Abstract
As predominant intestinal symbiotic bacteria, Bacteroides are essential in maintaining the health of the normal mammalian host; in return, the host provides a niche with plentiful nutrients for the symbionts. However, the intestinal environment is replete with chemical, physical, and biological challenges that require mechanisms for prompt and adept sensing of and responses to stress if the bacteria are to survive. Herein we propose that to persist in the intestine Bacteroides take advantage of their unusual bacterial sphingolipids to mediate signaling pathways previously known to be available only to higher organisms. Sphingolipids convey diverse signal transduction and stress response pathways and have profound physiological impacts demonstrated in a variety of eukaryotic cell types. We propose a mechanism by which the formation of specific sphingolipid membrane microdomains initiates signaling cascades that facilitate survival strategies within the bacteria. Our preliminary data suggest that sphingolipid signaling plays an important role in Bacteroides physiology, enabling these bacteria to persist in the intestine and to perform other functions related to symbiosis.
Keywords: stress response, bacterial sphingolipids, lipid rafts
The Bacteroides species make up a predominant genus of bacteria residing within the mammalian intestine. Organisms belonging to this genus have been the subject of extensive research and possess many distinctive features contributing to their ability to flourish in the intestine. For example, Bacteroides have sophisticated ability to degrade polysaccharides (for energy acquisition) (1), pronounced phase variation in surface polysaccharide synthesis (for antigenic versatility) (2, 3), and extensive surface decoration with host glycans (for molecular mimicry) (4). These bacteria display another extremely rare structural feature that has been recognized for decades but remains largely unstudied: membrane sphingolipids. Sphingolipids were generally thought to exist exclusively in eukaryotes until they were discovered in a handful of bacterial and viral lineages (5, 6). What is the function of sphingolipids in bacteria? Are they important in maintaining Bacteroides in their ecological niches? Herein we provide first evidence supporting an important role for sphingolipids in the ability of Bacteroides to survive stress, and we discuss the potential role of sphingolipids in maintaining intestinal symbiotic bacterial populations.
Results and Discussion
Are Sphingolipids Important for Bacteroides Growth?
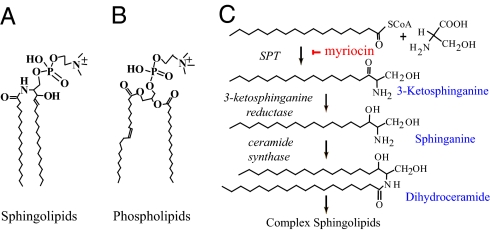
Sphingolipids are characterized by an aliphatic amino alcohol sphingoid backbone called long-chain base (LCB). LCB is attached via an amide bond to a fatty acid and via an ester linkage to a polar head group (Fig. 1A). Sphingolipids are always present in eukaryotic cells but are absent in most bacteria, whose membranes comprise only glycerol-based phospholipids (Fig. 1B). However, a few bacterial species possess both phospholipids and sphingolipids. These sphingolipid-containing bacteria are highly represented by genera in the phylum Bacteroidetes, including gut-associated Bacteroides (5, 7), oral cavity–associated Porphyromonas (8), gut and oral cavity–associated Prevotella (9), and soil-associated Sphingobacterium (10). Other prokaryotes constituted with sphingolipids include the genus Sphingomonas in the class Alphaproteobacterium, Bdellovibrio species in the class Deltaproteobacterium, and Fusobacterium species (11). This is by no means a complete list, and it is very likely additional bacterial species contain sphingolipids, especially in the Bacteroidetes phylum. Apart from the fact that the glycosphinogolipids in Sphingomonas are potent antigens in the activation of invariant natural killer T cell populations during infection (12, 13), little is known about bacterial sphingolipid function.
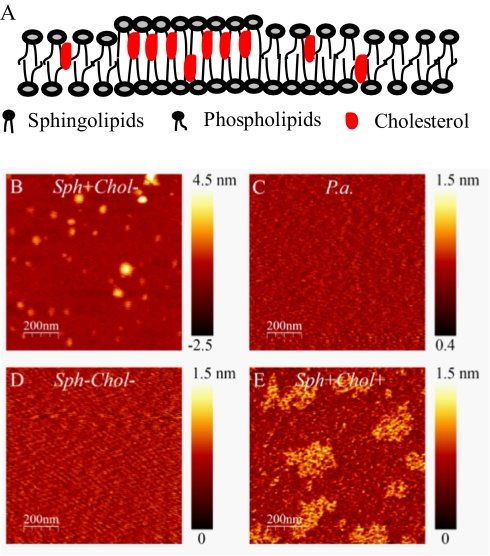
Fig. 1.
Typical structures of (A) sphingolipids and (B) phospholipids. (C) Initial steps in eukaryotic sphingolipid biosynthetic pathway.
