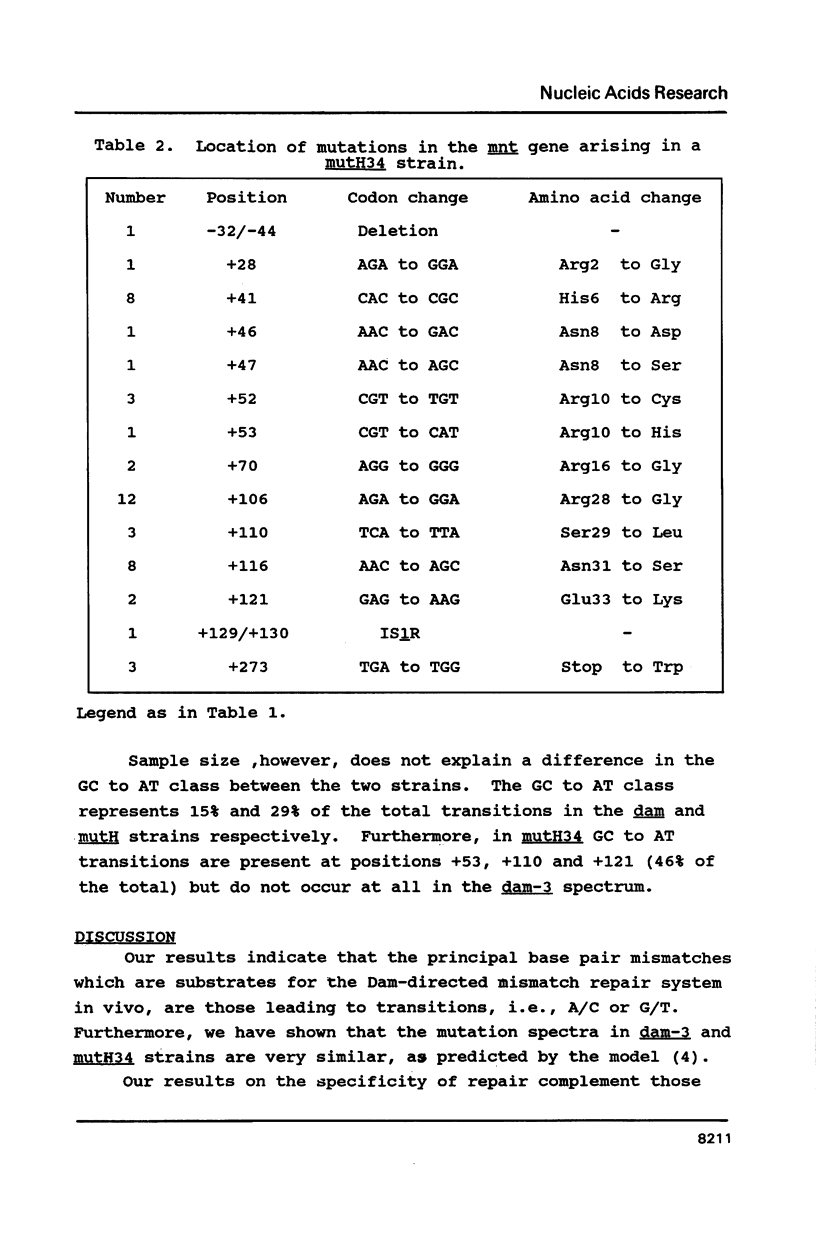
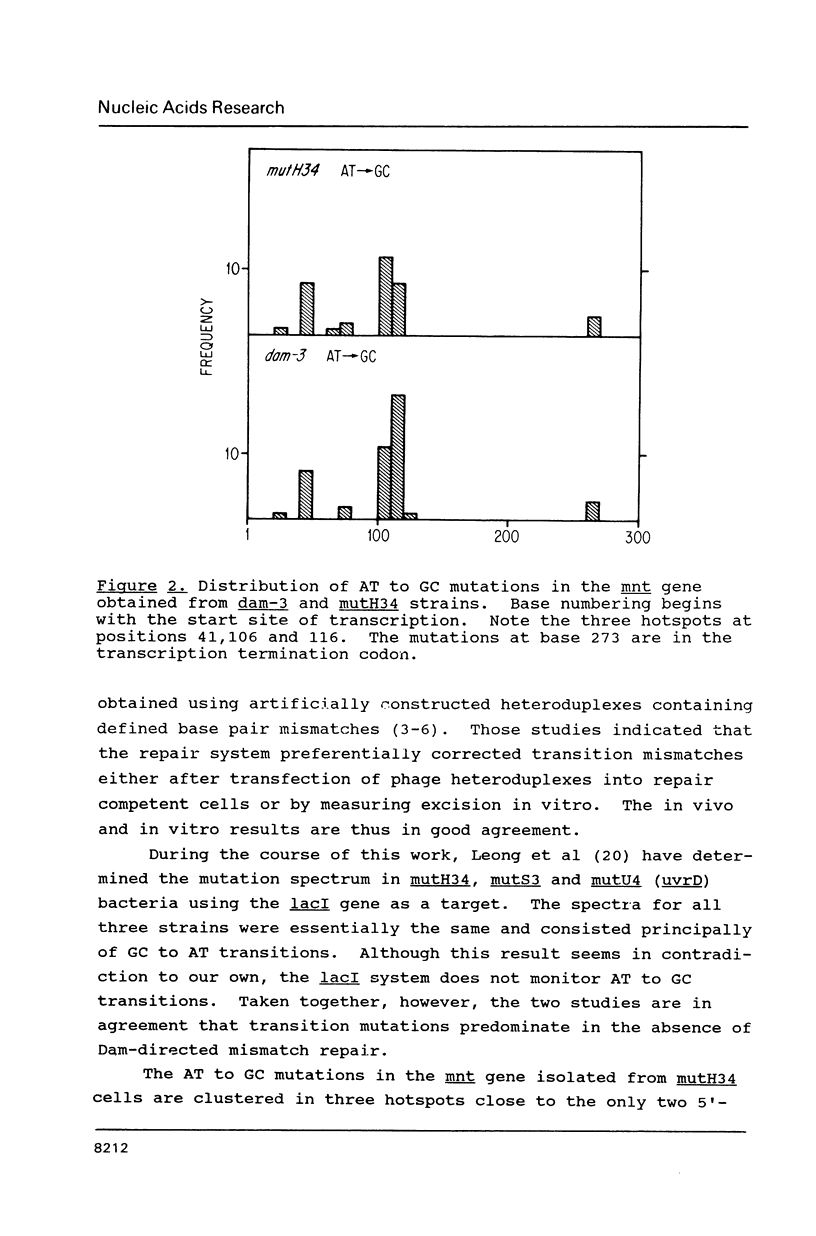
Abstract
The Dam-directed post-replicative mismatch repair system of Escherichia coli removes base pair mismatches from DNA. The products of the mutH, mutL and mutS genes, among others, are required for efficient mismatch repair. Absence of any of these gene products leads to persistence of mismatches in DNA with a resultant increase in spontaneous mutation rate. To determine the specificity of the mismatch repair system in vivo we have isolated and characterized 47 independent mutations from a mutH strain in the plasmid borne mnt repressor gene. The major class of mutations comprises AT to GC transitions that occur within six base pairs of the only two 5'-GATC-3' sequences in the mnt gene. In the wild type control strain, insertion of the IS1 element was the major spontaneous mutational event. A prediction of the Dam-directed mismatch repair model, that the mutation spectra of dam and mutH strains should be the same, was confirmed.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway M., Youderian P., Marinus M. G. Spontaneous mutations occur near dam recognition sites in a dam- Escherichia coli host. Genetics. 1987 Jul;116(3):343–347. doi: 10.1093/genetics/116.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B., Kramer W., Fritz H. J. Different base/base mismatches are corrected with different efficiencies by the methyl-directed DNA mismatch-repair system of E. coli. Cell. 1984 Oct;38(3):879–887. doi: 10.1016/0092-8674(84)90283-6. [DOI] [PubMed] [Google Scholar]
