Abstract
Alterations in the human intestinal microbiota are linked to conditions including inflammatory bowel disease, irritable bowel syndrome, and obesity. The microbiota also undergoes substantial changes at the extremes of life, in infants and older people, the ramifications of which are still being explored. We applied pyrosequencing of over 40,000 16S rRNA gene V4 region amplicons per subject to characterize the fecal microbiota in 161 subjects aged 65 y and older and 9 younger control subjects. The microbiota of each individual subject constituted a unique profile that was separable from all others. In 68% of the individuals, the microbiota was dominated by phylum Bacteroides, with an average proportion of 57% across all 161 baseline samples. Phylum Firmicutes had an average proportion of 40%. The proportions of some phyla and genera associated with disease or health also varied dramatically, including Proteobacteria, Actinobacteria, and Faecalibacteria. The core microbiota of elderly subjects was distinct from that previously established for younger adults, with a greater proportion of Bacteroides spp. and distinct abundance patterns of Clostridium groups. Analyses of 26 fecal microbiota datasets from 3-month follow-up samples indicated that in 85% of the subjects, the microbiota composition was more like the corresponding time-0 sample than any other dataset. We conclude that the fecal microbiota of the elderly shows temporal stability over limited time in the majority of subjects but is characterized by unusual phylum proportions and extreme variability.
Keywords: etagenomics, gerontology, inflammation, Faecalibacterium, Firmicutes
Recent advances in culture-independent techniques for microbial community analysis (1, 2) have led to major progress in understanding the complexity of the human intestinal ecosystem (reviewed in ref. 3). High-throughput amplicon pyrosequencing is emerging as the method of choice for determining the microbial phylotypes present in a sample (4). Total shotgun sequencing of the microbial metagenome is a robust approach to determine community composition with low amplification bias (5), but the cost is currently prohibitive for routine application to large numbers of samples.
The initial infant gut microbiota is a simple structure usually dominated by bifidobacteria, and through a series of successions and replacements, it migrates to a more complex adult pattern (2, 6). The adult intestinal microbiota has been shown to be relatively stable over time (7) and is sufficiently similar between individuals to allow identification of a core microbiome comprising 66 dominant operational taxonomic units (OTUs) that corresponds to 38% of sequence reads from 17 individuals (8). Estimates of the numbers of organisms present in the human gut range from 1,183 bacterial phylotypes based on a meta-analysis (9) to 3,180 OTUs (8). Changes in the adult gut microbiota have been linked to undesirable health conditions including obesity (10), inflammatory bowel disease (IBD) (11), and irritable bowel syndrome (IBS) (12). The link between obesity and the microbiota is likely to be more sophisticated than the simple phylum-level Bacteroidetes: Firmicutes ratio initially identified (13), and it is likely to involve microbiota–diet interaction (14–16). More subtle alterations in the levels of other bacteria in the gut may also impact on health. Some of the microbiota members most responsive to diet include the Clostridium clusters IV and XIVa; many of these species are producers of butyrate, which is an important energy source for intestinal cells (17, 18). Clostridium cluster IV also includes Faecalibacterium prausnitzii, which is depleted in IBD patients (11, 19, 20) and has antiinflammatory activity in vitro and in mouse models (21). In contrast, increases in the numbers of adherent Escherichia coli cells have been associated with IBD (22, 23).
The proportion of people over the age of 65 y is increasing in many countries and will exceed the proportion of infants under 5 y of age on a global basis in 10 y (24). The aging intestine is subject to a number of physiological changes that impact on food digestion and absorption (25, 26) and immune function (27). Accompanied by a loss in the proportions of naïve CD4+ T cells, the inflamm-aging process is characterized by persistent activation of innate immunity mediated by the NF-ĸB transcription factor (28). Given the potential for elements of the microbiota to impact on these dynamic processes, there is considerable interest in determining the composition of the gut microbiota of elderly subjects and characterizing its variation as a potential determinant of health (3). The gut microbiota composition of elderly subjects is expected to be in a state of flux, a theory supported by culture-dependent methods (29). More recent studies have suggested that the Bacteroides species diversity was marginally increased in the feces of elderly subjects and that of bifidobacteria reduced (30), whereas a further study suggested that both Bacteroides numbers and species diversity decline in the elderly (31, 32). The proportions of the phyla Firmicutes, Bacteroidetes, and Proteobacteria were 58%, 15%, and 27%, respectively (33) in three elderly subjects in Japan, using 16S rRNA gene-clone library sequencing. In a study based in four European countries using labeled probe technology, significant variations in the proportions of major bacteria phyla were recorded between countries, emphasizing the danger of generalization of findings from one study location to another (34) and illustrating the likelihood of diet–microbiota interactions being involved.
