Abstract
Copy-number variants (CNVs) are a major source of genetic variation in human health and disease. Previous studies have implicated replication stress as a causative factor in CNV formation. However, existing data are technically limited in the quality of comparisons that can be made between human CNVs and experimentally induced variants. Here, we used two high-resolution strategies—single nucleotide polymorphism (SNP) arrays and mate-pair sequencing—to compare CNVs that occur constitutionally to those that arise following aphidicolin-induced DNA replication stress in the same human cells. Although the optimized methods provided complementary information, sequencing was more sensitive to small variants and provided superior structural descriptions. The majority of constitutional and all aphidicolin-induced CNVs appear to be formed via homology-independent mechanisms, while aphidicolin-induced CNVs were of a larger median size than constitutional events even when mate-pair data were considered. Aphidicolin thus appears to stimulate formation of CNVs that closely resemble human pathogenic CNVs and the subset of larger nonhomologous constitutional CNVs.
IN recent years, submicroscopic structural variants (SVs) have been found to be widely distributed throughout the human genome where they represent an important component of genetic variation and phenotypic diversity (Iafrate et al. 2004; Sebat et al. 2004; Sharp et al. 2005; Conrad et al. 2010b). These include deletions, duplications, insertions, and inversions, with the majority being copy-number variations (CNVs) discovered in systematic studies using microarrays (Conrad et al. 2010b; Park et al. 2010). More than 10,000 CNVs have now been described in healthy individuals that represent gains or losses of ∼1 kb to >1 Mb. CNVs can alter gene expression in affected regions, confer redundancy, and provide substrates for evolution. Spontaneous CNVs are also known to be a major cause of genetic and developmental disorders, including mental retardation, autism, schizophrenia, epilepsy, skeletal defects, and many others (Stankiewicz and Beaudet 2007; Cook and Scherer 2008; Kirov et al. 2009; Tam et al. 2009; Zhang et al. 2009; Miller et al. 2010). Systematic studies of human population CNVs have provided further correlation to human conditions including Crohn's disease, rheumatoid arthritis, and diabetes (Craddock et al. 2010). Related systematic efforts have finally also revealed a high degree of submicroscopic chromosomal structural alterations in cancer (Stratton et al. 2009; Bignell et al. 2010).
Despite their importance, there is limited understanding of how SVs arise (Hastings et al. 2009b; Stankiewicz and Lupski 2010). The exceptions are local genome rearrangements that occur by unequal recombination between neighboring low-copy repeated sequences or segmental duplications, a process known as non-allelic homologous recombination (NAHR) (Sasaki et al. 2010). Such events are well described and underlie the specific recurrent alterations responsible for a variety of human microdeletion syndromes (Sasaki et al. 2010; Stankiewicz and Lupski 2010). However, the majority of both nonrecurrent pathogenic CNVs and those observed in the normal population do not appear to proceed by NAHR but instead show, at most, limited microhomology at the breakpoint junctions (Vissers et al. 2009; Conrad et al. 2010a). Multiple pathways might catalyze the formation of such junctions, including the best-described nonhomologous end-joining (NHEJ) pathway of DNA double-strand break repair (Lieber 2010; Lieber and Wilson 2010), alternative end-joining pathways recently implicated in chromosomal translocations (McVey and Lee 2008; Boboila et al. 2010; Simsek and Jasin 2010), and entirely distinct pathways in which stalled replication structures are processed by mechanisms variably known as template switching or microhomology-mediated break-induced replication (MMBIR) (Lee et al. 2007; Hastings et al. 2009a).
To date, these mechanisms have largely been inferred by examination of human CNV breakpoint sequences (Korbel et al. 2007; Vissers et al. 2009; Conrad et al. 2010a). To begin to explore CNV mechanisms experimentally, we recently reported a system in which normal human fibroblasts were treated with the replication inhibitor aphidicolin (Arlt et al. 2009). Treatment was associated with a substantially increased frequency of new CNVs in subclones, as detected by array comparative genome hybridization (aCGH). The observed CNVs were generally consistent with many normal and pathogenic human CNVs and suggested either template switching or nonhomologous repair formation mechanisms (Lee et al. 2007; Hastings et al. 2009a; Lieber 2010; Lieber and Wilson 2010). However, the resolution of the aCGH method used left uncertainty as to the full spectrum of CNVs that are induced by aphidicolin as compared to those observed in the human germline.
To address these issues, we have explored and optimized various methods for detecting CNVs and other SVs with a focus on those with sufficient power and low-enough cost for routine experimentation. We report an in-depth analysis of two complementary technologies—high-density SNP arrays and whole-genome mate-pair sequencing—and use them to compare aphidicolin-induced CNVs to the baseline constitutional CNVs in the same experimental samples. Our software platform, VAMP (Birkeland et al. 2010), was expanded to support the many bioinformatics aspects of the study. We found a surprisingly low correspondence between array and sequencing methods in the detection of constitutional SVs and accordingly identified >600 SVs by mate-pair analysis that were not previously known from systematic array-based studies (Conrad et al. 2010b; Park et al. 2010). A much higher method correspondence was observed for aphidicolin-induced CNVs mainly because these events were consistently larger than most constitutional CNVs even when the higher resolution of mate-pair sequencing was brought to bear in the analysis.
MATERIALS AND METHODS
Human cell lines:
All experiments were performed with normal human fibroblast cell line HGMDFN090 (090), which was obtained from the Progeria Research Foundation Cell and Tissue Bank (Peabody, MA). The source individual is a female of European descent with a normal 46,XX karyotype who does not carry the mutation for Hutchinson-Gilford progeria. Two aphidicolin-treated subclones of 090 that contain novel induced CNVs, called A3A2 and A1A1, which were obtained prior to immortalization of 090, have also been described (Arlt et al. 2009). More recently, 090 was immortalized by stable transfection with vector pBABE-Hygro-hTERT (Counter et al. 1998). A hygromycin-resistant clone was isolated, expanded, and called 090D2. Parental (i.e., not aphidicolin-treated) SNP array and mate-pair analyses were performed with 090D2 when specified. Genomic DNA was prepared from cell lines using the Blood and Cell Culture DNA Mini Kit (Qiagen).
