Abstract
In all eukaryotes, the Golgi apparatus is the main site of protein glycosylation. It is widely accepted that the glycosidases and glycosyltransferases involved in N-glycan processing are found concentrated within the Golgi stack where they provide their function. This means that enzymes catalyzing early steps in the processing pathway are located mainly at the cis-side, whereas late-acting enzymes mostly locate to the trans-side of the stacks, creating a non-uniform distribution along the cis–trans axis of the Golgi. There is compelling evidence that the information for their sorting to specific Golgi cisternae depends on signals encoded in the proteins themselves as well as on the trafficking machinery that recognizes these signals and it is believed that cisternal sub-compartmentalization is achieved and maintained by a combination of retention and retrieval mechanisms. Yet, the signals, mechanism(s), and molecular factors involved are still unknown. Here, we address recent findings and summarize the current understanding of this fundamental process in plant cell biology.
Keywords: Endomembrane system, Golgi apparatus, glycosylation, glycosyltransferase
THE PLANT GOLGI APPARATUS: AN IMPRESSIVE CARBOHYDRATE FACTORY
The Golgi apparatus occupies a central position in the secretory pathway and serves as the central site of protein glycosylation and cell wall polysaccharide synthesis. From a very basic point of view, this organelle consists of three compartments: the cis-, medial-, and trans-Golgi. It receives newly synthesized secretory cargo from the endoplasmic reticulum (ER), which enters the Golgi at the cis-face. On its passage towards the trans-face, cargo is exposed to the sequential action of numerous glycosidases and glycosyltransferases that are predicted to be arranged in an assembly line for the processing of oligo- and polysaccharide chains. In addition to these enzymes, proper glycosylation requires the import of nucleotide sugar substrates into the Golgi and export of metabolites like inorganic phosphate to the cytoplasm (Reyes and Orellana, 2008; Cubero et al., 2009).
The uniqueness of the plant Golgi apparatus has been highlighted in several recent reviews (Robinson et al., 2007; Faso et al., 2009; Hawes et al., 2010). One peculiar feature of the Golgi apparatus in plant cells is its existence in the form of numerous discrete stacks that display an actin-myosin-driven motility throughout the cytoplasm in vacuolated cells. The lack of an intermediate compartment between ER exit sites and the cis-Golgi and absence of Golgi stack fragmentation or disassembly during cell division are additional features that may reflect the major functional characteristic of the plant Golgi, which is the synthesis and heavy secretion of many of the complex cell wall polysaccharides to the cell wall, which represents the richest source of biomass on Earth. In fact, the biosynthesis of non-cellulosic polysaccharides such as hemicelluloses and pectin in the Golgi apparatus is invaluable for countless commercial and industrial processes (Driouich et al., 1993; Geisler et al., 2008; Pauly and Keegstra, 2010). Despite the functional and morphological differences between animal and plant cells, there seems to be a strong conservation of basic protein transport mechanisms and Golgi organization across species.
N-GLYCOSYLATION: AN EVOLUTIONARY CONSERVED AND HIGHLY COORDINATED PROCESS
In all eukaryotes, the N-glycosylation of proteins starts in the ER by transfer of an oligosaccharide precursor to asparagine side chains in Asn–X–Ser/Thr sequences of nascent polypeptide chains (Helenius and Aebi, 2001; Pattison and Amtmann, 2009). Subsequently, glucose and mannose residues are cleaved off from the N-glycan by ER-resident glucosidases and mannosidases. This process is highly regulated and links N-glycosylation to protein folding and protein quality control in the ER. The best characterized processes are the calnexin/calreticulin cycle and the ER-associated degradation (ERAD) of glycoproteins, where specific N-glycan moieties and ER-lectins are involved (Caramelo and Parodi, 2008; Vembar and Brodsky, 2008). The importance of these glycan-dependent processes for plant development and defence mechanisms has been shown recently (Jin et al., 2007; Hong et al., 2008, 2009; Li et al., 2009; Liebminger et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009; Häweker et al., 2010).
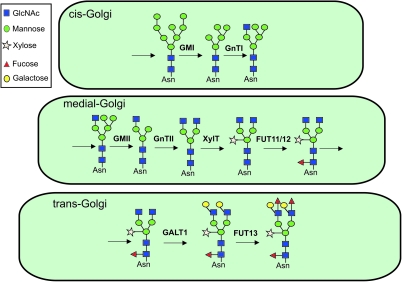
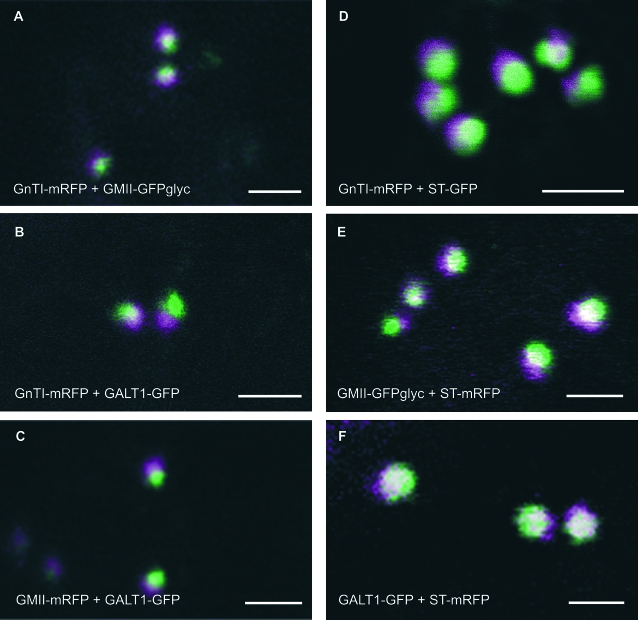
Further processing of the N-linked glycans to Man5GlcNAc2 is performed by Golgi-α-mannosidase I (GMI, Figure 1). This trimming reaction is the first committed step in the Golgi N-glycan processing pathway and has to be finished in order to enable subsequent N-glycan modifications (Strasser et al., 1999; Helenius and Aebi, 2001; Liebminger et al., 2009). Further processing within the Golgi is performed by the enzymes N-acetylglucosaminyltransferase I (GnTI), Golgi-α-mannosidase II (GMII), N-acetylglucosaminyltransferase II (GnTII), β1,2-xylosyltransferase (XylT), and core α1,3-fucosyltransferase (FUT11/12) (Strasser et al., 2006). Based on their in vitro and in vivo substrate specificity, no strict sub-compartmentalization of these processing enzymes is required to generate the characteristic complex N-glycan GnGnXF from the precursor substrate Man5GlcNAc2 (Strasser et al., 2004; Bencúr et al., 2005; Strasser et al., 2006). However, sub-cellular localization studies, mainly performed with fluorescent protein fusions, suggest that enzymes acting early in the N-glycan processing pathway tend to reside in cis- and medial-compartments of the Golgi, whereas later-acting enzymes seem to locate to the trans-Golgi cisternae and the trans-Golgi network (TGN). For example, Golgi live-cell imaging of fluorescent processing enzyme reporters revealed that the fluorescent signals derived from different enzymes overlapped only partially (Saint-Jore-Dupas et al., 2006; Schoberer et al., 2010) (Figure 2). This indicates a differential distribution of N-glycan processing enzymes in distinct Golgi cisternae and raises the question of how the sub-Golgi concentration of glycosylation enzymes is established and maintained in plant cells.
