Summary
The nonrandom distribution of meiotic recombination shapes patterns of inheritance and genome evolution, but chromosomal features governing this distribution are poorly understood. Formation of the DNA double-strand breaks (DSBs) that initiate recombination results in accumulation of Spo11 protein covalently bound to small DNA fragments. We show here that sequencing these fragments provides a genome-wide DSB map of unprecedented resolution and sensitivity. We use this map to explore the influence of large-scale chromosome structures, chromatin, transcription factors, and local sequence composition on DSB distributions. Our analysis supports the view that the recombination terrain is molded by combinatorial and hierarchical interaction of factors that work on widely different size scales. Mechanistic aspects of DSB formation and early processing steps are also uncovered. This map illuminates the occurrence of DSBs in repetitive DNA elements, repair of which can lead to chromosomal rearrangements. We discuss implications for evolutionary dynamics of recombination hotspots.
Introduction
Most sexual species induce homologous recombination in meiosis via a developmentally programmed pathway that forms numerous DNA double-strand breaks (DSBs) (Keeney, 2007). Recombination helps homologous chromosomes pair and become physically connected by crossovers, which promote accurate chromosome segregation at Meiosis I. Recombination also alters genome structure by disrupting linkage of sequence polymorphisms on the same DNA molecule (Kauppi et al., 2004). Thus, meiotic recombination is a powerful determinant of genome diversity and evolution.
Meiotic DSBs are formed by the conserved Spo11 protein, a topoisomerase relative, via a reaction in which a tyrosine severs the DNA backbone and attaches covalently to the 5′ end of the cleaved strand (Keeney, 2007) (Figure 1A). Two Spo11 molecules work in concert to cut both strands of a duplex. Endonucleolytic cleavage adjacent to the covalent protein-DNA complex liberates Spo11 bound to a short oligonucleotide (oligo) (Neale et al., 2005). In S. cerevisiae, there are two major oligo subpopulations differing in length. The longer (mostly ~21–37 nt) and shorter oligos (<12 nt) are equally abundant and may reflect asymmetry of DSB processing (see below). Further resection of 5′ DSB termini yields 3′-single stranded DNA (ssDNA) that is a substrate for strand exchange proteins.
Figure 1. Mapping DSBs by sequencing Spo11 oligos.
A. Formation and processing of meiotic DSBs.
B. Amplification of adaptor-ligated Spo11 oligos and mock immunoprecipitation processed in parallel.
C. Reproducibility of Spo11 oligo maps. Oligo counts are in hits per million mapped reads (hpM).
D. Proportions of reads mapped to unique regions and repetitive elements.
E. Quantitative agreement of Spo11 oligos and DSBs. Oligo counts in 19 hotspots were compared with DSBs (mean ± sd, 3 cultures) assayed by Southern blot of DNA from dmc1 and sae2 mutants.
F. The Spo11 oligo map agrees with dmc1 ssDNA microarray analysis (Buhler et al., 2007), but has higher resolution.
G. Agreement of the Spo11 oligo map with direct DSB detection in sae2 genomic DNA (spo11yf=DSB-deficient mutant spo11-Y135F. P=parental band, asterisk=cross-hybridization). Oligo counts in C,F,G were smoothed with a 201 bp sliding Hann window.
Recombination is more likely to occur in some genomic regions than others, largely because of nonrandom DSB distributions (Petes, 2001; Kauppi et al., 2004). DSBs in S. cerevisiae show many levels of spatial organization. There are large (tens of kb) DSB-hot and cold domains, within which are short regions, called hotspots, where DSBs form preferentially. Important determinants of this organization include open chromatin structure, presence of certain histone modifications, and, at some loci, binding of sequence-specific transcription factors (TFs) (Petes, 2001; Lichten, 2008). However, detailed understanding is lacking of how these and other factors influence DSB locations.
Prior studies of genome-wide DSB distributions used either covalent Spo11-DSB complexes that accumulate in rad50S-like mutants or ssDNA generated by DSB resection as microarray hybridization probes (e.g., Gerton et al., 2000; Blitzblau et al., 2007; Buhler et al., 2007). These studies provided considerable insight, but had limited quantitative and spatial resolution due to microarray design, dynamic range of hybridization signal, and the large size of DSB-associated DNA used as probes.
We overcame these limitations by using each Spo11 oligo as a tag that records precisely where a break was made. Sequencing these oligos allowed us to quantitatively map DSBs genome-wide at nucleotide resolution, with high sensitivity. This map elucidated chromosome features that govern DSB distributions; allowed us to test longstanding hypotheses concerning influence of TFs, chromatin, and other factors; and uncovered mechanistic details of the formation and early nucleolytic processing of DSBs.
Results and Discussion
A nucleotide-resolution map of meiotic DSBs
Spo11 oligos were purified from meiotic cultures and adaptors were added (Figure S1A). Because shorter oligos are difficult to map uniquely, longer ones were enriched by size fractionation. PCR yielded products of the anticipated size that were absent in controls from mock precipitation of the meiotic extracts (Figure 1B). We deep-sequenced three replicates from one culture and one from an independent culture, obtaining 2.19 million reads that were mapped to the genome of strain S288C and to a draft genome of SK1 (Liti et al., 2009), the source of Spo11 oligos (Table S1). More than 95% mapped to one or both genomes, mostly to unique sites. The maps agreed well: <0.8% of oligos mapped to different positions in the two strains (Table S1 and data not shown). The SK1 genome assembly is incomplete, so the S288C map was used for most analyses. Mapped reads matched sizes expected for longer oligos (Figure S1B). Replicates were highly reproducible (Pearson's r = 0.95–0.99) (Figure 1C, S1C), so data were pooled.
Sequenced DNA was highly specific for bona fide Spo11 oligos. The rDNA cluster, 100- 200 copies of a 9.1-kb repeat on Chr XII, is strongly repressed for meiotic recombination (Petes and Botstein, 1977). Only 0.15% of mappable reads were from rDNA (Figure 1D; other repeats are discussed below). Supposing that none of the rDNA reads are true Spo11 oligos, then the Spo11-independent background is 0.0011 hits per million mapped reads (hpM) per bp (assuming 150 rDNA repeats). This is likely an overestimate, as meiotic DSBs probably do form in the rDNA. Even so, this value is 75-fold below genome average (0.083 hpM/bp), and is 146- to 6,646-fold below oligo densities in hotspots (see below).
The Spo11 oligo map showed spatial and quantitative agreement with direct assays of DSB in genomic DNA (Figure 1E,G), and matched or exceeded sensitivity of DSB detection from rad50S-like mutants (e.g., note weak signals in the YCR048w ORF, Figure 1G). This agreement allows us to convert oligo counts to percentage of DNA broken (Figure 1E), from which we estimate that ~160 DSBs form in nonrepetitive sequences per meiotic cell in wild type (see Supplemental Experimental Procedures). This value agrees with prior estimates (Buhler et al., 2007) and can account for detectable crossovers and noncrossovers (mean = 136.7 recombination events per meiosis (Mancera et al., 2008)).