SNP microarrays:
Microarrays were the Illumina HumanOmni1-Quad BeadChip, which has both SNP and non-SNP probes selected by the vendor to optimize the detection of human CNVs. One microgram of genomic DNA was submitted to the University of Michigan DNA Sequencing Core for labeling, array hybridization, and scanning according to the manufacturer's instructions. X, Y, log-R ratio and B allele-frequency values were obtained using Illumina BeadStudio.
Mate-pair sequencing:
Genomic DNA (20–40 μg) was used to construct mate-pair libraries using the Illumina Mate Pair Library Prep Kit followed by paired end sequencing by the University of Michigan DNA Sequencing Core according to the manufacturer's instructions. Image analysis and base-calling were performed using the Illumina programs Firecrest and Bustard, respectively.
Data analysis:
All further data analysis was performed using an expanded version of our VAMP software platform (Birkeland et al. 2010), which is available for download at http://tewlab.path.med.umich.edu/vamp.html. See supporting information, File S1, Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, Figure S6, Figure S7, and Figure S8 for a description of the platform, logic, and parameters. Human genome Build 36 (hg18) served as the reference genome.
SNP microarray analysis was performed using moving average windows of 5, 10, 20, and 50 probes and a threshold of 5 standard deviations (SD) from the array mean. Candidate genome regions identified with these parameters were subjected to further filtering during visualization that required the best segment call within a region to have either (i) a change in the log 2 of the intensity ratio (log2R) of at least 0.15 or (ii) a change in the B allele frequency of informative probes of at least 0.083, as well as (iii) a Z-statistic of at least 7. The Z-statistic is the deviation of the average value of a contiguous segment of probes relative to the average value over all probes in the array, expressed as the number of standard errors of the array mean. Thus, Z is influenced by the absolute deviation of a segment, the number of probes within it, and the noise level of the array. All passing genome regions were individually examined and CNVs were manually adjusted and committed. Constitutional CNV analysis was performed on 090D2 array data alone. A3A2 was analyzed using 090D2 as the normalization reference.
For mate-pair sequencing, mapping filters allowed up to five mismatches relative to hg18, including indels, and up to 10 initial genome map positions per read. The combined 090 data, used to detect constitutional CNVs and as the reference for detecting induced CNVs in A3A2 and A1A1, merged four sequencing lanes—two derived from 090 libraries and two derived from 090D2 libraries. Candidate genome regions were identified by seeking sets of anomalous fragments as described in Birkeland et al. (2010) and in File S1. Sets were subjected to filtering during visualization that required them to have (i) no more than 40% promiscuously mapped fragments, (ii) a fractional overlap of no more than 90%, (iii) an average fragment size deviation, Δ, of no more than 3 population SD, (iv) no more than 40% of fragments where Δ exceeded 2 SD, and (iv) no more than 10% of fragments in the region contributed by the reference sample (comparative studies only). For insertions, an additional filter required that the set contain at least five fragments. All passing sets were individually examined and manually committed.
CNV segments predicted by SNP arrays and mate-pair sequencing were finally compared to each other, to an analysis of 090 performed using PennCNV (Wang et al. 2007), and to CNVs from published compendia (Conrad et al. 2010b; Park et al. 2010). Two events were declared as matching if the overlap of the two spans was at least 5% of the larger of the two spans. Conclusions were not substantially different when calculated at a match threshold of 33%, given that most events showed either no match or a strong match of >50% (Figure S9; Figure S10).
Breakpoint analysis:
For a subset of CNVs detected by mate pairs, a single PCR primer pair that flanked the anomalous junction predicted by the analysis in Figure S3 was designed. Occasionally, the first primer pair failed to give a product, but in most such cases products were obtained by moving the primers to different positions that were nonetheless consistent with the same junction. All products were then subjected to standard sequencing.
Data availability:
Array data have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession GSE26121, mate-pair sequence data have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) under study no. SRP003289, and called SVs have been deposited in NCBI dbVar (http://www.ncbi.nlm.nih.gov/dbvar).
RESULTS
Detection of aphidicolin-induced CNVs:
We first sought to compare the ability of SNP microarrays and mate-pair sequencing (Table 1) to detect induced CNVs in A3A2 and A1A1, two aphidicolin-treated subclones of the normal human fibroblast cell line 090, including CNVs previously identified by aCGH (Arlt et al. 2009) as well as potentially unknown events. All CNVs shown in Table 2 were detected by at least one of the three methods and subsequently validated—in two cases by confirming loss of heterozygosity of informative SNPs in the deleted region (A3A2, chromosome 7; A1A1, chromosome 13 (Arlt et al. 2009) and in all other cases by flanking PCR and sequencing of the junction (Table S1). Although concordance was high, no one technique identified all novel CNVs in the samples. As a trend, microarrays were least robust at detecting the smallest events because discovery algorithms are strongly influenced by the probe count. Not limited by probe density, mate-pair sequencing detected three new events not called by either the SNP array or aCGH (A3A2, chromosome 4; A1A1, chromosome 1; and A1A1, chromosome 16; Table 2), even though these could be appreciated in array data once attention was directed to the region. All were copy-number gains, which can be difficult to detect by microarray due to their smaller signal deviation, a limitation not applicable to mate-pair sequencing as it detects novel junctions directly. Mate-pair sequencing in turn failed to detect a pericentromeric deletion on A3A2, chromosome 7, which, because of repeat content, had too many artifactual mappings for reliable event calling. The A1A1 deletion on chromosome 13 was also missed, almost certainly as a result of the lower sequencing coverage of this sample (Table 1).