The ELDERMET consortium (http://eldermet.ucc.ie) was established in 2007 to investigate the role of the intestinal microbiota in elderly Irish subjects as an agent and indicator of health. The initial phase of the ELDERMET project is to establish the baseline microbiota composition in several hundred elderly subjects, for whom extensive clinical, health, and dietary intake information has been obtained. This data will allow identification of individuals and cohorts for full shotgun fecal microbiome sequencing to investigate functional linkages to health status. Here, we report the data for the microbiota composition of 161 elderly subjects at time 0, 26 of which were also sampled 3 mo later. The data are providing unprecedented insights into the composition and interindividual variability of the gut microbiota in the elderly and identifying several organisms whose high variability suggests they may be amenable to therapeutic modulation by dietary or other means.
Results
Study Design and Clinical Analysis of an Elderly Subject Cohort.
Full ethical approval for this study was obtained from the local Research Ethics Committee. One hundred sixty-one subjects entered the study: 82 female and 79 male (SI Materials and Methods); 26 subjects, 13 female and 13 male, returned after 3 mo for fecal microbiota-composition analysis. Nine healthy subjects, five female and four male, were also included as young controls, with ages between 28 and 46 y. Table S1 describes the antibiotic use and age of subjects at time 0 (T0) and those subjects who returned at 3 mo (T3).
Bacteroidetes-Dominant Aggregate Intestinal Microbiota in Elderly Irish Subjects.
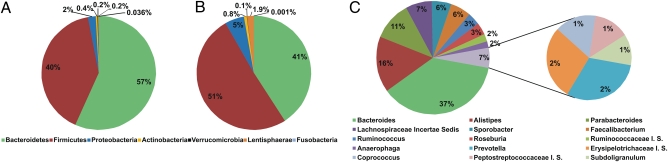
The composition of the fecal microbiota in each of the enrolled subjects was determined by sequencing ca. 40,000 16S rRNA gene V4 region amplicons as described in Materials and Methods and validated by comparison with a phylogenetic profiling microarray (35). Individual reads were identified to the lowest possible phylogenetic level using the Ribosomal Database Project (RDP) pipeline (36). The aggregate microbiota for 161 baseline samples plus 26 3-mo follow-up samples, based on a total of 6.3 million reads, indicated that the phylum Bacteroidetes was dominant at 57% (mean = 56.2 ± 1.1% SEM) compared with 40% (41.1 ± 1.1%) for phylum Firmicutes (Fig. 1A). Excluding either those 43 baseline subjects that had been treated with antibiotics or the 3-mo follow-up samples had little impact on the Bacteroidetes: Firmicutes ratio (Fig. S1). The fecal microbiota composition in nine younger adults, determined by the same protocol, was more typical for published studies of younger adults (Firmicutes-dominant) (Fig. 1B). The Bacteroidetes-dominant character of the microbiota in elderly subjects was also apparent from the genus assignments, despite the fact that the RDP classifier does not efficiently classify as many amplicon sequences at genus level as it does at phylum level; on average, 78 ± 1.1% SEM of all reads are assigned at genus level compared with 99 ± 0.1% at phylum level. Noteworthy among the well-characterized bacterial genera were Faecalibacterium spp. at 6% (6.1 ± 0.3%) of the 15 most abundant phyla, Ruminococcus spp. at 4% (3.9 ± 0.3%), Roseburia spp. at 3% (3.2 ± 0.2%) (Fig. 1C), and Bifidobacterium spp. at 0.41% (0.4 ± 0.07%). Exclusion of the subjects who had been treated with antibiotics in the 4 wk before the time-0 sample (n = 43) or exclusion of the 26 3-mo follow-up samples had no impact on the taxon assignment proportions (Fig. S1).
Fig. 1.
Aggregate microbiota composition at phylum level in (A) 187 (161 time-0 and 26 time 3-mo) fecal samples from elderly subjects, (B) 9 healthy younger adult controls, and (C) at genus level in the 187 fecal samples from elderly subjects (the smaller pie chart to right is inserted for clarity). Only major taxonomic groups are shown; these cover 95% of all reads assigned to genus level.