To determine whether sphingolipids are essential for maintaining cell integrity and normal growth, we studied the model organism Bacteroides fragilis, which is prevalent in the intestine and is of substantial clinical significance. In conducting this work, we took advantage of years of research on eukaryotic sphingolipids and employed myriocin, a widely used, potent, specific inhibitor of sphingolipid biosynthesis, to create sphingolipid-deficient B. fragilis cultures. Although genetic approaches (e.g., gene knockout and complementation) are necessary to fully address the function of sphingolipids, chemical methods have the advantage of producing fast and reliable results in initial studies. In the eukaryotic sphingolipid biosynthetic pathway (14, 15) depicted in Fig. 1C, myriocin chemically knocks out sphingolipid synthesis by disrupting the activity of serine palmitoyl transferase (SPT), the first enzyme of committed sphingolipid synthesis (16, 17) that condenses palmitoyl-CoA and serine to 3-ketosphinganine. The genome of wild-type B. fragilis strain NCTC 9343 has an SPT ortholog (encoded by BF2461) with a high degree of homology to the human SPT enzyme subunits SPTLC1, SPTLC2, and SPTLC3 [E values: ≤E−44 by standard BLASTP search (18); http://blast.ncbi.nlm.nih.gov]. In addition, SPT orthologs have been reported in several other sphingolipid-producing bacteria (19, 20) and are well conserved in Bacteroides and related species (Table S1). In the following steps, 3-ketosphinganine is converted to sphinganine by 3-ketosphinganine reductase, and sphinganine is further transformed to dihydroceramide by ceramide synthase. Dihydroceramide and its modifications (such as ceramide with a double bond between C4 and C5 on LCB and phytoceramide with an extra hydroxyl group at C5 on LCB) are the central precursors for more complex sphingolipids and themselves are often important components of membrane lipids. Because myriocin inhibition is specific to SPT but not other enzymes in the pathway, the quantity of ceramide-like molecules can be regarded as an indicator of the entire sphingolipid population in cells. By using electrospray ionization tandem mass spectrometry (ESI/MS/MS) (21), we found that addition of 5 μM myriocin to B. fragilis can chemically inhibit more than two thirds of ceramide-like structures (Figs. S1, S2 and S3). Thus the myriocin-treated B. fragilis cells are sphingolipid-deficient.
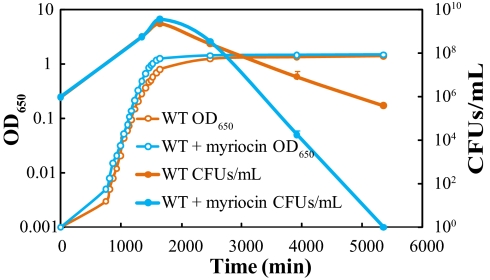
An overnight culture of the B. fragilis wild-type strain was diluted 1:1,000 in fresh rich medium (recipe in SI Materials and Methods) in the presence or absence of 5 μM myriocin, and the diluted cultures were incubated under anaerobic conditions at 37 °C in test tubes. Optical density at 650 nm (OD650) and colony-forming units (CFUs) per milliliter were monitored as measures of bacterial growth and death. The growth curves in Fig. 2 show that inhibition of sphingolipid synthesis resulted in no B. fragilis growth defect; the two cultures had analogous growth kinetics in the log phase and similar cell densities in the early stationary phase. However, the sphingolipid-deficient culture was completely wiped out within 60 h after entering the stationary phase, whereas ≈4 × 105 CFUs/mL of the sphingolipid-proficient cells were still alive at the same time point. These results suggest that sphingolipids probably are not essential for the active growth of B. fragilis but are involved in long-term survival of cells in the stressful stationary phase.
Fig. 2.
Sphingolipids affect the survival of Bacteroides in stationary phase but not the growth of the bacteria in log phase. Growth curves are shown for the wild-type B. fragilis culture in which sphingolipids were intact (WT) and the myriocin-treated B. fragilis culture in which sphingolipid synthesis was inhibited (WT + myriocin). Optical density curves are representative of three replications. Error bars for CFUs/mL curves show the SD of triplicate samples in one representative experiment.
Are Sphingolipids Important in the Bacteroides Stress Response?
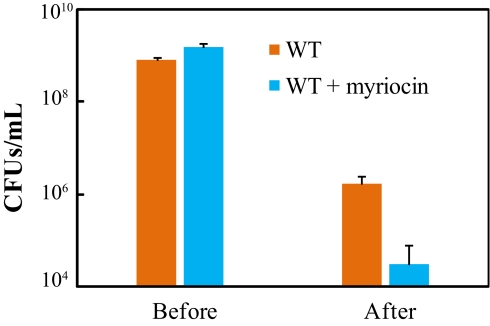
Seeking further evidence that B. fragilis sphingolipids are important for bacterial stress survival, we applied common types of stress encountered by intestinal microbes to stationary-phase cells grown in the presence or absence of myriocin, and we then monitored cell numbers. The stress challenge consisted of heat shock accompanied by oxidative stress whereby cultures were incubated at 42 °C in air for 14 h. Although cell death was significant in both cultures after such treatment, bacterial survival in the sphingolipid-deficient culture was approximately 2 logs lower than that in the sphingolipid-proficient culture (Fig. 3). Thus Bacteroides sphingolipids seem to play an important role in bacterial survival under stress.
Fig. 3.
Sphingolipids are important in the response of Bacteroides to stress. Rich-culture cells proficient with sphingolipids (WT) were compared with those deficient in sphingolipids (WT + myriocin) in terms of survival under heat shock and oxidative stress. Error bars show the SD of triplicate samples in one representative experiment.