As expected from prior studies (Petes, 2001; Lichten, 2008), most Spo11 oligos were from intergenic regions containing promoters, but a significant number mapped in ORFs (Figure 1G, S1D; discussed below). The oligo map agreed with microarray hybridization of ssDNA from dmc1 mutants (Blitzblau et al., 2007; Buhler et al., 2007; Borde et al., 2009), but gave much higher resolution (Figure 1F, S1E).
Thus, sequencing Spo11 oligos provides a genome-wide DSB map with unprecedented spatial and quantitative accuracy in recombination-proficient strains (Figure S2). Below, we explore this map at increasingly finer scale, from whole chromosome to single nucleotide. This analysis defines factors that interact in a hierarchical and combinatorial manner to shape DSB distributions.
Chromosome size-correlated variation in DSB frequencies
We exploited the quantitative nature of our data to address mechanisms behind chromosome size-associated variation in recombination. Small chromosomes cross over more often per kb than longer chromosomes (Kaback et al., 1992) (Figure 2A). Previously proposed mechanisms include smaller chromosomes having higher hotspot density, having more DSBs, favoring a crossover instead of noncrossover recombination outcome, and/or having less crossover interference (Kaback et al., 1992; Gerton et al., 2000; Martini et al., 2006; Blitzblau et al., 2007).
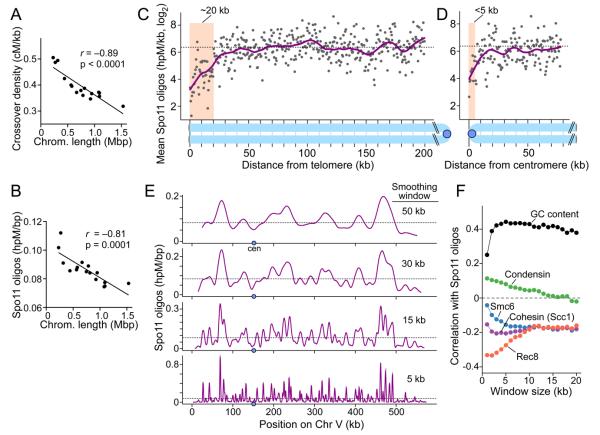
Figure 2. Large-scale DSB patterns.
A, B. Smaller chromosomes are hotter for crossing over (A, centimorgans (cM) per kb; data of Mancera et al., 2008) and DSBs (B).
C, D. Telomere-proximal and pericentric DSB suppression. Points are oligo densities in 500 bp segments averaged across all 32 chromosome arms. Dashed line, genome average; magenta line, smoothed fit (lowess); shading, DSB suppression zones.
E. Multiple levels of spatial organization of the DSB landscape. Oligo distributions on Chr V were smoothed with different sized Hann windows. Dashed lines, genome average.
F. Correlation (Pearson's r) between Spo11 oligo density and GC content or binding of chromosome structure proteins, analyzed in variable sized windows. Rec8 data were from 2 hr in meiosis (Kugou et al., 2009); others were from vegetative cells (Lindroos et al., 2006; D'Ambrosio et al., 2008). See Supplemental Experimental Procedures.
Similar to crossovers, more Spo11 oligos per kb were recovered from smaller chromosomes (Figure 2B, S3A), so crossover density correlated strongly with oligo density (r = 0.79, Figure S3B). In contrast, there appeared to be little difference between large and small chromosomes for either the crossover vs. noncrossover decision (Figure S3C), or the choice of homolog vs. sister chromatid as partner for recombination (Figure S3D). We infer that smaller chromosomes tend to experience more DSBs per kb, accounting for much of the crossover density variation. Spo11 oligo hotspots (described below) occurred at similar density on all chromosomes (Figure S3E), so the greater DSB density on smaller chromosomes is not simply because of a higher density of favorable DSB sites.
Sub-chromosomal domains of suppressed or enhanced DSB formation
Telomere-proximal regions
Spo11 oligos were less frequent in the 20 kb closest to each telomere (Figure 2C), matching DSB suppression zones seen by ssDNA mapping (Blitzblau et al., 2007; Buhler et al., 2007). Telomere structures in SK1 are not well defined, but inferring from the S288C map, oligo counts were 3.5-fold lower than genome average in the telomere-proximal 20 kb (p < 10−15 compared to a random sample, Mann-Whitney). Although most oligos from subtelomeric repeats do not map uniquely, their aggregate contribution can be estimated. If repeats were omitted, oligo counts appeared even more reduced (6.5-fold; data not shown). Notwithstanding this suppression, 1.5% of oligos mapped within 20 kb of a telomere, suggesting that meiotic cells experience 2–3 such DSBs on average, consistent with crossover rates near chromosome ends (Barton et al., 2008).
Pericentric regions
DSBs are suppressed near centromeres, but, as resolution of whole-genome methods has increased, size estimates for suppressed zones have decreased from ~20 kb (Gerton et al., 2000) to ~8–10 kb (Buhler et al., 2007). In our study, strong reduction extended only a short distance compared to telomeres: Spo11 oligo density was 7-fold lower in the 3 kb surrounding centromeres compared with a randomized sample (p < 10−4, Mann-Whitney), while segments further away were 2-3-fold lower than random but within genome-wide variation (Figure 2D, S3F). We observed hotspots near centromeres in agreement with Blitzblau et al. (2007) (Table S2; hotspot identification discussed below), but hotspot density was lower than expected within 10 kb of centromeres (60%, p < 0.02), and hotspot strength within 5 kb tended to be weaker (mean = 3.3-fold, p < 0.01) (Figure S3G,H). We infer that DSBs are rare within 1–3 kb of centromeres and that ~5–10 kb on either side is below average but not exceptionally cold. In total, 0.4% of oligos mapped within 5 kb of centromeres, equivalent to ~0.6 DSB per meiosis. Interestingly, pericentric oligo density varied 6.5-fold between chromosomes (Figure S3I), suggesting that different chromosomes may have different propensity toward missegregation caused by recombination disrupting pericentric cohesion (Rockmill et al., 2006; Chen et al., 2008).
Interstitial regions
Chromosomes show alternating domains of inherently higher or lower DSB frequency (Borde et al., 1999; Petes, 2001; Blat et al., 2002), which can be visualized by smoothing Spo11 oligo distributions with windows of increasing size (Figure 2E). Analyzed this way, peak spacing and peak-to-valley ratios varied substantially between regions (Figure 2E and data not shown), so the domains do not alternate in a highly regular fashion.
To explore mechanisms underlying these domains, we compared oligo distributions to several higher order chromosome structural features. Consistent with prior studies (Gerton et al., 2000; Blat et al., 2002), Spo11 oligos correlated positively with GC content. However, additional patterns emerged when correlations were evaluated using data binned in windows of varying sizes, such that the correlation with GC content was weak over short distances (~1 kb), but was uniformly strong at longer ranges (Figure 2F). This pattern reflects superposition of at least two levels of spatial organization: DSBs occur more often in relatively GC-rich domains, but at finer scale are mostly in intergenic regions (Figure S1D), which tend to be more AT-rich than their surroundings.
