L., M. E. Pique, and J. A. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363-378. [DOI] [PubMed] [Google Scholar]
- 113.Creuzenet, C. 2004. Characterization of CJ1293, a new UDP-GlcNAc C6 dehydratase from Campylobacter jejuni. FEBS Lett. 559:136-140. [DOI] [PubMed] [Google Scholar]
- 114.Creuzenet, C., M. J. Schur, J. Li, W. W. Wakarchuk, and J. S. Lam. 2000. FlaA1, a new bifunctional UDP-GlcNAc C6 dehydratase/C4 reductase from Helicobacter pylori. J. Biol. Chem. 275:34873-34880. [DOI] [PubMed] [Google Scholar]
- 115.Crosa, J. H. 1989. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev. 53:517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cullen, T. W., and M. S. Trent. 2010. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U. S. A. 107:5160-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Danielli, A., et al. 2009. Growth phase and metal-dependent transcriptional regulation of the fecA genes in Helicobacter pylori. J. Bacteriol. 191:3717-3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Danielli, A., et al. 2006. In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis. J. Bacteriol. 188:4654-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Danner, D. B., R. A. Deich, K. L. Sisco, and H. O. Smith. 1980. An eleven-base-pair sequence determines the specificity of DNA uptake in Haemophilus transformation. Gene 11:311-318. [DOI] [PubMed] [Google Scholar]
- 120.Darwin, K. H., and V. L. Miller. 1999. Molecular basis of the interaction of Salmonella with the intestinal mucosa. Clin. Microbiol. Rev. 12:405-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dasgupta, N., S. K. Arora, and R. Ramphal. 2000. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 182:357-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dasgupta, N., and R. Ramphal. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J. Bacteriol. 183:6636-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Davidsen, T., et al. 2004. Biased distribution of DNA uptake sequences towards genome maintenance genes. Nucleic Acids Res. 32:1050-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Debellis, L., E. Papini, R. Caroppo, C. Montecucco, and S. Curci. 2001. Helicobacter pylori cytotoxin VacA increases alkaline secretion in gastric epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G1440-G1448. [DOI] [PubMed] [Google Scholar]
- 125.de Boer, P., et al. 2002. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 44:351-359. [DOI] [PubMed] [Google Scholar]
- 126.Deiwick, J., et al. 2006. The translocated Salmonella effector proteins SseF and SseG interact and are required to establish an intracellular replication niche. Infect. Immun. 74:6965-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.de Jonge, R., et al. 2004. Role of the Helicobacter pylori outer-membrane proteins AlpA and AlpB in colonization of the guinea pig stomach. J. Med. Microbiol. 53:375-379. [DOI] [PubMed] [Google Scholar]
- 128.Delany, I., et al. 2002. Autoregulation of Helicobacter pylori Fur revealed by functional analysis of the iron-binding site. Mol. Microbiol. 46:1107-1122. [DOI] [PubMed] [Google Scholar]
- 129.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2003. An anti-repression Fur operator upstream of the promoter is required for iron-mediated transcriptional autoregulation in Helicobacter pylori. Mol. Microbiol. 50:1329-1338. [DOI] [PubMed] [Google Scholar]
- 130.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297-1309. [DOI] [PubMed] [Google Scholar]
- 131.De Lorenzo, V., M. Herrero, F. Giovannini, and J. B. Neilands. 1988. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur. J. Biochem. 173:537-546. [DOI] [PubMed] [Google Scholar]
- 132.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.DeVinney, R., J. L. Puente, A. Gauthier, D. Goosney, and B. B. Finlay. 2001. Enterohaemorrhagic and enteropathogenic Escherichia coli use a different Tir-based mechanism for pedestal formation. Mol. Microbiol. 41:1445-1458. [DOI] [PubMed] [Google Scholar]
- 134.Dhaenens, L., F. Szczebara, S. Van Nieuwenhuyse, and M. O. Husson. 1999. Comparison of iron uptake in different Helicobacter species. Res. Microbiol. 150:475-481. [DOI] [PubMed] [Google Scholar]
- 135.Dillingham, M. S., and S. C. Kowalczykowski. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72:642-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dillingham, M. S., and S. C. Kowalczykowski. 2001. A step backward in advancing DNA replication: rescue of stalled replication forks by RecG. Mol. Cell 8:734-736. [DOI] [PubMed] [Google Scholar]
- 137.Doig, P., N. Kinsella, P. Guerry, and T. J. Trust. 1996. Characterization of a posttranslational modification of Campylobacter flagellin: identification of a sero-specific glycosyl moiety. Mol. Microbiol. 19:379-387. [DOI] [PubMed] [Google Scholar]
- 138.Donnenberg, M. S. 2000. Pathogenic strategies of enteric bacteria. Nature 406:768-774. [DOI] [PubMed] [Google Scholar]
- 139.Dorer, M. S., J. Fero, and N. R. Salama. 2010. DNA damage triggers genetic exchange in Helicobacter pylori. PLoS Pathog. 6:e1001026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dorsey, C. W., M. C. Laarakker, A. D. Humphries, E. H. Weening, and A. J. Baumler. 2005. Salmonella enterica serotype Typhimurium MisL is an intestinal colonization factor that binds fibronectin. Mol. Microbiol. 57:196-211. [DOI] [PubMed] [Google Scholar]
- 141.Drake, S. L., and M. Koomey. 1995. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol. Microbiol. 18:975-986. [DOI] [PubMed] [Google Scholar]
- 142.Drake, S. L., S. A. Sandstedt, and M. Koomey. 1997. PilP, a pilus biogenesis lipoprotein in Neisseria gonorrhoeae, affects expression of PilQ as a high-molecular-mass multimer. Mol. Microbiol. 23:657-668. [DOI] [PubMed] [Google Scholar]
- 143.Drummond, M. H., A. Contreras, and L. A. Mitchenall. 1990. The function of isolated domains and chimaeric proteins constructed from the transcriptional activators NifA and NtrC of Klebsiella pneumoniae. Mol. Microbiol. 4:29-37. [DOI] [PubMed] [Google Scholar]
- 144.Dubnau, D. 1999. DNA uptake in bacteria. Annu. Rev. Microbiol. 53:217-244. [DOI] [PubMed] [Google Scholar]
- 145.Dubnau, D., and R. Provvedi. 2000. Internalizing DNA. Res. Microbiol. 151:475-480. [DOI] [PubMed] [Google Scholar]
- 146.Duckett, D. R., et al. 1988. The structure of the Holliday junction, and its resolution. Cell 55:79-89. [DOI] [PubMed] [Google Scholar]
- 147.Duffin, P. M., and H. S. Seifert. 2010. DNA uptake sequence-mediated enhancement of transformation in Neisseria gonorrhoeae is strain dependent. J. Bacteriol. 192:4436-4444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Dunderdale, H. J., et al. 1991. Formation and resolution of recombination intermediates by E. coli RecA and RuvC proteins. Nature 354:506-510. [DOI] [PubMed] [Google Scholar]
- 149.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dupuy, B., M. K. Taha, O. Possot, C. Marchal, and A. P. Pugsley. 1992. PulO, a component of the pullulanase secretion pathway of Klebsiella oxytoca, correctly and efficiently processes gonococcal type IV prepilin in Escherichia coli. Mol. Microbiol. 6:1887-1894. [DOI] [PubMed] [Google Scholar]
- 151.Eaton, K. A., D. R. Morgan, and S. Krakowka. 1992. Motility as a factor in the colonisation of gnotobiotic piglets by Helicobacter pylori. J. Med. Microbiol. 37:123-127. [DOI] [PubMed] [Google Scholar]
- 152.Eaton, K. A., S. Suerbaum, C. Josenhans, and S. Krakowka. 1996. Colonization of gnotobiotic piglets by Helicobacter pylori deficient in two flagellin genes. Infect. Immun. 64:2445-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Elkins, C., C. E. Thomas, H. S. Seifert, and P. F. Sparling. 1991. Species-specific uptake of DNA by gonococci is mediated by a 10-base-pair sequence. J. Bacteriol. 173:3911-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ellermeier, C. D., and J. M. Slauch. 2003. RtsA and RtsB coordinately regulate expression of the invasion and flagellar genes in Salmonella enterica serovar Typhimurium. J. Bacteriol. 185:5096-5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Elsinghorst, E. A., and J. A. Weitz. 1994. Epithelial cell invasion and adherence directed by the enterotoxigenic Escherichia coli tib locus is associated with a 104-kilodalton outer membrane protein. Infect. Immun. 62:3463-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Engel, A. S., et al. 2003. Filamentous “Epsilonproteobacteria” dominate microbial mats from sulfidic cave springs. Appl. Environ. Microbiol. 69:5503-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Enz, S., H. Brand, C. Orellana, S. Mahren, and V. Braun. 2003. Sites of interaction between the FecA and FecR signal transduction proteins of ferric citrate transport in Escherichia coli K-12. J. Bacteriol. 185:3745-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Erhardt, M., and K. T. Hughes. 2010. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol. Microbiol. 75:376-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ernst, F. D., et al. 2005. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology 151:533-546. [DOI] [PubMed] [Google Scholar]
- 161.Ernst, F. D., et al. 2005. Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J. Bacteriol. 187:3687-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ernst, P. B., and B. D. Gold. 2000. The disease spectrum of Helicobacter pylori: the immunopathogenesis of gastroduodenal ulcer and gastric cancer. Annu. Rev. Microbiol. 54:615-640. [DOI] [PubMed] [Google Scholar]
- 163.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1998. Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283:537-547. [DOI] [PubMed] [Google Scholar]
- 164.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Evans, D. G., D. J. Evans, Jr., and D. Y. Graham. 1992. Adherence and internalization of Helicobacter pylori by HEp-2 cells. Gastroenterology 102:1557-1567. [DOI] [PubMed] [Google Scholar]
- 166.Evans, D. J., Jr., et al. 1995. Characterization of a Helicobacter pylori neutrophil-activating protein. Infect. Immun. 63:2213-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ewing, C. P., E. Andreishcheva, and P. Guerry. 2009. Functional characterization of flagellin glycosylation in Campylobacter jejuni 81-176. J. Bacteriol. 191:7086-7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Facius, D., M. Fussenegger, and T. F. Meyer. 1996. Sequential action of factors involved in natural competence for transformation of Neisseria gonorrhoeae. FEMS Microbiol. Lett. 137:159-164. [DOI] [PubMed] [Google Scholar]
- 169.Facius, D., and T. F. Meyer. 1993. A novel determinant (comA) essential for natural transformation competence in Neisseria gonorrhoeae and the effect of a comA defect on pilin variation. Mol. Microbiol. 10:699-712. [DOI] [PubMed] [Google Scholar]
- 170.Falk, P., et al. 1993. An in vitro adherence assay reveals that Helicobacter pylori exhibits cell lineage-specific tropism in the human gastric epithelium. Proc. Natl. Acad. Sci. U. S. A. 90:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Falush, D., et al. 2001. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: estimates of clock rates, recombination size, and minimal age. Proc. Natl. Acad. Sci. U. S. A. 98:15056-15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fecker, L., and V. Braun. 1983. Cloning and expression of the fhu genes involved in iron(III)-hydroxamate uptake by Escherichia coli. J. Bacteriol. 156:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ferguson, D. A., Jr., et al. 1993. Isolation of Helicobacter pylori from saliva. J. Clin. Microbiol. 31:2802-2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fiedorek, S. C., et al. 1991. Factors influencing the epidemiology of Helicobacter pylori infection in children. Pediatrics 88:578-582. [PubMed] [Google Scholar]
- 175.Field, L. H., V. L. Headley, S. M. Payne, and L. J. Berry. 1986. Influence of iron on growth, morphology, outer membrane protein composition, and synthesis of siderophores in Campylobacter jejuni. Infect. Immun. 54:126-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Filloux, A., G. Michel, and M. Bally. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev. 22:177-198. [DOI] [PubMed] [Google Scholar]
- 177.Finkel, S. E., and R. Kolter. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183:6288-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Finlay, B. B., and S. Falkow. 1988. Comparison of the invasion strategies used by Salmonella cholerae-suis, Shigella flexneri and Yersinia enterocolitica to enter cultured animal cells: endosome acidification is not required for bacterial invasion or intracellular replication. Biochimie 70:1089-1099. [DOI] [PubMed] [Google Scholar]
- 180.Fischer, W., D. Hofreuter, and R. Hass. 2001. Natural transformation, recombination, and repair, p. 249-257. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori physiology and genetics. ASM Press, Washington, DC. [PubMed]
- 181.Fischer, W., and R. Haas. 2004. The RecA protein of Helicobacter pylori requires a posttranslational modification for full activity. J. Bacteriol. 186:777-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fitzmaurice, W. P., R. C. Benjamin, P. C. Huang, and J. J. Scocca. 1984. Characterization of recognition sites on bacteriophage HP1c1 DNA which interact with the DNA uptake system of Haemophilus influenzae Rd. Gene 31:187-196. [DOI] [PubMed] [Google Scholar]