Figure 1.
Assembly-Line Model for N-Glycan Processing in Plant Golgi Stacks.
While the spatial separation of cis- from medial-located enzymes is less well established, the steady-state distribution of GALT1 and FUT13 is in the trans-Golgi.
Figure 2.
Localization of Fluorescently Tagged Glycosylation Enzymes within Golgi Stacks Using a Confocal Microscopy Approach.
Each two-colored image (A–F) shows highly magnified Golgi stacks double-labeled by two different enzymes that were transiently co-expressed in N. benthamiana leaf epidermal cells. Note the shift in the overlapping signals with the central region being white and the non-overlapping regions being green and magenta. (A) GnTI–mRFP (magenta) and GMII–GFPglyc (green) as well as (F) GALT1–GFP (green) and ST–mRFP (magenta) show larger areas of white, whilst (B) GnTI–mRFP (magenta) and GALT1–GFP (green), (C) GMII–mRFP (magenta) and GALT1–GFP (green), (D) GnTI–mRFP (magenta) and ST–GFP (green), and (E) GMII–GFPglyc (green) and ST–mRFP (magenta), respectively, display larger non-overlapping areas (magenta and green) indicating distinct intra-Golgi distributions. Scale bars = 2 μm.
SUB-COMPARTMENTALIZATION OF PLANT N-GLYCAN PROCESSING ENZYMES IN THE GOLGI APPARATUS
All of the Golgi-resident plant N-glycan processing enzymes are type II membrane proteins. The N-terminal region consists of a short cytoplasmic part, a single transmembrane domain (TMD), and a stem region (together, the CTS region, Table 1), which orients the catalytic domain into the Golgi lumen. The CTS region seems to contain all the targeting information for sub-Golgi localization (Saint-Jore-Dupas et al., 2006; Schoberer et al., 2009). A fluorescent protein fusion to the CTS region of rat α2,6-sialyltransferase (ST, Table 1) has become one of the most commonly used Golgi markers and, by using immuno-electron microscopy, was localized to the trans-half of Golgi stacks in leaves of Nicotiana clevelandii (Boevink et al., 1998). The same trans-Golgi labeling was found when a tagged full-length version of ST was expressed in Arabidopsis callus tissues (Wee et al., 1998). Moreover, the expression of a chimeric protein consisting of the CTS region of ST fused to the catalytic domain of human β1,4-galactosyltransferase (β1,4-GALT) resulted in the efficient production of di-galactosylated N-glycan structures in N. benthamiana (Strasser et al., 2009). In contrast, the CTS region of XylT fused to the catalytic domain of β1,4-GALT resulted in the formation of mainly mono-galactosylated and hybrid N-glycan structures (Bakker et al., 2006). Since galactosylation of N-glycans blocks further processing by GMII and GnTII, these incompletely processed N-glycans show that the ST–CTS region confers targeting to a later Golgi sub-compartment than the XylT–CTS region in leaf epidermal cells.
Table 1.
A Comparison of Putative CTS Regions from Different cis/medial- and trans-Golgi enzymes from Plants and Rat.
| Golgi enzymes | Cytoplasmic-transmembrane-stem region (the transmembrane domain is underlined and the length is indicated) | Total length of CTS region(aa)a |
| Cis/medial | ||
| 16 aa | ||
| GMIb | MARGSRSVGSSSSKWRYCNPSYYLKRPKRLALLFIVFVCVSFVFWDRQT | 49 aa |
| 18 aa | ||
| GnTIc | MRGYKFCCDFRYLLILAAVAFIYIQMRLFATQSEYADRLAAAIEAENHCTSQTRLLIDQISQQQGRIVALEEQMKRQ | 77 aa |
| 20 aa | ||
| XT1d | MIEKCIGAHRFRRLQRFMRQGKVTILCLVLTVIVLRGTIGAGKFGTPEKDIEEIREHFFYTRKRGEPHRV | 70 aaj |
| Medial | ||
| 23 aa | ||
| MUR3e | MFPRVSMRRRSAEVSPTEPMEKGNGKNQTNRICLLVALSLFFWALLLYFHFVVLGTSNIDKQLQLQPSYA | 70 aaj |
| Trans | ||
| 18 aa | ||
| GALT1f | MKRFYGGLLVVSMCMFLTVYRYVDLNTPVEKPYITAAASVVVTPNTTLPMEWLRITLPDF | 60 aa |
| 23 aa | ||
| FUT13g | MPMRYLNAMAALLMMFFTLLILSFTGILEFPSASTSMEHSIDPEPKLSDSTS | 52 aa |
| 23 aa | ||
| FUT1h | MDQNSYRRRSSPIRTTTGGSKSVNFSELLQMKYLSSGTMKLTRTFTTCLIVFSVLVAFSMIFHQHPSDSN | 70 aaj |
| 17 aa | ||
| STi | MIHTNLKKKFSLFILVFLLFAVICVWKKGSDYEALTLQAKEFQMPKSQEKVA | 52 aa |
Amino acids.
