Abstract
Protein components of the Drosophila male ejaculate are critical modulators of reproductive success, several of which are known to evolve rapidly. Recent evidence of adaptive evolution in female reproductive tract proteins suggests this pattern may reflect sexual selection at the molecular level. Here we explore the evolutionary dynamics of a five-paralog gene family of female reproductive proteases within geographically isolated subspecies of Drosophila mojavensis. Remarkably, four of five paralogs show exceptionally low differentiation between subspecies and unusually structured haplotypes that suggest the retention of old polymorphisms. These gene genealogies are accompanied by deviations from neutrality consistent with diversifying selection. While diversifying selection has been observed among the reproductive molecules of mammals and marine invertebrates, our study provides the first evidence of this selective regime in any Drosophila reproductive protein, male or female.
THE molecular interface that underlies sexual reproduction is extraordinarily dynamic. In a host of biologically diverse organisms, proteins with reproductive functions evolve rapidly through changes in coding sequence (reviewed in Swanson and Vacquier 2002a,b; Clark et al. 2006; Panhuis et al. 2006), lineage-specific gene duplications, evolution of novel proteins, and regulatory changes (Begun and Lindfors 2005; Mueller et al. 2005; Begun et al. 2006; Findlay et al. 2008). Within species, reproductive proteins show evidence of two contrasting selective regimes. Reduced polymorphism or elevated divergence at many reproductive protein loci indicates they have experienced positive directional selection (Lee et al. 1995; Aguadé 1998, 1999; Calkins et al. 2007; Clark et al. 2007). In contrast, other reproductive proteins exhibit remarkable intraspecific diversity generated by balancing selection (Metz and Palumbi 1996; Gasper and Swanson 2006; Levitan and Ferrell 2006; Hamm et al. 2007; Clark et al. 2009), alternative splicing (Moy et al. 2008; Springer et al. 2008), copy number variation (Dopman and Hartl 2007), and gene conversion (Kelleher and Markow 2009).
The exceptional dynamics of reproductive proteins are thought to result from sexual selection that occurs after copulation or gamete release (reviewed in Swanson and Vacquier 2002a,b; Clark et al. 2006; Panhuis et al. 2006). Sperm competition predicts an ongoing arms race between the abundant gametes of multiple males to achieve fertilization of more limited female gametes (Birkhead and Møller 1998). Cryptic female choice, a postcopulatory equivalent of traditional sexual selection, suggests that females can bias fertilization success toward certain males on the basis of qualities of the male ejaculate (Eberhard 1996). Finally, sexual conflict, or a difference in the reproductive interests of the two sexes, proposes that an evolutionary arms race between males and females over control of reproductive outcomes can cause rapid evolution of the proteins involved (Parker 1979). Although both cryptic female choice and sexual conflict predict a role for female reproductive proteins in driving the dynamics of their male interactors, when compared to the extensive literature on the evolution of male reproductive molecules, examination of the female side remains lacking.
The promiscuous mating system and unique reproductive biology of the cactus breeding fruit fly, Drosophila mojavensis, presents an exciting system in which to explore the evolution of female reproductive proteins. These females remate daily (reviewed in Markow 1996), and broods of wild-caught females have been demonstrated to have up to six different sires, implying strong postcopulatory sexual selection (Good et al. 2006). D. mojavensis females, furthermore, are known to incorporate male-derived molecules into somatic tissues and oocytes (Markow and Ankney 1984), a nutritional benefit to copulation that dramatically contrasts the “cost of mating” incurred by the females of their congener D. melanogaster (Chapman et al. 1995; Pitnick and García-González 2002; Kuijper et al. 2006; Barnes et al. 2008). Finally, D. mojavensis females exhibit an insemination reaction, an opaque mass of unknown composition that occupies the uterus after every copulation (Patterson 1946; Alonso-Pimentel et al. 1994). This reaction mass is thought to protect the male's nutritional investment from cuckoldry by competing ejaculates (Markow and Ankney 1984, 1988; Pitnick et al. 1997), and furthermore, may coevolve antagonistically between the sexes (Knowles and Markow 2001).