- Leong P. M., Hsia H. C., Miller J. H. Analysis of spontaneous base substitutions generated in mismatch-repair-deficient strains of Escherichia coli. J Bacteriol. 1986 Oct;168(1):412–416. doi: 10.1128/jb.168.1.412-416.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki H., Kornberg A. Proofreading by DNA polymerase III of Escherichia coli depends on cooperative interaction of the polymerase and exonuclease subunits. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4389–4392. doi: 10.1073/pnas.84.13.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968 Mar 23;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler A., Blöcker H., Frank R., Kahmann R. The mom gene of bacteriophage Mu: the mechanism of methylation-dependent expression. EMBO J. 1986 Oct;5(10):2719–2728. doi: 10.1002/j.1460-2075.1986.tb04556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youderian P., Vershon A., Bouvier S., Sauer R. T., Susskind M. M. Changing the DNA-binding specificity of a repressor. Cell. 1983 Dec;35(3 Pt 2):777–783. doi: 10.1016/0092-8674(83)90110-1. [DOI] [PubMed] [Google Scholar]
- Zagursky R. J., Berman M. L. Cloning vectors that yield high levels of single-stranded DNA for rapid DNA sequencing. Gene. 1984 Feb;27(2):183–191. doi: 10.1016/0378-1119(84)90139-2. [DOI] [PubMed] [Google Scholar]