Glycine max Golgi-α-mannosidase I.
Nicotiana tabacum N-acetylglucosaminyltransferase I.
Arabidopsis thaliana α1,6-xylosyltransferase.
A. thaliana β1,2-galactosyltransferase.
A. thaliana β1,3-galactosyltransferase.
A. thaliana α1,4-fucosyltransferase.
A. thaliana α1,2-fucosyltransferase.
Rattus norvegicus α2,6-sialyltransferase.
the first 70 amino acids are shown for these enzymes, since the N-terminal targeting regions have not been determined.
The formation of the Lewis a carbohydrate structure on N-glycans involves the sequential action of β1,3-galactosyltransferase (GALT1) and α1,4-fucosyltransferase (FUT13) (Strasser et al., 2007) (Figure 1). It has been shown that these reactions are late Golgi events and hence take place in the trans-Golgi (Fitchette et al., 1999). Moreover, fluorescent fusions of the GALT1–CTS region to GFP resulted in a trans-Golgi distribution as observed by co-expression experiments with ST and other Golgi markers (Schoberer et al., 2010). Together, these data are clearly in favor of a sequential distribution of N-glycan processing enzymes across the Golgi stack, with a clear separation of early- (GMI, GnTI, GMII, GnTII, XylT, and FUT11/12) from late-acting enzymes (GALT1, FUT13). As a consequence, the existence of a distinct medial-Golgi or intermediate location can be questioned. However, GFP-tagged forms of GnTI (Reichardt et al., 2007) and XylT (Pagny et al., 2003) were found to display an increased labeling of medial stacks in electron micrographs and GMI located nearly exclusively to medial-Golgi cisternae in a 3D-tomographic model obtained by using an antibody that detects the native Arabidopsis GMI proteins (Staehelin and Kang, 2008). Although the latter finding is in clear contrast with two other studies that detected GMI–GFP in cis-cisternae (Nebenführ et al., 1999; Saint-Jore-Dupas, 2006), these results support the idea that a distinct concentration of enzymes in medial-Golgi cisternae exists in plants. The still limited dataset indicates that a cis/medial location significantly overlaps.
As mentioned earlier, the plant Golgi apparatus contains numerous glycosyltransferases involved in the formation and remodeling of cell wall polysaccharides. For most of these proteins, the sub-cellular targeting regions have not been analyzed in detail. However, recently, the cis/medial- and trans-Golgi location of three GFP-tagged Arabidopsis glycosyltransferases—α1,6-xylosyltransferase (XT1), β1,2-galactosyltransferase (MUR3), and α1,2-fucosyltransferase (FUT1)—involved in the biosynthesis of xyloglucan side chains has been shown by immunogold electron microscopy in tobacco BY2 cells (Chevalier et al., 2010). Similarly to the aforementioned N-glycan processing enzymes, all three glycosyltransferases are type II membrane proteins with a short N-terminal cytoplasmic region (Table 1). Moreover, analysis of the domain structure of other Golgi-located plant glycosyltransferases indicates that the vast majority of them are type II membrane proteins (Dunkley et al., 2006; Strasser, unpublished). These data suggest that plant N-glycan processing enzymes and glycosyltransferases for cell wall polysaccharides utilize similar conserved mechanisms and signals for sequential distribution in the different Golgi cisternae.
MODELS FOR DISTINCT SUB-GOLGI LOCALIZATION OF GLYCOSYLATION ENZYMES
Models for sorting and concentration of glycosylation enzymes are inevitably dependent on the nature of protein transport through the Golgi (Munro, 1998). The two prevailing models are the cisternal maturation and the vesicular transport model (Pelham and Rothman, 2000; Rabouille and Klumperman, 2005; Jackson, 2009; Tu and Banfield, 2010). In the cisternal maturation model, an efficient retrieval of resident proteins from late Golgi cisternae back to early cisternae is required to fine-balance the spatial organization of the glycosylation enzymes across the stack. This mechanism is based on discrimination between cis/medial- and trans-located enzymes in order to achieve their non-homogeneous distribution throughout the Golgi stack. Moreover, the selective enrichment of late-Golgi-resident proteins could also be achieved by continuous and highly efficient recycling from post-Golgi organelles, like the TGN, back to the trans-Golgi. The vesicular transport model requires trafficking of glycosylation enzymes to specific cisternae and a mechanism to retain them by preventing their anterograde transport. While the debate on protein transport models is still ongoing, the data from plant cells are more in favor of the cisternal maturation model (Hawes and Satiat-Jeunemaitre, 2005; Hawes et al., 2010). Dense vesicles, for example, which transport proteins to protein storage vacuoles, form at the rims of cis-Golgi cisternae and proceed and mature towards the trans-face of the Golgi stack while staying attached to the cisternae (Hillmer et al., 2001; Robinson et al., 2005). A similar observation has been made for many scale types of green algae, which progress through the stack in a cis-to-trans direction within maturing cisternae, even though algal scales are small enough to fit into coat protein (COP)I-coated vesicles (Becker and Melkonian, 1996).