Our previous expressed sequence tag (EST) analysis of the lower reproductive tracts of D. arizonae, a closely related sister species of D. mojavensis [most recent common ancestor (MRCA) ∼0.7 MYA] (Reed et al. 2007; Matzkin 2008), identified 241 candidate female reproductive tract proteins that potentially interact with the male ejaculate (Kelleher et al. 2007). The most intriguing of these candidates were five recently duplicated endoprotease families, exclusively expressed in the lower female reproductive tract (Kelleher et al. 2007). The current study explores the evolutionary history of one of the two largest protease families in four ecologically and geographically isolated subspecies of D. mojavensis; Baja Peninsula (D. m. baja), Catalina Island (D. m. wrigleyi), mainland Sonora (D. m. sonorensis), and the Mojave Desert (D. m. mojavensis) (Pfeiler et al. 2009). The family encodes five serine-endoprotease paralogs (Figure 1, protease gene family 1; Kelleher et al. 2007), all of which contain signal peptides suggesting they are secreted into the female reproductive tract lumen, creating the potential for biochemical interaction and coevolution with male seminal proteins (Kelleher et al. 2007). Consistent with a hypothesis of molecular coevolution, the protease gene family examined here exhibits evidence of positive selection at some codons, as inferred from divergence between species and paralogs (Kelleher et al. 2007).
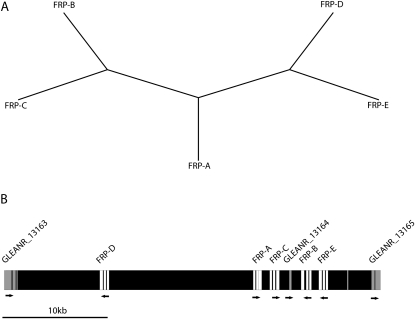
Figure 1.—
Phylogenetic relationships and genomic arrangement of the paralogs examined in this study. (A) Evolutionary relationships between paralogs adapted from Kelleher et al. (2007). (B) Genomic arrangement of the protease gene family on D. mojavensis chromosome 4 (scaffold_6680 bp 10216565-10169309, http://rana.lbl.gov/drosophila/). Open blocks indicate individual exons of duplicated paralogs, while shaded blocks indicate individual exons of flanking and interspersed coding sequences. Arrows indicate the direction of transcription.
Previous studies have demonstrated that the four subspecies of D. mojavnesis are genetically and morphologically differentiated (Machado et al. 2007; Reed et al. 2007; Matzkin 2008; reviewed in Pfeiler et al. 2009), and that male and female contributions to reproductive outcomes are coadapted within them (Knowles and Markow 2001; Pitnick et al. 2003; Knowles et al. 2005; Kelleher and Markow 2007). Consistent with these phenotypic observations, we find considerable evidence for population-specific selection in the female reproductive proteins examined here. We furthermore find that in contrast to D. melanogaster, which exhibits a high frequency of positive directional selection among its female reproductive tract proteins (Swanson et al. 2004; Panhuis and Swanson 2006; Lawniczak and Begun 2007), D. mojavensis female reproductive tract proteins display deviations from neutrality consistent with balancing selection. We discuss our results in terms of sexual selection theory and the unique reproductive biology of D. mojavensis.
MATERIALS AND METHODS
Fly strains:
D. mojavensis were collected from Catalina Island (2001), Mojave Desert (2002), Baja Peninsula (2002), and mainland Sonora (2007) by J. Bono, L. Reed, and L. Matzkin. D. arizonae, the sister species of D. mojavensis, were collected in Tucson, Arizona (2000) by L. Matzkin. All flies used in population analyses were maintained as isofemale lines. Between seven and eight isofemale lines were sampled from each population for each locus. A third, closely related species, D. navojoa, was obtained from the Tucson Drosophila Stock Center.