TABLE 1.
Mate-pair sample summary
| Aphidicolin treated |
|||
|---|---|---|---|
| Untreated 090 | A3A2 | A1A1 | |
| Lanes | 4 | 3 | 2 |
| Libraries | 4 | 2 | 2 |
| Read length | 35–39 | 35–36 | 35–36 |
| Mean fragment length (kb) | 2.9–4.4 | 2.8–3.4 | 2.9–3.0 |
| SD fragment length (kb) | 0.22–0.42 | 0.24–0.55 | 0.29–0.35 |
| Mate pairs | 5.9 × 107 | 2.9 × 107 | 1.7 × 107 |
| Unique mate pairs | 4.4 × 107 (74%) | 2.1 × 107 (74%) | 1.2 × 107 (73%) |
| Fraction mapped reads | 84% | 81% | 83% |
| ∼3-kb fragments | 2.4 × 107 (56%) | 8.3 × 106 (39%) | 6.0 × 106 (48%) |
| Haploid fragment coverage | 24.9 | 8.7 | 5.8 |
| Diploid fragment coverage | 12.4 | 4.3 | 2.9 |
| Haploid read coverage | 0.6 | 0.2 | 0.1 |
| Diploid read coverage | 0.3 | 0.1 | 0.1 |
TABLE 2.
Aphidicolin-induced CNVs
| SNP array |
||||||||
|---|---|---|---|---|---|---|---|---|
| Type | Chromosome | Start | End | Size (kb) | aCGH log2R | log2R | B frequency | Mate pairs |
| A3A2 | ||||||||
| Deletion | 7 | 61,379,057 | 61,440,507 | 61 | −0.28 (26)*** | 0.04 (11) | (0) | 0 |
| Deletion | 11 | 6,476,303 | 6,564,800 | 88 | −0.51 (73)**** | −0.33 (51)**** | 1.00 (1) | 6 |
| Deletion | 15 | 69,469,199 | 69,499,819 | 31 | −0.30 (23)*** | −0.33 (11)**** | 1.00 (4)*** | 6 |
| Deletion | 18 | 7,630,270 | 7,864,089 | 234 | −0.31 (183)**** | −0.38 (85)**** | 1.00 (22)**** | 5 |
| Bi-allelic deletion | 3 | 117,988,930 | 118,751,078 | 762 | — | — | — | 2 |
| 118,056,525 | 118,673,931 | 617 | — | — | — | 2 | ||
| 117,988,930 | 118,056,525 | 68 | −0.43 (58)**** | −0.40 (19)**** | 1.00 (10)**** | — | ||
| 118,056,525 | 118,673,931 | 617 | −1.23 (516)**** | −6.63 (232)**** | 0.90 (74)**** | — | ||
| 118,673,931 | 118,751,078 | 77 | −0.46 (66)**** | −0.38 (31)**** | 1.00 (9)**** | — | ||
| Duplication | 1 | 172,806,440 | 172,861,505 | 55 | 0.27 (23)** | 0.24 (7) | (0) | 2 |
| Duplication | 9 | 130,303,541 | 130,699,329 | 396 | 0.20 (232)**** | 0.16 (126)**** | 0.62 (43)**** | 5 |
| Duplication | 10 | 111,820,163 | 114,253,039 | 2433 | 0.29 (1971)**** | 0.19 (879)**** | 0.64 (303)**** | 9 |
| Inversion | 4 | 123,973,228 | 124,044,943 | 72 | — | — | — | 4 |
| 123,999,958 | 124,068,805 | 69 | — | — | — | 9 | ||
| 123,973,228 | 123,999,958 | 27 | 0.26 (24)**** | 0.15 (14)* | 0.62 (5)**** | — | ||
| 124,044,943 | 124,068,805 | 24 | 0.28 (20)*** | 0.15 (14)** | 0.63 (2) | — | ||
| A1A1 | ||||||||
| Deletion | 13 | 93,393,314 | 93,545,246 | 152 | −0.24 (129)**** | ND | ND | 0 |
| Duplication | 1 | 29,268,937 | 29,423,288 | 154 | 0.12 (101)**** | ND | ND | 2 |
| Duplication | 16 | 81,847,144 | 81,882,508 | 35 | 0.15 (28)*** | ND | ND | 3 |
Columns “log2R” and “B frequency” show the average log2 of the sample to reference ratio or the average B allele frequency, respectively, followed by the number of probes in parentheses, followed by a significance value calculated by using the t-test comparing all event probes to the mean value for the array: ****P < 0.00001; ***P < 0.0001; **P < 0.001; *P < 0.01. Boldface type indicates that the event was called by the detection algorithm. For the complex A3A2 bi-allelic deletion (chromosome 3) and inversion with flanking duplications (chromosome 4), rows show either the anomalous junctions with corresponding mate-pair counts or the CNV spans created by these junctions with corresponding array parameters. ND, not done; —, not applicable.
When using optimized filtering parameters (see materials and methods), the list of candidate new CNVs returned from A3A2 SNP array data did not include any false positives. A larger number of false-positive CNVs were returned as candidates from mate-pair data (∼20 per sample). These false events all failed to show the expected change in coverage of the normal ∼3-kb fragments within the putative CNV and were not corroborated by array data, in contrast to real events (Figure 1; Figure S11). They also consistently had only two crossing mate pairs and thus likely correspond to stochastic artifactual fragments or mappings. These artifacts were easy to dismiss with experience, but visual inspection of all candidate events was necessary and CNV calling was most reliable when parallel array and mate-pair data could be compared.