Recently, a new model, the so-called cisternal progenitor model, has been proposed (Pfeffer, 2010). It postulates the existence of stable compartments that have the capability to generate subsequent compartments of the Golgi apparatus in a process that involves homotypic fusion and fission of Golgi membranes driven by Golgi-located Rab family GTPases acting together in so-called Rab cascades (Grosshans et al., 2006; Rivera-Molina and Novick, 2009). As a consequence of the model, the presence and activation of certain Rabs would create membrane microdomains and, in the context of the Golgi apparatus, biochemically define a Golgi sub-compartment by regulating the specific retention of glycosyltransferases in such domains. Although this alternative concept does not preclude the existence of transport vesicles, it is not clear how the concentration of Golgi enzymes like GMII in COPI vesicles can be combined with the cisternal progenitor model (Martinez-Menárguez et al., 2001; Nilsson et al., 2009). Until now, a clear support for the progenitor model is missing in plants, but it has been shown that Rab proteins determine membrane identity and membrane traffic in the early endomembrane system of plants (Woollard and Moore, 2008). It has become clear that Rabs are key players in a highly coordinated ‘interaction network’ consisting of effector molecules such as soluble N-ethylmaleimide-sensitive factor attachment receptor (SNARE) proteins, coat proteins, and a cohort of tethers with well established roles in membrane trafficking and organization of the Golgi stack. So-called tethering factors include multi-protein complexes, such as the conserved oligomeric Golgi (COG) complex, and coiled-coil proteins of the golgin family. The latter are regulated by Rabs, which help to bring vesicles closer to their targets and act in concert with subsets of SNAREs that facilitate docking and fusion (Robinson et al., 2007; Hawes et al., 2008). Although Arabidopsis homologs of all eight COG complex subunits have been identified in silico by database mining (Latijnhouwers et al., 2005), the molecular function of this complex in plants is still unknown. In mammals and yeast, on the other hand, there is compelling evidence that this tethering complex controls vesicular, and more specifically retrograde, trafficking within the Golgi apparatus (Smith and Lupashin, 2008).
In mammals, four different models have been proposed for the sub-Golgi localization of glycosylation enzymes. In the ‘kin recognition model’, it has been proposed that especially cis/medial-Golgi enzymes such as GnTI and GMII oligomerize to form higher-order complexes that are excluded from forward transport to downstream compartments (Nilsson et al., 1993, 1994). In mammalian cells, it was recently proposed that in addition to the oligomerization of cis/medial-resident processing enzymes, also medial/trans enzymes such as β1,4-GALT and ST form protein complexes (Hassinen et al., 2010), which might be required for correct and efficient processing of N-glycans. However, evidence for oligomerization of early and/or late Golgi-resident glycosylation enzymes is still lacking in plants and the CTS regions of β1,4-GALT and ST, respectively, target different Golgi regions in N. benthamiana leaf epidermal cells (Strasser et al., 2009), suggesting that these two mammalian proteins do not reside within the same Golgi cisternae and therefore do not form a protein complex in plant cells. Arguing against a potential oligomerization-based retention mechanism are studies using fluorescence recovery after photobleaching (FRAP) techniques. The expression and subsequent photobleaching of the GFP-tagged signal anchor sequences of various Golgi-resident N-glycan processing enzymes such as GnTI, GALT1, and ST (Brandizzi et al., 2002a) have revealed that these integral Golgi membrane proteins recover rapidly (within minutes) after bleaching and have little or no immobile fractions (Schoberer et al., 2010). This indicates that, first, these enzymes have unhindered mobility in the Golgi membranes and hence do not seem to be physically restrained by a potential complex formation, and, second, appear to associate only transiently with this organelle before recycling to the ER rather than being stable resident components of the Golgi (Ward and Brandizzi, 2004).
The ‘bilayer thickness model’ suggests that the membrane thickness gradually increases from the ER to the plasma membrane and this change excludes proteins with a shorter transmembrane domain (TMD) from forward transport (Bretscher and Munro, 1993; Munro, 1995). Since the composition of the lipid bilayer changes in the Golgi, this process could be involved in trafficking and retention of membrane proteins within different cisternae. Indeed, the TMD length plays a role in the targeting of type I transmembrane proteins in plants (Brandizzi et al., 2002b) and an increase in TMD length of the early enzyme GMI from 16 to 23 amino acids resulted in trans-Golgi targeting (Saint-Jore-Dupas et al., 2006). However, the TMD lengths of different ER- and Golgi-resident glycosylation enzymes vary significantly, which indicates that this model cannot explain the segregation of glycosylation enzymes into different Golgi cisternae. ST, for example, has a rather short membrane-spanning region with a length of 17 amino acids, which is in a similar range to the predicted TMD lengths of cis/medial-located enzymes such as GMI and GnTI (Saint-Jore-Dupas et al., 2006; Liebminger et al., 2009; Schoberer et al., 2009) (Table 1). On the other hand, these enzymes’ TMD lengths have not been determined experimentally and the TMD length predictions vary, dependent on the used algorithm (Davis et al., 2006). In addition, TMDs might not only be integrated into the membrane in a linear manner, but could also be subjected to adaptation by tilting or oligomerization of transmembrane segments (Killian and Nyholm, 2006). Recently, the systematic investigation of TMDs of integral membrane proteins from fungi and mammals revealed organelle-specific differences in length of the TMDs as well as in composition (Sharpe et al., 2010). No changes between cis- and medial-Golgi proteins were identified when the mean residue hydrophobicity or the amino acid volume were compared. However, due to the limited number of proteins with confirmed sub-Golgi location, the dataset to analyze specific features of transmembrane segments was rather small.