- 183.Flanagan, R. C., J. M. Neal-McKinney, A. S. Dhillon, W. G. Miller, and M. E. Konkel. 2009. Examination of Campylobacter jejuni putative adhesins leads to the identification of a new protein, designated FlpA, required for chicken colonization. Infect. Immun. 77:2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Follett, E. A. C., and J. Gordon. 1963. An electron microscope study of Vibrio flagella. J. Gen. Microbiol. 32:235-239. [DOI] [PubMed] [Google Scholar]
- 185.Fouts, D. E., et al. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Foynes, S., et al. 2000. Helicobacter pylori possesses two CheY response regulators and a histidine kinase sensor, CheA, which are essential for chemotaxis and colonization of the gastric mucosa. Infect. Immun. 68:2016-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Francez-Charlot, A., et al. 2003. RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol. Microbiol. 49:823-832. [DOI] [PubMed] [Google Scholar]
- 188.Francis, N. R., G. E. Sosinsky, D. Thomas, and D. J. DeRosier. 1994. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261-1270. [DOI] [PubMed] [Google Scholar]
- 189.Freitag, N. E., H. S. Seifert, and M. Koomey. 1995. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16:575-586. [DOI] [PubMed] [Google Scholar]
- 190.Friedman, C. R., J. Neimann, H. C. Wegener, and R. V. Tauxe. 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industralized nations, p. 121-138. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 191.Fu, Y., and J. E. Galan. 1999. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401:293-297. [DOI] [PubMed] [Google Scholar]
- 192.Fussenegger, M., D. Facius, J. Meier, and T. F. Meyer. 1996. A novel peptidoglycan-linked lipoprotein (ComL) that functions in natural transformation competence of Neisseria gonorrhoeae. Mol. Microbiol. 19:1095-1105. [DOI] [PubMed] [Google Scholar]
- 193.Gaasbeek, E. J., et al. 2009. Functional characterization of excision repair and RecA-dependent recombinational DNA repair in Campylobacter jejuni. J. Bacteriol. 191:3785-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 195.Galindo, M. A., W. A. Day, B. H. Raphael, and L. A. Joens. 2001. Cloning and characterization of a Campylobacter jejuni iron-uptake operon. Curr. Microbiol. 42:139-143. [DOI] [PubMed] [Google Scholar]
- 196.Galkin, V. E., et al. 2008. Divergence of quaternary structures among bacterial flagellar filaments. Science 320:382-385. [DOI] [PubMed] [Google Scholar]
- 197.Gancz, H., S. Censini, and D. S. Merrell. 2006. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74:602-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Garmendia, J., et al. 2004. TccP is an enterohaemorrhagic Escherichia coli O157:H7 type III effector protein that couples Tir to the actin-cytoskeleton. Cell. Microbiol. 6:1167-1183. [DOI] [PubMed] [Google Scholar]
- 199.Gehring, A. M., K. A. Bradley, and C. T. Walsh. 1997. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry 36:8495-8503. [DOI] [PubMed] [Google Scholar]
- 200.Geis, G., H. Leying, S. Suerbaum, U. Mai, and W. Opferkuch. 1989. Ultrastructure and chemical analysis of Campylobacter pylori flagella. J. Clin. Microbiol. 27:436-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Geis, G., S. Suerbaum, B. Forsthoff, H. Leying, and W. Opferkuch. 1993. Ultrastructure and biochemical studies of the flagellar sheath of Helicobacter pylori. J. Med. Microbiol. 38:371-377. [DOI] [PubMed] [Google Scholar]
- 202.Gerlach, R. G., et al. 2008. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell. Microbiol. 10:2364-2376. [DOI] [PubMed] [Google Scholar]
- 203.Gerlach, R. G., et al. 2007. Salmonella pathogenicity island 4 encodes a giant non-fimbrial adhesin and the cognate type 1 secretion system. Cell. Microbiol. 9:1834-1850. [DOI] [PubMed] [Google Scholar]
- 204.Ghelardi, E., et al. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424-6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gibbs, C. P., et al. 1989. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature 338:651-652. [DOI] [PubMed] [Google Scholar]
- 207.Glover, K. J., E. Weerapana, M. M. Chen, and B. Imperiali. 2006. Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry 45:5343-5350. [DOI] [PubMed] [Google Scholar]
- 208.Glover, K. J., E. Weerapana, and B. Imperiali. 2005. In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc. Natl. Acad. Sci. U. S. A. 102:14255-14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Glover, K. J., E. Weerapana, S. Numao, and B. Imperiali. 2005. Chemoenzymatic synthesis of glycopeptides with PglB, a bacterial oligosaccharyl transferase from Campylobacter jejuni. Chem. Biol. 12:1311-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Golden, N. J., and D. W. Acheson. 2002. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect. Immun. 70:1761-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Goldmark, P. J., and S. Linn. 1970. An endonuclease activity from Escherichia coli absent from certain rec− strains. Proc. Natl. Acad. Sci. U. S. A. 67:434-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Gonzalez-Lopez, M. A., and J. J. Olivares-Trejo. 2009. The gene frpB2 of Helicobacter pylori encodes an hemoglobin-binding protein involved in iron acquisition. Biometals 22:889-894. [DOI] [PubMed] [Google Scholar]
- 213.Gonzalez-Pedrajo, B., T. Minamino, M. Kihara, and K. Namba. 2006. Interactions between C ring proteins and export apparatus components: a possible mechanism for facilitating type III protein export. Mol. Microbiol. 60:984-998. [DOI] [PubMed] [Google Scholar]
- 214.Goodgal, S. H., and R. M. Herriott. 1961. Studies on transformations of Haemophilus influenzae. I. Competence. J. Gen. Physiol. 44:1201-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Goodman, S. D., and J. J. Scocca. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 85:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Goodwin, C. S., R. K. McCulloch, J. A. Armstrong, and S. H. Wee. 1985. Unusual cellular fatty acids and distinctive ultrastructure in a new spiral bacterium (Campylobacter pyloridis) from the human gastric mucosa. J. Med. Microbiol. 19:257-267. [DOI] [PubMed] [Google Scholar]
- 217.Goon, S., et al. 2006. A σ28-regulated nonflagella gene contributes to virulence of Campylobacter jejuni 81-176. Infect. Immun. 74:769-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Goon, S., J. F. Kelly, S. M. Logan, C. P. Ewing, and P. Guerry. 2003. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 50:659-671. [DOI] [PubMed] [Google Scholar]
- 219.Gorbalenya, A. E., and E. V. Koonin. 1993. Helicases: amino acid sequence conservation comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419-429. [Google Scholar]
- 220.Gradia, S., S. Acharya, and R. Fishel. 1997. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell 91:995-1005. [DOI] [PubMed] [Google Scholar]
- 221.Grant, C. C. R., M. E. Konkel, W. Cieplak, Jr., and L. S. Tompkins. 1993. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect. Immun. 61:1764-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Grass, G. 2006. Iron transport in Escherichia coli: all has not been said and done. Biometals 19:159-172. [DOI] [PubMed] [Google Scholar]
- 223.Grass, G., et al. 2005. FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Arch. Microbiol. 183:9-18. [DOI] [PubMed] [Google Scholar]
- 224.Grass, S., C. F. Lichti, R. R. Townsend, J. Gross, and J. W. St. Geme III. 2010. The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 6:e1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 226.Green, J. C., et al. 2009. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J. Mol. Biol. 391:679-690. [DOI] [PubMed] [Google Scholar]
- 227.Gruenheid, S., et al. 2001. Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3:856-859. [DOI] [PubMed] [Google Scholar]
- 228.Grys, T. E., M. B. Siegel, W. W. Lathem, and R. A. Welch. 2005. The StcE protease contributes to intimate adherence of enterohemorrhagic Escherichia coli O157:H7 to host cells. Infect. Immun. 73:1295-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Guerry, P., R. A. Alm, M. E. Power, S. M. Logan, and T. J. Trust. 1991. Role of two flagellin genes in Campylobacter motility. J. Bacteriol. 173:4757-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Guerry, P., et al. 1996. Identification and characterization of genes required for post-translational modification of Campylobacter coli VC167 flagellin. Mol. Microbiol. 19:369-378. [DOI] [PubMed] [Google Scholar]
- 231.Guerry, P., et al. 2006. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol. Microbiol. 60:299-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Guerry, P., S. M. Logan, S. Thornton, and T. J. Trust. 1990. Genomic organization and expression of Campylobacter flagellin genes. J. Bacteriol. 172:1853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Guerry, P., J. Perez-Casal, R. Yao, A. McVeigh, and T. J. Trust. 1997. A genetic locus involved in iron utilization unique to some Campylobacter strains. J. Bacteriol. 179:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Guerry, P., et al. 1994. Development and characterization of recA mutants of Campylobacter jejuni for inclusion in attenuated vaccines. Infect. Immun. 62:426-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Guerry, P., and C. M. Szymanski. 2008. Campylobacter sugars sticking out. Trends Microbiol. 16:428-435. [DOI] [PubMed] [Google Scholar]
- 236.Guo, B., et al. 2008. CmeR functions as a pleiotropic regulator and is required for optimal colonization of Campylobacter jejuni in vivo. J. Bacteriol. 190:1879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Gupta, R. S. 2000. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol. Rev. 24:367-402. [DOI] [PubMed] [Google Scholar]
- 238.Guy, R. L., L. A. Gonias, and M. A. Stein. 2000. Aggregation of host endosomes by Salmonella requires SPI2 translocation of SseFG and involves SpvR and the fms-aroE intragenic region. Mol. Microbiol. 37:1417-1435. [DOI] [PubMed] [Google Scholar]
- 239.Hamilton, H. L., and J. P. Dillard. 2006. Natural transformation of Neisseria gonorrhoeae: from DNA donation to homologous recombination. Mol. Microbiol. 59:376-385. [DOI] [PubMed] [Google Scholar]
- 240.Hamoen, L. W., G. Venema, and O. P. Kuipers. 2003. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology 149:9-17. [DOI] [PubMed] [Google Scholar]
- 241.Hansen-Wester, I., and M. Hensel. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549-559. [DOI] [PubMed] [Google Scholar]
- 242.Hantke, K. 1984. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 197:337-341. [DOI] [PubMed] [Google Scholar]
- 243.Hantke, K. 1997. Ferrous iron uptake by a magnesium transport system is toxic for Escherichia coli and Salmonella typhimurium. J. Bacteriol. 179:6201-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hantke, K. 1982. Negative control of iron uptake systems in Escherichia coli. FEMS Microbiol. Lett. 15:83-86. [Google Scholar]
- 245.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 246.Hardt, W. D., L. M. Chen, K. E. Schuebel, X. R. Bustelo, and J. E. Galan. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell 93:815-826. [DOI] [PubMed] [Google Scholar]
- 247.Harle, C., I. Kim, A. Angerer, and V. Braun. 1995. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 14:1430-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hayashi, F., et al. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 249.Hayward, R. D., and V. Koronakis. 1999. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Hazell, S. L., A. Lee, L. Brady, and W. Hennessy. 1986. Campylobacter pyloridis and gastritis: association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J. Infect. Dis. 153:658-663. [DOI] [PubMed] [Google Scholar]
- 251.Hendrixson, D. R. 2006. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol. Microbiol. 61:1646-1659. [DOI] [PubMed] [Google Scholar]
- 252.Hendrixson, D. R. 2008. Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni. Mol. Microbiol. 70:519-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Hendrixson, D. R., B. J. Akerley, and V. J. DiRita. 2001. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol. Microbiol. 40:214-224. [DOI] [PubMed] [Google Scholar]
- 254.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 255.Hendrixson, D. R., and V. J. DiRita. 2003. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol. Microbiol. 50:687-702. [DOI] [PubMed] [Google Scholar]
- 256.Hernandez, L. D., K. Hueffer, M. R. Wenk, and J. E. Galan. 2004. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science 304:1805-1807. [DOI] [PubMed] [Google Scholar]
- 257.Herriott, R. M., E. M. Meyer, and M. Vogt. 1970. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J. Bacteriol. 101:517-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Higashi, H., et al. 2002. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science 295:683-686. [DOI] [PubMed] [Google Scholar]
- 259.Ho, T. D., B. M. Davis, J. M. Ritchie, and M. K. Waldor. 2008. Type 2 secretion promotes enterohemorrhagic Escherichia coli adherence and intestinal colonization. Infect. Immun. 76:1858-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Hofreuter, D., A. Karnholz, and R. Haas. 2003. Topology and membrane interaction of Helicobacter pylori ComB proteins involved in natural transformation competence. Int. J. Med. Microbiol. 293:153-165. [DOI] [PubMed] [Google Scholar]