Loci and primer design:
The genomic arrangement and phylogenetic relationships of the protease gene family examined in this study are presented in Figure 1. Although the genes remain unannotated, our previous study showed that they were expressed in D. arizonae, and that orthologous sequences were present in the D. mojavensis genome (Kelleher et al. 2007). Intron–exon splice sites were inferred from D. arizonae ESTs in Kelleher et al. (2007). For simplicity, we refer to these genes as female reproductive proteases A–E, or FRP-A–E, where FRP-A corresponds to Dari\anon-EST:Kelleher9, FRP-B corresponds to Dari\anon-EST:Kelleher8, FRP-C corresponds to Dari\anon-EST:Kelleher7, FRP-D corresponds to Dari\anon-EST:Kelleher6, and FRP-E corresponds to anon-EST:Kelleher-5. D. mojavensis (http://rana.lbl.gov/drosophila/) orthologs further were aligned to available D. arizonae ESTs to generate paralog-specific primers that amplified the majority of the coding sequence for each locus. All paralogs were reciprocally monophyletic (not shown). Further, heterozygosity in sampled isofemale lines was quite low, except for flies that recently had been introduced to the lab (fewer than five generations). We are confident, therefore, that each set of primers amplified a unique genomic location.
Sequencing:
Genomic DNA was isolated from whole flies using the DNeasy it (Qiagen) according to manufacturer instructions. Standard PCR was performed using internal, paralog-specific primers (Figure 1). All sequencing was performed on an ABI 3700 DNA sequencer with Big Dye Terminator chemistry. Primers and PCR conditions are available from the authors upon request. Base calling and assembly were performed in Sequencher 4.8.
Polymorphism analyses:
Haplotypes were phased in Arlequin (http://cmpg.unibe.ch/software/arlequin3/), and a single haplotype for each individual was retained for subsequent analyses. Polymorphism analyses, estimation of population parameters, and tests of selection were performed in DNAsp (Rozas et al. 2003). Sample sizes, sequence lengths, and estimates of polymorphism are presented in supporting information, Table S1. Significance of site frequency spectra statistics and measurements of linkage disequilibrium were assessed by coalescent simulations under the conservative assumption of no recombination. For tests requiring an outgroup, one or more D. arizonae orthologs were used for FRP-A–FRP-C. For FRP-D we used an allele of FRP-E as the outgroup, and vice versa, due to uncertainty surrounding the identity of the D. arizonae ortholog. Using closely related paralogs as an outgroup sequence, in place of sibling species, is known to be a conservative approach for McDonald–Kreitman (MK) tests (McDonald and Kreitman 1991; Thornton and Long 2005; Thornton 2007). We note, however, that using the putative D. arizonae ortholog had no effect on the outcome of the test. Tests were polarized with the appropriate sequence from D. navojoa.
Gene conversion was detected by GENECONV within an alignment of all unique haplotypes for all paralogs using the method of Sawyer (1989). Briefly, gene conversion tracts between pairs of sequences are identified by considerable stretches of complete identity interspersed between two regions of considerable mismatch or one region of mismatch and the end of the alignment. Statistical significance of these fragments is determined by permutation tests. Neighbor-joining gene trees (Saitou and Nei 1987) were constructed in Paup*4.0b10 (Swofford 2000).
Three-dimensional modeling:
Serine endoprotease catalytic sites (reviewed in Polgar 2005), and protease inhibitor sites (reviewed in Srinivasan et al. 2006), as well as previously identified Bayes empirical Bayes positively selected sites (Yang et al. 2005; Kelleher et al. 2007), were mapped to a predicted three-dimensional (3D) model for FRP-C obtained from Swiss-Model (Schwede et al. 2003).
To test for an association between positively selected sites and protease inhibitor sites, we implemented a permutation analysis previously described in Clark et al. (2007). Briefly, the distance from each selected site to the nearest inhibitor site was calculated, and its mean value compared to a distribution of distances between random pairs of sites. Buried, core sites with ≤10% surface exposure per residue, as calculated by GETAREA (Fraczkiewicz and Braun 1998), were not considered for random sets. This exclusion makes the test more conservative, because these sites evolve slowly relative to surface sites and rarely are inferred as positively selected. Statistical significance was determined as the fraction of random permutations with a mean distance equal to or lower than the observed mean distance between selected and inhibitor sites.