Spo11 oligos correlated negatively with presence of meiosis-specific cohesin subunit Rec8, as expected (Kugou et al., 2009), but anticorrelation was strongest at short range (<5 kb) and was weaker at larger scales (Figure 2F). This pattern also likely reflects superposition of different levels of spatial organization. Anticorrelation in larger windows is consistent with the hypothesis that a fundamental organizing principle of DSB distributions is the arrangement of chromosomes as ~10–20 kb chromatin loops emanating from a cohesin-enriched axis, with DSBs forming preferentially in cohesin-poor loops (Blat et al., 2002; Kleckner, 2006). Why anticorrelation is even stronger at short range is unknown, but may reflect a tendency for Rec8 to be especially depleted in promoters.
We also compared our data with mitotic distributions of other chromosome structure proteins (Lindroos et al., 2006; D'Ambrosio et al., 2008). Spo11 oligos showed only a weak correlation at short distances with mitotic condensin, but there was a strong anticorrelation with the G2/M distribution of Smc6 and, as previously noted, the mitotic cohesin subunit Scc1/Mcd1 (Blat and Kleckner, 1999) (Figure 2F). This is consistent with the known correlation between Scc1 and the Smc5/6 complex (Lindroos et al., 2006), but the anticorrelation of Spo11 oligos with the two proteins had different size dependence.
Taken together, these patterns point to existence of multiple levels of spatial organization of the DSB terrain, supporting the view that DSB distributions are shaped by numerous high order chromosome structures that vary over different size scales, and that intersect in complex combinations (Petes, 2001; Kleckner, 2006; Keeney, 2007).
Comprehensive identification of DSB hotspots
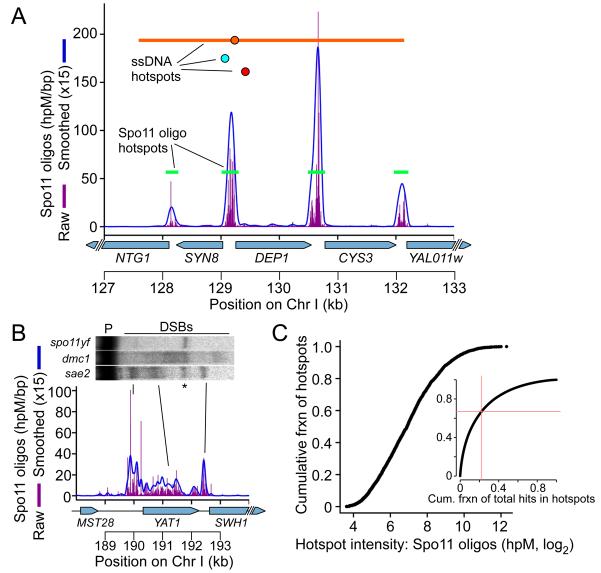
We defined 3,604 DSB hotspots as clusters of Spo11 oligos (Figure 3A, Table S2; see Supplemental Experimental Procedures). These hotspots agreed well with direct DSB detection both spatially (e.g., Figure 1G) and quantitatively (Figure 1E), and included 94 hotspots previously documented in SK1 by Southern blot (Supplemental Experimental Procedures). Spo11 oligo hotspots account for nearly all hotspots identified by ssDNA mapping (Blitzblau et al., 2007; Buhler et al., 2007; Borde et al., 2009), if allowance is made for spatial ambiguity from DSB hyperresection in dmc1 mutants (Figure 3A, S4A). However, spatial precision for Spo11 oligo hotspots was better (Figure 3A, S4A), and the oligo map resolved hotspots that were merged in microarray data by overlapping ssDNA resection tracts (Figure 3A). Thus Spo11 oligos provide the highest resolution and most complete compilation of DSB hotspots available to date in a recombination-proficient organism.
Figure 3. Identification and properties of DSB hotspots.
A. Hotspots near CYS3. Green bars are hotspots from this study. Hotspots from ssDNA mapping are also indicated (red and teal circles, peaks only (Buhler et al. 2007; Borde et al. 2009); orange circle and line, peak and region with signal over threshold (Blitzblau et al. 2007)).
B. Verification of an unusually broad hotspot by Southern blot of genomic DNA (labels as in Figure 1G). DSB signal in dmc1 spreads out because of hyperresection.
C. Hotspot intensities vary over a smooth continuum. Inset: cumulative fraction of hotspots vs. cumulative fraction of Spo11 oligos. Red lines mark the fraction of hotspot oligos in the weakest 67% of hotspots.
Canonical and noncanonical characteristics of DSB hotspots
Apparent hotspot traits emerged from prior studies, such as a narrow width (~50–250 bp) and a tendency to overlap promoters (Petes, 2001; Lichten, 2008). However, because few hotspots have been studied in detail and current whole-genome data do not resolve individual hotspots (see above), the full range of variability was unknown. Spo11 oligos address this issue and reveal additional features.
Oligo hotspots had a median width of 189 bp, and 73.4% were 50–300 bp wide (Figure S4B). Most (88.2%) overlapped with promoters (Table S2), agreeing with studies of Chr III (Baudat and Nicolas, 1997). Thus, most hotspots conform to stereotypical patterns inferred from direct mapping of a small subset. Nonetheless, there were many exceptions. For example, 10.4% of hotspots were ≥500 bp wide, and nine were >1.5 kb wide (Figure S4B). Where tested, direct assays verified anomalously wide hotspots, some of which overlapped ORFs (Figure 3B, S4C; see also below). Moreover, non-promoter hotspots accounted for 4.8% of uniquely mapped Spo11 oligos.
When hotspots were rank-ordered, their oligo counts followed a smooth continuum over a 410-fold range (Figure 3C). This pattern has several implications. First, lack of an obvious break in the continuum indicates that the cutoff is arbitrary between sites that are hotspots and those that are not. Second, most hotspots were very similar to neighbors on the continuum. Thus, small changes in measured DSB activity can cause large changes in hotspot rank, which may contribute to variability of hotspot compilations that implicitly rely on rank ordering. Third, the strongest 33% of hotspots contained >75% of all hotspot-associated oligos (Figure 3C, inset). Thus, most DSBs occur in a small subset of hotspots, while many different sites account for a smaller (but still substantial) fraction of DSBs. Strikingly, 11% of uniquely mapped Spo11 oligos fell outside of clear hotspots. A purely hotspot-centric view thus misses a considerable fraction of total recombination events.
Nucleosome-depleted regions as windows of opportunity for DSB formation
Of the few hotspots tested to date, most are nuclease-hypersensitive sites in chromatin of meiotic and vegetative cells (Petes, 2001; Lichten, 2008). An open chromatin structure is thus inferred to be necessary for Spo11 to access its DNA substrate, but the genome-wide relationship between DSBs and nucleosome occupancy remains unexplored. We therefore generated high resolution maps of micrococcal nuclease (MNase)-resistant mononucleosomes during meiosis and compared them to Spo11 oligos.