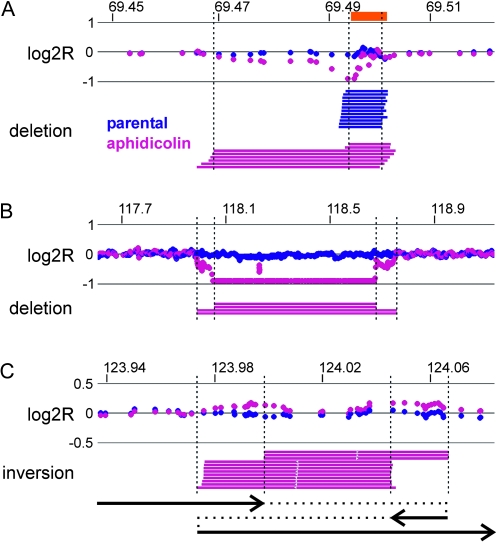
Figure 1.—
Examples of aphidicolin-induced CNVs. Intensity data (log2R) from SNP arrays are at the top of each panel with mate pairs of the indicated type below. Array data are averaged over a five-probe window. Individual mate pairs are drawn as bars connecting the two reads as mapped to the reference genome. Blue denotes the reference 090 sample and magenta the aphidicolin-treated A3A2 sample. Chromosome coordinates are in megabases. (A) An induced deletion on chromosome 15 that overlaps a constitutional CNV. The orange box denotes the position of a known human population CNV. (B) A bi-allelic deletion on chromosome 3. (C) A complex inversion on chromosome 4 that creates two associated copy-number gains. One potential underlying allele is drawn as an event chain similar to Figure S7.
Detection of constitutional CNVs:
We next sought to explore the content of constitutional CNVs in 090 for comparison to induced CNVs and to better understand the capabilities of the methods. Table 3 shows the number of copy-number gains and losses called from SNP array and mate-pair sequence data, grouped according to their correspondence to the other detection method (see Table S2 for a complete list). Consistent with other human individuals (Korbel et al. 2007; Wheeler et al. 2008; Conrad et al. 2010b; Pang et al. 2010; Park et al. 2010), >103 CNVs were discovered, with more losses than gains (1513 vs. 146, respectively). Unlike induced CNVs, a surprisingly large number of constitutional CNVs were called by only one of the two detection methods (1329, or 80% of all CNVs; Table 3, rows A–C; see Figure 2 for examples). A small number of events were judged to be false-negative calls (Table 3, row D). Other failed detections were attributable to technical limitations, in particular to having too few array probes (Table 3, last column) or too small an event size for mate-pair detection. However, even after these factors were considered, many CNVs that might have been detected by both methods were not.
TABLE 3.
Constitutional CNVs
| Detection method: | SNP array |
Mate pair |
||||
|---|---|---|---|---|---|---|
| Detected by other method: | Yes | No | Yes | No | <5 probes | |
| A | Gains | 4 | 98 | 4 | 22 | 22 |
| B | Losses | 326 | 120 | 322 | 382 | 685 |
| C | Total events | 330 | 218 | 326 | 404 | 707 |
| D | False-negative calls | — | 1 (0.5%) | — | 27 (7%) | — |
| E | Matches known CNV | 326 (99%) | 202 (93%) | 319 (98%) | 376 (93%) | 92 (13%) |
| F | Median size (kb) | 1.8 | 1.9 | 3.1 | 2.6 | 2.6 |
| G | Median fold probe density vs. array | 20.8 | 18.4 | 12.2 | 12.7 | 0.0 |
| H | Median (log2R) | 0.48 | 0.33 | 0.45 | 0.13 | — |
| I | Median (Z statistic) | 13.4 | 8.9 | 13.1 | 3.0 | — |
| J | Median mate pairs | — | — | 13 | 9 | 8 |
| K | Median fraction mismatched bases | — | — | 0.007 | 0.008 | 0.008 |
| L | Created by inversion | 1 (0.3%) | ? | 1 (0.3%) | 3 (0.7%) | 66 (9%) |
| M | Tandem repeat (losses only) | 112 (34%) | 82 (68%) | 170 (53%) | 291 (76%) | 458 (67%) |
?, cannot be determined; —, not applicable.
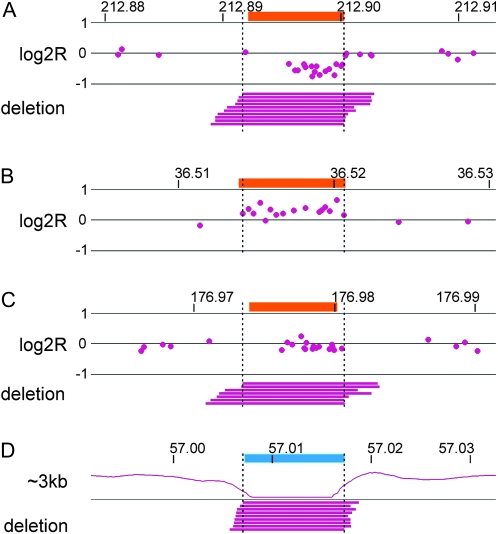
Figure 2.—
Examples of constitutional CNVs. Major classes of constitutional CNVs observed in 090 are depicted similarly to Figure 1. Orange boxes highlight known human CNVs. (A) An 8.2-kb deletion on chromosome 2 evident in both array and mate-pair data. (B) A 6.5-kb gain on chromosome 18 evident in array but not mate-pair data. (C) A 6.5-kb deletion on chromosome 2 evident in mate-pair but not array data. (D) A 10.2-kb deletion on chromosome 12 evident in mate-pair data in a region with no array probes. A trace shows the relative density of normal ∼3-kb mate-pair mappings, and a cyan box highlights a HERVK element in the reference genome.