RESULTS
Multiple paralogs evolve nonindependently through gene conversion:
Gene conversion and nonallelic homologous recombination result in nonindependent evolutionary histories of paralogous loci. Describing this process is critical, as it leads to complex genealogies and unusual patterns of polymorphism not seen for single copy genes (Innan 2003; Thornton 2007). In our FRPs, gene conversion tracts between pairs of paralogous haplotypes were identified as fragments of complete identity flanked by regions of significant mismatch, using the method of Sawyer (1989). No evidence of gene conversion was observed between FRP-A and any other paralog, indicating this locus evolves independently (Figure 2). In contrast, there is evidence of gene conversion in at least one pairwise comparison between all other paralogs in the examined gene family (Figure 2). The lack of detectable gene conversion between FRP-A and the other paralogs may suggest that gene conversion does not occur, but could also indicate that it is deleterious. Indeed, all sampled haplotypes of this paralog exhibit the same replacement changes in the three residues of the catalytic triad (reviewed in Polgar 2005), suggesting it has acquired a divergent, nonproteolytic function. In contrast, all other paralogs have retained a catalytic triad, indicating that their biochemical functions may be more similar.
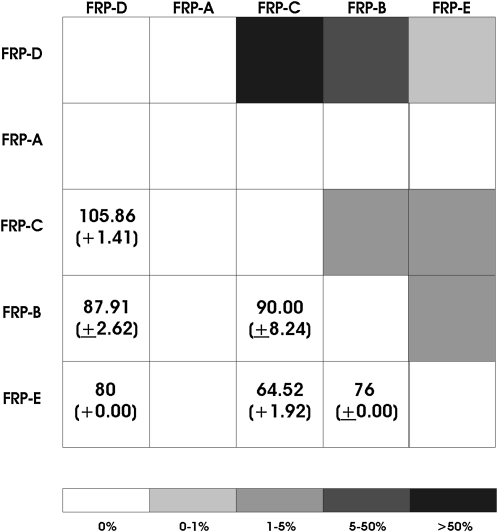
Figure 2.—
Ectopic recombination. An alignment of all unique haplotypes was used to detect significant fragments of complete identity in GENECONV, on the basis of the method of Sawyer (1989). The percentage of pairwise comparisons between paralogs that show evidence of gene conversion is shaded in the upper right. The average length of identified conversion tracts between paralogs, and the standard deviation of this estimate, are indicated in the lower left.
In terms of both tract length and frequency, the most extensive gene conversion was observed between paralogs FRP-C and FRP-D. These paralogs are neither physically adjacent, nor are they genetically more similar to each other than to the remainder of the gene family. Conversely, minimal gene conversion was observed between adjacent paralogs or between genetically similar pairs FRP-D and FRP-E, and FRP-B and FRP- C. There is no evidence, therefore for an association between phylogenetic or physical distance and gene conversion.
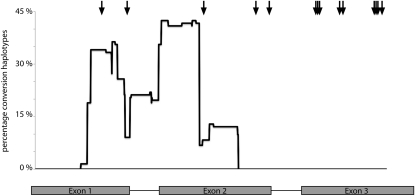
Examination of the frequency that a given site is found within a significant fragment reveals that gene conversion is nonrandomly distributed along the chromosome (Figure 3). Gene conversion is frequent in the 5′ end of the gene, peaks near the center, and is entirely absent from the 3′ end. Intriguingly, previously described selected sites (Kelleher et al. 2007) are highly concentrated at the 3′ end of the gene (Figure 3), suggesting a possible negative association between gene conversion and adaptive evolution. Consistent with this hypothesis, we observed that positively selected sites exhibited a significantly lower frequency of gene conversion than nonselected sites (Wilcoxon rank sum test, P = 0.0016).