- 261.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 262.Hofreuter, D., S. Odenbreit, G. Henke, and R. Haas. 1998. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol. Microbiol. 28:1027-1038. [DOI] [PubMed] [Google Scholar]
- 263.Hofreuter, D., et al. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74:4694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Holliday, R. 1964. A mechanism for gene conversion in fungi. Genet. Res. 5:282-304. [DOI] [PubMed] [Google Scholar]
- 265.Holmes, K., et al. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151:243-257. [DOI] [PubMed] [Google Scholar]
- 266.Homma, M., Y. Komeda, T. Iino, and R. M. Macnab. 1987. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J. Bacteriol. 169:1493-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Horii, Z., and A. J. Clark. 1973. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J. Mol. Biol. 80:327-344. [DOI] [PubMed] [Google Scholar]
- 268.Howard, S. L., et al. 2009. Campylobacter jejuni glycosylation island important in cell charge, legionaminic acid biosynthesis, and colonization of chickens. Infect. Immun. 77:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Hu, L., and D. J. Kopecko. 1999. Campylobacter jejuni 81-176 associates with microtubules and dynein during invasion of human intestinal cells. Infect. Immun. 67:4171-4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hughes, D. T., M. B. Clarke, K. Yamamoto, D. A. Rasko, and V. Sperandio. 2009. The QseC adrenergic signaling cascade in enterohemorrhagic E. coli (EHEC). PLoS Pathog. 5:e1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Hughes, K. T., K. L. Gillen, M. J. Semon, and J. E. Karlinsey. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277-1280. [DOI] [PubMed] [Google Scholar]
- 273.Humphries, A. D., et al. 2003. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol. Microbiol. 48:1357-1376. [DOI] [PubMed] [Google Scholar]
- 274.Husson, M. O., D. Legrand, G. Spik, and H. Leclerc. 1993. Iron acquisition by Helicobacter pylori: importance of human lactoferrin. Infect. Immun. 61:2694-2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Ilver, D., et al. 1998. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science 279:373-377. [DOI] [PubMed] [Google Scholar]
- 276.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ishikawa, T., et al. 2003. The iron-binding protein DPS confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol. 185:1010-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Israel, D. A., A. S. Lou, and M. J. Blaser. 2000. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol. Lett. 186:275-280. [DOI] [PubMed] [Google Scholar]
- 279.Israel, D. A., et al. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. U. S. A. 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Iwasaki, H., M. Takahagi, A. Nakata, and H. Shinagawa. 1992. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 6:2214-2220. [DOI] [PubMed] [Google Scholar]
- 281.Iwasaki, H., M. Takahagi, T. Shiba, A. Nakata, and H. Shinagawa. 1991. Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J. 10:4381-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Janssen, R., et al. 2008. Host-pathogen interactions in Campylobacter infections: the host perspective. Clin. Microbiol. Rev. 21:505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Janvier, B., et al. 1998. Characterization and gene sequencing of a 19-kDa periplasmic protein of Campylobacter jejuni/coli. Res. Microbiol. 149:95-107. [DOI] [PubMed] [Google Scholar]
- 284.Jeon, B., W. Muraoka, O. Sahin, and Q. Zhang. 2008. Role of Cj1211 in natural transformation and transfer of antibiotic resistance determinants in Campylobacter jejuni. Antimicrob. Agents Chemother. 52:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Jeon, B., and Q. Zhang. 2007. Cj0011c, a periplasmic single- and double-stranded DNA-binding protein, contributes to natural transformation in Campylobacter jejuni. J. Bacteriol. 189:7399-7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Jeong, J. Y., et al. 2000. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Jervis, A. J., et al. 2010. Characterization of N-linked protein glycosylation in Helicobacter pullorum. J. Bacteriol. 192:5228-5236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Jiang, X., et al. 2004. The related effector proteins SopD and SopD2 from Salmonella enterica serovar Typhimurium contribute to virulence during systemic infection of mice. Mol. Microbiol. 54:1186-1198. [DOI] [PubMed] [Google Scholar]
- 289.Jin, S., et al. 2001. JlpA, a novel surface-exposed lipoprotein specific to Campylobacter jejuni, mediates adherence to host epithelial cells. Mol. Microbiol. 39:1225-1236. [DOI] [PubMed] [Google Scholar]
- 290.Jin, S., Y. C. Song, A. Emili, P. M. Sherman, and V. L. Chan. 2003. JlpA of Campylobacter jejuni interacts with surface-exposed heat shock protein 90alpha and triggers signalling pathways leading to the activation of NF-kappaB and p38 MAP kinase in epithelial cells. Cell. Microbiol. 5:165-174. [DOI] [PubMed] [Google Scholar]
- 291.Johnson, D. C., D. R. Dean, A. D. Smith, and M. K. Johnson. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74:247-281. [DOI] [PubMed] [Google Scholar]
- 292.Jones, A. C., et al. 1997. A flagellar sheath protein of Helicobacter pylori is identical to HpaA, a putative N-acetylneuraminyllactose-binding hemagglutinin, but is not an adhesin for AGS cells. J. Bacteriol. 179:5643-5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Jones, B. D., H. F. Paterson, A. Hall, and S. Falkow. 1993. Salmonella typhimurium induces membrane ruffling by a growth factor-receptor-independent mechanism. Proc. Natl. Acad. Sci. U. S. A. 90:10390-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Jones, C. J., R. M. Macnab, H. Okino, and S. Aizawa. 1990. Stoichiometric analysis of the flagellar hook-(basal-body) complex of Salmonella typhimurium. J. Mol. Biol. 212:377-387. [DOI] [PubMed] [Google Scholar]
- 295.Jonsson, A. B., G. Nyberg, and S. Normark. 1991. Phase variation of gonococcal pili by frameshift mutation in pilC, a novel gene for pilus assembly. EMBO J. 10:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Josenhans, C., R. L. Ferrero, A. Labigne, and S. Suerbaum. 1999. Cloning and allelic exchange mutagenesis of two flagellin genes of Helicobacter felis. Mol. Microbiol. 33:350-362. [DOI] [PubMed] [Google Scholar]
- 297.Josenhans, C., A. Labigne, and S. Suerbaum. 1995. Comparative ultrastructural and functional studies of Helicobacter pylori and Helicobacter mustelae flagellin mutants: both flagellin subunits, FlaA and FlaB, are necessary for full motility in Helicobacter species. J. Bacteriol. 177:3010-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Josenhans, C., et al. 2002. Functional characterization of the antagonistic flagellar late regulators FliA and FlgM of Helicobacter pylori and their effects on the H. pylori transcriptome. Mol. Microbiol. 43:307-322. [DOI] [PubMed] [Google Scholar]
- 299.Josenhans, C., L. Vossebein, S. Friedrich, and S. Suerbaum. 2002. The neuA/flmD gene cluster of Helicobacter pylori is involved in flagellar biosynthesis and flagellin glycosylation. FEMS Microbiol. Lett. 210:165-172. [DOI] [PubMed] [Google Scholar]
- 300.Joslin, S. N., and D. R. Hendrixson. 2009. Activation of the Campylobacter jejuni FlgSR two-component system is linked to the flagellar export apparatus. J. Bacteriol. 191:2656-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Joslin, S. N., and D. R. Hendrixson. 2008. Analysis of the Campylobacter jejuni FlgR response regulator suggests integration of diverse mechanisms to activate an NtrC-like protein. J. Bacteriol. 190:2422-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kabir, S. 2004. Detection of Helicobacter pylori DNA in feces and saliva by polymerase chain reaction: a review. Helicobacter 9:115-123. [DOI] [PubMed] [Google Scholar]
- 303.Kadner, R. J., K. Heller, J. W. Coulton, and V. Braun. 1980. Genetic control of hydroxamate-mediated iron uptake in Escherichia coli. J. Bacteriol. 143:256-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Kakuda, T., and V. J. DiRita. 2006. Cj1496c encodes a Campylobacter jejuni glycoprotein that influences invasion of human epithelial cells and colonization of the chick gastrointestinal tract. Infect. Immun. 74:4715-4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kalmokoff, M., et al. 2006. Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188:4312-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Kang, J., and M. J. Blaser. 2006. Bacterial populations as perfect gases: genomic integrity and diversification tensions in Helicobacter pylori. Nat. Rev. Microbiol. 4:826-836. [DOI] [PubMed] [Google Scholar]
- 308.Kang, J., and M. J. Blaser. 2008. Repair and antirepair DNA helicases in Helicobacter pylori. J. Bacteriol. 190:4218-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Kang, J., S. Huang, and M. J. Blaser. 2005. Structural and functional divergence of MutS2 from bacterial MutS1 and eukaryotic MSH4-MSH5 homologs. J. Bacteriol. 187:3528-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kang, J., D. Tavakoli, A. Tschumi, R. A. Aras, and M. J. Blaser. 2004. Effect of host species on recG phenotypes in Helicobacter pylori and Escherichia coli. J. Bacteriol. 186:7704-7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Kanipes, M. I., L. C. Holder, A. T. Corcoran, A. P. Moran, and P. Guerry. 2004. A deep-rough mutant of Campylobacter jejuni 81-176 is noninvasive for intestinal epithelial cells. Infect. Immun. 72:2452-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Karlinsey, J. E., et al. 2000. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220-1231. [DOI] [PubMed] [Google Scholar]
- 313.Karlyshev, A. V., et al. 2004. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology 150:1957-1964. [DOI] [PubMed] [Google Scholar]
- 314.Karlyshev, A. V., J. M. Ketley, and B. W. Wren. 2005. The Campylobacter jejuni glycome. FEMS Microbiol. Rev. 29:377-390. [DOI] [PubMed] [Google Scholar]
- 315.Karlyshev, A. V., D. Linton, N. A. Gregson, and B. W. Wren. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148:473-480. [DOI] [PubMed] [Google Scholar]
- 316.Karnholz, A., et al. 2006. Functional and topological characterization of novel components of the comB DNA transformation competence system in Helicobacter pylori. J. Bacteriol. 188:882-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Karudapuram, S., X. Zhao, and G. J. Barcak. 1995. DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. J. Bacteriol. 177:3235-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Kazmierczak, M. J., M. Wiedmann, and K. J. Boor. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69:527-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Kelly, A., et al. 2004. A global role for Fis in the transcriptional control of metabolism and type III secretion in Salmonella enterica serovar Typhimurium. Microbiology 150:2037-2053. [DOI] [PubMed] [Google Scholar]
- 320.Kelly, J., et al. 2006. Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 188:2427-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Kenny, B. 2001. The enterohaemorrhagic Escherichia coli (serotype O157:H7) Tir molecule is not functionally interchangeable for its enteropathogenic E. coli (serotype O127:H6) homologue. Cell. Microbiol. 3:499-510. [DOI] [PubMed] [Google Scholar]
- 322.Kenny, B., et al. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 323.Khan, I. H., T. S. Reese, and S. Khan. 1992. The cytoplasmic component of the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 89:5956-5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Kim, I., A. Stiefel, S. Plantor, A. Angerer, and V. Braun. 1997. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol. Microbiol. 23:333-344. [DOI] [PubMed] [Google Scholar]
- 325.Kim, J. S., J. H. Chang, S. I. Chung, and J. S. Yum. 1999. Molecular cloning and characterization of the Helicobacter pylori fliD gene, an essential factor in flagellar structure and motility. J. Bacteriol. 181:6969-6976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Kingsley, R. A., R. L. Santos, A. M. Keestra, L. G. Adams, and A. J. Baumler. 2002. Salmonella enterica serotype Typhimurium ShdA is an outer membrane fibronectin-binding protein that is expressed in the intestine. Mol. Microbiol. 43:895-905. [DOI] [PubMed] [Google Scholar]
- 327.Kingsley, R. A., K. van Amsterdam, N. Kramer, and A. J. Baumler. 2000. The shdA gene is restricted to serotypes of Salmonella enterica subspecies I and contributes to efficient and prolonged fecal shedding. Infect. Immun. 68:2720-2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Kinsella, N., P. Guerry, J. Cooney, and T. J. Trust. 1997. The flgE gene of Campylobacter coli is under the control of the alternative σ factor σ54. J. Bacteriol. 179:4647-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kirby, A. E., D. J. Metzger, E. R. Murphy, and T. D. Connell. 2001. Heme utilization in Bordetella avium is regulated by RhuI, a heme-responsive extracytoplasmic function sigma factor. Infect. Immun. 69:6951-6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Klose, K. E., and J. J. Mekalanos. 1998. Differential regulation of multiple flagellins in Vibrio cholerae. J. Bacteriol. 180:303-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 332.Knudsen, S. K., A. Stensballe, M. Franzmann, U. B. Westergaard, and D. E. Otzen. 2008. Effect of glycosylation on the extracellular domain of the Ag43 bacterial autotransporter: enhanced stability and reduced cellular aggregation. Biochem. J. 412:563-577. [DOI] [PubMed] [Google Scholar]