Can the Stress Response Be Mediated by Sphingolipid Signaling in Bacteroides?
Sphingolipids, mostly enriched in the outer leaflet of the eukaryotic plasma membrane as well as in endosomes, are key signal transduction molecules with high specificity in response to various cell stresses, such as cytokines, growth factors, death factors, heat, and UV radiation (15, 22). The biological outcomes of sphingolipid signaling are profound and diverse, including cell apoptosis, proliferation, differentiation, mitogenesis, and migration; angiogenesis; and inflammation. Cholesterol is indispensable for sphingolipid signaling in mammals, facilitating formation of sphingolipid-enriched platforms (“lipid rafts”) in membranes to function as membrane-reactive centers. In the mammalian intestine, Bacteroides species have access to abundant cholesterol—both provided by food intake and secreted by the host (23). Therefore, under stressful conditions, Bacteroides have both sphingolipids and cholesterol at their disposal to assemble a unique stress-response signaling mechanism previously known to be available only to higher organisms.
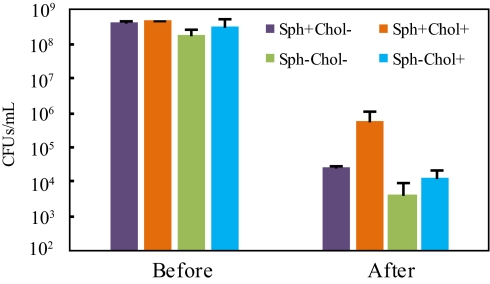
If Bacteroides use a eukaryotic-like sphingolipid- and cholesterol-dependent mechanism to respond to stress, this mechanism will be most efficient only when both sphingolipids and cholesterol are present. To test this hypothesis, we prepared sphingolipid-proficient cultures with and without cholesterol (Sph+Chol+ and Sph+Chol−) and sphingolipid-deficient cultures with and without cholesterol (Sph−Chol+ and Sph−Cho−). Because the rich medium used in the studies depicted in Figs. 2 and 3 contains a yeast product (ergosterol) that can functionally substitute for cholesterol in sphingolipid signaling (24), we prepared the cholesterol-free cultures in a glucose-supplemented minimal medium (recipe in SI Materials and Methods). Cholesterol (30 μg/mL) was added to formulate cholesterol-rich cultures; the addition of cholesterol at this concentration had no remarkable impact on growth. Moreover, we purposefully chose a distinct stress type—DNA crosslinking—in this experiment. DNA crosslinking is common and can be induced in living organisms by both exogenous and endogenous agents, such as alkylating agents, nitrous acids, and other reactive species (25). Mitomycin C—a potent chemotherapeutic compound commonly used to treat a broad range of tumors, including gastrointestinal cancers (26)—served as the crosslinking agent here. When both sphingolipids and cholesterol were available to B. fragilis, the survival rate was the highest, with more than 1.4 logs more cells alive than in any of the other three cultures (Fig. 4). Deficiency of either cholesterol or sphingolipids caused a lower survival rate, and loss of both only made the bacteria slightly less viable than loss of either, suggesting cholesterol and sphingolipids function by related mechanisms. Using a different stress challenge under different growth conditions, this experiment confirmed the conclusion suggested by data in Fig. 3 and proposed that Bacteroides organisms probably use the eukaryotic-like sphingolipid signaling mechanism for various stress survival.
Fig. 4.
Sphingolipids and cholesterol are both needed for an efficient stress response. Cells grown in minimal medium with different combinations of sphingolipids and cholesterol were compared in terms of survival after DNA crosslinking and oxidative stress. Error bars show the SD of duplicate samples in one representative experiment.
Is Sphingolipid Signaling Feasible in Bacteroides?
From a genomic viewpoint, conservation of SPT in sphingolipid-producing bacteria demonstrates that these bacteria are committed to sphingolipid metabolism and suggests that they may share other enzymes in the sphingolipid biosynthetic pathway (Fig. 1C) with eukaryotes. However, a standard BLASTP search in Bacteroides genomes does not return dependable 3-ketosphinganine reductase and ceramide synthase homologs. Another important class of enzymes is sphingomyelinase, which degrades sphingomyelin to ceramide. Bacteroides largely lack these enzymes, although they are found in many pathogens [e.g., Staphylococcus aureus (27)]. It is possible that Bacteroides have structural homologs to these enzymes, which would not be revealed by standard BLASTP searches. Nonetheless, complex and simple sphingolipid molecules are detected in cultures of Bacteroides species. For example, in addition to our data in Fig. S1, it was reported that as much as 10% of total lipid content was (dihydro)ceramide in B. fragilis (7). The discrepancy between genomic predictions and chemical analyses indicates either that a more in-depth search of the genome is needed or that bacteria and eukaryotes use different pathways to synthesize sphingolipids.