In a related model, the rapid partitioning model, which was proposed by the Lippincott-Schwartz group, glycosylation enzymes are supposed to be partitioned from the export machinery using a lipid-based sorting mechanism (Patterson et al., 2008). This model suggests a bidirectional transport between different cisternae and proposes a role for self-organizing lipid-domains in partitioning of cargo and resident proteins into domains of different lipid compositions. Although this model might explain the mono-exponential kinetics of cargo exit from the Golgi in mammalian cells, it also proposes the formation of incompletely processed N-glycans, which are normally not observed, and is in conflict with established data for the transport of large cargo through the Golgi (Nilsson et al., 2009). Moreover, also for that model, the intrinsic protein signals for lipid-driven sorting of Golgi-resident processing enzymes have not been identified and the relevance of this model for plant cells has not been tested yet. Recent data from mammalian cells have highlighted the importance of lipid membrane composition in the organization of Golgi-resident glycosylation enzymes. Changes in Golgi phosphatidylinositol-4-phosphate content resulted in mislocalization of GnTI and GMII accompanied by defects in glycosylation (Cheong et al., 2010). These data show that spatial regulation of phospholipids is a key regulator of Golgi organization. In plants, similar experiments have not been performed and functional data on phospholipids are still limited, but imaging of phosphatidylinositol-4-phosphate showed a clear co-location with ST in the Golgi (Vermeer et al., 2009). Yet, in another study, it was shown that inhibition of glucosylceramide biosynthesis disturbed transport of membrane and secretory proteins but remarkably had no effect on the Golgi targeting of ST in tobacco leaf epidermal cells (Melser et al., 2010). However, differences in sub-Golgi targeting were not investigated in this study.
In the last model, it has been proposed that the cytoplasmic tails of different glycosyltransferases contain important information for their sub-Golgi targeting and concentration. Exchange of the cytoplasmic protein parts resulted in alterations in sub-Golgi targeting of two glycosyltransferases involved in glycolipid biosynthesis in mammalian cells (Uliana et al., 2006). In yeast, the Vps74p protein has been found to directly bind to a conserved motif in the cytoplasmic tail of glycosyltransferases and by doing so might act as an adaptor for COPI-mediated retrieval leading to the dynamic localization of glycosylation enzymes (Schmitz et al., 2008; Tu et al., 2008). While this model is quite appealing and motifs in cytoplasmic regions of plant membrane proteins have been identified as signals for ER-to-Golgi targeting or retrieval (Hanton et al., 2005; Yuasa et al., 2005; Chatre et al., 2009; Schoberer et al., 2009), no obvious Vps74p homolog is present in plants and a Vps74p-like binding motif is not unequivocally recognizable in the cytoplasmic region of plant enzymes (Table 1). Therefore, it remains to be shown whether a similar mechanism that involves binding to the short cytoplasmic regions and results in either retrieval from late Golgi compartments or retention within distinct cisternae exists in plant cells.
COMPARTMENTALIZATION OF GLYCOSYLATION ENZYMES IN THE GOLGI: AN UNSOLVED MYSTERY?
In conclusion, it becomes obvious that none of the proposed models can so far explain the sub-Golgi distribution of a given N-glycan processing enzyme in plant cells. Most likely, the signals and mechanisms for the localization to their correct place in the Golgi have to be determined for each enzyme individually. Molecular determinants such as proteins that bind to cytoplasmic regions or recognize amino acid motifs in TMDs are still elusive in plants. The sequence heterogeneity observed in the rather short N-terminal CTS targeting regions of different glycosylation enzymes (Table 1) suggests that features within this region probably function in a combinatorial manner and may dictate the non-uniform distribution of different enzymes in different ways, which is strongly in favor of a contribution of multiple mechanisms to the spatial concentration of these proteins. A combination of lipid- (Patterson et al., 2008) and protein-based sorting (Schmitz et al., 2008; Tu et al., 2008), with an important role for the TMDs and flanking regions of glycosylation enzymes for establishing the polar distribution in the Golgi, might be essential determinants of sub-cellular targeting. Despite intense research over the past decade, the underlying mechanisms are still widely unsolved, but recent data emphasize that sorting/partitioning of proteins and lipids into different Golgi cisternae are highly dependent processes (Lippincott-Schwartz and Phair, 2010), which are accomplished by vesicular transport, organelle maturation, and continual fusion and fission of compartments. In the context of glycosylation enzymes, the existence of plant homologs of the COG complex subunits could be one possible answer to the question of how the Golgi apparatus maintains its non-uniform distribution of enzymes in the face of heavy secretion of transiting cargo (Latijnhouwers et al., 2005; Faso et al., 2009). Studies of the yeast and mammalian COG proteins imply that this complex is crucial for glycosylation enzyme homeostasis, as it orchestrates the recycling of cis- and medial-Golgi enzymes by tethering vesicles that constantly retrieve these proteins from trans-Golgi compartments back to their site of function (Smith and Lupashin, 2008; Foulquier, 2009). Mutations in components of the hetero-octameric COG complex severely affect protein N-glycosylation and in humans can cause so-called congenital disorders of glycosylation (Wu et al., 2004; Foulquier et al., 2006; Kranz et al., 2007). Our current research focuses on the identification of the respective plant COG component homologs and determination of their role in retrograde intra-Golgi transport of Golgi glycosylation enzymes in plants.