- 333.Knutton, S., D. R. Lloyd, and A. S. McNeish. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect. Immun. 55:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Ko, M., and C. Park. 2000. H-NS-dependent regulation of flagellar synthesis is mediated by a LysR family protein. J. Bacteriol. 182:4670-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Konforti, B. B., and R. W. Davis. 1987. 3′ homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 84:690-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Konkel, M. E., et al. 2005. Identification of a fibronectin-binding domain within the Campylobacter jejuni CadF protein. Mol. Microbiol. 57:1022-1035. [DOI] [PubMed] [Google Scholar]
- 337.Konkel, M. E., S. G. Garvis, S. L. Tipton, D. E. Anderson, Jr., and W. Cieplak, Jr. 1997. Identification and molecular cloning of a gene encoding a fibronectin-binding protein (CadF) from Campylobacter jejuni. Mol. Microbiol. 24:953-963. [DOI] [PubMed] [Google Scholar]
- 338.Konkel, M. E., S. F. Hayes, L. A. Joens, and W. Cieplak, Jr. 1992. Characteristics of the internalization and intracellular survival of Campylobacter jejuni in human epithelial cell cultures. Microb. Pathog. 13:357-370. [DOI] [PubMed] [Google Scholar]
- 339.Konkel, M. E., B. J. Kim, V. Rivera-Amill, and S. G. Garvis. 1999. Bacterial secreted proteins are required for the internalization of Campylobacter jejuni into cultured mammalian cells. Mol. Microbiol. 32:691-701. [DOI] [PubMed] [Google Scholar]
- 340.Konkel, M. E., et al. 2004. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J. Bacteriol. 186:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Konkel, M. E., C. L. Larson, and R. C. Flanagan. 2010. Campylobacter jejuni FlpA binds fibronectin and is required for maximal host cell adherence. J. Bacteriol. 192:68-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Koomey, M. 1998. Competence for natural transformation in Neisseria gonorrhoeae: a model system for studies of horizontal gene transfer. APMIS Suppl. 84:56-61. [DOI] [PubMed] [Google Scholar]
- 343.Koster, W., and V. Braun. 1990. Iron(III) hydroxamate transport of Escherichia coli: restoration of iron supply by coexpression of the N- and C-terminal halves of the cytoplasmic membrane protein FhuB cloned on separate plasmids. Mol. Gen. Genet. 223:379-384. [DOI] [PubMed] [Google Scholar]
- 344.Kostrzynska, M., J. D. Betts, J. W. Austin, and T. J. Trust. 1991. Identification, characterization, and spatial localization of two flagellin species in Helicobacter pylori flagella. J. Bacteriol. 173:937-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Kowalczykowski, S. C., and R. A. Krupp. 1987. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J. Mol. Biol. 193:97-113. [DOI] [PubMed] [Google Scholar]
- 347.Kowarik, M., et al. 2006. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314:1148-1150. [DOI] [PubMed] [Google Scholar]
- 348.Kowarik, M., et al. 2006. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25:1957-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Kuhle, V., G. L. Abrahams, and M. Hensel. 2006. Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic 7:716-730. [DOI] [PubMed] [Google Scholar]
- 350.Kuhle, V., and M. Hensel. 2002. SseF and SseG are translocated effectors of the type III secretion system of Salmonella pathogenicity island 2 that modulate aggregation of endosomal compartments. Cell. Microbiol. 4:813-824. [DOI] [PubMed] [Google Scholar]
- 351.Kustu, S., E. Santero, J. Keener, D. Popham, and D. Weiss. 1989. Expression of sigma 54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol. Rev. 53:367-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Kusumoto, A., et al. 2006. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J. Biochem. 139:113-121. [DOI] [PubMed] [Google Scholar]
- 353.Kusumoto, A., N. Nishioka, S. Kojima, and M. Homma. 2009. Mutational analysis of the GTP-binding motif of FlhF which regulates the number and placement of the polar flagellum in Vibrio alginolyticus. J. Biochem. 146:643-650. [DOI] [PubMed] [Google Scholar]
- 354.Kusumoto, A., et al. 2008. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154:1390-1399. [DOI] [PubMed] [Google Scholar]
- 355.Kutsukake, K. 1994. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella typhimurium. Mol. Gen. Genet. 243:605-612. [DOI] [PubMed] [Google Scholar]
- 356.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Kuzminov, A. 1993. RuvA, RuvB and RuvC proteins: cleaning-up after recombinational repairs in E. coli. Bioessays 15:355-358. [DOI] [PubMed] [Google Scholar]
- 358.Kwok, T., S. Backert, H. Schwarz, J. Berger, and T. F. Meyer. 2002. Specific entry of Helicobacter pylori into cultured gastric epithelial cells via a zipper-like mechanism. Infect. Immun. 70:2108-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Langman, L., I. G. Young, G. E. Frost, H. Rosenberg, and F. Gibson. 1972. Enterochelin system of iron transport in Escherichia coli: mutations affecting ferric-enterochelin esterase. J. Bacteriol. 112:1142-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Lanka, E., and B. M. Wilkins. 1995. DNA processing reactions in bacterial conjugation. Annu. Rev. Biochem. 64:141-169. [DOI] [PubMed] [Google Scholar]
- 361.Larsen, J. C., C. Szymanski, and P. Guerry. 2004. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J. Bacteriol. 186:6508-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Lee, S. H., and J. E. Galan. 2004. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol. Microbiol. 51:483-495. [DOI] [PubMed] [Google Scholar]
- 363.Lehnen, D., et al. 2002. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol. Microbiol. 45:521-532. [DOI] [PubMed] [Google Scholar]
- 364.Lennox, E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 365.Leung, W. K., et al. 1999. Isolation of Helicobacter pylori from vomitus in children and its implication in gastro-oral transmission. Am. J. Gastroenterol. 94:2881-2884. [DOI] [PubMed] [Google Scholar]
- 366.Leunk, R. D., P. T. Johnson, B. C. David, W. G. Kraft, and D. R. Morgan. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 26:93-99. [DOI] [PubMed] [Google Scholar]
- 367.Levine, S. M., et al. 2007. Plastic cells and populations: DNA substrate characteristics in Helicobacter pylori transformation define a flexible but conservative system for genomic variation. FASEB J. 21:3458-3467. [DOI] [PubMed] [Google Scholar]
- 368.Leying, H., S. Suerbaum, G. Geis, and R. Haas. 1992. Cloning and genetic characterization of a Helicobacter pylori flagellin gene. Mol. Microbiol. 6:2863-2874. [DOI] [PubMed] [Google Scholar]
- 369.Linden, S. K., C. Wickstrom, G. Lindell, K. Gilshenan, and I. Carlstedt. 2008. Four modes of adhesion are used during Helicobacter pylori binding to human mucins in the oral and gastric niches. Helicobacter 13:81-93. [DOI] [PubMed] [Google Scholar]
- 370.Lindenthal, C., and E. A. Elsinghorst. 1999. Identification of a glycoprotein produced by enterotoxigenic Escherichia coli. Infect. Immun. 67:4084-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Linton, D., E. Allan, A. V. Karlyshev, A. D. Cronshaw, and B. W. Wren. 2002. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol. Microbiol. 43:497-508. [DOI] [PubMed] [Google Scholar]
- 372.Linton, D., et al. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55:1695-1703. [DOI] [PubMed] [Google Scholar]
- 373.Linton, D., et al. 2000. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol. Microbiol. 35:1120-1134. [DOI] [PubMed] [Google Scholar]
- 374.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Lloyd, R. G., C. Buckman, and F. E. Benson. 1987. Genetic analysis of conjugational recombination in Escherichia coli K12 strains deficient in RecBCD enzyme. J. Gen. Microbiol. 133:2531-2538. [DOI] [PubMed] [Google Scholar]
- 376.Lloyd, R. G., and G. J. Sharples. 1992. Genetic analysis of recombination in prokaryotes. Curr. Opin. Genet. Dev. 2:683-690. [DOI] [PubMed] [Google Scholar]
- 377.Logan, S. M. 2006. Flagellar glycosylation—a new component of the motility repertoire? Microbiology 152:1249-1262. [DOI] [PubMed] [Google Scholar]
- 378.Logan, S. M., L. A. Harris, and T. J. Trust. 1987. Isolation and characterization of Campylobacter flagellins. J. Bacteriol. 169:5072-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Logan, S. M., et al. 2009. Identification of novel carbohydrate modifications on Campylobacter jejuni 11168 flagellin using metabolomics-based approaches. FEBS J. 276:1014-1023. [DOI] [PubMed] [Google Scholar]
- 380.Logan, S. M., J. F. Kelly, P. Thibault, C. P. Ewing, and P. Guerry. 2002. Structural heterogeneity of carbohydrate modifications affects serospecificity of Campylobacter flagellins. Mol. Microbiol. 46:587-597. [DOI] [PubMed] [Google Scholar]
- 381.Logan, S. M., T. J. Trust, and P. Guerry. 1989. Evidence for posttranslational modification and gene duplication of Campylobacter flagellin. J. Bacteriol. 171:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Long, C. D., D. M. Tobiason, M. P. Lazio, K. A. Kline, and H. S. Seifert. 2003. Low-level pilin expression allows for substantial DNA transformation competence in Neisseria gonorrhoeae. Infect. Immun. 71:6279-6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Louwen, R., et al. 2008. The sialylated lipooligosaccharide outer core in Campylobacter jejuni is an important determinant for epithelial cell invasion. Infect. Immun. 76:4431-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Lovett, S. T., and R. D. Kolodner. 1989. Identification and purification of a single-stranded-DNA-specific exonuclease encoded by the recJ gene of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 86:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Luke, C. J., and C. W. Penn. 1995. Identification of a 29 kDa flagellar sheath protein in Helicobacter pylori using a murine monoclonal antibody. Microbiology 141:597-604. [DOI] [PubMed] [Google Scholar]
- 386.Lundrigan, M. D., and R. J. Kadner. 1986. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB. J. Biol. Chem. 261:10797-10801. [PubMed] [Google Scholar]
- 387.Lundstrom, A. M., K. Blom, V. Sundaeus, and I. Bolin. 2001. HpaA shows variable surface localization but the gene expression is similar in different Helicobacter pylori strains. Microb. Pathog. 31:243-253. [DOI] [PubMed] [Google Scholar]
- 388.MacFadyen, L. P., et al. 2001. Competence development by Haemophilus influenzae is regulated by the availability of nucleic acid precursors. Mol. Microbiol. 40:700-707. [DOI] [PubMed] [Google Scholar]
- 389.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 390.Macnab, R. M. 2004. Type III flagellar protein export and flagellar assembly. Biochim. Biophys. Acta 1694:207-217. [DOI] [PubMed] [Google Scholar]
- 391.Mademidis, A., et al. 1997. ATP-dependent ferric hydroxamate transport system in Escherichia coli: periplasmic FhuD interacts with a periplasmic and with a transmembrane/cytoplasmic region of the integral membrane protein FhuB, as revealed by competitive peptide mapping. Mol. Microbiol. 26:1109-1123. [DOI] [PubMed] [Google Scholar]
- 392.Madiraju, M. V., A. Templin, and A. J. Clark. 1988. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc. Natl. Acad. Sci. U. S. A. 85:6592-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Magariyama, Y., S. Yamaguchi, and S. Aizawa. 1990. Genetic and behavioral analysis of flagellar switch mutants of Salmonella typhimurium. J. Bacteriol. 172:4359-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Mahdavi, J., et al. 2002. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 297:573-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Mahren, S., and V. Braun. 2003. The FecI extracytoplasmic-function sigma factor of Escherichia coli interacts with the beta′ subunit of RNA polymerase. J. Bacteriol. 185:1796-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Marlovits, T. C., W. Haase, C. Herrmann, S. G. Aller, and V. M. Unger. 2002. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc. Natl. Acad. Sci. U. S. A. 99:16243-16248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Marshall, B. J., J. A. Armstrong, D. B. McGechie, and R. J. Glancy. 1985. Attempt to fulfil Koch's postulates for pyloric Campylobacter. Med. J. Aust. 142:436-439. [DOI] [PubMed] [Google Scholar]
- 398.Marsin, S., et al. 2010. Genetic dissection of Helicobacter pylori AddAB role in homologous recombination. FEMS Microbiol. Lett. 311:44-50. [DOI] [PubMed] [Google Scholar]
- 399.Marsin, S., A. Mathieu, T. Kortulewski, R. Guerois, and J. P. Radicella. 2008. Unveiling novel RecO distant orthologues involved in homologous recombination. PLoS Genet. 4:e1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 401.Masse, E., C. K. Vanderpool, and S. Gottesman. 2005. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 187:6962-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Mathis, L. S., and J. J. Scocca. 1982. Haemophilus influenzae and Neisseria gonorrhoeae recognize different specificity determinants in the DNA uptake step of genetic transformation. J. Gen. Microbiol. 128:1159-1161. [DOI] [PubMed] [Google Scholar]