The clostridia constitute a large class in phylum Firmicutes that has been divided into 19 groups or clusters (37). These clusters include organisms that are dominant or subdominant organisms in the human intestinal microbiota (9), and variations in some of them have been reported in stool of IBD patients (38); F. prausnitzii, a prominent member of Clostridium cluster IV, is also depleted in IBD (39). To assist in the assignment of the short V4 amplicon sequences to Clostridium groups, we developed an approach based on an association table of V4 region sequences from species previously assigned to Clostridium clusters (Materials and Methods). Thus, some 25% of 6.3 million amplicon reads were assigned to Clostridium clusters. Clostridium clusters IV and XIVa represented the dominant assignments, with 56% (56.6 ± 1.1%) and 35% (34.7 ± 1.1%) of the assigned sequences, respectively. Clusters XVI and XI comprised 3% (3.4 ± 0.4%) and 2% (1.7 ± 0.2%), respectively, of the total assignments. In the younger adults, Clostridium clusters IV and XIVa were also the dominant clusters with similar proportions, at 56% (56.0 ± 1.6%) and 36% (35.3 ± 1.9%), respectively.
Dramatic Interindividual Variability of the Elderly Gut Microbiota.
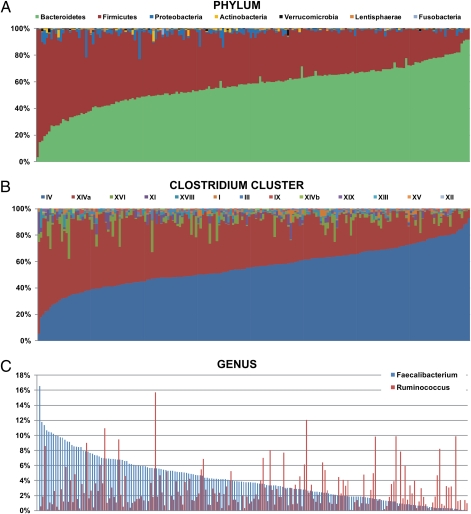
Whereas the aggregate microbiota was dominated by the Bacteroidetes, the individual composition datasets showed extraordinary variation, with the proportion of the Bacteroidetes ranging from 3% to 92% and the proportion of the phylum Firmicutes ranging from 7% to 94% (Fig. 2A). Exclusion of the 43 antibiotic-treated subjects or 3-mo samples had no impact on the degree of variability (Fig. S2). The proportions of five other major phyla also varied substantially between individuals. For example, in several microbiota communities, the phylum Proteobacteria was unusually abundant, comprising between 11% and 23% of total reads, whereas the average was 2% (Fig. S2A). Although the average proportion of the phylum Actinobacteria was under 1% (0.44%) (Fig. 2), some samples harbored unusually high levels, at 8%, 5%, and 4%. Similarly, considerable variation was detected for the proportions of the Clostridium clusters (Fig. 2B). Proportions of the total number of reads for clusters IV and XIVa ranged from 1.7% to 34% and 1.1% to 22%, respectively. None of the extreme values were from subjects treated with antibiotics or those returning after 3 mo (Fig. S3). Faecalibacterium and Ruminococcus varied most in their proportions, reaching 16.5% and 15.7% in some subjects, respectively, (Fig. 2C). The two highest proportions were, perhaps surprisingly, detected in individuals who had been administered antibiotics (Fig. S4B), but unusually high proportions of ca. 10–12% of both Faecalibacterium and Ruminococcus were also recorded in subjects not taking antibiotics (Fig. S4 A and C).
Fig. 2.
Interindividual variation in the proportion of major phyla (A), Clostridium clusters (B), and the genera Faecalibacterium and Ruminococcus (C) in 187 fecal samples from elderly subjects. Only the seven largest phyla are shown in A. Samples are ordered, from left, according to their Bacteroidetes proportion (A), cluster IV proportion of all reads assigned to Clostridium clusters (B), and Faecalibacterium proportion (C) to illustrate the dramatic interindividual variation.
Different Fecal Core Microbiota in Younger and Older Adults.