Nucleosome occupancy was determined as previously described (Kaplan et al., 2009) (Figure S5A and Supplemental Experimental Procedures). Chromatin digestion and deep sequencing were carried out in two laboratories at 0, 1, 2, and 3 hr (dataset N1) and 0 and 4 hr in meiosis (dataset N2), respectively. Dataset N2 samples were more heavily MNase-digested, but the two datasets nonetheless agreed well for patterns described below. (See Supplemental Experimental Procedures and Figures S5B,C,D for additional discussion.) Relatively few differences were observed when comparing premeiotic (0 hr) with meiotic samples, indicating that steady-state nucleosome occupancy changes little during early meiosis. Moreover, patterns agreed well with prior studies in vegetatively growing haploids of different strains (Jiang and Pugh, 2009), attesting to the conserved structure of yeast chromatin (Radman-Livaja and Rando, 2010; Tsankov et al., 2010).
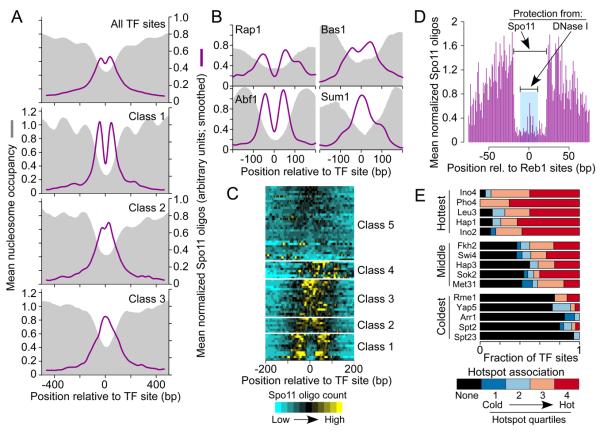
Most S. cerevisiae promoters exhibit a short (~130 bp) nucleosome-depleted region (NDR) flanked by well-positioned nucleosomes, with the transcription start site (TSS) in the first (+1) nucleosome (Radman-Livaja and Rando, 2010) (Figure 4A). Substantial overlap of DSBs with promoters is largely explained by DNA accessibility in this stereotypical structure (Lichten, 2008). As expected, Spo11 oligos mapped preferentially in promoter NDRs (Figure 4B,C).
Figure 4. DSB formation and local chromatin structure.
A. Stereotypical chromatin structure of most yeast promoters.
B. Spo11 oligos cluster in promoter NDRs. Normalized nucleosome occupancy is the number of sequence reads covering a base pair divided by genome average coverage (Kaplan et al., 2009).
C. Genome average nucleosome occupancy and Spo11 oligo density around all annotated TSS and separately for TATA-containing and TATA-less genes.
D. Hotspots are nucleosome depleted (n=3600, omitting exogenous hotspots).
E. DSB activity is not correlated with degree of nucleosome depletion at hotspot centers. Hotspots were divided into quintiles by oligo count.
F. Low nucleosome occupancy is not sufficient for strong DSB activity. Mean profiles are shown for 3′ ends of convergent genes.
Spo11 oligos in C, D, F were smoothed with a 75 bp Hann window.
Most Pol II promoters (~80%) lack a TATA sequence; this class is enriched for constitutively expressed genes, while TATA-containing promoters are more common for inducible genes (Basehoar et al., 2004). These classes differed in average chromatin structure: TATA-less promoters had a narrower average NDR and well positioned +1 and −1 nucleosomes, whereas TATA-containing promoters had a wider average NDR (Figure 4C) (Mavrich et al., 2008). Spo11 oligo distributions matched this difference (Figure 4C). TATA-containing promoters also had a 1.5-fold higher mean oligo count (Figure 4C,S5E), which may account for amino acid biosynthetic genes being enriched in hotspots in a prior study (Gerton et al., 2000). TATA-containing promoters have a higher level of histone turnover, potentially providing opportunities for increased access by Spo11 (Tirosh and Barkai, 2008).
The conclusion that DSBs occur nearly exclusively on non-nucleosomal DNA is reinforced by the fact that essentially all Spo11 oligo hotspots had low nucleosome occupancy (Figure 4D, S5F). However, hotspot oligo counts did not correlate with quantitative scores for nucleosome occupancy (Figure 4E). Moreover, low nucleosome occupancy is not sufficient for robust DSB formation. For example, NDRs are also prominent at 3′ ends of genes (Kaplan et al., 2009) (Figure S5D,G), but these are not strong DSB sites unless they coincide with the promoter NDR of a downstream gene (Figure 4F).
The hottest fifth of hotspots showed a wider average zone of low nucleosome occupancy (red line, Figure 4E). Because stronger hotspots tended to be wider on average (Figure S4B), we examined chromatin separately for “normal” width hotspots and unusually wide ones. Indeed, wider hotspots tended to have wider regions of nucleosome depletion (Figure S5H), suggesting that chromatin structure is a primary determinant of hotspot width. This conclusion is further supported by the exceptionally wide hotspots at YAT1, NAR1, and WHI5, where overall nucleosome occupancy was low and nucleosomes appeared relatively disordered (Figure S4D), suggesting that stably bound nucleosomes are sparse and variably positioned among cells.
Our findings support the view that stable nucleosomes occlude Spo11 access to DNA in vivo, in turn suggesting that variability of nucleosome occupancy contributes to variation in the DSB landscape between individual cells or between strains. However, although lack of a nucleosome is a prerequisite for DSB formation, other factors play a more dominant role in determining the probability of DNA cleavage.
Influence of TFs on DSB spatial patterns
The effect on DSB formation of a few TFs—Bas1, Bas2 and Rap1—has been explored (reviewed in Petes, 2001). It was hypothesized that TF binding (but not transcription) promotes DSBs nearby by influencing chromatin structure and/or interacting with the DSB machinery (Petes, 2001). It was also hypothesized that TFs compete with Spo11 for DNA access, occluding DSB formation at their binding sites (Xu and Petes, 1996; Petes, 2001). Spo11 oligos allowed us to test these hypotheses genome-wide.
Spo11 oligos mapped frequently near 4,233 binding sites of 77 TFs annotated based on chromatin immunoprecipitation and conservation (MacIsaac et al., 2006) (Figure 5A,B), which is not surprising since TF sites are enriched in promoters. We examined fine-scale patterns by grouping TFs based on local oligo distributions (Figure 5C and Table S3). We discuss three of these groups below. Other TFs are not considered further because they showed little spatial correlation with Spo11 oligos, having either local oligo enrichment offset from the TF sites (Class 4 in Figure 5C) or evenly distributed oligos (Class 5).
Figure 5. DSB patterns around TF binding sites.
A, B. Mean normalized Spo11 oligo profiles (see Supplemental Experimental Procedures), smoothed with 75 bp window, and nucleosome occupancy (3 hr sample) around TF sites.
C. TFs clustered according to local spatial pattern of Spo11 oligos. Each horizontal line on the heat map shows the mean profile for binding sites of a single TF, grouped by k-means clustering. Because locally normalized oligo counts were used, color coding reflects DSB spatial pattern, not total DSB intensity.