- 404.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 405.Mattick, J. S., and R. A. Alm. 1995. Response from Mattick and Alm: common architecture of type 4 fimbriae and complexes involved in macromolecular traffic. Trends Microbiol. 3:411-413. [Google Scholar]
- 406.Maughan, H., and R. J. Redfield. 2009. Extensive variation in natural competence in Haemophilus influenzae. Evolution 63:1852-1866. [DOI] [PubMed] [Google Scholar]
- 407.McGee, D. J., et al. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 73:1820-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.McHugh, J. P., et al. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 409.McNally, D. J., et al. 2007. Targeted metabolomics analysis of Campylobacter coli VC167 reveals legionaminic acid derivatives as novel flagellar glycans. J. Biol. Chem. 282:14463-14475. [DOI] [PubMed] [Google Scholar]
- 410.McNally, D. J., et al. 2006. Functional characterization of the flagellar glycosylation locus in Campylobacter jejuni 81-176 using a focused metabolomics approach. J. Biol. Chem. 281:18489-18498. [DOI] [PubMed] [Google Scholar]
- 411.McNally, D. J., et al. 2007. Commonality and biosynthesis of the O-methyl phosphoramidate capsule modification in Campylobacter jejuni. J. Biol. Chem. 282:28566-28576. [DOI] [PubMed] [Google Scholar]
- 412.McPartland, A., L. Green, and H. Echols. 1980. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell 20:731-737. [DOI] [PubMed] [Google Scholar]
- 413.McSweegan, E., and R. I. Walker. 1986. Identification and characterization of two Campylobacter jejuni adhesins for cellular and mucous substrates. Infect. Immun. 53:141-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Megraud, F. 2007. Helicobacter pylori and antibiotic resistance. Gut 56:1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Meresse, S., et al. 2001. Remodelling of the actin cytoskeleton is essential for replication of intravacuolar Salmonella. Cell. Microbiol. 3:567-577. [DOI] [PubMed] [Google Scholar]
- 416.Merrell, D. S., et al. 2003. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect. Immun. 71:6510-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 418.Miao, E. A., and S. I. Miller. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc. Natl. Acad. Sci. U. S. A. 97:7539-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Miller, C. E., J. D. Rock, K. A. Ridley, P. H. Williams, and J. M. Ketley. 2008. Utilization of lactoferrin-bound and transferrin-bound iron by Campylobacter jejuni. J. Bacteriol. 190:1900-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Miller, C. E., P. H. Williams, and J. M. Ketley. 2009. Pumping iron: mechanisms for iron uptake by Campylobacter. Microbiology 155:3157-3165. [DOI] [PubMed] [Google Scholar]
- 421.Mills, S. D., S. R. Ruschkowski, M. A. Stein, and B. B. Finlay. 1998. Trafficking of porin-deficient Salmonella typhimurium mutants inside HeLa cells: ompR and envZ mutants are defective for the formation of Salmonella-induced filaments. Infect. Immun. 66:1806-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Minamino, T., and K. Namba. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485-488. [DOI] [PubMed] [Google Scholar]
- 423.Misawa, N., and M. J. Blaser. 2000. Detection and characterization of autoagglutination activity by Campylobacter jejuni. Infect. Immun. 68:6168-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Mitra, K., D. Zhou, and J. E. Galan. 2000. Biophysical characterization of SipA, an actin-binding protein from Salmonella enterica. FEBS Lett. 482:81-84. [DOI] [PubMed] [Google Scholar]
- 425.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell. Biol. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 426.Monteville, M. R., J. E. Yoon, and M. E. Konkel. 2003. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology 149:153-165. [DOI] [PubMed] [Google Scholar]
- 427.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Moormann, C., I. Benz, and M. A. Schmidt. 2002. Functional substitution of the TibC protein of enterotoxigenic Escherichia coli strains for the autotransporter adhesin heptosyltransferase of the AIDA system. Infect. Immun. 70:2264-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Moran, A. P. 2008. Relevance of fucosylation and Lewis antigen expression in the bacterial gastroduodenal pathogen Helicobacter pylori. Carbohydr. Res. 343:1952-1965. [DOI] [PubMed] [Google Scholar]
- 430.Morgan, E., et al. 2004. Identification of host-specific colonization factors of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 54:994-1010. [DOI] [PubMed] [Google Scholar]
- 431.Morimatsu, K., and S. C. Kowalczykowski. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11:1337-1347. [DOI] [PubMed] [Google Scholar]
- 432.Morris, A., and G. Nicholson. 1987. Ingestion of Campylobacter pyloridis causes gastritis and raised fasting gastric pH. Am. J. Gastroenterol. 82:192-199. [PubMed] [Google Scholar]
- 433.Morris, A. J., M. R. Ali, G. I. Nicholson, G. I. Perez-Perez, and M. J. Blaser. 1991. Long-term follow-up of voluntary ingestion of Helicobacter pylori. Ann. Intern. Med. 114:662-663. [DOI] [PubMed] [Google Scholar]
- 434.Muller, B., C. Jones, B. Kemper, and S. C. West. 1990. Enzymatic formation and resolution of Holliday junctions in vitro. Cell 60:329-336. [DOI] [PubMed] [Google Scholar]
- 435.Muller, B., I. R. Tsaneva, and S. C. West. 1993. Branch migration of Holliday junctions promoted by the Escherichia coli RuvA and RuvB proteins. I. Comparison of RuvAB- and RuvB-mediated reactions. J. Biol. Chem. 268:17179-17184. [PubMed] [Google Scholar]
- 436.Muller, B., I. R. Tsaneva, and S. C. West. 1993. Branch migration of Holliday junctions promoted by the Escherichia coli RuvA and RuvB proteins. II. Interaction of RuvB with DNA. J. Biol. Chem. 268:17185-17189. [PubMed] [Google Scholar]
- 437.Murray, T. S., and B. I. Kazmierczak. 2006. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J. Bacteriol. 188:6995-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Nachamkin, I., X.-H. Yang, and N. J. Stern. 1993. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl. Environ. Microbiol. 59:1269-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Naikare, H., K. Palyada, R. Panciera, D. Marlow, and A. Stintzi. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74:5433-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Nakagawa, S., et al. 2005. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7:1619-1632. [DOI] [PubMed] [Google Scholar]
- 441.Nandal, A., et al. 2009. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe(2+)-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol. Microbiol. 75:637-657. [DOI] [PubMed] [Google Scholar]
- 442.Natrajan, G., et al. 2003. Structures of Escherichia coli DNA mismatch repair enzyme MutS in complex with different mismatches: a common recognition mode for diverse substrates. Nucleic Acids Res. 31:4814-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Navaneethan, U., and R. A. Giannella. 2008. Mechanisms of infectious diarrhea. Nat. Clin. Pract. Gastroenterol. Hepatol. 5:637-647. [DOI] [PubMed] [Google Scholar]
- 444.Nedenskov-Sorensen, P., G. Bukholm, and K. Bovre. 1990. Natural competence for genetic transformation in Campylobacter pylori. J. Infect. Dis. 161:365-366. [DOI] [PubMed] [Google Scholar]
- 445.Neilands, J. B. 1995. Siderophores: structure and function of microbial iron transport compounds. J. Biol. Chem. 270:26723-26726. [DOI] [PubMed] [Google Scholar]
- 446.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 447.Niehus, E., et al. 2004. Genome-wide analysis of transcriptional hierarchy and feedback regulation in the flagellar system of Helicobacter pylori. Mol. Microbiol. 52:947-961. [DOI] [PubMed] [Google Scholar]
- 448.Nies, D. H. 1999. Microbial heavy-metal resistance. Appl. Microbiol. Biotechnol. 51:730-750. [DOI] [PubMed] [Google Scholar]
- 449.Nikaido, H., and E. Y. Rosenberg. 1990. Cir and Fiu proteins in the outer membrane of Escherichia coli catalyze transport of monomeric catechols: study with beta-lactam antibiotics containing catechol and analogous groups. J. Bacteriol. 172:1361-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Nita-Lazar, M., M. Wacker, B. Schegg, S. Amber, and M. Aebi. 2005. The N-X-S/T consensus sequence is required but not sufficient for bacterial N-linked protein glycosylation. Glycobiology 15:361-367. [DOI] [PubMed] [Google Scholar]
- 451.North, A. K., K. E. Klose, K. M. Stedman, and S. Kustu. 1993. Prokaryotic enhancer-binding proteins reflect eukaryote-like modularity: the puzzle of nitrogen regulatory protein C. J. Bacteriol. 175:4267-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Nothaft, H., X. Liu, D. J. McNally, J. Li, and C. M. Szymanski. 2009. Study of free oligosaccharides derived from the bacterial N-glycosylation pathway. Proc. Natl. Acad. Sci. U. S. A. 106:15019-15024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Nougayrede, J. P., P. J. Fernandes, and M. S. Donnenberg. 2003. Adhesion of enteropathogenic Escherichia coli to host cells. Cell. Microbiol. 5:359-372. [DOI] [PubMed] [Google Scholar]
- 454.Novik, V., D. Hofreuter, and J. E. Galan. 2010. Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect. Immun. 78:3540-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Nuijten, P. J., F. J. van Asten, W. Gaastra, and B. A. van der Zeijst. 1990. Structural and functional analysis of two Campylobacter jejuni flagellin genes. J. Biol. Chem. 265:17798-17804. [PubMed] [Google Scholar]
- 456.Nuijten, P. J. M., N. M. C. Bleumink-Pluym, W. Gaastra, and B. A. M. van der Zeijst. 1989. Flagellin expression in Campylobacter jejuni is regulated at the transcriptional level. Infect. Immun. 57:1084-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Obhi, R. K., and C. Creuzenet. 2005. Biochemical characterization of the Campylobacter jejuni Cj1294, a novel UDP-4-keto-6-deoxy-GlcNAc aminotransferase that generates UDP-4-amino-4,6-dideoxy-GalNAc. J. Biol. Chem. 280:20902-20908. [DOI] [PubMed] [Google Scholar]
- 458.Odenbreit, S., G. Faller, and R. Haas. 2002. Role of the AlpAB proteins and lipopolysaccharide in adhesion of Helicobacter pylori to human gastric tissue. Int. J. Med. Microbiol. 292:247-256. [DOI] [PubMed] [Google Scholar]
- 459.Odenbreit, S., et al. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 460.Oelschlaeger, T. A., P. Guerry, and D. J. Kopecko. 1993. Unusual microtubule-dependent endocytosis mechanisms triggered by Campylobacter jejuni and Citrobacter freundii. Proc. Natl. Acad. Sci. U. S. A. 90:6884-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Ohnishi, K., K. Kutsukake, H. Suzuki, and T. Iino. 1990. Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221:139-147. [DOI] [PubMed] [Google Scholar]
- 462.Ohnishi, K., K. Kutsukake, H. Suzuki, and T. Iino. 1992. A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an anti-sigma factor inhibits the activity of the flagellum-specific sigma factor, σF. Mol. Microbiol. 6:3149-3157. [DOI] [PubMed] [Google Scholar]
- 463.Olivier, N. B., M. M. Chen, J. R. Behr, and B. Imperiali. 2006. In vitro biosynthesis of UDP-N,N′-diacetylbacillosamine by enzymes of the Campylobacter jejuni general protein glycosylation system. Biochemistry 45:13659-13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.O'Toole, P. W., M. Kostrzynska, and T. J. Trust. 1994. Non-motile mutants of Helicobacter pylori and Helicobacter mustelae defective in flagellar hook production. Mol. Microbiol. 14:691-703. [DOI] [PubMed] [Google Scholar]
- 465.Ottemann, K. M., and A. C. Lowenthal. 2002. Helicobacter pylori uses motility for initial colonization and to attain robust infection. Infect. Immun. 70:1984-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Otto, B. R., S. J. van Dooren, J. H. Nuijens, J. Luirink, and B. Oudega. 1998. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J. Exp. Med. 188:1091-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Palyada, K., D. Threadgill, and A. Stintzi. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186:4714-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Pandza, S., et al. 2000. The G-protein FlhF has a role in polar flagellar placement and general stress response induction in Pseudomonas putida. Mol. Microbiol. 36:414-423. [DOI] [PubMed] [Google Scholar]
- 469.Panina, E. M., A. A. Mironov, and M. S. Gelfand. 2001. Comparative analysis of FUR regulons in gamma-proteobacteria. Nucleic Acids Res. 29:5195-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Papini, E., et al. 1998. Selective increase of the permeability of polarized epithelial cell monolayers by Helicobacter pylori vacuolating toxin. J. Clin. Invest. 102:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Park, S. F., D. Purdy, and S. Leach. 2000. Localized reversible frameshift mutation in the flhA gene confers phase variability to flagellin gene expression in Campylobacter coli. J. Bacteriol. 182:207-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Park, S. F., and P. T. Richardson. 1995. Molecular characterization of a Campylobacter jejuni lipoprotein with homology to periplasmic siderophore-binding proteins. J. Bacteriol. 177:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Parkhill, J., et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 474.Parsonnet, J., et al. 1991. Helicobacter pylori infection and the risk of gastric carcinoma. N. Engl. J. Med. 325:1127-1131. [DOI] [PubMed] [Google Scholar]