Figure 3.—
Sliding window analysis of gene conversion. Y-axis denotes the percentage of haplotypes from the full alignment, including all haplotypes of all subspecies and all paralogs that show evidence of gene conversion at a particular site. The intron–exon structure is shown to scale along the x-axis, with a total length of ∼1 kb. Solid arrows denote selected sites from Kelleher et al. (2007).
Female reproductive proteases exhibit unusually low population structure:
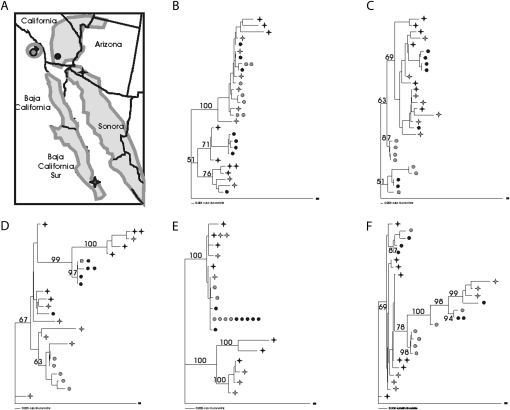
A previous examination of genome-wide variation in D. mojavensis shows that haplotypes are structured between the four geographically isolated subspecies, D. m. baja, D. m. wrigleyi, D. m. sonorensis, and D. m. mojavensis (Machado et al. 2007; Figure 4A). Specifically, a concatenated genealogy of 10 nuclear loci revealed a well-supported (>86%) reciprocally monophyletic clade for each subspecies (Machado et al. 2007). To explore the relationships between female reproductive protease haplotypes sampled here, gene genealogies were constructed for each of the five paralogs (Figure 4 B–F). Well-supported clades (bootstrap values >95%) containing individuals from multiple subspecies were observed for FRP-A and FRP-C– FRP-E, providing little evidence for a geographical structuring of sampled haplotypes. The genealogies of FRP-A and FRP-D are particularly unusual, as haplotypes sort into two divergent clades, suggesting ancestral polymorphism. Recombination, or gene conversion between paralogs is expected to lengthen terminal branch lengths and reduce internal branch lengths (Schierup and Hein 200). Thus, while gene conversion may affect the phylogenetic structure of duplicated loci, it is unlikely to give rise to the deep bifurcations we observe in FRP-A and FRP-D. To determine whether these results represent a genuine difference from the neutral loci examined in Machado et al. (2007), we constructed gene genealogies for each of the 10 loci sampled in that study individually. No well-supported clades (>95%) grouping haplotypes from multiple subspecies were observed (not shown), indicating that the genealogies of the duplicated proteases examined here are distinct from those of putatively neutral loci.
Figure 4.—
Female reproductive proteases exhibit unusual haplotype structures. (A) Geographic distribution and collection sites of four isolated subspecies of D. mojavensis: D. m. baja (Baja Peninsula, shaded star). D. m. wrigleyi (Catalina Island, shaded circle), D. m. sonorensis (mainland Sonora, solid star), and D. m. mojavensis (Mojave Desert, solid circle). Gene genealogies of FRP-A (B), FRP-B (C), FRP-C (D), FRP-D (E), and FRP-E (F). All genealogies were inferred by neighbor joining in Paup*4.0 b10 (Swofford 2000). Symbols indicate subspecies of origin, and number of symbols indicates individual sampled alleles that correspond to that haplotype. Bootstrap values are indicated.
To further quantify the disparity in population structure between neutral loci and our female reproductive proteases, we compared FST values between the two gene classes (Table 1). For all six pairwise combinations of subspecies, the mean FST value for FRPs was lower than that of putatively neutral loci (Machado et al. 2007), although the difference was only significant for three of these comparisons (Table 1). Furthermore, 21 individual combinations of loci and population pairs exhibit FST values that are significantly lower than the neutral distribution from Machado et al. (2007), 13 of which remain significant after correction for multiple testing (Table 1). Finally, the global FST value across all four subspecies was significantly lower than those of neutral loci for every FRP examined except FRP-A (Table 1). Collectively these comparisons suggest that FRPs exhibit reduced population structure, consistent with a regime of natural selection that acts to maintain genetic variation within subspecies.