While the fundamental processes of targeting, retention, and retrieval of Golgi-resident glycan processing enzymes seem to be conserved between different phyla, it remains to be shown whether plant glycosylation enzymes also exist in protein complexes as proposed for mammalian and yeast cells (Nilsson et al., 1994; Jungmann and Munro, 1998; Opat et al., 2000; Hassinen et al., 2010) or whether the unique organization and function of the plant Golgi as a biosynthetic factory for cell wall polysaccharides (Driouich et al., 1993; Hawes and Satiat-Jeunemaitre, 2005; Robinson et al., 2007) resulted in different targeting/retention mechanisms. Clearly, a better understanding of the determinants and mechanisms that are essential to establish and maintain a non-uniform distribution of these Golgi enzymes and hence are important for the integrity of N-glycosylation and very likely also for cell wall polysaccharide synthesis will help us to harness this fundamental process optimally in terms of production of plant food and non-food products. In the future, emerging imaging technologies, such as three-dimensional structured illumination microscopy (3D-SIM) (Fitzgibbon et al., 2010), unbiased genetics screens (Boulaflous et al., 2008), the development of proteomic approaches to unravel protein–protein and protein–lipid interactions involved in membrane trafficking (Osterrieder et al., 2010), and a detailed characterization of the protein domains of Golgi-resident glycosylation enzymes, should help us to decipher one of the enigmas of this fascinating organelle.
FUNDING
Work in the Strasser group is funded by the Austrian Science Fund (P19494 and 20817). Some of the work reported in this review was funded by a research fellowship from the Austrian Science Fund (J2981-B20) awarded to J.S.
Acknowledgments
We would like to thank Prof. Chris Hawes for critical reading of the manuscript. No conflict of interest declared.
References
- Bakker H, et al. An antibody produced in tobacco expressing a hybrid beta-1,4-galactosyltransferase is essentially devoid of plant carbohydrate epitopes. Proc. Natl Acad. Sci. U S A. 2006;103:7577–7582. doi: 10.1073/pnas.0600879103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Melkonian M. The secretory pathway of protists: spatial and functional organization and evolution. Microbiol. Rev. 1996;60:697–721. doi: 10.1128/mr.60.4.697-721.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencúr P, et al. Arabidopsis thaliana beta1,2-xylosyltransferase: an unusual glycosyltransferase with the potential to act at multiple stages of the plant N-glycosylation pathway. Biochem. J. 2005;388:515–525. doi: 10.1042/BJ20042091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C. Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 1998;15:441–447. doi: 10.1046/j.1365-313x.1998.00208.x. [DOI] [PubMed] [Google Scholar]
- Boulaflous A, Faso C, Brandizzi F. Deciphering the Golgi apparatus: from imaging to genes. Traffic. 2008;9:1613–1617. doi: 10.1111/j.1600-0854.2008.00769.x. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C. Membrane protein transport between the endoplasmic reticulum and the golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. Plant Cell. 2002a;14:1293–1309. doi: 10.1105/tpc.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus J, Paris N. The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell. 2002b;14:1077–1092. doi: 10.1105/tpc.000620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher M, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Caramelo J, Parodi A. Getting in and out from calnexin/calreticulin cycles. J. Biol. Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatre L, Wattelet-Boyer V, Melser S, Maneta-Peyret L, Brandizzi F, Moreau P. A novel di-acidic motif facilitates ER export of the syntaxin SYP31. J. Exp. Bot. 2009;60:3157–3165. doi: 10.1093/jxb/erp155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong FY, et al. Spatial regulation of Golgi phosphatidylinositol-4-phosphate is required for enzyme localization and glycosylation fidelity. Traffic. 2010;11:1180–1190. doi: 10.1111/j.1600-0854.2010.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier L, et al. Subcompartment localization of the side chain xyloglucan-synthesizing enzymes within Golgi stacks of tobacco suspension-cultured cells. Plant J. 2010;64:977–989. doi: 10.1111/j.1365-313X.2010.04388.x. [DOI] [PubMed] [Google Scholar]
- Cubero B, et al. The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol. Plant. 2009;2:535–552. doi: 10.1093/mp/ssp013. [DOI] [PubMed] [Google Scholar]
- Davis M, Zhang F, Yuan Z, Teasdale R. MemO: a consensus approach to the annotation of a protein's membrane organization. Silico Biol. 2006;6:387–399. [PubMed] [Google Scholar]
- Driouich A, Faye L, Staehelin L. The plant Golgi apparatus: a factory for complex polysaccharides and glycoproteins. Trends Biochem. Sci. 1993;18:210–214. doi: 10.1016/0968-0004(93)90191-o. [DOI] [PubMed] [Google Scholar]
- Dunkley T, et al. Mapping the Arabidopsis organelle proteome. Proc. Natl Acad. Sci. U S A. 2006;103:6518–6523. doi: 10.1073/pnas.0506958103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faso C, Boulaflous A, Brandizzi F. The plant Golgi apparatus: last 10 years of answered and open questions. FEBS Lett. 2009;583:3752–3757. doi: 10.1016/j.febslet.2009.09.046. [DOI] [PubMed] [Google Scholar]
- Fitchette AC, Cabanes-Macheteau M, Marvin L, Martin B, Satiat-Jeunemaitre B, Gomord V, Crooks K, Lerouge P, Faye L. Biosynthesis and immunolocalization of Lewis a-containing N-glycans in the plant cell. Plant Physiol. 1999;121:333–344. doi: 10.1104/pp.121.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbon J, Bell K, King E, Oparka K. Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiol. 2010;153:1453–1463. doi: 10.1104/pp.110.157941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulquier F. COG defects, birth and rise! Biochim. Biophys. Acta. 2009;1792:896–902. doi: 10.1016/j.bbadis.2008.10.020. [DOI] [PubMed] [Google Scholar]