- 475.Parsonnet, J., et al. 1994. Helicobacter pylori infection and gastric lymphoma. N. Engl. J. Med. 330:1267-1271. [DOI] [PubMed] [Google Scholar]
- 476.Parsonnet, J., H. Shmuely, and T. Haggerty. 1999. Fecal and oral shedding of Helicobacter pylori from healthy infected adults. JAMA 282:2240-2245. [DOI] [PubMed] [Google Scholar]
- 477.Parsons, C. A., A. Stasiak, R. J. Bennett, and S. C. West. 1995. Structure of a multisubunit complex that promotes DNA branch migration. Nature 374:375-378. [DOI] [PubMed] [Google Scholar]
- 478.Patel, J. C., and J. E. Galan. 2005. Manipulation of the host actin cytoskeleton by Salmonella—all in the name of entry. Curr. Opin. Microbiol. 8:10-15. [DOI] [PubMed] [Google Scholar]
- 479.Paul, K., M. Erhardt, T. Hirano, D. F. Blair, and K. T. Hughes. 2008. Energy source of flagellar type III secretion. Nature 451:489-492. [DOI] [PubMed] [Google Scholar]
- 480.Pead, P. J. 1979. Electron microscopy of Campylobacter jejuni. J. Med. Microbiol. 12:383-385. [DOI] [PubMed] [Google Scholar]
- 481.Peck, B., M. Ortkamp, K. D. Diehl, E. Hundt, and B. Knapp. 1999. Conservation, localization and expression of HopZ, a protein involved in adhesion of Helicobacter pylori. Nucleic Acids Res. 27:3325-3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Pecqueur, L., et al. 2006. Structural changes of Escherichia coli ferric uptake regulator during metal-dependent dimerization and activation explored by NMR and X-ray crystallography. J. Biol. Chem. 281:21286-21295. [DOI] [PubMed] [Google Scholar]
- 483.Pesci, E. C., D. L. Cottle, and C. L. Pickett. 1994. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect. Immun. 62:2687-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Piao, H., et al. 2011. Tissue binding patterns and in vitro effects of Campylobacter jejuni DNA-binding protein from starved cells. Neurochem. Res. 36:58-66. [DOI] [PubMed] [Google Scholar]
- 485.Piao, H., et al. 2010. Induction of paranodal myelin detachment and sodium channel loss in vivo by Campylobacter jejuni DNA-binding protein from starved cells (C-Dps) in myelinated nerve fibers. J. Neurol. Sci. 288:54-62. [DOI] [PubMed] [Google Scholar]
- 486.Pickett, C. L., T. Auffenberg, E. C. Pesci, V. L. Sheen, and S. S. Jusuf. 1992. Iron acquisition and hemolysin production by Campylobacter jejuni. Infect. Immun. 60:3872-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Pinto, A. V., et al. 2005. Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol. Cell 17:113-120. [DOI] [PubMed] [Google Scholar]
- 488.Poh, J., et al. 2008. SteC is a Salmonella kinase required for SPI-2-dependent F-actin remodelling. Cell. Microbiol. 10:20-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Poly, F., et al. 2007. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect. Immun. 75:3859-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Porter, M. L., and A. S. Engel. 2008. Diversity of uncultured Epsilonproteobacteria from terrestrial sulfidic caves and springs. Appl. Environ. Microbiol. 74:4973-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Pounder, R. E., and D. Ng. 1995. The prevalence of Helicobacter pylori infection in different countries. Aliment. Pharmacol. Ther. 9(Suppl. 2):33-39. [PubMed] [Google Scholar]
- 492.Power, M. E., P. Guerry, W. D. McCubbin, C. M. Kay, and T. J. Trust. 1994. Structural and antigenic characteristics of Campylobacter coli FlaA flagellin. J. Bacteriol. 176:3303-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Pressler, U., H. Staudenmaier, L. Zimmermann, and V. Braun. 1988. Genetics of the iron dicitrate transport system of Escherichia coli. J. Bacteriol. 170:2716-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Prouty, M. G., N. E. Correa, and K. E. Klose. 2001. The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39:1595-1609. [DOI] [PubMed] [Google Scholar]
- 495.Pugsley, A. P., and B. Dupuy. 1992. An enzyme with type IV prepilin peptidase activity is required to process components of the general extracellular protein secretion pathway of Klebsiella oxytoca. Mol. Microbiol. 6:751-760. [DOI] [PubMed] [Google Scholar]
- 496.Rafferty, J. B., et al. 1996. Crystal structure of DNA recombination protein RuvA and a model for its binding to the Holliday junction. Science 274:415-421. [DOI] [PubMed] [Google Scholar]
- 497.Ramsden, A. E., L. J. Mota, S. Munter, S. L. Shorte, and D. W. Holden. 2007. The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cell. Microbiol. 9:2517-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Raphael, B. H., and L. A. Joens. 2003. FeoB is not required for ferrous iron uptake in Campylobacter jejuni. Can. J. Microbiol. 49:727-731. [DOI] [PubMed] [Google Scholar]
- 499.Raymond, K. N., and E. A. Dertz. 2004. Biochemical and physical properties of siderophores, p. 3-17. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 500.Raymond, K. N., E. A. Dertz, and S. S. Kim. 2003. Enterobactin: an archetype for microbial iron transport. Proc. Natl. Acad. Sci. U. S. A. 100:3584-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Rendon, M. A., et al. 2007. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. U. S. A. 104:10637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Richardson, P. T., and S. F. Park. 1995. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology 141:3181-3191. [DOI] [PubMed] [Google Scholar]
- 503.Ridley, K. A., J. D. Rock, Y. Li, and J. M. Ketley. 2006. Heme utilization in Campylobacter jejuni. J. Bacteriol. 188:7862-7875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Roca, A. I., and M. M. Cox. 1990. The RecA protein: structure and function. Crit. Rev. Biochem. Mol. Biol. 25:415-456. [DOI] [PubMed] [Google Scholar]
- 505.Roman, L. J., D. A. Dixon, and S. C. Kowalczykowski. 1991. RecBCD-dependent joint molecule formation promoted by the Escherichia coli RecA and SSB proteins. Proc. Natl. Acad. Sci. U. S. A. 88:3367-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Rosenshine, I., et al. 1996. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 15:2613-2624. [PMC free article] [PubMed] [Google Scholar]
- 507.Rostas, K., S. J. Morton, S. M. Picksley, and R. G. Lloyd. 1987. Nucleotide sequence and LexA regulation of the Escherichia coli recN gene. Nucleic Acids Res. 15:5041-5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Rothman, R. H., and A. J. Clark. 1977. The dependence of postreplication repair on uvrB in a recF mutant of Escherichia coli K-12. Mol. Gen. Genet. 155:279-286. [DOI] [PubMed] [Google Scholar]
- 509.Rudel, T., et al. 1995. Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 92:7986-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Russell, R. G., and D. C. Blake, Jr. 1994. Cell association and invasion of Caco-2 cells by Campylobacter jejuni. Infect. Immun. 62:3773-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Rust, M., et al. 2009. The Helicobacter pylori anti-sigma factor FlgM is predominantly cytoplasmic and cooperates with the flagellar basal body protein FlhA. J. Bacteriol. 191:4824-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Saadat, I., et al. 2007. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature 447:330-333. [DOI] [PubMed] [Google Scholar]
- 513.Salcedo, S. P., and D. W. Holden. 2003. SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 22:5003-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Saldana, Z., et al. 2009. The Escherichia coli common pilus and the bundle-forming pilus act in concert during the formation of localized adherence by enteropathogenic E. coli. J. Bacteriol. 191:3451-3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Samatey, F. A., et al. 2001. Structure of the bacterial flagellar protofilament and implications for a switch for supercoiling. Nature 410:331-337. [DOI] [PubMed] [Google Scholar]
- 516.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 517.Santos, R. L., et al. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect. 3:1335-1344. [DOI] [PubMed] [Google Scholar]
- 518.Satin, B., et al. 2000. The neutrophil-activating protein (HP-NAP) of Helicobacter pylori is a protective antigen and a major virulence factor. J. Exp. Med. 191:1467-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Sauer, M., K. Hantke, and V. Braun. 1987. Ferric-coprogen receptor FhuE of Escherichia coli: processing and sequence common to all TonB-dependent outer membrane receptor proteins. J. Bacteriol. 169:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Schirm, M., I. C. Schoenhofen, S. M. Logan, K. C. Waldron, and P. Thibault. 2005. Identification of unusual bacterial glycosylation by tandem mass spectrometry analyses of intact proteins. Anal. Chem. 77:7774-7782. [DOI] [PubMed] [Google Scholar]
- 521.Schirm, M., et al. 2003. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 48:1579-1592. [DOI] [PubMed] [Google Scholar]
- 522.Schlumberger, M. C., and W. D. Hardt. 2006. Salmonella type III secretion effectors: pulling the host cell's strings. Curr. Opin. Microbiol. 9:46-54. [DOI] [PubMed] [Google Scholar]
- 523.Schmidt, H., and M. Hensel. 2004. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 17:14-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Schmitt, W., S. Odenbreit, D. Heuermann, and R. Haas. 1995. Cloning of the Helicobacter pylori recA gene and functional characterization of its product. Mol. Gen. Genet. 248:563-572. [DOI] [PubMed] [Google Scholar]
- 525.Schmitz, A., C. Josenhans, and S. Suerbaum. 1997. Cloning and characterization of the Helicobacter pylori flbA gene, which codes for a membrane protein involved in coordinated expression of flagellar genes. J. Bacteriol. 179:987-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Schoenhofen, I. C., et al. 2006. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J. Biol. Chem. 281:8907-8916. [DOI] [PubMed] [Google Scholar]
- 527.Schoenhofen, I. C., D. J. McNally, J. R. Brisson, and S. M. Logan. 2006. Elucidation of the CMP-pseudaminic acid pathway in Helicobacter pylori: synthesis from UDP-N-acetylglucosamine by a single enzymatic reaction. Glycobiology 16:8C-14C. [DOI] [PubMed] [Google Scholar]
- 528.Schoenhofen, I. C., et al. 2006. Functional characterization of dehydratase/aminotransferase pairs from Helicobacter and Campylobacter: enzymes distinguishing the pseudaminic acid and bacillosamine biosynthetic pathways. J. Biol. Chem. 281:723-732. [DOI] [PubMed] [Google Scholar]
- 529.Schreiber, S., et al. 2004. The spatial orientation of Helicobacter pylori in the gastric mucus. Proc. Natl. Acad. Sci. U. S. A. 101:5024-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Schwarz-Linek, U., M. Hook, and J. R. Potts. 2004. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol. Microbiol. 52:631-641. [DOI] [PubMed] [Google Scholar]
- 531.Scocca, J. J., R. L. Poland, and K. C. Zoon. 1974. Specificity in deoxyribonucleic acid uptake by transformable Haemophilus influenzae. J. Bacteriol. 118:369-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Sellars, M. J., S. J. Hall, and D. J. Kelly. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184:4187-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Senkovich, O., S. Ceaser, D. J. McGee, and T. L. Testerman. 2010. Unique host iron utilization mechanisms of Helicobacter pylori revealed with iron-deficient chemically defined media. Infect. Immun. 78:1841-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Sharma, C. M., et al. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250-255. [DOI] [PubMed] [Google Scholar]
- 535.Sharples, G. J., S. M. Ingleston, and R. G. Lloyd. 1999. Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J. Bacteriol. 181:5543-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Sheridan, P. P., K. K. Freeman, and J. E. Brenchley. 2003. Estimated minimal divergence times of the major bacterial and archaeal phyla. Geomicrobiol. J. 20:1-14. [Google Scholar]
- 537.Sherlock, O., U. Dobrindt, J. B. Jensen, R. Munk Vejborg, and P. Klemm. 2006. Glycosylation of the self-recognizing Escherichia coli Ag43 autotransporter protein. J. Bacteriol. 188:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Sheu, B. S., S. M. Sheu, H. B. Yang, A. H. Huang, and J. J. Wu. 2003. Host gastric Lewis expression determines the bacterial density of Helicobacter pylori in babA2 genopositive infection. Gut 52:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Shevchik, V. E., J. Robert-Baudouy, and G. Condemine. 1997. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 16:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Shin, S., and C. Park. 1995. Modulation of flagellar expression in Escherichia coli by acetyl phosphate and the osmoregulator OmpR. J. Bacteriol. 177:4696-4702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Singleton, M. R., S. Scaife, and D. B. Wigley. 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 107:79-89. [DOI] [PubMed] [Google Scholar]
- 542.Sisco, K. L., and H. O. Smith. 1979. Sequence-specific DNA uptake in Haemophilus transformation. Proc. Natl. Acad. Sci. U. S. A. 76:972-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. U. S. A. 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Skirrow, M. B., and M. J. Blaser. 1995. Campylobacter jejuni, p. 825-848. In M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, New York, NY.