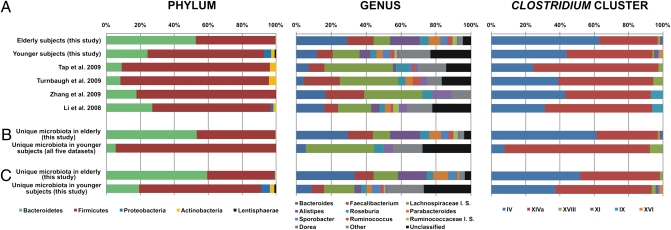
We identified and compared the core microbiota of younger adults [derived from previously published studies (8, 13, 40, 41) and ELDERMET subjects] (Table S2). Because procedures differ between individual studies, the bioinformatic analysis was harmonized by extracting V4 sequences from published full-length datasets from younger adults. Selection was based on full-length sequence availability (V4 extraction possible), being derived from healthy nonelderly adults and information on the subject from which they came. Although this comparison of high-coverage pyrosequencing data with lower coverage clone-library Sanger reads is complicated by the potential sampling bias of the latter, previous studies have shown similar distributions of the major taxa when measured by both full-length and partial pyrosequencing reads (4, 13, 41). We excluded from this comparison all subjects that had been treated with antibiotics, were subjected to gastric bypass (subjects gb1–3 in ref. 41), and were aged between 18 and 58 y among the healthy adults (subjects GG and BB in ref. 40). The extracted V4 region sequences were binned into unique sequences and grouped by subject. As had also been observed at the 97% OTU level (8), the subject specificity (proportion of unique sequences present in only one sample) was high in all datasets (Table S2). We subsequently defined the core microbiota as all of the unique sequences present in at least one-half of the subjects, following an approach applied in a recent study (8). Assignments to phylum, genus, and Clostridium cluster for each of these datasets revealed large differences between the elderly and younger adults (Fig. 3). More than one-half of the core microbiota (53%) in the elderly subjects-comprised Bacteroidetes were made up by the genera Bacteroides (29%), Alistipes (17%), and Parabacteroides (7%) compared with 8–27% in the younger adults. Although less pronounced, assignments to the two major Clostridium clusters were also unequal, even more than when comparing aggregate microbiota, with cluster IV being the more predominant cluster among subjects older than 65 y, whereas cluster XIVa was more prevalent in the younger cohort.
Fig. 3.
(A) The core fecal microbiota of elderly subjects at the levels of phylum, genus and Clostridium cluster compared with similarly defined core microbiota from four studies of younger adults and nine young adult controls. (B) Unique microbiota defined by comparing elderly and younger adults (published datasets). (C) Unique microbiota defined by comparing elderly and younger adults (this study).
Because the same variable region was used in this comparison of five studies, we were able to identify and classify core sequences that were unique to subjects either at least 65 y of age (400 sequences) or between 18 and 58 y (18 sequences) (Tables S3 and S4). The sequences unique to the elderly subjects correspond to a wide array of Bacteroides, Alistipes, and Parabacteroides species, whereas younger subjects had one unique Bacteroides species, most similar to Bacteroides vulgatus. The elderly subjects also had a much higher abundance of Faecalibacterium spp. and consequently, Clostridium cluster IV. The composition of the (Bacteroidetes-dominated) core microbiota that was unique to elderly subjects was not markedly different when it was defined by either comparison with the datasets of younger subjects from this study combined with the four published datasets (Fig. 3B) or comparison exclusively with the data from the nine younger subjects generated in this study (Fig. 3C). The latter comparison resulted in 167 and 1,177 core sequences unique to the elderly and younger adults in this study, respectively. This shows that the low sampling size of the Sanger data in the former comparison did not compromise the outcome.
Intraindividual Fecal Microbiota Stability in a Majority of Elderly Subjects.
To analyze changes in the microbiota over 3 mo in the same subject, we first analyzed the relatedness of the total pool of reads from all samples using UniFrac, as detailed in Materials and Methods. Briefly, a phylogenetic tree was built from the alignment of all ∼1 million unique V4 sequence reads, from which UniFrac distances between all samples were calculated. These distances were used for hierarchical clustering, whereby relative abundances of unique sequences were factored (weighted UniFrac tree) (Fig. S5A) or only presence/absence data were factored (unweighted UniFrac tree) (Fig. S5B). In the unweighted tree for 161 time-0 and 26 time 3-mo samples, 85% (22/26) of the microbiota compositions for the time 3-mo samples localize most closely to their time-0 sample. This figure drops to 62% based on the weighted tree, presumably because of discrimination based on phylotypes being overwhelmed by weighting values of fewer taxa that each have more reads. However, by either approach, a given microbiota after 3 mo was more like to localize to its original composition than to any other sample analyzed.
Fig. S6 shows the fecal microbiota composition for the 26 pairs of time-0/time 3-mo fecal samples at phylum (Fig. S6A) and genus assignment levels (Fig. S6B). Four of six temporal pairs that matched only in the unweighted tree were still proximal in the weighted tree (Fig. S5B). It is evident that those temporal pairs that are separated in both weighted and unweighted trees also show larger variations of phyla and genus proportions. However, even for some temporal pairs that are each other's closest neighbors in the unweighted tree only (e.g., EM047 and EM053), there are also relatively large phylum and genus variations.