- Foulquier F, et al. Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc. Natl Acad. Sci. U S A. 2006;103:3764–3769. doi: 10.1073/pnas.0507685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler DA, Sampathkumar A, Mutwil M, Persson S. Laying down the bricks: logistic aspects of cell wall biosynthesis. Curr. Opin. Plant Biol. 2008;11:647–652. doi: 10.1016/j.pbi.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl Acad. Sci. U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton S, Renna L, Bortolotti L, Chatre L, Stefano G, Brandizzi F. Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell. 2005;17:3081–3093. doi: 10.1105/tpc.105.034900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassinen A, Rivinoja A, Kauppila A, Kellokumpu S. Golgi N-glycosyltransferases form both homo- and heterodimeric enzyme complexes in live cells. J. Biol. Chem. 2010;285:17771–17777. doi: 10.1074/jbc.M110.103184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häweker H, et al. Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 2010;285:4629–4636. doi: 10.1074/jbc.M109.063073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C, Satiat-Jeunemaitre B. The plant Golgi apparatus: going with the flow. Biochim. Biophys. Acta. 2005;1744:466–480. doi: 10.1016/j.bbamcr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Hawes C, Osterrieder A, Hummel E, Sparkes I. The plant ER–Golgi interface. Traffic. 2008;9:1571–1580. doi: 10.1111/j.1600-0854.2008.00773.x. [DOI] [PubMed] [Google Scholar]
- Hawes C, Schoberer J, Hummel E, Osterrieder A. Biogenesis of the plant Golgi apparatus. Biochem. Soc. Trans. 2010;38:761–767. doi: 10.1042/BST0380761. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- Hillmer S, Movafeghi A, Robinson DG, Hinz G. Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J. Cell Biol. 2001;152:41–50. doi: 10.1083/jcb.152.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, et al. Mutations of an alpha1,6 mannosyltransferase inhibit endoplasmic reticulum-associated degradation of defective brassinosteroid receptors in Arabidopsis. Plant Cell. 2009;21:3792–3802. doi: 10.1105/tpc.109.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell. 2008;20:3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson C. Mechanisms of transport through the Golgi complex. J. Cell Sci. 2009;122:443–452. doi: 10.1242/jcs.032581. [DOI] [PubMed] [Google Scholar]
- Jin H, Yan Z, Nam K, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell. 2007;26:821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J, Munro S. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killian J, Nyholm T. Peptides in lipid bilayers: the power of simple models. Curr. Opin. Struct. Biol. 2006;16:473–479. doi: 10.1016/j.sbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Kranz C, et al. COG8 deficiency causes new congenital disorder of glycosylation type IIh. Hum. Mol. Genet. 2007;16:731–741. doi: 10.1093/hmg/ddm028. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M, Hawes C, Carvalho C. Holding it all together? Candidate proteins for the plant Golgi matrix. Curr. Opin. Plant Biol. 2005;8:632–639. doi: 10.1016/j.pbi.2005.09.014. [DOI] [PubMed] [Google Scholar]
- Li J, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl Acad. Sci. U S A. 2009;106:15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebminger E, et al. Class I alpha-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell. 2009;21:3850–3867. doi: 10.1105/tpc.109.072363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Phair R. Lipids and cholesterol as regulators of traffic in the endomembrane system. Annu. Rev. Biophys. 2010;39:559–578. doi: 10.1146/annurev.biophys.093008.131357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Menárguez J, et al. Peri-Golgi vesicles contain retrograde but not anterograde proteins consistent with the cisternal progression model of intra-Golgi transport. J. Cell Biol. 2001;155:1213–1224. doi: 10.1083/jcb.200108029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melser S, et al. Glucosylceramide biosynthesis is involved in Golgi morphology and protein secretion in plant cells. Traffic. 2010;11:479–490. doi: 10.1111/j.1600-0854.2009.01030.x. [DOI] [PubMed] [Google Scholar]
- Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A, et al. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 1999;121:1127–1142. doi: 10.1104/pp.121.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, et al. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 2009;28:3428–3438. doi: 10.1038/emboj.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Au C, Bergeron J. Sorting out glycosylation enzymes in the Golgi apparatus. FEBS Lett. 2009;583:3764–3769. doi: 10.1016/j.febslet.2009.10.064. [DOI] [PubMed] [Google Scholar]
- Nilsson T, et al. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe M, Warren G. Kin recognition: a model for the retention of Golgi enzymes. FEBS Lett. 1993;330:1–4. doi: 10.1016/0014-5793(93)80906-b. [DOI] [PubMed] [Google Scholar]
- Opat A, Houghton F, Gleeson P. Medial Golgi but not late Golgi glycosyltransferases exist as high molecular weight complexes: role of luminal domain in complex formation and localization. J. Biol. Chem. 2000;275:11836–11845. doi: 10.1074/jbc.275.16.11836. [DOI] [PubMed] [Google Scholar]
- Osterrieder A, Hummel E, Carvalho C, Hawes C. Golgi membrane dynamics after induction of a dominant-negative mutant Sar1 GTPase in tobacco. J. Exp. Bot. 2010;61:405–422. doi: 10.1093/jxb/erp315. [DOI] [PubMed] [Google Scholar]
- Pagny S, et al. Structural requirements for Arabidopsis beta1,2-xylosyltransferase activity and targeting to the Golgi. Plant J. 2003;33:189–203. doi: 10.1046/j.0960-7412.2002.01604.x. [DOI] [PubMed] [Google Scholar]
- Patterson G, Hirschberg K, Polishchuk R, Gerlich D, Phair R, Lippincott-Schwartz J. Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell. 2008;133:1055–1067. doi: 10.1016/j.cell.2008.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattison RJ, Amtmann A. N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 2009;14:92–99. doi: 10.1016/j.tplants.2008.11.008. [DOI] [PubMed] [Google Scholar]