Several bacterial sphingolipid structures have been characterized (7–9, 28). In bacteria, both LCBs and fatty acid acyl chains are typically saturated with 17–19 carbons in length (dihydroceramide- or phytoceramide-based) and sometimes the fatty acids have an extra 3-hydroxyl group. They are also usually branched with a methyl group in either the iso or the anteiso position. These characteristics are different from those of eukaryotic sphingolipids, which usually have a double bond between C4 and C5 on the LCBs (ceramide-based) and have unbranched acyl chains 18–20 carbons in length. Another notable feature is the polar head group. Eukaryotes most often have phosphorylcholine as the head group to form sphingomyelin, whereas bacteria are largely devoid of sphingomyelin but instead characteristically have ceramide phosphorylethanolamine (CPE), ceramide phosphorylglycerol (CPG), ceramide phosphoryl-myo-inositol, ceramide phosphorylmannose, and other complex glycosphingolipids (5, 10, 29–31). The major sphingolipid species in B. fragilis include CPE and CPG. These structural analyses suggest that bacterial sphingolipids largely resemble their eukaryotic counterparts and probably behave similarly in cell membrane bilayers.
Because of differences in chemical structures and physical properties, phospholipids and sphingolipids form separate domains in model membranes (32). The distinction between the two membrane domains is strengthened by the presence of cholesterol or cholesterol-like lipid molecules in eukaryotic cells. The heterogeneity in membrane structure is thought to provide a physical and chemical basis for sphingolipid signaling. Although this topic is controversial and unresolved in many respects, a popular view is that membrane signaling is associated with the sphingolipid domains, which are also enriched in cholesterol and specific proteins. These domains are more tightly packed and have a higher melting temperature because of the highly hydrophobic interactions of the sphingolipid acyl chains with cholesterol and the extensive hydrogen bonding facilitated by the amide linkage and the hydroxyl group. Sphingolipid-enriched domains are thus in the liquid-ordered phase and have greater lateral mobility than the surrounding liquid-disordered phospholipid domains (Fig. 5A). When signaling is triggered by stresses, considerable ceramide is generated through sphingomyelin hydrolysis or de novo synthesis (33–35). The increase in ceramide can significantly promote the liquid-ordered state and result in merging of adjacent microdomains and formation of bigger platforms. These spatially organized platforms can effectively concentrate and recruit specific proteins, generate signaling molecules, and initiate downstream events (36–39). In this process, sphingolipids are thought to be important in two ways: they are the central components of the bioactive membrane domains, and they can be signaling molecules themselves.
Fig. 5.
Bacteroides lipids form sphingolipid- and cholesterol-dependent heterogeneous domains in supported bilayers. (A) Schematic sketch shows formation of heterogeneous membrane domains. (B–E) Representative AFM height images (1 μm × 1 μm) of reconstituted bilayers using lipid extractions from B. fragilis Sph+Chol− (B), P. aeruginosa (P.a.; C), B. fragilis Sph−Chol− (D), and B. fragilis Sph+Chol+ (E). Color bars along AFM images indicate height ranges. The extended height range in sample Sph+Chol− was due to assembly of multiple lipid bilayers instead of single bilayers. Areas of multiple lipid bilayers were excluded from calculations shown in text.
If Bacteroides organisms are capable of sphingolipid-mediated signaling, the lipid membrane presumably will have distinct microdomains. Atomic force microscopy (AFM), a widely used method, has shown that eukaryotic phospholipid and sphingolipid species can form separate domains when these lipids are mixed and reconstituted in lipid bilayers on inert surfaces (40). The two domains are characterized by two discrete phases with a small variation (<2 nm) in the “height image.” Of these two phases, sphingolipid domains are taller. Furthermore, AFM clearly shows the impact of cholesterol, revealing that its addition considerably enlarges the microdomains. To investigate whether such heterogeneity exists in Bacteroides lipid bilayers, we extracted lipids from stationary-phase B. fragilis cultures grown under several conditions. Each lipid sample was then reconstituted on a mica surface to form bilayers and was inspected by AFM.
We first grew B. fragilis cells in minimal medium without exogenously added cholesterol (Sph+Chol−). After reconstitution in bilayers, a two-phase pattern was observed by AFM. These two phases represented phospholipid- and sphingolipid-enriched domains with an average height difference of 0.5 nm. The average size (diameter) of the taller sphingolipid domain was 44 ± 15 nm (Fig. 5B). We next applied the same procedure to a lipid preparation from Pseudomonas aeruginosa, a bacterium that produces phospholipids but not sphingolipids. Serving as a negative control, lipids in this sample were homogeneously distributed, with one single height and phase across the examined surface (Fig. 5C). To investigate whether the heterogeneity shown in Fig. 5B was due to the presence of sphingolipids, we prepared lipids from myriocin-treated B. fragilis culture (Sph−Chol−) and found that the extracted lipids became homogeneously distributed on the surface bilayer in a manner resembling that seen with the P. aeruginosa sample (Fig. 5D). Finally, to examine whether the presence of cholesterol changes lipid domain organization, we extracted lipids from a B. fragilis culture grown in the presence of 30 μg/mL cholesterol (Sph+Chol+). As predicted, the sphingolipid domains were considerably enlarged, with an average size of ≈168 ± 45 nm (Fig. 5E). The average height difference of the two phases in this sample was 0.3 nm. These observations provide strong evidence that the lipid bilayer organization in Bacteroides is characterized by heterogeneity and bears a resemblance to that of eukaryotic plasma membrane lipids. Therefore, from a physicochemical point of view, Bacteroides sphingolipids can form membrane functional units for signaling.