- 545.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection, p. 69-88. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.
- 546.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Smeets, L. C., J. J. Bijlsma, E. J. Kuipers, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. The dprA gene is required for natural transformation of Helicobacter pylori. FEMS Immunol. Med. Microbiol. 27:99-102. [DOI] [PubMed] [Google Scholar]
- 548.Smeets, L. C., and J. G. Kusters. 2002. Natural transformation in Helicobacter pylori: DNA transport in an unexpected way. Trends Microbiol. 10:159-162. [DOI] [PubMed] [Google Scholar]
- 549.Smith, G. R. 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35:243-274. [DOI] [PubMed] [Google Scholar]
- 550.Smith, H. O., M. L. Gwinn, and S. L. Salzberg. 1999. DNA uptake signal sequences in naturally transformable bacteria. Res. Microbiol. 150:603-616. [DOI] [PubMed] [Google Scholar]
- 551.Smith, H. O., J. F. Tomb, B. A. Dougherty, R. D. Fleischmann, and J. C. Venter. 1995. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science 269:538-540. [DOI] [PubMed] [Google Scholar]
- 552.Smith, K. D., et al. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4:1247-1253. [DOI] [PubMed] [Google Scholar]
- 553.Smith, M. A., G. L. Mendz, M. A. Jorgensen, and S. L. Hazell. 1999. Fumarate metabolism and the microaerophily of Campylobacter species. Int. J. Biochem. Cell Biol. 31:961-975. [DOI] [PubMed] [Google Scholar]
- 554.Snelling, W. J., et al. 2007. HorB (HP0127) is a gastric epithelial cell adhesin. Helicobacter 12:200-209. [DOI] [PubMed] [Google Scholar]
- 555.Sockett, H., S. Yamaguchi, M. Kihara, V. M. Irikura, and R. M. Macnab. 1992. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J. Bacteriol. 174:793-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Sogin, M. L., et al. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Solomon, J. M., and A. D. Grossman. 1996. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 12:150-155. [DOI] [PubMed] [Google Scholar]
- 558.Sommerlad, S. M., and D. R. Hendrixson. 2007. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J. Bacteriol. 189:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Song, Y. C., et al. 2004. FlaC, a protein of Campylobacter jejuni TGH9011 (ATCC43431) secreted through the flagellar apparatus, binds epithelial cells and influences cell invasion. Mol. Microbiol. 53:541-553. [DOI] [PubMed] [Google Scholar]
- 560.Soo, E. C., et al. 2004. Selective detection and identification of sugar nucleotides by CE-electrospray-MS and its application to bacterial metabolomics. Anal. Chem. 76:619-626. [DOI] [PubMed] [Google Scholar]
- 561.Soutourina, O., et al. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Sparling, P. F. 1966. Genetic transformation of Neisseria gonorrhoeae to streptomycin resistance. J. Bacteriol. 92:1364-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 564.Spohn, G., and V. Scarlato. 1999. Motility of Helicobacter pylori is coordinately regulated by the transcriptional activator FlgR, an NtrC homolog. J. Bacteriol. 181:593-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Sprencel, C., et al. 2000. Binding of ferric enterobactin by the Escherichia coli periplasmic protein FepB. J. Bacteriol. 182:5359-5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Staudenmaier, H., B. Van Hove, Z. Yaraghi, and V. Braun. 1989. Nucleotide sequences of the fecBCDE genes and locations of the proteins suggest a periplasmic-binding-protein-dependent transport mechanism for iron(III) dicitrate in Escherichia coli. J. Bacteriol. 171:2626-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 567.Steer, H. W. 1984. Surface morphology of the gastroduodenal mucosa in duodenal ulceration. Gut 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Stein, M., R. Rappuoli, and A. Covacci. 2000. Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc. Natl. Acad. Sci. U. S. A. 97:1263-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Stein, M. A., K. Y. Leung, M. Zwick, F. Garcia-del Portillo, and B. B. Finlay. 1996. Identification of a Salmonella virulence gene required for formation of filamentous structures containing lysosomal membrane glycoproteins within epithelial cells. Mol. Microbiol. 20:151-164. [DOI] [PubMed] [Google Scholar]
- 570.Stender, S., et al. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206-1221. [DOI] [PubMed] [Google Scholar]
- 571.Sterzenbach, T., et al. 2008. Role of the Helicobacter hepaticus flagellar sigma factor FliA in gene regulation and murine colonization. J. Bacteriol. 190:6398-6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 572.Stevens, M. P., P. M. van Diemen, G. Frankel, A. D. Phillips, and T. S. Wallis. 2002. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70:5158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Stingl, K., S. Muller, G. Scheidgen-Kleyboldt, M. Clausen, and B. Maier. 2010. Composite system mediates two-step DNA uptake into Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 107:1184-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 574.Stintzi, A., A. H. M. van Vliet, and J. M. Ketley. 2008. Iron metabolism, transport, and regulation, p. 591-610. In I. Nachamkin, C. M. Szymanski, and M. J. Blaser (ed.), Campylobacter, 3rd ed. ASM Press, Washington, DC.
- 575.Stintzi, A., C. Barnes, J. Xu, and K. N. Raymond. 2000. Microbial iron transport via a siderophore shuttle: a membrane ion transport paradigm. Proc. Natl. Acad. Sci. U. S. A. 97:10691-10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Stojiljkovic, I., A. J. Baumler, and K. Hantke. 1994. Fur regulon in Gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 577.Stojiljkovic, I., M. Cobeljic, and K. Hantke. 1993. Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 108:111-115. [DOI] [PubMed] [Google Scholar]
- 578.Stojiljkovic, I., and K. Hantke. 1995. Functional domains of the Escherichia coli ferric uptake regulator protein (Fur). Mol. Gen. Genet. 247:199-205. [DOI] [PubMed] [Google Scholar]
- 579.Su, B., et al. 1999. Signal transduction-mediated adherence and entry of Helicobacter pylori into cultured cells. Gastroenterology 117:595-604. [DOI] [PubMed] [Google Scholar]
- 580.Suerbaum, S., and C. Josenhans. 2007. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 5:441-452. [DOI] [PubMed] [Google Scholar]
- 581.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Sukupolvi, S., et al. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 65:5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Swisher, S. C., and A. J. Barbati. 2007. Helicobacter pylori strikes again: gastric mucosa-associated lymphoid tissue (MALT) lymphoma. Gastroenterol. Nurs. 30:348-354. [DOI] [PubMed] [Google Scholar]
- 584.Szabo, I., et al. 1999. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. EMBO J. 18:5517-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Szymanski, C. M., D. H. Burr, and P. Guerry. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70:2242-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Szymanski, C. M., S. M. Logan, D. Linton, and B. W. Wren. 2003. Campylobacter—a tale of two protein glycosylation systems. Trends Microbiol. 11:233-238. [DOI] [PubMed] [Google Scholar]
- 587.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 588.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 589.Takata, T., T. Ando, D. A. Israel, T. M. Wassenaar, and M. J. Blaser. 2005. Role of dprA in transformation of Campylobacter jejuni. FEMS Microbiol. Lett. 252:161-168. [DOI] [PubMed] [Google Scholar]
- 590.Talley, N. J., et al. 1991. Gastric adenocarcinoma and Helicobacter pylori infection. J. Natl. Cancer Inst. 83:1734-1739. [DOI] [PubMed] [Google Scholar]
- 591.Tan, S., L. S. Tompkins, and M. R. Amieva. 2009. Helicobacter pylori usurps cell polarity to turn the cell surface into a replicative niche. PLoS Pathog. 5:e1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Tauxe, R. V. 1992. Epidemiology of Campylobacter jejuni infections in the United States and other industrial nations, p. 9-12. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter jejuni: current and future trends. ASM Press, Washington, DC.