D. Zones of protection from Spo11 or DNase I (Hesselberth et al. 2009) around Reb1 binding sites (n=156).
E. TFs that are most or least associated with frequent DSB formation nearby. TFs were rank ordered by mean oligo count ±500 bp from their binding sites. A subset of TFs is shown, others are in Figure S6B. Bars depict the fraction of sites not in hotspots (black) and divide the remainder according to hotspot quartile.
For 12 TFs, there was strong evidence for DSB occlusion at their binding sites (Class 1, Figure 5A,C). The two most striking examples were Abf1 and Reb1, whose binding sites were often located in Spo11 oligo hotspots (Table S3). Both proteins showed strong oligo enrichment adjacent to their sites but depletion in the central ~40 bp (Figure 5B, 5D and S6A). Abf1 or Reb1 binding promotes nucleosome exclusion nearby (Badis et al., 2008; Kaplan et al., 2009), and both bind chromatin in meiosis (Schlecht et al., 2008), so it is likely that they influence hotspot activity at least indirectly by providing favorable chromatin structure. Class 1 also includes Rap1 (Figure 5B and S6A), whose binding sites occluded DSB formation in an altered HIS4 hotspot (Xu and Petes, 1996). Our results show that Spo11 tends to be prevented from cutting in natural Rap1 binding sites genome-wide.
Abf1, Reb1, and Rap1 footprints of protection against Spo11 cleavage (40~42 bp) were larger than for protection from DNase I cleavage in chromatin (19~24 bp, Hesselberth et al., 2009) (Figure 5D, S6A). We infer that Spo11 (and associated proteins) has a larger effective size than DNase I for cleaving DNA, and that steric constraints place the Spo11 active site ≥10 bp (30~40 Å) away from surfaces of competing DNA binding proteins. The findings also suggest that it is unlikely that Spo11-associated proteins must form an extensive DNA binding surface prior to DNA binding by Spo11 itself.
Seven TFs showed only weak DSB occlusion at their binding sites (Class 2, Figure 5A, C). A good example is Bas1. Consistent with prior studies (Mieczkowski et al., 2006), 32/37 (86.5%) analyzed Bas1 sites were in 18 Spo11 oligo hotspots (Table S3). However, Bas1 sites showed only modest depression of oligo counts in their immediate vicinity (Figure 5B). Thus, not all TFs that affect DSBs can block Spo11 access to DNA. Class 2 TFs may have low steady-state occupancy of their binding sites when DSBs form (e.g., because of short dwell time on DNA or because they act earlier), or Spo11 and/or accessory factors can displace them.
Binding sites for another 17 TFs showed local oligo enrichment but, unlike Classes 1 or 2, no detectable DSB occlusion (Class 3, Figure 5A,C). An example is Sum1 (Figure 5B), which represses meiotic genes in vegetative cells and is displaced during meiosis (Ahmed et al., 2009). Thus, some Class 3 TFs may not block Spo11 simply because they are not chromatin-bound at the relevant time. Nevertheless, some may influence DSBs by prior hit-and-run action on nucleosome occupancy or histone modifications.
Correlating TF binding sites with DSB frequency
We also examined whether TFs can be linked, positively or negatively, to DSB hotspot activity. TFs differed widely when we compared the average total oligo counts near their binding sites (Figure 5E, S6B). TF classes defined above did not correlate with oligo counts (Figure S6C), thus DSB frequency and DSB spatial distribution are separate features of the interplay between TFs and DSBs. The five TFs with the highest mean oligo counts (the Ino2/Ino4 complex, Pho4, Leu3, and Hap1; Figure 5E) are not known to influence meiotic recombination, but enrichment of Pho4 sites in hotspots was noted before (Gerton et al., 2000). It is not yet clear whether these TFs are active players or innocent bystanders in hotspot activity, but their known properties are consistent with their being bound to target promoters during sporulation (Figure S6 legend). On the other end of the scale, several TFs had low oligo counts near their binding sites (Figure 5E). Most have not been characterized in meiosis, but it may be that they do not exist in SK1 (Figure S6 legend), are not expressed or not bound to targets during meiosis, or are linked to formation of closed chromatin that inhibits DSBs.
These findings thus reveal TFs whose binding sites are predictive of hotspot activity or lack thereof. However, for most TFs, oligo counts varied widely between individual binding sites. For example, Fkh2 and Swi4 sites were about equally likely to be in a hotspot as not, and the hotspots they were associated with ran the gamut from weak to strong (Figure 5E). Most TFs had similar characteristics (Figure 5E, S6B), so presence of these binding sites is a poor predictor of DSB frequency. This pattern reinforces the view that promoters provide windows of opportunity for Spo11, but that DSB frequency is more strongly dictated by other factors.
Fine-scale analysis of DSB sites
Spo11 displays biases for which phosphodiester bonds are cleaved, but it has been difficult to discern patterns behind these preferences (Keeney, 2007; Murakami and Nicolas, 2009). Our data address this issue by providing a large library of individual cleavage sites. Importantly, the fine-scale Spo11 oligo distribution agreed well with direct DSB mapping (r = 0.77, Figure S7A) (Murakami and Nicolas, 2009), after accounting for spatial ambiguity of oligos whose 5′ ends map next to C residues (Figure S1A).
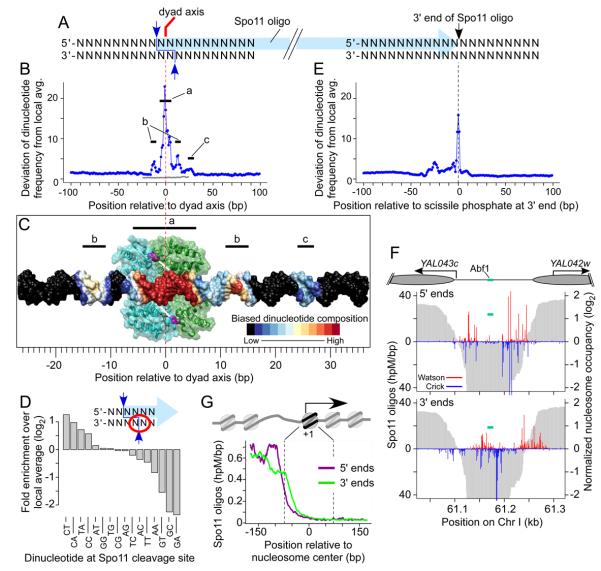
To explore Spo11 preferences, we aligned DNA sequences around each uniquely mapped oligo, using the SK1 genome sequence (Figure 6A, S7B). All mapped oligos were included, but conclusions discussed below were also obtained if we used only oligos without 5′-C ambiguity (data not shown). No consensus was apparent, supporting the idea that Spo11 is flexible in terms of DNA sequences it can cleave (Murakami and Nicolas, 2009). Nonetheless, base composition was highly nonrandom from −16 to +30 relative to the predicted dyad axis of cleavage (Figure S7B). Figure 6B summarizes this pattern. The strongest bias encompassed 10–12 bp centered on the dyad axis (segment “a”, Figure 6B), a region predicted to contact Spo11 based on docking DNA against Top6A, the archaeal Spo11 homolog (Nichols et al., 1999) (Figure 6C). This biased composition likely reflects DNA properties promoting Spo11 binding and/or catalysis, so we examined this region in detail. Overall, it is AT-enriched (64.6% vs. 60.3% local average), and the dinucleotide composition is consistent with a preference for relatively narrow, deep grooves on the side of the DNA facing Spo11 (Figure S7D). G was enriched and C was depleted at the third base in Spo11 oligos (Figure S7B), which is the complement of the base 5′ of the scissile phosphate on the opposite strand (Figure 6D). Thus, Spo11 cleavage is favored 3′ of C and, as previously shown (Murakami and Nicolas, 2009), cleavage 3′ of G is disfavored. Dinucleotide frequencies further refined Spo11 preference at the scissile phosphate: 5′-C[A/C/T] and TA were favored, whereas G[A/C/T] and AA were disfavored (Figure 6D).