Proteomic studies from the literature provide additional information suggesting the existence of bacterial sphingolipid microdomains. Proteins with an SPFH (stomatin, prohibitin, flotillin, HflK/C) domain play important roles in scaffolding sphingolipid-enriched, detergent-resistant eukaryotic membrane microdomains and have become the hallmarks of the “lipid raft” (41). Remarkably, prokaryotic SPFH orthologs are widely distributed in the phylogenetic tree, including Bacteroides species, and are proposed as the origins of their eukaryotic counterparts (42). It is tempting to speculate that Bacteroides SPFH orthologs interact with sphingolipids and cholesterol to facilitate the formation of signaling microdomains. Moreover, the presence of SPFH orthologs in bacterial species that do not produce sphingolipids is intriguing. It is well known that bacterial phospholipids have highly diverse structures generated by lipid homeostasis mechanisms (43). One hypothesis is that these SPFH orthologs interact with a subset of phospholipids to form special functional membrane units for signal transduction. Confirmation of this hypothesis would indicate that membrane heterogeneity is a common theme in all bacteria.
Does Bacteroides Sphingolipid Signaling Have Ecological Significance?
Sphingolipid signaling in eukaryotes is an exceedingly vibrant process characterized by a high turnover rate of lipid species and extremely versatile signaling functions (22, 44, 45). The magnitude of the complexity has just begun to be elucidated, but what is already known suggests that eukaryotic cells craft an enormously dynamic environment for themselves, commanding signaling processes with a high degree of sophistication. As single-celled prokaryotes, bacteria usually do not encounter such fierce challenges except if their survival depends on close association with eukaryotic hosts as in the case of intestinal symbionts. These symbiotic microbes thus are under tremendous selection pressure and must deal efficiently with the ever-changing host environment.
Born sterile, mammals become colonized with microbes immediately after birth, at which point their life-long relationship with microbial partners (the microbiota) commences. The colonization of many sites in and on mammals is most strikingly exemplified in the large intestine, where the density of microbial organisms can be as high as 1012/g luminal content. This intense colonization evolves into a mutually beneficial relationship. In exchange for niches with the right growth conditions, the microbiota endows mammals with abilities they have not developed on their own. Consequently, symbiotic bacteria are implicated in a number of major health issues. In a sense, the intestinal microbiota serves as an acquired multifunctional eukaryotic organ (46–48).
As the “acquired organ” develops postnatally, the intestinal microbiota is continuously challenged by the host environment. Along with the normal peristaltic propulsion of the intestine, the renewal of intestinal epithelial cells at estimated rates of 20–50 million and 2–5 million cells per minute in the small intestine and the colon, respectively (49), constantly generates a fresh interface between epithelial cells and microbial cells; these changes require immediate recognition and tolerance. In addition, thermal instability results from fluctuations in the local intestinal temperature in response to various cues, including food intake, inflammation, pharmacologic agents, or emotional stresses. Moreover, the intestine presents microbes with an extraordinarily complex chemical environment that includes various organic molecules (e.g., polysaccharides, proteins, amino acids, and lipids) as well as inorganic ions (e.g., calcium and phosphate at high levels). Although unimaginably rich in nutrients, this chemical environment is full of antimicrobial peptides, reactive oxidative species, hormones, cytokines, and neurotransmitters whose impact on the microbiota has yet to be comprehensively elucidated (50, 51). Resident microbes also constantly compete for niches with foreign microbial species that have traveled from the upper gastrointestinal tract. Furthermore, the aging of the host is usually accompanied by dampened lymphocyte function and increased risk of infection with pathogens such as Clostridium difficile; the result is more frequent occurrence of chronic inflammation and carcinogenesis in elderly hosts (52). In addition, intestinal diseases that are not related to the host's age (e.g., cancers and inflammatory bowel diseases) induce heightened cytokine production, uncontrolled inflammation, and tissue damage that also render the intestinal environment extremely hostile to the microbiota (53). Finally, oral administration of medications directly subjects the intestine and the intestinal microbiota to the effects of drugs like antibiotics and nonsteroidal anti-inflammatory agents. Therefore, the mammalian intestine selects for microbial colonizers that not only are metabolically adaptable but also possess efficient sensing and responding mechanisms for stress survival. How symbiotic intestinal bacteria manifest their stress response and persist in the intestinal environment remains largely unknown but deserves extensive study.
As one of the most dominant Gram-negative bacterial genera in the intestine, Bacteroides must excel at dealing with stresses. Given its known functions in eukaryotic cells, sphingolipid-mediated signaling is an excellent candidate mechanism for the Bacteroides stress response. The highly flexible and dynamic nature of lipids guarantees timely transmission of signals. Situated at the membrane interface between cells and their external environments, sphingolipids are ideally positioned to broadcast information from outside the cell to the cytoplasm through lipid–lipid, protein–lipid, and protein–protein interactions. It is not surprising that genomic analyses of Bacteroides have revealed an unusually large number of extracytoplasmic function-type σ factors and hybrid two-component systems whose purposes are likely to be sensing and response (54). One possibility is that these membrane proteins are specifically and functionally associated with the sphingolipid-enriched domains. Downstream of these interactions is a profound transcriptional response. For example, B. fragilis can transcriptionally adjust 45% of its genome expression to withstand oxidative stress and become aerotolerant (55).