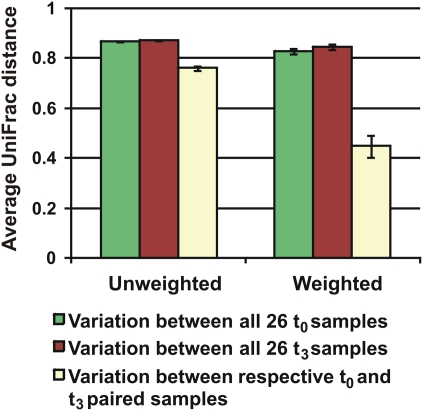
Microbiota compositional differences were greater between individuals than within individuals (Fig. 4). The average unweighted UniFrac distances between 26 fecal microbiota communities were 0.87 ± 0.0016 SEM for the time-0 samples and 0.87 ± 0.0014 for the time 3-mo samples. The average unweighted UniFrac distance within the time-0 to time 3-mo samples was lower, at 0.76 ± 0.0104. The unweighted UniFrac distance within the 26 individuals from time 0 to time 3 mo was significantly lower than the between individual unweighted UniFrac distances at both time 0 (robust t test; P < 0.001) and time 3 mo (robust t test; P < 0.001). The weighted UniFrac analysis show a similar trend with average UniFrac distances between time-0 samples of 0.83 ± 0.0102 SEM, an average distance of 0.85 ± 0.0115 between all time 3-mo samples, and a much lower average UniFrac distance of 0.45 ± 0.0434 for between time-0 and time 3-mo samples. The weighted UniFrac distance within the 26 individuals from time 0 to time 3 mo was significantly lower than the between individual weighted UniFrac distances at both time 0 (robust t test; P < 0.001) and time 3 mo (robust t test; P < 0.001).
Fig. 4.
Variation in the microbiota between time-0 and time 3-mo samples from 26 subjects. Mean (± SEM) unweighted and weighted UniFrac distances between the samples as indicated.
In some paired samples (e.g., EM059), the loss of phylogenetic proximity was not readily apparent at phylum level but was evident at genus level (Fig. S6B). Analysis of the genus assignments for these pair-wise comparisons showed that relative increases in the Firmicutes proportions, particularly Sporobacter spp. (order Clostridiales, family Ruminococcaceae), and the Lachnospiraceae family (order Clostridiales), were often present in microbiota pairs that diverged. Changes in the proportions of Alistipes spp. (order Bacteroidales, family Rikenellaceae), Parabacteroides (order Bacteroidales), or Anaerophaga (order Bacteroidales) were also responsible for other divergent paired samples.
Effect of Antibiotics on the Microbiota.
The impact of antibiotics usage was not readily evident from high-level comparisons of aggregate data (Fig. S1 B and C); a modest but significant effect of antibiotic use on the abundance of both phyla and genera was recorded. The average proportion of total reads assigned to the different phyla and genera were compared according to antibiotic use for the 161 baseline samples. Of the seven largest phyla in terms of composition proportion, a significantly increased relative abundance of Bacteroidetes (Mann–Whitney test; P = 0.0297) and a significant increase in the Bacteroidetes: Firmicutes ratio (Mann–Whitney test; P = 0.0363) were recorded in antibiotic users, with decreases in the relative abundance of both Firmicutes (Mann–Whitney test; P = 0.0445) and Proteobacteria [significant using the Mann–Whitney (P = 0.0232) but not the t test (P > 0.05)]. At the level of genus, only the relative abundance of Parabacteroides was significantly increased (Mann–Whitney test; P = 0.0465) with antibiotic use.
Five of twenty-six subjects that provided both time-0 and time 3-mo fecal samples had been administered antibiotics between these visits, one belonging to the first group in Fig. S6 (EM059), three belonging to the second group (EM047, EM053, and EM077), and one belonging to the last group (EM090). There were significant differences in the time-0 and time 3-mo fecal microbiota profiles of these subjects according to antibiotic use. Antibiotic use significantly decreased the relative abundance of Actinobacteria (Mann–Whitney test; P = 0.0472). At genus level, Alistipes abundance was significantly increased in subjects who had taken antibiotics using the t test (P < 0.05) but not the Mann–Whitney test (P = 0.1263). Only two of these five subjects had been prescribed the same antibiotic, albeit at different dosages. A low, 250-mg dose of clarithromycin decreased the proportion of Bacteroidetes (Bacteroides and Parabacteroides) and equivalently increased the proportion of Firmicutes (Alistipes). In contrast, a higher, 500-mg dose of clarithromycin increased the proportion of Bacteroidetes (Parabacteroides) and nonequivalently decreased the proportion of Firmicutes (Alistipes), indicating changes in the proportions of other phyla.