TABLE 1.
Comparison of FST values for neutral and FRP loci
| wr. and mo. | ba. and wr. | so. and wr. | ba. and mo. | so. and mo. | so. and ba. | All | |
|---|---|---|---|---|---|---|---|
| Neutral mean FST | 0.60 (±0.24) | 0.44 (±0.32) | 0.42 (±0.32) | 0.46 (±0.25) | 0.53 (±0.19) | 0.19 (±0.21) | 0.44 (±0.24) |
| FRP-A | 0.45* | 0.13** | 0.45 | 0.12***a | −0.02***a | 0.11 | 0.3** |
| FRP-B | 0.26***a | 0.32 | 0.23 | 0.19** | 0.13***a | 0.02** | 0.22**a |
| FRP-C | 0.67 | 0.08**a | 0.45 | 0.28 | 0.25***a | 0.14 | 0.26 |
| FRP-D | 0.00***a | 0.30 | 0.25* | 0.30 | 0.25***a | 0.00**a | 0.18***a |
| FRP-E | 0.26***a | 0.25 | 0.65 | −0.05***a | 0.26***a | 0.23 | 0.2* |
| FRP mean Fst | 0.33 (±0.25)* | 0.21 (±0.11) | 0.41 (±0.17) | 0.16 (±0.14)* | 0.18 (±0.12)***a | 0.11 (± 0.094) | 0.24 (±0.05)*a |
Neutral mean Fst is calculated from loci in Machado et al. (2007). Values in parentheses represent the standard deviation. For individual loci and and subspecies, we determined whether FST values were significantly different from the neutral distribution using a Z-test. Furthermore, we compared the distribution of FST values between neutral and FRP loci using a two-sample t-test. All FST values were calculated in DnaSP (Rozas et al. 2003). wr., D. m. wrigleyi; ba., D. m. baja; so., D. m. sonorensis; mo., D. m. mojavensis. *P < 0.05, **P < 0.01, ***P < 0.001.
Values that remain significant after a conservative Bonferroni correction for multiple measures.
Female reproductive protein haplotypes exhibit nonneutral levels of linkage disequilibrium:
To further characterize the unusual haplotype structure of FRPs, we estimated linkage disequilibrium within each locus and subspecies, using Zns (Kelly 1997) and Za (Rozas et al. 2001) (Table 2). Zns measures the correlation of pairwise linkage disequilibrium, r2, for all polymorphic sites within a locus (Kelly 1997), while Za estimates the correlation in r2 values between adjacent polymorphic sites only (Rozas et al. 2001). For both statistics, values that approach 1 indicate high linkage disequilibrium and low asymmetry in the frequency of polymorphic sites, a pattern that could result either from cryptic population structure, or from natural selection acting on a polymorphism that is closely linked to neutral sites in this region. For example, alcohol dehydrogenase (Adh), a frequently cited example of balancing selection in D. melanogaster (Hudson et al. 1987) exhibits significantly elevated values of Zns within the region that has undergone nonneutral evolution (Kelly 1997). After correction for multiple testing, four of five paralogs exhibit a degree of linkage disequilibrium that is inconsistent with a standard neutral model in at least one subspecies, as assessed by coalescent simulations (Table 2). These significant values may result from natural selection, as no evidence for cryptic population structure within subspecies has been seen for other loci (Machado et al. 2007; Reed et al. 2007; Matzkin 2008).
TABLE 2.