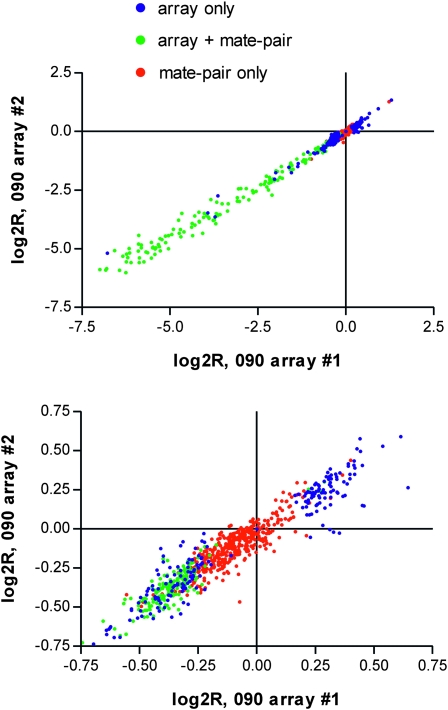
We entertained many explanations for the low concordance between SNP arrays and mate pairs. First, VAMP might give poor detection of CNVs from SNP array data. Use of other CNV calling algorithms such as PennCNV (Wang et al. 2007) did not improve the concordance, however (Figure S9). Moreover, the array data in genomic regions called as mate-pair-specific CNVs were not statistically different from the array average (Table 3, rows H and I; Figure 3). This dichotomy could not be attributed to a specific array run (Figure 3). An alternative would be that VAMP substantially overcalled CNVs from mate-pair data. Many observations suggest that this is not the case. The three categories of mate-pair CNVs showed little or no quality difference in the number of associated mate-pair mappings or rate of base mismatches in associated reads (Table 3, row J and K; Figure S12). Also, when mate-pair-specific CNVs had adequate array probe coverage, the fraction that matched a known human CNV remained very high (Table 3, row E).
Figure 3.—
Array run correlation by CNV category. Scatter plots show the correlation of two different SNP array runs of 090. where the top panel shows all data and the bottom panel is magnified to the region near the origin, corresponding to predicted heterozygous events. Green circles are the array log2R values of CNV regions detected by both array and mate pairs, blue circles are CNVs detected only by array, and red circles are CNVs detected only by mate pairs for which array probes were present.
Interestingly, the category of mate-pair-specific SVs having too few SNP probes for array detection (Table 3, last column, row G) was notably different in having a low rate of correspondence to human CNVs detected by ultra-high-density aCGH (Table 3, row E) (Conrad et al. 2010b; Park et al. 2010). We looked for systematic factors that might make these events amenable to mate-pair as opposed to array detection. Most CNVs created by underlying inversions were not well sampled by the SNP array (66, or 94% of all inversion CNVs; Table 3, row L), perhaps because they were relatively small (median size: 1.6 kb; range: 0.1–20 kb). A majority (67%) of mate-pair events with insufficient array probe coverage contained short tandem repeat elements (Benson 1999) whose contraction might lead to size loss by a non-CNV mechanism (Table 3, row M). Some mate-pair-specific events also likely represent simple deletions of mobile repetitive elements, such as long interspersed nuclear elements (LINEs) and human endogenous retroviruses (HERVs), for which array probes often cannot be meaningfully designed (e.g., Figure 2D).
Finally, we examined the smaller number of CNVs identified by SNP arrays but not by mate pairs. The array statistics for these events were consistently robust (Table 3, rows H and I; Figure 3), and they once again showed a high concordance with known human CNVs (Table 3, row E).
In summary, most constitutional event calls could be validated either internally by virtue of detection by both methods or externally by correspondence to known human CNVs, despite the low concordance between SNP arrays and mate pairs. Mate-pair-specific SVs were the largest unvalidated group, so we randomly selected 35 such events and attempted to amplify the anomalous junctions by PCR (Table S1). These SVs encompassed most observed event types, including CNVs with and without sufficient probes for array detection. Only two SVs had been previously described by systematic array studies (Conrad et al. 2010b; Park et al. 2010). Nearly all showed a PCR product consistent with the predicted SV (31/35, or 89%, Table S1), confirming the high reliability of mate-pair calls.
Fine structure of aphidicolin-induced CNVs:
Mate-pair sequencing was very robust at describing SV structures. With regard to aphidicolin-induced CNVs, this included recognition that one breakpoint of a deletion on chromosome 15 fell 0.4 kb from a known human population CNV, as well as the unambiguous characterization of the two alleles underlying a homozygous deletion on chromosome 3 (Figure 1, A and B). Most strikingly, mate pairs described the precise structure of a complex inversion that created two duplication segments on chromosome 4, an event difficult to detect and impossible to describe from array data alone (Figure 1C). Similar observations held true for constitutional events, including the identification of CNVs associated with 38 inversions with a median inverted segment size of 2.3 kb (range: 0.1–28 kb, Table S2).
Aphidicolin-induced CNVs are large compared to many constitutional CNVs:
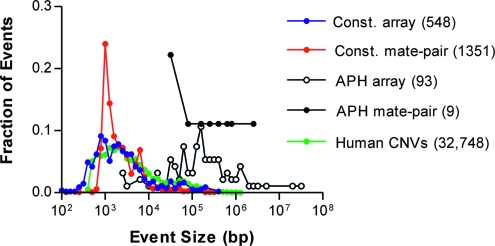
Our last and most important goal was to compare constitutional and aphidicolin-induced SVs to explore the hypothesis that replication stress induces events typical of human polymorphic CNVs and whether the detection method had any effect on this conclusion. As expected, the size distribution of constitutional 090 SVs matched the pattern described for other human individuals and the human population (Korbel et al. 2007; Wheeler et al. 2008; Conrad et al. 2010b; Pang et al. 2010; Park et al. 2010), with an inverse correlation between size and frequency down to a lower detection limit of ∼1 kb (Figure 4). Array CNVs matched many known human CNVs particularly closely, as expected since the SNP array was designed to target these events. Mate-pair events showed a slightly different pattern in being very sensitive for small gains and losses in regions not as easily sampled by arrays, but overall the methods gave a similar result that most constitutional CNVs (∼94%) were <10 kb.