Discussion
This study constitutes the largest and deepest sampling of the composition of the elderly gut microbiota reported to date. The fecal microbiota of younger adults was previously shown by temperature gradient gel electrophoresis (TGGE) and phylochip (HitChip) analysis to be stable and individual-specific (7, 42). In the present study, all 161 of the time-0 microbiota samples could be clearly separated by UniFrac analysis (Fig. S5), and fine-detail analysis of genus confirmed individual-specific microbiota configurations. The HitChip study also found that temporally paired samples from elderly subjects were more similar than randomly compared samples from different subjects. However, the similarity of paired samples decreased from time 1 mo to time 2 mo. In the five elderly subjects that they investigated, the decrease in microbiota similarity was more pronounced in the Actinobacteria (42), whereas in younger adults, greatest divergence was recorded for Clostridium cluster IV. We observed a similar phenomenon in the 3-mo microbiota comparisons, suggesting that these levels of bacteria are commonly in flux in elderly subjects.
Subjects over 65 y of age often have morbidities ranging from mild to severe that increase in complexity and seriousness with advancing age. These conditions are treated with a complex repertoire of medications that are likely to have individual or combined effects on the microbiota. For the 4 of 26 (∼15%) of the time 3-mo microbiota samples that were not stable over 3 mo in the unweighted UniFrac tree, no association was identified with the potentially influential factors of nonantibiotic prescription medication change or illness. Because antibiotic treatment is relatively common in the elderly, we did not exclude enrolling such subjects in the study but rather, treated them as a separate group for most analyses. Antibiotic treatment has been shown by culture-dependent methods (32), TGGE (43), and an elegant molecular method (4) to dramatically disturb the composition of the fecal microbiota. In three subjects treated with ciprofloxacin (4), the antibiotic caused a decrease in the taxonomic richness, diversity, and evenness of the community. Intriguingly, the magnitude of the changes and the taxa affected were different in the three subjects, and the overall community structure had been restored by 4 wk posttreatment. Stochastic or individual-specific effects on the microbiota were also described in another study (43). In the simplest comparison of antibiotic treatment that we could make, different doses of clarithromycin resulted in counterintuitive impacts on the microbiota, which could clearly be complicated and confounded by factors other than antibiotic use.
The gut microbiota of the elderly revealed here (by analysis of a fecal surrogate sample, similar to the majority of other studies) was substantially different from that of younger adults (13, 40, 44–46), particularly with regard to the lower proportion of phylum Firmicutes detected. Over 65% of the time-0 microbiota communities (Fig. 3) had atypical Bacteroidetes: Firmicutes ratios compared with those described for younger adults. In a previous pilot study using the same primers, we showed that V4-based phylogenetic assignments correlated well with microbiota composition data produced by hybridizing full-length 16S rRNA gene amplicons to a phylochip (35). This indicates lack of a primer-based bias in favor of the Bacteroidetes. Furthermore, the aggregate composition of the nine young adults analyzed using our DNA extraction, amplification, sequencing, and phylogenetic methodologies was similar to that found in other studies of younger adults (45) and was not Bacteroidetes-dominant. The diversity and range of phylum proportions that we identified herein mean that comparison of aggregate figures must be interpreted judiciously. Nevertheless, some top-level comparisons are informative. The range of phylum Firmicutes in our study, across all 161 time-0 samples, varied from 8% to 80%, whereas phylum Bacteroidetes varied from 14% to 92%. The phylum Actinobacteria includes Bifidobacterium spp., which are among the best characterized of the commensal organisms considered beneficial or probiotic in nature (47). This phylum ranged from 0% to 8% in the total dataset from 161 fecal samples, comparing with values of 0.4% (41) to 11.4% (46) in other studies of younger adults. In our analysis of the core microbiota, we observed a clear shift to a more Clostridium cluster IV-dominated community in the elderly. These elderly-specific core sequences all belonged to the Ruminococcaceae family and essentially, comprised Faecalibacterium, Sporobacter, and Ruminococcus species. In a recent study, albeit using lower-resolution molecular methods (48), the proportion of Bacteroidetes in the fecal microbiota of 17 institutionalized elderly subjects was significantly higher than in younger adults. That study also noted relatively low Bifidobacterium proportions in elderly subjects, lower proportions of Clostridium cluster IV, and lower diversity overall (48).