Figure 6. High resolution view of DSB sites.
A. Schematic of a Spo11 DSB. Staggered cuts (blue arrows) by a Spo11 dimer generate a 2-nt 5′ overhang, the middle of which is a two-fold rotational symmetry axis. Nucleolytic cleavage (black arrow) forms the oligo 3′ end.
B. Nonrandom dinucleotide composition around Spo11 cleavage sites. At each position, deviation of dinucleotide frequencies from local average was summed (Supplemental Experimental Procedures). Gray line, deviation for a randomized sample of 50mers.
C. Biased DNA composition relative to predicted Spo11 binding. Dimeric structure is shown of a fragment of a Spo11 homolog, archaeal Top6A (Nichols et al., 1999), with monomers in green and blue, catalytic tyrosines in magenta. White bars indicate scissile phosphates in DNA docked on the dimer and color-coded by composition bias in B.
D. Spo11 preference at the scissile phosphate (dinucleotide indicated by the red circle).
E. Nonrandom dinucleotide composition around 3′ ends, as in B.
F, G. Nucleosomes and TFs are not a barrier to 3′ end formation. F, Spo11 oligos in an intergenic region. Cartoon depicts annotated nucleosomes (elipses), TSS (arrows), and Abf1 binding site. Graphs show 5′ and 3′ ends of oligos from top (red) and bottom (blue) strands. Nucleosome occupancy (3 hr) is in gray. G, Oligos around +1 nucleosomes (n=5,036). Genome averages were smoothed with 5-bp sliding window.
Bias was also observed at 11–16 bp symmetrically to the right and left of the dyad axis, outside the predicted Spo11 footprint (“b” segments, Figures 6B,C). These zones, which are modestly GC-enriched (41.8% vs. 39.7% local average), likely reflect preference of a Spo11-associated protein or a Spo11 domain not modeled by the Top6A structure. Another region that was asymmetric relative to the dyad axis (segment “c”, Figure 6B,C) probably reflects bias for oligo 3′-end formation, discussed below.
For the central 32 bp, dinucleotide composition on the right correlated with the reverse complementary composition on the left (Figure S7E,F), as predicted for two-fold rotational symmetry around the theoretical dyad axis. This symmetry does not imply that individual Spo11 cleavage sites are palindromic. Instead, it appears that left and right half-sites contribute separately, because sites with a favored base composition on one side were less likely to show favored composition on the other side (data not shown). Importantly, DNA 5′ of each oligo was engaged by Spo11 and accessory factors in vivo, but was never encountered by enzymes used in vitro to manipulate the oligos. Thus, left:right symmetry demonstrates that observed biases are inherent to DSB formation and cannot be artifacts of methods to sequence Spo11 oligos.
Formation of Spo11 oligo 3′ ends
Because reads on the 454 platform are relatively long, we could define the 3′ end of each oligo, which is likely formed by nuclease activity of Mre11, or possibly Sae2 (Figure 6A) (Keeney, 2007). Base composition around 3′ ends was nonrandom in a pattern distinct from 5′ ends (Figure 6E, S7C). Strong bias was limited to 3~4 bp centered on the 3′ ends, most notably a small but significant enrichment for T. Mre11 (or Sae2) activity thus appears to be only modestly affected by DNA composition around the scissile phosphate, with a slight preference for homopolymeric T runs.
Unlike 5′ ends, 3′ ends frequently mapped within boundaries of positioned nucleosomes and in TF binding sites where 5′ ends were rare (Figure 6F,G, S6A). Thus, neither nucleosomes nor the TFs that block Spo11 appear to be a barrier to endonucleolytic DSB processing. Possibly, Mre11-dependent cleavage can occur on DNA still bound by histones or TFs, but it seems more likely that these protein-DNA interactions are disrupted before cleavage, either through normal dynamics of histone- or TF-DNA interactions or via active displacement by Spo11 and/or accessory proteins. In principle, disruption of these protein-DNA interactions could occur either prior to or as a consequence of DSB formation. However, Mre11 associates with DSB sites independent of DSB formation (Borde et al., 2004), and is required for DSBs (Keeney, 2007). Thus, we propose that assembly of Spo11-containing pre-DSB complexes on DNA competitively replaces or actively displaces other proteins. This scenario can explain increased MNase sensitivity observed in hotspots prior to DSBs (Ohta et al., 1994), much of which is in NDRs themselves and is not accompanied by loss of positioned nucleosomes (Lichten, 2008).
Evidence for asymmetry in an early DSB processing step
We proposed three models to explain the 1:1 ratio of prominent oligo subpopulations (Neale et al., 2005): each DSB could be processed asymmetrically to yield one short and one long oligo (Figure 7A.i); each DSB could be processed symmetrically to yield two long or two short oligos, with the two outcomes equally likely genome-wide (Figure 7A.ii); or nucleolytic cleavage could occur either near or far from each DSB, with independent positions on the left and right and a ~50% chance for near vs. far (Figure 7A.iii). The latter model predicts a mix of DSBs with oligos that are asymmetric, symmetric long, or symmetric short.
Figure 7. Spo11 oligo asymmetry.
A. Models for origin of long and short oligos. i. Nicks (red arrows) positioned asymmetrically relative to Spo11. ii. Two DSB classes with symmetric nicks. iii. Variable nick placement, independent on left and right.
B. Asymmetry at DSB sites in the YCR048w hotspot (Chr III, 211, 744-211, 764 bp). Arrows show two known DSB sites that cannot be resolved due to ambiguity of oligos with 5′-C. Gray brackets, regions pooled to tally oligos for each strand.
C. Substantial asymmetry at Spo11 cleavage sites genome wide. Strand-specific counts were tallied for sites with ≥8 oligos total (n=49, 115). Points in red are significantly asymmetric (Poisson test, p≤0.05 after correction for false discovery). Green lines, 95% confidence intervals for Poisson sampling with equal frequencies on Watson and Crick.
D. Net asymmetry in the CYS3 hotspot.
E. Net asymmetry of hotspots genome wide (n=3, 604, colors as in C).
Because we recovered mostly the longer oligos, symmetry can be evaluated by asking whether oligos were recovered equally from the top (“Watson”) and bottom (“Crick”) strands at individual DSB positions. Figure 7B shows two strong cleavage sites in the YCR048w hotspot (Murakami and Nicolas, 2009). These sites are not resolved in our study because of 5′-C ambiguity, but total oligos from these sites can be tallied for the two strands (gray brackets, Figure 7B). We recovered 1,615 Crick oligos but only 44 Watson oligos, revealing strong asymmetry (p < 2.2 × 10−16, Poisson test). We infer that equal oligo numbers were formed on both strands in vivo, but that Watson oligos often escaped detection, perhaps because they were too short.