The data presented in this paper strongly suggest that Bacteroides organisms have the capacity to use sphingolipid-mediated mechanisms to respond to stress. This possibility raises several important questions. First, where are the sphingolipids located in bacterial membranes? The complicated envelope structures of Bacteroides, with double lipid membranes, lipopolysaccharides, extracellular polysaccharides, and capsular polysaccharides, certainly cloud the issue. The current model would predict that these lipids are likely enriched in the outer leaflet of the outer membrane, together with cholesterol molecules taken up from the intestinal environment. Second, what are the specific genetic components in Bacteroides sphingolipid signal transduction? Although tremendous progress has been made in understanding the protein constituents in eukaryotic sphingolipid signaling functional units, debate remains heated about a reliable protocol to separate “raft” domains and associated proteins from other membrane structures (56–58). Given the ability of Bacteroides to grow in a cholesterol-free environment where effective membrane organization and sphingolipid signaling are not possible, these bacteria present a unique opportunity to study this question. Cells grown under the cholesterol-free condition can be compared in terms of proteomic or transcriptional profiles with those grown in the presence of cholesterol and engaged in active signaling. This comparison allows one possible way to gain valuable information about important genes and proteins in signaling. Third, what are the signaling molecules? The enormous structural capacity and the functional specificity of sphingolipids make them ideal signaling molecules; yet the actual signals are known to be only transiently present at low concentrations and therefore difficult to analyze. Nevertheless, knowledge and methodology gained from studies of eukaryotic sphingolipids will be helpful in tackling this question in bacteria. Last, what is the biological significance of Bacteroides sphingolipid signaling? For example, does this mechanism mediate important functions in vivo to make Bacteroides species successful as symbionts in mammalian hosts? Can similar mechanisms help other sphingolipid-containing symbiotic bacteria to survive (e.g., Porphyromonas, Prevotella)? What impact does this signaling have on microbial populations in the intestine that do not produce sphingolipids (e.g., Firmicutes)? Does this mechanism provide a common platform on which the host and the bacteria can share important information? Answers to these questions will help to better understand host–bacteria interactions.
Concluding Remarks.
Past research has greatly advanced our appreciation of Bacteroides as symbionts; however, the survival of these bacteria in a challenging intestinal environment is poorly understood. We propose that Bacteroides species use a unique sphingolipid-mediated mechanism to establish a foothold in the enormously stressful intestinal environment, so that they are able to deliver important beneficial functions to the host. Further investigation of this proposal will likely reveal previously unknown metabolic and signaling pathways in Bacteroides and help to illuminate their relationship with the host.
Materials and Methods
Bacterial Stress Challenges.
For challenges using rich-medium cultures, bacteria were grown to stationary phase in the presence or absence of 5 μM myriocin. Each sample was then pipette in an Eppendorf tube with 0.5 mL in volume and incubated for 14 h in a 42 °C water bath under aerobic condition. For challenges using minimal-medium cultures, bacteria were grown with 2 × 2 combinations of myriocin (5 μM; Sigma-Aldrich) and cholesterol (30 μg/mL; Sigma-Aldrich). Each sample, 0.5 mL in volume in an Eppendorf tube, was added with 1 μg/mL mitomycin C (Sigma-Aldrich) and followed by aerobic incubation for 2 h at 37 °C. Before and after treatments, CFUs/mL were counted. Each experiment was repeated at least three times.
Bacterial Lipid Purification and AFM Studies.
Stationary-phase bacterial cells (50 mL) were centrifuged, and the cell pellets were resuspended in a mixture of methanol (20 mL) and chloroform (10 mL) in glass tubes. After the addition of 8 mL of water, each sample was extensively vortexed and centrifuged for phase separation. The lower organic phase was transferred to a new glass tube, and the water extraction step was repeated. Finally, the organic phase in a clean glass tube was dried with a gentle stream of N2. Lipids were then weighed and resuspended in chloroform to a concentration of 0.5 mg/mL (59). For AFM studies, lipid samples were reconstituted into bilayers on a mica surface by a previously described vesicle fusion method (60, 61). In brief, the lipid sample was dried and resuspended in water by prolonged sonication. After all lipid molecules were dissolved, 20 μL of the solution was placed on a clean mica surface at 60 °C. The mica plate was then cooled down slowly to room temperature in a closed chamber so that the surface retained moisturized. The lipid bilayers were then examined by AFM (Veeco Multimode Nanoscope IIIA). The microscope was operated in the tapping mode according to standard procedure. Rastering of a probing tip (Olympus OMCL-AC160TS-W2) across a given area provided a height image that recorded the topography on top of the lipid bilayers. At least five images were taken for each sample.
Supplementary Material
Acknowledgments
We thank the D.L.K. laboratory and Dr. Laurie Comstock at Harvard Medical School for helpful discussions; and Dr. Scot Martin at Harvard University for providing equipment for AFM studies. C.N. thanks the University of Notre Dame Faculty Scholarship Award Program (FASP) for financial support.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Microbes and Health,” held November 2–3, 2009, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA. The complete program and audio files of most presentations are available on the NAS Web site at http://www.nasonline.org/SACKLER_Microbes_and_Health.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1001501107/-/DCSupplemental.