This microbiota composition profiling provides the basis for a follow-up phase focusing on the function of the elderly gut microbiota. By first investigating the microbiota composition in a large panel of subjects of defined community setting, health status, and diet, we can rationally select samples for fecal shotgun metagenomics by reference to both host parameters and microbiota diversity. This approach is validated by the remarkable interindividual microbiota diversity that was evident in 161 time-0 fecal samples from phylum down to unique OTU level. Prior knowledge of microbiota composition will allow for more cost-effective application of shotgun microbiome sequencing to samples selected for ranges or types of phylogenetic diversity.
Materials and Methods
Subject Recruitment and Sample Collection.
This study was approved by the Clinical Research Ethics Committee of the Cork Teaching Hospitals. Subjects, aged 65 y and older, were recruited and clinically investigated in two local hospitals. Exclusion criteria were a history of alcohol abuse, participation in an investigational drug evaluation within the last 30 days, or advanced organic disease. Informed consent was obtained from all subjects or in cases of cognitive impairment, next of kin in accordance with the local research ethics committee guidelines. Data collected included anthropometric measurements, clinical history and status, and medication history. Antibiotic use was also recorded for each subject. Nine younger adult subjects of ages ranging from 28 to 46 y that were not antibiotic-treated were also recruited by informed consent.
Molecular Methods and Bioinformatics.
DNA was extracted from fecal samples, and the V4 region of the 16S rRNA gene was amplified, sequenced, and analyzed as described in SI Materials and Methods and ref. 35. In addition, a phylogenetic tree was built from an alignment of unique sequences (i.e., 100% identity OTUs) using FastTree (49). Based on this tree and abundance/presence/absence data of the unique sequences, Fast UniFrac (50) was used for hierarchical Unweighted Pair Group Method with Arithmetic Mean clustering and principal coordinate analysis. The resulting trees were visualized with Dendroscope (51). To assign reads to Clostridium clusters, 58,000 V4 sequences were extracted from full-length 16S rRNA genes with complete species assignments (RDP release 10.16). An association table was derived by comparing scores from an all-against-all BLAST search of the extracted sequences with a list of species previously assigned to Clostridium clusters. Subsequently, V4 sequence reads could be assigned to clusters if their BLAST scores against the custom database were above the threshold for the best-hit species in the association table. Sequences were deposited in the Short Read Archive.
Statistical Methods.
Statistical analyses were carried out using Stata SE Release 11.0 (StataCorp 2009 Stata: Release 11. Statistical Software; StataCorp). Detailed statistical analysis is in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Catherine Lozupone for UniFrac advice, Jennifer Deane, Nessa Gallwey, Karen O'Donovan, and Ann O'Neill for technical and clinical help, and the staffs at St. Finbarr's Hospital, Cork, and Cork University Hospital for facilitating subject recruitment. This work is supported by the Goverment of Ireland Department of Agriculture Fisheries and Food/Health Research Board Food for Health Research Initiative award to the ELDERMET project as well as by a Science Foundation Ireland award to the Alimentary Pharmabiotic Centre. M.J.C. is now funded by a fellowship from the Health Research Board of Ireland.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
Data deposition: The sequence data reported in this paper have been deposited in the National Center for Biotechnology Information Short Read Archive (accession no. SRA021022).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1000097107/-/DCSupplemental.