Many DSB sites analyzed showed significant asymmetry (39.3%, Figure 7C), which is incompatible with the obligate symmetry model (Figure 7A.ii) and with versions of the mixed model (Figure 7A.iii) in which 3′ end positions are random every time a DSB is made. Instead, our findings are compatible with the obligate asymmetry model (Figure 7A.i). Oligo counts are a population average, thus the fact that not all DSB sites showed asymmetry could mean that every DSB is processed asymmetrically, with different sites showing greater or lesser propensity for the direction to be the same in different cells. The results are also compatible with the mixed cleavage model (Figure 7A.iii) if degree of asymmetry is particular to individual DSB sites rather than being the same genome-wide.
We also evaluated hotspot asymmetry. On a population basis, asymmetric DSBs in a hotspot can either reinforce or cancel one another. For example, the CYS3 hotspot had net 1.3-fold asymmetry in favor of Watson (p < 10−14) (Figure 7D). Of the 3,604 hotspots identified, 1,754 (48.7%) showed net asymmetry (Figure 7E). Direction or presence of asymmetry does not correlate with orientation of adjacent transcription units (data not shown). It remains to be determined whether hotspot asymmetry is simply the aggregate of independent DSB sites, or if directionality is influenced by as yet unknown local chromosomal features. Nonetheless, the findings provide candidate sites to test the proposal that DSB asymmetry might influence later recombination steps, such as which end is first to invade the unbroken homolog (Neale et al., 2005).
DSBs at risk for genome rearrangements
DSBs in repetitive DNA can lead to genome rearrangement if non-allelic homologous sequences are used as recombination templates (reviewed in Sasaki et al., 2010). Our data allowed us to examine these “at-risk” DSBs. Only 1.53% of Spo11 oligos mapped to two or more positions in the genome, and an additional 0.23% mapped to unique positions within boundaries of repetitive elements (Figure 1D). High copy number repeats (rDNA, telomeres, retrotransposons, and tRNA genes) accounted for only 1.16% of total oligos, despite these repeats occupying ~14% of the genome. Remaining oligos that mapped to multiple locations were from low copy repeats such as multigene families or from regions with low sequence complexity. As noted above, few oligos were from rDNA; these mapped fairly uniformly across the repeat unit (Figure S1F).
Subtelomeric X and Y′ elements accounted for 0.43% of mappable reads, or ~0.7 DSB per meiosis assuming all recovered sequences were from bona fide Spo11 oligos (Figure 1D). Most of these oligos were from Y′ elements (Figure S1F), consistent with frequent rearrangement of chromosome ends through Y′ recombination in meiosis (Horowitz et al., 1984).
S288C has 50 full Ty retrotransposons and many more solo long terminal repeats (LTRs) or LTR fragments, totaling ~3% of its genome, but SK1 has only ca. half as many full-length Tys and insertion sites are not conserved (Gabriel et al., 2006; Liti et al., 2009) (M.S., unpublished data). Meiotic recombination was rare in artificial Ty constructs and meiotic DSBs were not detected in a full Ty element, likely reflecting its relatively closed chromatin structure (Ben-Aroya et al., 2004). Correspondingly, only 0.28% of Spo11 oligos were from full Tys or LTRs (Figure 1D, S1F), indicating that meiotic DSBs tend to be somewhat suppressed in natural Ty elements.
These findings demonstrate that DSB formation is suppressed within repetitive DNA genome-wide, albeit to various degrees for different repeat families. Nonetheless, the total burden of such DSBs is substantial: from the number of Spo11 oligos recovered, we estimate an average of ~2–3 DSBs per meiosis. Not all types of repeats have the same potential to generate lethal chromosome rearrangements through non-allelic homologous recombination, but all have potential to contribute to genome plasticity and evolution, and all have potential to adversely affect meiotic chromosome pairing and disjunction. Thus, as yet poorly understood mechanisms that control non-allelic recombination are clearly critical in nearly every meiosis to maintain genome integrity (Sasaki et al., 2010).
Conclusions
Our findings reinforce the view that the DSB landscape in S. cerevisiae is shaped by combinatorial action of many factors (Petes, 2001; Kleckner, 2006; Keeney, 2007; Lichten, 2008). These factors operate over many size scales and include whole chromosome variation, large subchromosomal domains, chromosome structure proteins, chromatin structure (including nucleosomes and sequence-specific DNA binding proteins), and local DNA composition. Moreover, these factors work hierarchically, with the general trend that level in hierarchy is proportional to scale. Thus, for example, two DNA segments that are equally free of nucleosomes may have substantially different DSB probability depending on their locations relative to loop-axis chromosome organization or telomeres.
These studies also lead to reexamination of the definition of DSB hotspots. From the earliest proposals that recombination initiates preferentially at defined sites (Holliday, 1964), the concept of hotspots has been useful for describing how recombination is distributed and for cataloging preferred sites. However, we suggest that this concept also tends to lead, intentionally or not, to the view that hotspots are discrete functional entities. We show here that many base pairs—and maybe all—are potential DSB sites, with a continuous distribution of cleavage probability. We also show that hotspot definition is arbitrary, i.e., that it is difficult to draw a biologically meaningful boundary between what is and what is not a hotspot. Moreover, while most hotspots comprise a narrow cluster of DSB sites within a promoter, many do not fit this stereotype and many DSBs occur outside of hotspots entirely. These findings argue against a granular, hotspot-centric view of DSB distributions. Instead, the DSB landscape is better seen as a quantitative, probabilistic distribution across the entire genome. A hotspot is a group of phosphodiester bonds that share a high local likelihood of cleavage, i.e., simply one spatial organization level among many. In this view, Spo11 is an opportunistic cutter and hotspots are its windows of opportunity (Lichten, 2008).
As shown here and in prior studies, chromatin structure is a key determinant of these windows. Low nucleosome occupancy in promoters is conserved in S. cerevisiae strains and related species (this work and Tsankov et al., 2010), likely because of selection to maintain appropriate chromatin structure for gene expression. In fact, chromosome structures at all size scales are probably under evolutionary constraints because of functions important for DNA replication, compaction, and segregation. Hence, many factors that govern Spo11 activity are under selective pressures independent of meiosis or recombination per se (Nicolas et al., 1989). This predicts that some features of DSB landscapes will show significant similarity between related yeast species: quantitative values are likely to differ substantially, but many hotspots in one strain or species are likely to be hotspots in another. In this regard, S. cerevisiae and its relatives may contrast with mammals, where a meiosis-specific factor, PRDM9, appears to specifically target SPO11 to sites that may not have another intrinsic function (Neale, 2010), and which might thus be easier to evolve toward hotspot extinction because of different selective constraints. It will be interesting to compare Spo11 oligo maps between species, such as mammals and budding and fission yeasts, whose chromosomes have distinct architectures and evolutionary dynamics.