- Pauly M, Keegstra K. Plant cell wall polymers as precursors for biofuels. Curr. Opin. Plant Biol. 2010;13:305–312. doi: 10.1016/j.pbi.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Pelham H, Rothman J. The debate about transport in the Golgi: two sides of the same coin? Cell. 2000;102:713–719. doi: 10.1016/s0092-8674(00)00060-x. [DOI] [PubMed] [Google Scholar]
- Pfeffer SR. How the Golgi works: a cisternal progenitor model. Proc. Natl Acad. Sci. U S A. 2010;107:19614–19618. doi: 10.1073/pnas.1011016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Klumperman J. Opinion: the maturing role of COPI vesicles in intra-Golgi transport. Nat. Rev. Mol. Cell Biol. 2005;6:812–817. doi: 10.1038/nrm1735. [DOI] [PubMed] [Google Scholar]
- Reichardt I, et al. Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr. Biol. 2007;17:2047–2053. doi: 10.1016/j.cub.2007.10.040. [DOI] [PubMed] [Google Scholar]
- Reyes F, Orellana A. Golgi transporters: opening the gate to cell wall polysaccharide biosynthesis. Curr. Opin. Plant Biol. 2008;11:244–251. doi: 10.1016/j.pbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- Rivera-Molina FE, Novick PJ. A Rab GAP cascade defines the boundary between two Rab GTPases on the secretory pathway. Proc. Natl Acad. Sci. U S A. 2009;106:14408–14413. doi: 10.1073/pnas.0906536106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Herranz M-C, Bubeck J, Pepperkok R, Ritzenthaler C. Membrane dynamics in the early secretory pathway. Critical Reviews in Plant Sciences. 2007;26:199–225. [Google Scholar]
- Robinson DG, Oliviusson P, Hinz G. Protein sorting to the storage vacuoles of plants: a critical appraisal. Traffic. 2005;6:615–625. doi: 10.1111/j.1600-0854.2005.00303.x. [DOI] [PubMed] [Google Scholar]
- Saijo Y, et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C, et al. Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell. 2006;18:3182–3200. doi: 10.1105/tpc.105.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz K, et al. Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev. Cell. 2008;14:523–534. doi: 10.1016/j.devcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, et al. Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic. 2009;10:101–115. doi: 10.1111/j.1600-0854.2008.00841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Runions J, Steinkellner H, Strasser R, Hawes C, Osterrieder A. Sequential depletion and acquisition of proteins during Golgi stack disassembly and reformation. Traffic. 2010;11:1429–1444. doi: 10.1111/j.1600-0854.2010.01106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell. 2010;142:158–169. doi: 10.1016/j.cell.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RD, Lupashin VV. Role of the conserved oligomeric Golgi (COG) complex in protein glycosylation. Carbohydr. Res. 2008;343:2024–2031. doi: 10.1016/j.carres.2008.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staehelin L, Kang B. Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol. 2008;147:1454–1468. doi: 10.1104/pp.108.120618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Altmann F, Mach L, Glössl J, Steinkellner H. Generation of Arabidopsis thaliana plants with complex N-glycans lacking beta1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett. 2004;561:132–136. doi: 10.1016/S0014-5793(04)00150-4. [DOI] [PubMed] [Google Scholar]
- Strasser R, et al. A unique beta1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell. 2007;19:2278–2292. doi: 10.1105/tpc.107.052985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, et al. Improved virus neutralization by plant-produced anti-HIV antibodies with a homogeneous β1,4-galactosylated N-glycan profile. J. Biol. Chem. 2009;284:20479–20485. doi: 10.1074/jbc.M109.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Mucha J, Schwihla H, Altmann F, Glössl J, Steinkellner H. Molecular cloning and characterization of cDNA coding for beta1,2N-acetylglucosaminyltransferase I (GlcNAc-TI) from Nicotiana tabacum. Glycobiology. 1999;9:779–785. doi: 10.1093/glycob/9.8.779. [DOI] [PubMed] [Google Scholar]
- Strasser R, Schoberer J, Jin C, Glössl J, Mach L, Steinkellner H. Molecular cloning and characterization of Arabidopsis thaliana Golgi alpha-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 2006;45:789–803. doi: 10.1111/j.1365-313X.2005.02648.x. [DOI] [PubMed] [Google Scholar]
- Tu L, Banfield D. Localization of Golgi-resident glycosyltransferases. Cell Mol. Life Sci. 2010;67:29–41. doi: 10.1007/s00018-009-0126-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Tai W, Chen L, Banfield D. Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science. 2008;321:404–407. doi: 10.1126/science.1159411. [DOI] [PubMed] [Google Scholar]
- Uliana A, Giraudo C, Maccioni H. Cytoplasmic tails of SialT2 and GalNAcT impose their respective proximal and distal Golgi localization. Traffic. 2006;7:604–612. doi: 10.1111/j.1600-0854.2006.00413.x. [DOI] [PubMed] [Google Scholar]
- Vembar S, Brodsky J. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer J, Thole J, Goedhart J, Nielsen E, Munnik T, Gadella TJ. Imaging phosphatidylinositol 4-phosphate dynamics in living plant cells. Plant J. 2009;57:356–372. doi: 10.1111/j.1365-313X.2008.03679.x. [DOI] [PubMed] [Google Scholar]
- Ward TH, Brandizzi F. Dynamics of proteins in Golgi membranes: comparisons between mammalian and plant cells highlighted by photobleaching techniques. Cell Mol. Life Sci. 2004;61:172–185. doi: 10.1007/s00018-003-3355-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee EG, Sherrier DJ, Prime TA, Dupree P. Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell. 1998;10:1759–1768. doi: 10.1105/tpc.10.10.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard AA, Moore I. The functions of Rab GTPases in plant membrane traffic. Curr. Opin. Plant Biol. 2008;11:610–619. doi: 10.1016/j.pbi.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Wu X, et al. Mutation of the COG complex subunit gene COG7 causes a lethal congenital disorder. Nat. Med. 2004;10:518–523. doi: 10.1038/nm1041. [DOI] [PubMed] [Google Scholar]
- Yuasa K, Toyooka K, Fukuda H, Matsuoka K. Membrane-anchored prolyl hydroxylase with an export signal from the endoplasmic reticulum. Plant J. 2005;41:81–94. doi: 10.1111/j.1365-313X.2004.02279.x. [DOI] [PubMed] [Google Scholar]