Figure 4.—
Size distribution of SVs. Histograms show the fraction of events by event size within each of four groups. “Const. array”: constitutional CNVs observed in 090D2 by SNP array (blue circles). “Const. mate-pair”: constitutional SVs observed in 090 and/or 090D2 by mate-pair sequencing (red circles). “APH array”: new CNVs observed in a series of 23 independent aphidicolin-treated subclones of 090 or 090D2 by SNP array and/or aCGH (open circles). “APH mate-pair”: new CNVs observed in aphidicolin-treated subclones A3A2 and A1A1 by mate-pair sequencing (black circles). Also shown is a trace of the CNVs detected by microarray from two published compendia (green circles) (Conrad et al. 2010b; Park et al. 2010). Numbers in parentheses indicate the total number of events in each group. Array traces combine both gains and losses, while mate-pair traces combine deletion and duplication sets.
In marked contrast to the constitutional CNVs, 93% of aphidicolin-induced CNVs were >10 kb (median 148 kb; Figure 4), a data set that includes all aphidicolin-induced CNVs that we have described previously (Arlt et al. 2009), in the current study, and in ongoing unpublished analyses using Illumina 1M SNP arrays. We have noted this different size distribution previously but had not known whether it might simply reflect a bias of arrays against detecting small induced events (Arlt et al. 2009). In contrast to the deliberate oversampling of constitutional CNV regions, most induced CNVs are sampled at the average density of 0.37 probes per kilobase, so that ∼14 kb are required to cross the five probes needed for CNV calling. Mate pairs are not subject to this limit and indeed were readily able to detect constitutional changes as small as 1 kb (Figure 4). Despite this, mate pairs did not reveal a new and more frequent class of small aphidicolin-induced CNVs in two different subclones. Some new aphidicolin-induced CNVs were discovered by mate pairs that were indeed smaller than those previously known from aCGH, but these were nonetheless >10 kb (Table 2).
Aphidicolin-induced and constitutional CNVs show similar nonhomologous junctions:
To assess the mechanisms likely to underlie formation of the observed CNVs, we sequenced a subset of breakpoint junctions (Table S1). Extending previous observations (Arlt et al. 2009), all seven aphidicolin-induced events showed microhomologies, blunt joints, and insertions and thus were inconsistent with NAHR. Constitutional CNV junctions revealed a mixture of apparent mechanisms typical of previous reports (Korbel et al. 2007; Vissers et al. 2009; Conrad et al. 2010a), with 2 of 14 (14%) showing extended homology indicative of NAHR and 12 of 14 (86%) being homology independent. We further observed that only 337 of 1351 090 mate-pair deletions and duplications (25%) had a homology segment (at least 75 bp and 80% identity within 500 bp of the predicted junction) available to support NAHR (24 of 80 (30%) and 313 of 1271 (25%) for events larger than and <10 kb, respectively). Thus, nonhomologous mechanisms must also account for most observed constitutional CNVs regardless of size.
Despite the different size distribution, examining the exact breakpoint structure of all available sequenced aphidicolin-induced and human constitutional homology-independent CNVs did not reveal an obvious difference when comparing either aphidicolin-induced CNVs to constitutional CNVs or large CNVs to small CNVs (Table S1; data not shown). Thus, aphidicolin-induced CNVs appear typical of all larger nonhomologous CNVs.
DISCUSSION
Much recent work has been devoted to the description of CNVs and other SVs in the human genome. We approached this subject from the specific perspective of optimizing an experimental cell system being used to probe the environmental and genetic influences on SV formation (Arlt et al. 2009). The combined data provide a strong basis for comparing the properties of induced and constitutional CNVs as well as their methods of detection.
Constitutional compared to replication stress-induced CNVs:
We have described the constitutional SVs in a female of European descent who is not one of the commonly studied individuals. We nonetheless observed a very similar overall pattern of genetic changes as previous studies (Korbel et al. 2007; Wheeler et al. 2008; Conrad et al. 2010b; Pang et al. 2010; Park et al. 2010), including the number of SVs observed and the relatively small ∼2.5-kb median event size (Table 3; Figure 4; Table S2). Similar to human population studies (Conrad et al. 2010a), we infer that the majority (∼75%) of SV junctions were formed by homology-independent mechanisms regardless of event size and type, a fact confirmed in the 14 new junction sequences reported (Table S1). The potential impact of these changes on shaping inter-individual variation is evident in the 180 distinct genes having at least one exon affected by a SV (Table S3). Importantly, the event list here contained >600 previously undocumented SVs. Although some of these may have been missed in population studies due to methodological considerations below, some likely represent low-frequency population polymorphisms or private alleles in our individual.
Relative to the constitutional CNVs, aphidicolin-induced CNVs showed a much larger median size of 148 kb and exclusive utilization of homology-independent mechanisms (Figure 4; Table S1). Importantly, although most constitutional CNVs are <10 kb, larger homology-independent events are still readily observed in most individuals (Korbel et al. 2007; Wheeler et al. 2008; Conrad et al. 2010a,b; Pang et al. 2010; Park et al. 2010). Thus, aphidicolin-induced CNVs are best described as correlating mainly to the subset of germline SVs that are both larger and homology independent (Figure 4). Several factors might contribute to this pattern. First, we have analyzed only two aphidicolin-treated subclones and 11 observed CNVs by the higher-resolution mate-pair method. Nontechnical factors include that constitutional events in any human individual have been subjected to a much greater negative selection pressure than what we observe in cell culture. This almost certainly skews population polymorphisms toward smaller events inherently less likely to disturb gene function. An interesting corollary observation is that most human pathogenic CNVs are large, including nonhomologus CNVs (Vissers et al. 2009), but once again these have mostly been discovered using low-resolution arrays (Stankiewicz and Beaudet 2007; Cook and Scherer 2008; Kirov et al. 2009; Tam et al. 2009; Zhang et al. 2009; Miller et al. 2010).