- 593.Taylor, A., and G. R. Smith. 1980. Unwinding and rewinding of DNA by the RecBC enzyme. Cell 22:447-457. [DOI] [PubMed] [Google Scholar]
- 594.Taylor, A. F. 1992. Movement and resolution of Holliday junctions by enzymes from E. coli. Cell 69:1063-1065. [DOI] [PubMed] [Google Scholar]
- 595.Terebiznik, M. R., et al. 2002. Elimination of host cell PtdIns(4,5)P(2) by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat. Cell Biol. 4:766-773. [DOI] [PubMed] [Google Scholar]
- 596.Terry, K., S. M. Williams, L. Connolly, and K. M. Ottemann. 2005. Chemotaxis plays multiple roles during Helicobacter pylori animal infection. Infect. Immun. 73:803-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Thibault, P., et al. 2001. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J. Biol. Chem. 276:34862-34870. [DOI] [PubMed] [Google Scholar]
- 598.Thompson, S. A., and M. J. Blaser. 1995. Isolation of the Helicobacter pylori recA gene and involvement of the recA region in resistance to low pH. Infect. Immun. 63:2185-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Tomb, J. F., et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 600.Tombola, F., et al. 2001. The Helicobacter pylori VacA toxin is a urea permease that promotes urea diffusion across epithelia. J. Clin. Invest. 108:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Tonello, F., et al. 1999. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol. Microbiol. 34:238-246. [DOI] [PubMed] [Google Scholar]
- 602.Tonjum, T., N. E. Freitag, E. Namork, and M. Koomey. 1995. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 16:451-464. [DOI] [PubMed] [Google Scholar]
- 603.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 604.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 605.Tsaneva, I. R., B. Muller, and S. C. West. 1992. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell 69:1171-1180. [DOI] [PubMed] [Google Scholar]
- 606.Unsworth, K. E., and D. W. Holden. 2000. Identification and analysis of bacterial virulence genes in vivo. Philos. Trans. R. Soc. Lond. B Biol. Sci. 355:613-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Valdez, Y., R. B. Ferreira, and B. B. Finlay. 2009. Molecular mechanisms of Salmonella virulence and host resistance. Curr. Top. Microbiol. Immunol. 337:93-127. [DOI] [PubMed] [Google Scholar]
- 608.van Alphen, L. B., et al. 2008. A functional Campylobacter jejuni maf4 gene results in novel glycoforms on flagellin and altered autoagglutination behaviour. Microbiology 154:3385-3397. [DOI] [PubMed] [Google Scholar]
- 609.van der Velden, A. W., A. J. Baumler, R. M. Tsolis, and F. Heffron. 1998. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 66:2803-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.van Sorge, N. M., et al. 2009. N-glycosylated proteins and distinct lipooligosaccharide glycoforms of Campylobacter jejuni target the human C-type lectin receptor MGL. Cell. Microbiol. 11:1768-1781. [DOI] [PubMed] [Google Scholar]
- 611.van Vliet, A. H., J. M. Ketley, S. F. Park, and C. W. Penn. 2002. The role of iron in Campylobacter gene regulation, metabolism and oxidative stress defense. FEMS Microbiol. Rev. 26:173-186. [DOI] [PubMed] [Google Scholar]
- 612.van Vliet, A. H., J. D. Rock, L. N. Madeleine, and J. M. Ketley. 2000. The iron-responsive regulator Fur of Campylobacter jejuni is expressed from two separate promoters. FEMS Microbiol. Lett. 188:115-118. [DOI] [PubMed] [Google Scholar]
- 613.van Vliet, A. H. M., M.-L. A. Baillon, C. W. Penn, and J. M. Ketley. 1999. Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Velayudhan, J., et al. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 615.Wacker, M., et al. 2002. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298:1790-1793. [DOI] [PubMed] [Google Scholar]
- 616.Wadhams, G. H., and J. P. Armitage. 2004. Making sense of it all: bacterial chemotaxis. Nat. Rev. Mol. Cell. Biol. 5:1024-1037. [DOI] [PubMed] [Google Scholar]
- 617.Wagegg, W., and V. Braun. 1981. Ferric citrate transport in Escherichia coli requires outer membrane receptor protein FecA. J. Bacteriol. 145:156-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 618.Wai, S. N., K. Nakayama, K. Umene, T. Moriya, and K. Amako. 1996. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol. Microbiol. 20:1127-1134. [DOI] [PubMed] [Google Scholar]
- 619.Wai, S. N., T. Takata, A. Takade, N. Hamasaki, and K. Amako. 1995. Purification and characterization of ferritin from Campylobacter jejuni. Arch. Microbiol. 164:1-6. [DOI] [PubMed] [Google Scholar]
- 620.Waidner, B., et al. 2002. Essential role of ferritin Pfr in Helicobacter pylori iron metabolism and gastric colonization. Infect. Immun. 70:3923-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 621.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 622.Walsh, C. T., and C. G. Marshall. 2004. Siderophore biosynthesis in bacteria, p. 18-37. In J. H. Crosa, A. R. Mey, and S. M. Payne (ed.), Iron transport in bacteria. ASM Press, Washington, DC.
- 623.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 624.Wang, G., Y. Hong, A. Olczak, S. E. Maier, and R. J. Maier. 2006. Dual roles of Helicobacter pylori NapA in inducing and combating oxidative stress. Infect. Immun. 74:6839-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 625.Wang, G., and R. J. Maier. 2008. Critical role of RecN in recombinational DNA repair and survival of Helicobacter pylori. Infect. Immun. 76:153-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 626.Wang, Q., Y. Zhao, M. McClelland, and R. M. Harshey. 2007. The RcsCDB signaling system and swarming motility in Salmonella enterica serovar Typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J. Bacteriol. 189:8447-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 627.Wang, S., R. T. Fleming, E. M. Westbrook, P. Matsumura, and D. B. McKay. 2006. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 355:798-808. [DOI] [PubMed] [Google Scholar]
- 628.Wang, T. V., and K. C. Smith. 1984. recF-dependent and recF recB-independent DNA gap-filling repair processes transfer dimer-containing parental strands to daughter strands in Escherichia coli K-12 uvrB. J. Bacteriol. 158:727-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 629.Wang, Y., and D. E. Taylor. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172:949-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 630.Warawa, J., and B. Kenny. 2001. Phosphoserine modification of the enteropathogenic Escherichia coli Tir molecule is required to trigger conformational changes in Tir and efficient pedestal elongation. Mol. Microbiol. 42:1269-1280. [DOI] [PubMed] [Google Scholar]
- 631.Wassenaar, T. M., N. M. C. Bleumink-Pluym, D. G. Newell, P. J. Nuijten, and B. A. M. van der Zeijst. 1994. Differential flagellin expression in a flaA flaB+ mutant of Campylobacter jejuni. Infect. Immun. 62:3901-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 632.Wassenaar, T. M., N. M. C. Bleumink-Pluym, and B. A. M. van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 633.Wassenaar, T. M., B. A. M. van der Zeijst, R. Ayling, and D. G. Newell. 1993. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J. Gen. Microbiol. 139:1171-1175. [DOI] [PubMed] [Google Scholar]
- 634.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 635.Watson, R. O., and J. E. Galan. 2008. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 4:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Weening, E. H., et al. 2005. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect. Immun. 73:3358-3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Wei, B. L., et al. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245-256. [DOI] [PubMed] [Google Scholar]
- 638.West, S. C., E. Cassuto, and P. Howard-Flanders. 1981. Mechanism of E. coli RecA protein directed strand exchanges in post-replication repair of DNA. Nature 294:659-662. [DOI] [PubMed] [Google Scholar]
- 639.West, S. C., E. Cassuto, and P. Howard-Flanders. 1981. RecA protein promotes homologous-pairing and strand-exchange reactions between duplex DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 78:2100-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Whitby, M. C., L. Ryder, and R. G. Lloyd. 1993. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell 75:341-350. [DOI] [PubMed] [Google Scholar]
- 641.Wiesner, R. S., D. R. Hendrixson, and V. J. DiRita. 2003. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 185:5408-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Williams, S. M., et al. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 75:3747-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Wilson, D. J., et al. 2009. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol. Biol. Evol. 26:385-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Wolfgang, M., et al. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321-330. [DOI] [PubMed] [Google Scholar]
- 645.Wolfgang, M., J. P. van Putten, S. F. Hayes, and M. Koomey. 1999. The comP locus of Neisseria gonorrhoeae encodes a type IV prepilin that is dispensable for pilus biogenesis but essential for natural transformation. Mol. Microbiol. 31:1345-1357. [DOI] [PubMed] [Google Scholar]
- 646.Wood, M. W., R. Rosqvist, P. B. Mullan, M. H. Edwards, and E. E. Galyov. 1996. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a Sip-dependent mechanism and promotes bacterial entry. Mol. Microbiol. 22:327-338. [DOI] [PubMed] [Google Scholar]
- 647.Wooldridge, K. G., P. H. Williams, and J. M. Ketley. 1994. Iron-responsive genetic regulation in Campylobacter jejuni: cloning and characterization of a fur homolog. J. Bacteriol. 176:5852-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Worley, M. J., G. S. Nieman, K. Geddes, and F. Heffron. 2006. Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc. Natl. Acad. Sci. U. S. A. 103:17915-17920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 649.Worst, D. J., B. R. Otto, and J. de Graaff. 1995. Iron-repressible outer membrane proteins of Helicobacter pylori involved in heme uptake. Infect. Immun. 63:4161-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Wosten, M. M., et al. 2010. Temperature-dependent FlgM/FliA complex formation regulates Campylobacter jejuni flagella length. Mol. Microbiol. 75:1577-1591. [DOI] [PubMed] [Google Scholar]
- 651.Wosten, M. M. S. M., J. A. Wagenaar, and J. P. M. van Putten. 2004. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J. Biol. Chem. 279:16214-16222. [DOI] [PubMed] [Google Scholar]
- 652.Wozniak, C. E., C. Lee, and K. T. Hughes. 2009. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J. Bacteriol. 191:1498-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 653.Xiao, B., et al. 2009. Identification of small noncoding RNAs in Helicobacter pylori by a bioinformatics-based approach. Curr. Microbiol. 58:258-263. [DOI] [PubMed] [Google Scholar]
- 654.Xiao, B., et al. 2009. Screening and identification of natural antisense transcripts in Helicobacter pylori by a novel approach based on RNase I protection assay. Mol. Biol. Rep. 36:1853-1858. [DOI] [PubMed] [Google Scholar]
- 655.Xu, F., X. Zeng, R. D. Haigh, J. M. Ketley, and J. Lin. 2010. Identification and characterization of a new ferric enterobactin receptor, CfrB, in Campylobacter. J. Bacteriol. 192:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Yamaguchi, H., et al. 1996. Flow cytometric analysis of the heat shock protein 60 expressed on the cell surface of Helicobacter pylori. J. Med. Microbiol. 45:270-277. [DOI] [PubMed] [Google Scholar]
- 657.Yamaguchi, S., et al. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Yamaguchi, S., H. Fujita, A. Ishihara, S. Aizawa, and R. M. Macnab. 1986. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J. Bacteriol. 166:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 659.Yanagihara, S., S. Iyoda, K. Ohnishi, T. Iino, and K. Kutsukake. 1999. Structure and transcriptional control of the flagellar master operon of Salmonella typhimurium. Genes Genet. Syst. 74:105-111. [DOI] [PubMed] [Google Scholar]
- 660.Yao, R., et al. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 661.Yao, R., D. H. Burr, and P. Guerry. 1997. CheY-mediated modulation of Campylobacter jejuni virulence. Mol. Microbiol. 23:1021-1031. [DOI] [PubMed] [Google Scholar]
- 662.Yeh, Y. C., T. L. Lin, K. C. Chang, and J. T. Wang. 2003. Characterization of a ComE3 homologue essential for DNA transformation in Helicobacter pylori. Infect. Immun. 71:5427-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 663.Yonekura, K., S. Maki-Yonekura, and K. Namba. 2003. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 424:643-650. [DOI] [PubMed] [Google Scholar]
- 664.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. U. S. A. 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Young, N. M., et al. 2002. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277:42530-42539. [DOI] [PubMed] [Google Scholar]
- 666.Zeaiter, Z., et al. 2008. Analysis of detergent-resistant membranes of Helicobacter pylori infected gastric adenocarcinoma cells reveals a role for MARK2/Par1b in CagA-mediated disruption of cellular polarity. Cell. Microbiol. 10:781-794. [DOI] [PubMed] [Google Scholar]
- 667.Zheng, M., B. Doan, T. D. Schneider, and G. Storz. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Zhou, D., L. M. Chen, L. Hernandez, S. B. Shears, and J. E. Galan. 2001. A Salmonella inositol polyphosphatase acts in conjunction with other bacterial effectors to promote host cell actin cytoskeleton rearrangements and bacterial internalization. Mol. Microbiol. 39:248-259. [DOI] [PubMed] [Google Scholar]
- 669.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc. Natl. Acad. Sci. U. S. A. 96:10176-10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 670.Zhou, D., M. S. Mooseker, and J. E. Galan. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283:2092-2095. [DOI] [PubMed] [Google Scholar]
- 671.Ziprin, R. L., C. R. Young, L. H. Stanker, M. E. Hume, and M. E. Konkel. 1999. The absence of cecal colonization of chicks by a mutant of Campylobacter jejuni not expressing bacterial fibronectin-binding protein. Avian Dis. 43:586-589. [PubMed] [Google Scholar]