References
- 1.Sonnenburg JL, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 2.Krinos CM, et al. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature. 2001;414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 3.Cerdeño-Tárraga AM, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 4.Coyne MJ, Reinap B, Lee MM, Comstock LE. Human symbionts use a host-like pathway for surface fucosylation. Science. 2005;307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 5.Kato M, Muto Y, Tanaka-Bandoh K, Watanabe K, Ueno K. Sphingolipid composition in Bacteroides species. Anaerobe. 1995;1:135–139. doi: 10.1006/anae.1995.1009. [DOI] [PubMed] [Google Scholar]
- 6.Wilson WH, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 7.Miyagawa E, Azuma R, Suto T, Yano I. Occurrence of free ceramides in Bacteroides fragilis NCTC 9343. J Biochem. 1979;86:311–320. doi: 10.1093/oxfordjournals.jbchem.a132528. [DOI] [PubMed] [Google Scholar]
- 8.Nichols FC, et al. Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis. J Lipid Res. 2004;45:2317–2330. doi: 10.1194/jlr.M400278-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.LaBach JP, White DC. Identification of ceramide phosphorylethanolamine and ceramide phosphorylglycerol in the lipids of an anaerobic bacterium. J Lipid Res. 1969;10:528–534. [PubMed] [Google Scholar]
- 10.Naka T, et al. Structural analysis of sphingophospholipids derived from Sphingobacterium spiritivorum, the type species of genus Sphingobacterium. Biochim Biophys Acta. 2003;1635:83–92. doi: 10.1016/j.bbalip.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 11.Olsen I, Jantzen E. Sphingolipids in Bacteria and Fungi. Anaerobe. 2001;7:103–112. [Google Scholar]
- 12.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 13.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 14.Dickson RC. Thematic review series: Sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res. 2008;49:909–921. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartke N, Hannun YA. Bioactive sphingolipids: Metabolism and function. J Lipid Res. 2009;50(Suppl):S91–S96. doi: 10.1194/jlr.R800080-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyake Y, Kozutsumi Y, Nakamura S, Fujita T, Kawasaki T. Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem Biophys Res Commun. 1995;211:396–403. doi: 10.1006/bbrc.1995.1827. [DOI] [PubMed] [Google Scholar]
- 17.Hidari KIPJ, Ichikawa S, Fujita T, Sakiyama H, Hirabayashi Y. Complete removal of sphingolipids from the plasma membrane disrupts cell to substratum adhesion of mouse melanoma cells. J Biol Chem. 1996;271:14636–14641. doi: 10.1074/jbc.271.24.14636. [DOI] [PubMed] [Google Scholar]
- 18.Altschul SF, et al. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005;272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikushiro H, Hayashi H, Kagamiyama H. A water-soluble homodimeric serine palmitoyltransferase from Sphingomonas paucimobilis EY2395T strain. Purification, characterization, cloning, and overproduction. J Biol Chem. 2001;276:18249–18256. doi: 10.1074/jbc.M101550200. [DOI] [PubMed] [Google Scholar]
- 20.Ikushiro H, Islam MM, Tojo H, Hayashi H. Molecular characterization of membrane-associated soluble serine palmitoyltransferases from Sphingobacterium multivorum and Bdellovibrio stolpii. J Bacteriol. 2007;189:5749–5761. doi: 10.1128/JB.00194-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bielawski J, et al. Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography–tandem mass spectrometry. In: Armstrong D, editor. Methods in Molecular Biology: Lipidomics. Vol. 579. Totowa, NJ: Humana Press; 2009. pp. 443–467. [DOI] [PubMed] [Google Scholar]
- 22.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 23.Iqbal J, Hussain MM. Intestinal lipid absorption. Am J Physiol Endocrinol Metab. 2009;296:E1183–E1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dickson R, Lester R. Sphingolipid functions in Saccharomyces cerevisiae. Biochim Biophys Acta. 2002;1583:13–25. doi: 10.1016/s1388-1981(02)00210-x. [DOI] [PubMed] [Google Scholar]
- 25.McCabe KM, Olson SB, Moses RE. DNA interstrand crosslink repair in mammalian cells. J Cell Physiol. 2009;220:569–573. doi: 10.1002/jcp.21811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofheinz RD, Beyer U, Al-Batran SE, Hartmann JT. Mitomycin C in the treatment of gastrointestinal tumours: Recent data and perspectives. Onkologie. 2008;31:271–281. doi: 10.1159/000122590. [DOI] [PubMed] [Google Scholar]
- 27.Walev I, Weller U, Strauch S, Foster T, Bhakdi S. Selective killing of human monocytes and cytokine release provoked by sphingomyelinase (beta-toxin) of Staphylococcus aureus. Infect Immun. 1996;64:2974–2979. doi: 10.1128/iai.64.8.2974-2979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikuya Y, Sadao I, Ikuko T, Eiko Y. Separation and analysis of free ceramides containing 2-hydroxy fatty acids in Sphingobacterium species. FEMS Microbiol Lett. 1983;20:449–453. [Google Scholar]