Experimental Procedures
Detailed methods are in Supplemental Experimental Procedures. Strains are diploid derivatives of SK1 (Table S4). The Spo11-HA construct used here gives reduced DSBs (~80% of wild type), but does not appear to grossly alter DSB distributions (Figure S7). Spo11 oligos were sequenced as follows (Figure S1A). Cultures were harvested at 4 hr of synchronous meiosis, then denaturing extracts were prepared from nuclei of hypotonically lysed spheroplasts. Spo11-HA-oligo complexes were immunoprecipitated, size-fractionated by SDS-PAGE, then protease-digested. The 3′ ends of resulting Spo11 oligos were extended with GTP and terminal deoxynucleotidyl transferase, then ligated to a double-stranded DNA adaptor using T4 RNA ligase 2. The complementary strands of Spo11 oligos were synthesized, then gel-purified on denaturing PAGE and subjected to another round of 3′-GTP tailing, adaptor ligation, and strand synthesis. Sequencing primers were added by low cycle numbers of PCR, products were sequenced on the 454 platform (Roche) according to manufacturer's instructions, and reads were mapped to S288C and SK1 genomes. Nucleosome mapping was carried out by deep sequencing of MNase-digested chromatin according to an established method (Kaplan et al., 2009). Data analysis utilized R (http://www.r-project.org/) and Bioconductor (http://www.bioconductor.org/). Raw sequence data are available from the GEO repository (accession numbers GSE26449 and GSE26452). Map files in several formats are also available at http://www.cbio.mskcc.org/Public/Spo11.
Supplementary Material
Acknowledgments
We thank A. Viale (MSKCC Genomics Core Laboratory) for sequencing; S. Shuman and J. Nandakumar (MSKCC) for generous gifts of RNA ligase; and M. Lichten (NCI), I. Whitehouse (MSKCC), and G. Smith (Fred Hutchinson Cancer Res. Ctr.) for comments on the manuscript. This work was supported in part by NIH grants GM58673 (SK) and HD53855 (SK and MJ). MJN is supported by a Royal Society University Research Fellowship, an MRC New Investigator Grant, and an HFSP Career Development Award. JP was supported by a Leukemia and Lymphoma Society Fellowship. RK was supported by NIH training grant T32 GM008539. SK is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed NT, Bungard D, Shin ME, Moore M, Winter E. The Ime2 protein kinase enhances the disassociation of the Sum1 repressor from middle meiotic promoters. Mol. Cell. Biol. 2009;29:4352–4362. doi: 10.1128/MCB.00305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton AB, Pekosz MR, Kurvathi RS, Kaback DB. Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae. Genetics. 2008;179:1221–1235. doi: 10.1534/genetics.107.083493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast Chromosome III. Proc. Natl. Acad. Sci. USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S, Mieczkowski PA, Petes TD, Kupiec M. The compact chromatin structure of a Ty repeated sequence suppresses recombination hotspot activity in Saccharomyces cerevisiae. Mol. Cell. 2004;15:221–231. doi: 10.1016/j.molcel.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- Blitzblau HG, Bell GW, Rodriguez J, Bell SP, Hochwagen A. Mapping of Meiotic Single-Stranded DNA Reveals Double-Strand-Break Hotspots near Centromeres and Telomeres. Curr. Biol. 2007;17:2003–2012. doi: 10.1016/j.cub.2007.10.066. [DOI] [PubMed] [Google Scholar]
- Borde V, Lin W, Novikov E, Petrini JH, Lichten M, Nicolas A. Association of Mre11p with double-strand break sites during yeast meiosis. Mol. Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Wu T-C, Lichten M. Use of a recombination reporter insert to define meiotic recombination domains on Chromosome III of Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:4832–4842. doi: 10.1128/mcb.19.7.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler C, Borde V, Lichten M. Mapping Meiotic Single-Strand DNA Reveals a New Landscape of DNA Double-Strand Breaks in Saccharomyces cerevisiae. PLoS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, Vader G, Hochwagen A, Roeder GS, Fung JC. Global analysis of the meiotic crossover landscape. Dev. Cell. 2008;15:401–415. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes. Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel A, Dapprich J, Kunkel M, Gresham D, Pratt SC, Dunham MJ. Global mapping of transposon location. PLoS Genet. 2006;2:e212. doi: 10.1371/journal.pgen.0020212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Inaugural article: global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:11383–11390. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Horowitz H, Thorburn P, Haber JE. Rearrangements of highly polymorphic regions near telomeres of Saccharomyces cerevisiae. Mol. Cell. Biol. 1984;4:2509–2517. doi: 10.1128/mcb.4.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol. 2009;10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback DB, Guacci V, Barber D, Mahon JW. Chromosome size-dependent control of meiotic recombination. Science. 1992;256:228–232. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Proust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Lankenau DH, editor. Recombination and Meiosis. Springer-Verlag; Heidelberg: 2007. pp. 81–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–194. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- Kugou K, Fukuda T, Yamada S, Ito M, Sasanuma H, Mori S, Katou Y, Itoh T, Matsumoto K, Shibata T, et al. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol. Biol. Cell. 2009;20:3064–3076. doi: 10.1091/mbc.E08-12-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M. Meiotic chromatin: The substrate for recombination initiation. In: Egel R, Lankenau DH, editors. Recombination and Meiosis: Models, Means, and Evolution. Springer-Verlag; Berlin: 2008. pp. 165–193. [Google Scholar]
- Lindroos HB, Strom L, Itoh T, Katou Y, Shirahige K, Sjogren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126:285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski PA, Dominska M, Buck MJ, Gerton JL, Lieb JD, Petes TD. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:1014–1027. doi: 10.1128/MCB.26.3.1014-1027.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Nicolas A. Locally, meiotic double-strand breaks targeted by Gal4BD-Spo11 occur at discrete sites with a sequence preference. Mol. Cell. Biol. 2009;29:3500–3516. doi: 10.1128/MCB.00088-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ. PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11:104. doi: 10.1186/gb-2010-11-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD. Meiotic recombination hot spots and cold spots. Nat. Rev. Genet. 2001;2:360–369. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- Petes TD, Botstein D. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc. Natl. Acad. Sci. USA. 1977;74:5091–5095. doi: 10.1073/pnas.74.11.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. Nucleosome positioning: how is it established, and why does it matter. Dev. Biol. 2010;339:258–266. doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Voelkel-Meiman K, Roeder GS. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat. Rev. Mol. Cell. Biol. 2010;11:182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlecht U, Erb I, Demougin P, Robine N, Borde V, Nimwegen E, Nicolas A, Primig M. Genome-wide expression profiling, in vivo DNA binding analysis, and probabilistic motif prediction reveal novel Abf1 target genes during fermentation, respiration, and sporulation in yeast. Mol. Biol. Cell. 2008;19:2193–2207. doi: 10.1091/mbc.E07-12-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Petes TD. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics. 1996;143:1115–1125. doi: 10.1093/genetics/143.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.