References
- 1.Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut. 2008;57:1605–1615. doi: 10.1136/gut.2007.133603. [DOI] [PubMed] [Google Scholar]
- 2.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Toole PW, Claesson MJ. Gut microbiota: Changes throughout the lifespan from infancy to elderly. Int Dairy J. 2010;20:281–291. [Google Scholar]
- 4.Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raes J, Foerstner KU, Bork P. Get the most out of your metagenome: Computational analysis of environmental sequence data. Curr Opin Microbiol. 2007;10:490–498. doi: 10.1016/j.mib.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zoetendal EG, Akkermans AD, De Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol. 1998;64:3854–3859. doi: 10.1128/aem.64.10.3854-3859.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tap J, et al. Towards the human intestinal microbiota phylogenetic core. Environ Microbiol. 2009;11:2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x. [DOI] [PubMed] [Google Scholar]
- 9.Rajilić-Stojanović M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol. 2007;9:2125–2136. doi: 10.1111/j.1462-2920.2007.01369.x. [DOI] [PubMed] [Google Scholar]
- 10.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 11.Frank DN, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kassinen A, et al. The fecal microbiota of irritable bowel syndrome patients differs significantly from that of healthy subjects. Gastroenterology. 2007;133:24–33. doi: 10.1053/j.gastro.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan SH, et al. Human colonic microbiota associated with diet, obesity and weight loss. Int J Obes (Lond) 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- 15.Schwiertz A, et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 16.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6–14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 18.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- 19.Manichanh C, et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez-Medina M, Aldeguer X, Gonzalez-Huix F, Acero D, Garcia-Gil LJ. Abnormal microbiota composition in the ileocolonic mucosa of Crohn's disease patients as revealed by polymerase chain reaction-denaturing gradient gel electrophoresis. Inflamm Bowel Dis. 2006;12:1136–1145. doi: 10.1097/01.mib.0000235828.09305.0c. [DOI] [PubMed] [Google Scholar]
- 21.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darfeuille-Michaud A, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 23.Kotlowski R, Bernstein CN, Sepehri S, Krause DO. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. 2007;56:669–675. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinsella K, He W. US Census Bureau. An Aging World: 2008. Washington, DC: International Population Reports; 2009. [Google Scholar]
- 25.Kleessen B, Sykura B, Zunft HJ, Blaut M. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am J Clin Nutr. 1997;65:1397–1402. doi: 10.1093/ajcn/65.5.1397. [DOI] [PubMed] [Google Scholar]
- 26.Camilleri M, Lee JS, Viramontes B, Bharucha AE, Tangalos EG. Insights into the pathophysiology and mechanisms of constipation, irritable bowel syndrome, and diverticulosis in older people. J Am Geriatr Soc. 2000;48:1142–1150. doi: 10.1111/j.1532-5415.2000.tb04793.x. [DOI] [PubMed] [Google Scholar]
- 27.Franceschi C, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 28.Salminen A, et al. Activation of innate immunity system during aging: NF-kB signaling is the molecular culprit of inflamm-aging. Ageing Res Rev. 2008;7:83–105. doi: 10.1016/j.arr.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Mitsuoka T. Intestinal Bacteria and Health. Tokyo: Harcourt Brace Jovanovich; 1978. [Google Scholar]
- 30.Hopkins MJ, Macfarlane GT. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J Med Microbiol. 2002;51:448–454. doi: 10.1099/0022-1317-51-5-448. [DOI] [PubMed] [Google Scholar]
- 31.Woodmansey EJ. Intestinal bacteria and ageing. J Appl Microbiol. 2007;102:1178–1186. doi: 10.1111/j.1365-2672.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- 32.Woodmansey EJ, McMurdo ME, Macfarlane GT, Macfarlane S. Comparison of compositions and metabolic activities of fecal microbiotas in young adults and in antibiotic-treated and non-antibiotic-treated elderly subjects. Appl Environ Microbiol. 2004;70:6113–6122. doi: 10.1128/AEM.70.10.6113-6122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi H, Sakamoto M, Kitahara M, Benno Y. Molecular analysis of fecal microbiota in elderly individuals using 16S rDNA library and T-RFLP. Microbiol Immunol. 2003;47:557–570. doi: 10.1111/j.1348-0421.2003.tb03418.x. [DOI] [PubMed] [Google Scholar]
- 34.Mueller S, et al. Differences in fecal microbiota in different European study populations in relation to age, gender, and country: A cross-sectional study. Appl Environ Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Claesson MJ, et al. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE. 2009;4:e6669. doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole JR, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collins MD, et al. The phylogeny of the genus Clostridium: Proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 38.Sokol H, et al. Specificities of the fecal microbiota in inflammatory bowel disease. Inflamm Bowel Dis. 2006;12:106–111. doi: 10.1097/01.MIB.0000200323.38139.c6. [DOI] [PubMed] [Google Scholar]
- 39.Sokol H, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- 40.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajilić-Stojanović M, et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: Analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol. 2009;11:1736–1751. doi: 10.1111/j.1462-2920.2009.01900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De La Cochetière MF, et al. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson AF, et al. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ventura M, et al. Genome-scale analyses of health-promoting bacteria: Probiogenomics. Nat Rev Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 48.Zwielehner J, et al. Combined PCR-DGGE fingerprinting and quantitative-PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Exp Gerontol. 2009;44:440–446. doi: 10.1016/j.exger.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hamady M, Lozupone C, Knight R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007 doi: 10.1186/1471-2105-8-460. 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.