A nonexclusive possibility is that the mechanisms contributing to formation of constitutional CNVs are more diverse than those stimulated by aphidicolin. This is certainly true for the subset of small SVs manifest as changes in variable number tandem repeats (VNTRs) or mobile genetic elements. Beyond these special cases, we found no obvious structural signature that could distinguish the nonhomologous junctions that characterize most aphidicolin-induced and constitutive CNVs of any size, although sequence information is scant for constitutional CNVs >30 kb. Regardless of size or source, most CNVs are variably characterized by microhomologies, blunt ends, and/or short inserted sequences, features that might result from many mechanisms including NHEJ, alternative end joining, MMBIR, and template switching (Lee et al. 2007; McVey and Lee 2008; Hastings et al. 2009a; Lieber 2010; Lieber and Wilson 2010). It cannot be judged from current data whether aphidicolin induces just a subset of these mechanisms that are more diversely utilized in constitutional events or whether some unknown feature causes the same mechanism(s) to be used for joint formation but with a tendency toward larger segment jumps when inhibited replication is the underlying stimulus.
SNP arrays compared to mate-pair sequencing:
Overall, mate-pair sequencing had the best power for describing SVs as compared to 1M feature SNP arrays. This was apparent not only in the increased detection of bona fide induced and constitutional events, but also in the markedly superior descriptions of their underlying structure (Tables 2 and 3; Figures 1 and 2). Array analysis nonetheless had its advantages, among them a much greater simplicity and lower cost. Moreover, because arrays use an entirely different basis of detection, they often helped to clarify mate-pair data and uniquely detected a number of events. Indeed, the best descriptions of SVs were undoubtedly obtained when array and mate-pair data could be compared.
The failed correlations of arrays and mate-pair sequencing highlight the limitations of each technique. An overriding issue was the strong dependence on the genome locations sampled by the array design. Human population CNVs are often small (∼1 kb) but could be detected by the 1M feature SNP array because these regions were deliberately oversampled. This obviously limits arrays for detection of unknown small events sampled at the average array density, such as might be induced in our cell system or underlie a pathology of interest. Genomic regions that are difficult to sample will be further underrepresented. Indeed, the largest single category of method discrepancies was events detected by mate pairs in regions that had too few probes for array detection (Table 3). A less obvious problem is the potential negative impact of oversampling on probe quality, including the use of non-SNP probes and the increased frequency of probes placed in repetitive elements. This is evident in the large number of known human CNVs unambiguously detected by mate pairs in 090 for which the SNP array data were not statistically deviant from the array average (Figure 3).
For mate-pair sequencing, a main limitation was genome coverage, easily appreciated by comparing the A3A2 and A1A1 samples (Tables 1 and 2). However, present technology is substantially advanced over that used to obtain much of our data so that a single Illumina sequencing lane now provides sufficient coverage for three or more crossing fragments per junction. More challenging is mapping mate pairs and making event calls. False-negative calls are the most likely errors as a result of CNVs contained entirely within highly repetitive genome regions where accurate mapping is all but impossible. A special class of false-negative calls might occur when the reference genome itself carries a duplication common to the studied sample. Here, mate pairs would not detect an anomaly but array methods would still reveal the increased copy number. This phenomenon might help account for the bias of array-specific calls toward copy-number gains (Table 3; Figure 3).
False positive mate-pair calls are possible but much less likely when multiple independent fragments predict an event. In this context, many factors likely contribute to the seemingly large number of mate-pair-specific calls. First, all calls were relative to Build 36/hg18 and need not represent true SVs. Indeed, at least once, a sequenced SV mapped correctly to an alternative genome assembly. Copy-number neutrality might also be consistent with a mate-pair event if there are corresponding gains and losses on different alleles for which one allele escaped mate-pair detection. Most importantly, mate-pair analysis detects all manner of SVs that are invisible to arrays, including tandem repeat expansions and gain or loss of mobile genetic elements.
Recent descriptions of data from the 1000 Genomes Project and other high-coverage human genome sequences highlight final limitations of both SNP arrays and low-depth mate-pair sequencing (Durbin et al. 2010; Pang et al. 2010; Sudmant et al. 2010). First, it is now clear that the frequency of human population CNVs increases continuously with decreasing event size and does not show the rapid drop-off below ∼1 kb as observed here (Figure 4), which for SNP arrays and mate-pair sequencing reflect the limitations of probe density and fragment size distribution, respectively. Further, the new ability to compare deep sequencing of multiple human individuals has established the high frequency of human CNVs in repetitive genome segments such as segmental duplications (Sudmant et al. 2010), regions inherently difficult to study by either microarray or low-depth mate-pair sequencing. Nonetheless, results here demonstrate the robust detection of most induced CNVs in an experimental setting by either SNP arrays or low-depth mate-pair sequencing.
Acknowledgments
We thank Jun Li and Steve Qin for many helpful discussions regarding mate-pair sequencing in the early phases of this project. This work was supported by National Institutes of Health grant RCI-ES018672 to T.E.W. and T.W.G. and by a research grant from the March of Dimes Foundation to T.W.G.
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.124776/DC1.
Array data have been deposited in the NCBI Gene Expression Omnibus under accession no. GSE26121. Mate-pair sequence data have been deposited in the NCBI Sequence Read Archive under study no. SRP003289. Called SVs have been deposited in NCBI dbVar.