- 29.Steiner S, Conti SF, Lester RL. Occurrence of phosphonosphingolipids in Bdellovibrio bacteriovorus strain UKi2. J Bacteriol. 1973;116:1199–1211. doi: 10.1128/jb.116.3.1199-1211.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawahara K, et al. Chemical structure of glycosphingolipids isolated from Sphingomonas paucimobilis. FEBS Lett. 1991;292:107–110. doi: 10.1016/0014-5793(91)80845-t. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe Y, et al. A novel sphingophosphonolipid head group 1-hydroxy-2-aminoethyl phosphonate in Bdellovibrio stolpii. Lipids. 2001;36:513–519. doi: 10.1007/s11745-001-0751-3. [DOI] [PubMed] [Google Scholar]
- 32.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 33.Mathias S, et al. Activation of the sphingomyelin signaling pathway in intact EL4 cells and in a cell-free system by IL-1 beta. Science. 1993;259:519–522. doi: 10.1126/science.8424175. [DOI] [PubMed] [Google Scholar]
- 34.Haimovitz-Friedman A, et al. Ionizing radiation acts on cellular membranes to generate ceramide and initiate apoptosis. J Exp Med. 1994;180:525–535. doi: 10.1084/jem.180.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Li X, Becker KA, Gulbins E. Ceramide-enriched membrane domains—structure and function. Biochim Biophys Acta. 2009;1788:178–183. doi: 10.1016/j.bbamem.2008.07.030. [DOI] [PubMed] [Google Scholar]
- 36.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 37.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 38.Gulbins E, Dreschers S, Wilker B, Grassmé H. Ceramide, membrane rafts and infections. J Mol Med. 2004;82:357–363. doi: 10.1007/s00109-004-0539-y. [DOI] [PubMed] [Google Scholar]
- 39.Grassmé H, Riethmüller J, Gulbins E. Biological aspects of ceramide-enriched membrane domains. Prog Lipid Res. 2007;46:161–170. doi: 10.1016/j.plipres.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Connell SD, Smith DA. The atomic force microscope as a tool for studying phase separation in lipid membranes. Mol Membr Biol. 2006;23:17–28. doi: 10.1080/09687860500501158. [DOI] [PubMed] [Google Scholar]
- 41.Langhorst MF, Reuter A, Stuermer CA. Scaffolding microdomains and beyond: The function of reggie/flotillin proteins. Cell Mol Life Sci. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinderhofer M, et al. Evolution of prokaryotic SPFH proteins. BMC Evol Biol. 2009;9:10. doi: 10.1186/1471-2148-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang YM, Rock CO. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol. 2008;6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 44.Wymann MP, Schneiter R. Lipid signalling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 45.Pruett ST, et al. Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J Lipid Res. 2008;49:1621–1639. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu J, Gordon JI. Inaugural Article: Honor thy symbionts. Proc Natl Acad Sci USA. 2003;100:10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 48.Edelman SM, Kasper DL. Symbiotic commensal bacteria direct maturation of the host immune system. Curr Opin Gastroenterol. 2008;24:720–724. doi: 10.1097/MOG.0b013e32830c4355. [DOI] [PubMed] [Google Scholar]
- 49.Croft DN, Cotton PB. Gastro-intestinal cell loss in man. Its measurement and significance. Digestion. 1973;8:144–160. doi: 10.1159/000197310. [DOI] [PubMed] [Google Scholar]
- 50.Freestone PP, Sandrini SM, Haigh RD, Lyte M. Microbial endocrinology: How stress influences susceptibility to infection. Trends Microbiol. 2008;16:55–64. doi: 10.1016/j.tim.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 51.Furness JB. The enteric nervous system: Normal functions and enteric neuropathies. Neurogastroenterol Motil. 2008;20(Suppl 1):32–38. doi: 10.1111/j.1365-2982.2008.01094.x. [DOI] [PubMed] [Google Scholar]
- 52.Haynes L, Maue AC. Effects of aging on T cell function. Curr Opin Immunol. 2009;21:414–417. doi: 10.1016/j.coi.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgart DC, Carding SR. Inflammatory bowel disease: Cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 54.Xu J, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 55.Sund CJ, et al. The Bacteroides fragilis transcriptome response to oxygen and H2O2: The role of OxyR and its effect on survival and virulence. Mol Microbiol. 2008;67:129–142. doi: 10.1111/j.1365-2958.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- 56.Munro S. Lipid rafts: Elusive or illusive? Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 57.Hancock JF. Lipid rafts: Contentious only from simplistic standpoints. Nat Rev Mol Cell Biol. 2006;7:456–462. doi: 10.1038/nrm1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng YZ, Foster LJ. Contributions of quantitative proteomics to understanding membrane microdomains. J Lipid Res. 2009;50:1976–1985. doi: 10.1194/jlr.R900018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 60.Burns AR. Atomic force microscopy of lipid domains in supported model membranes. Methods Mol Biol. 2007;398:263–282. doi: 10.1007/978-1-59745-513-8_18. [DOI] [PubMed] [Google Scholar]
- 61.Lin WC, Blanchette CD, Ratto TV, Longo ML. Lipid domains in supported lipid bilayer for atomic force microscopy. Methods Mol Biol. 2007;400:503–513. doi: 10.1007/978-1-59745-519-0_34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.