References
- Arlt, M. F., J. G. Mulle, V. M. Schaibley, R. L. Ragland, S. G. Durkin et al., 2009. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am. J. Hum. Genet. 84 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell, G. R., C. D. Greenman, H. Davies, A. P. Butler, S. Edkins et al., 2010. Signatures of mutation and selection in the cancer genome. Nature 463 893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland, S. R., N. Jin, A. C. Ozdemir, R. H. Lyons, L. S. Weisman et al., 2010. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole genome sequencing. Genetics 186 1127–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila, C., M. Jankovic, C. T. Yan, J. H. Wang, D. R. Wesemann et al., 2010. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl. Acad. Sci. USA 107 3034–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, D. F., C. Bird, B. Blackburne, S. Lindsay, L. Mamanova et al., 2010. a Mutation spectrum revealed by breakpoint sequencing of human germline CNVs. Nat. Genet. 42 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad, D. F., D. Pinto, R. Redon, L. Feuk, O. Gokcumen et al., 2010. b Origins and functional impact of copy number variation in the human genome. Nature 464 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, E. H., Jr., and S. W. Scherer, 2008. Copy-number variations associated with neuropsychiatric conditions. Nature 455 919–923. [DOI] [PubMed] [Google Scholar]
- Counter, C. M., W. C. Hahn, W. Wei, S. D. Caddle, R. L. Beijersbergen et al., 1998. Dissociation among in vitro telomerase activity, telomere maintenance, and cellular immortalization. Proc. Natl. Acad. Sci. USA 95 14723–14728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock, N., M. E. Hurles, N. Cardin, R. D. Pearson, V. Plagnol et al., 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464 713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin, R. M., G. R. Abecasis, D. L. Altshuler, A. Auton, L. D. Brooks et al., 2010. A map of human genome variation from population-scale sequencing. Nature 467 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings, P. J., G. Ira and J. R. Lupski, 2009. a A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5 e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings, P. J., J. R. Lupski, S. M. Rosenberg and G. Ira, 2009. b Mechanisms of change in gene copy number. Nat. Rev. Genet. 10 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate, A. J., L. Feuk, M. N. Rivera, M. L. Listewnik, P. K. Donahoe et al., 2004. Detection of large-scale variation in the human genome. Nat. Genet. 36 949–951. [DOI] [PubMed] [Google Scholar]
- Kirov, G., D. Grozeva, N. Norton, D. Ivanov, K. K. Mantripragada et al., 2009. Support for the involvement of large copy number variants in the pathogenesis of schizophrenia. Hum. Mol. Genet. 18 1497–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel, J. O., A. E. Urban, J. P. Affourtit, B. Godwin, F. Grubert et al., 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318 420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J. A., C. M. Carvalho and J. R. Lupski, 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131 1235–1247. [DOI] [PubMed] [Google Scholar]
- Lieber, M. R., 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber, M. R., and T. E. Wilson, 2010. SnapShot: nonhomologous DNA end joining (NHEJ). Cell 142 496–496.e1 [DOI] [PubMed] [Google Scholar]
- McVey, M., and S. E. Lee, 2008. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet. 24 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, D. T., M. P. Adam, S. Aradhya, L. G. Biesecker, A. R. Brothman et al., 2010. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 86 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, A. W., J. R. MacDonald, D. Pinto, J. Wei, M. A. Rafiq et al., 2010. Towards a comprehensive structural variation map of an individual human genome. Genome Biol. 11 R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, H., J. I. Kim, Y. S. Ju, O. Gokcumen, R. E. Mills et al., 2010. Discovery of common Asian copy number variants using integrated high-resolution array CGH and massively parallel DNA sequencing. Nat. Genet. 42 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, M., J. Lange and S. Keeney, 2010. Genome destabilization by homologous recombination in the germ line. Nat. Rev. Mol. Cell Biol. 11 182–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat, J., B. Lakshmi, J. Troge, J. Alexander, J. Young et al., 2004. Large-scale copy number polymorphism in the human genome. Science 305 525–528. [DOI] [PubMed] [Google Scholar]
- Sharp, A. J., D. P. Locke, S. D. McGrath, Z. Cheng, J. A. Bailey et al., 2005. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek, D., and M. Jasin, 2010. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat. Struct. Mol. Biol. 17 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz, P., and A. L. Beaudet, 2007. Use of array CGH in the evaluation of dysmorphology, malformations, developmental delay, and idiopathic mental retardation. Curr. Opin. Genet. Dev. 17 182–192. [DOI] [PubMed] [Google Scholar]
- Stankiewicz, P., and J. R. Lupski, 2010. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 61 437–455. [DOI] [PubMed] [Google Scholar]
- Stratton, M. R., P. J. Campbell and P. A. Futreal, 2009. The cancer genome. Nature 458 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant, P. H., J. O. Kitzman, F. Antonacci, C. Alkan, M. Malig et al., 2010. Diversity of human copy number variation and multicopy genes. Science 330 641–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam, G. W., R. Redon, N. P. Carter and S. G. Grant, 2009. The role of DNA copy number variation in schizophrenia. Biol. Psychiatry 66 1005–1012. [DOI] [PubMed] [Google Scholar]
- Vissers, L. E., S. S. Bhatt, I. M. Janssen, Z. Xia, S. R. Lalani et al., 2009. Rare pathogenic microdeletions and tandem duplications are microhomology-mediated and stimulated by local genomic architecture. Hum. Mol. Genet. 18 3579–3593. [DOI] [PubMed] [Google Scholar]
- Wang, K., M. Li, D. Hadley, R. Liu, J. Glessner et al., 2007. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17 1665–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, D. A., M. Srinivasan, M. Egholm, Y. Shen, L. Chen et al., 2008. The complete genome of an individual by massively parallel DNA sequencing. Nature 452 872–876. [DOI] [PubMed] [Google Scholar]
- Zhang, F., W. Gu, M. E. Hurles and J. R. Lupski, 2009. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10 451–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Array data have been deposited in the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession GSE26121, mate-pair sequence data have been deposited in the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra) under study no. SRP003289, and called SVs have been deposited in NCBI dbVar (http://www.ncbi.nlm.nih.gov/dbvar).