Linkage disequilibrium
| All |
No conversion |
|||
|---|---|---|---|---|
| Zns | Za | Zns | Za | |
| FRP-A | ||||
| D. m. baja | 0.71 | 0.81** | NA | NA |
| D. m. wrigleyi | 0.42 | 0.52 | NA | NA |
| D. m. sonorensis | 0.47 | 0.53 | NA | NA |
| D. m. mojavensis | 0.81* | 0.90**a | NA | NA |
| FRP-B | ||||
| D. m. baja | 0.62 | 0.74 | 0.49 | 0.60 |
| D. m. wrigleyi | 0.77** | 0.96** | 0.78 | 0.92* |
| D. m. sonorensis | 0.39 | 0.6 | 0.35 | 0.67 |
| D. m. mojavensis | 0.77* | 0.97**a | 0.75 | 0.95** |
| FRP-C | ||||
| D. m. baja | 0.38 | 0.54 | 0.38 | 0.54 |
| D. m. wrigleyi | NA | NA | NA | |
| D. m. sonorensis | 0.73 | 0.75 | 0.71 | 0.69 |
| D. m. mojavensis | NA | NA | NA | |
| FRP-D | ||||
| D. m. baja | 0.91** | 0.96***a | 0.90***a | 0.96***a |
| D. m. wrigleyi | NA | NA | NA | NA |
| D. m. sonorensis | 0.67 | 0.80* | 0.39 | 0.53 |
| D. m. mojavensis | NA | NA | NA | NA |
| FRP-E | ||||
| D. m. baja | 0.85** | 0.85** | 0.84** | 0.85** |
| D. m. wrigleyi | 0.71 | 0.95**a | NA | NA |
| D. m. sonorensis | 0.38 | 0.47 | NA | NA |
| D. m. mojavensis | 0.91** | 0.93**a | NA | NA |
Zns (Kelly 1997) and Za (Rozas et al. 2001) were calculated for all combinations of subspecies and loci in DnaSP, and statistical significance was determined by coalescent simulations under the conservative assumption of no recombination (Rozas and Rozas 1995). Values are for all sites, as well as values where sites with evidence for gene conversion are reported. NA, incalculable or there was no evidence for gene conversion. *P < 0.05, **P < 0.01, ***P < 0.001.
Values that remain significant after a conservative Bonferroni correction for multiple measures.
If gene conversion introduces multiple linked polymorphisms within the same conversion tract, it could lead to strong linkage disequilibrium between polymorphic sites and significant values of Zns and Za. Because no gene conversion was detected between FRP-A and any other paralog, this phenomenon cannot explain the degree of linkage disequilibrium observed at this locus. To elucidate the contribution of gene conversion to linkage disequilibrium at the other four loci, we identified sites with evidence of gene conversion within each population and excluded them from the analysis. While in some cases, significant linkage disequilibrium was no longer detected when sites of gene conversion were excluded, the majority of values remained significant (Table 2). We cannot, however, rule out the possibility that GENECONV failed to detect older gene conversion events that nonetheless contribute to linkage disequilibrium.
Polymorphism and divergence analyses:
One explanation for the unusual patterns of haplotype structure and linkage disequilibrium observed in the female reproductive proteases examined here is that these loci are subject to balancing selection. Two statistics were used to detect skews in the site-frequency spectra indicative of nonneutral evolution. Tajima's D (Tajima 1989) detects an excess of intermediate or low-frequency polymorphisms suggestive of balancing or directional selection, respectively. Similarly, Fu and Li's F (Fu and Li 1993) identifies an excess of old polymorphisms indicative of balancing selection by assigning variation to branches on a gene genealogy. Although the site-frequency spectra can be affected by demographic processes, these statistics tend to be slightly negative and close to zero at putatively neutral loci for all four races of D. mojavensis (Machado et al. 2007; Kelleher and Markow 2009). Significantly positive or negative values, therefore, cannot be attributed to demographic history.
There was a general trend toward positive values of D and F among the five paralogs (Table 3). This result is not unexpected, as gene conversion is predicted to create a marginally positive skew in the site frequency spectra of duplicate loci (Innan 2003; Thornton 2007). Significantly positive values do not result from neutral gene conversion, however, as an overestimation of the variance makes these statistics quite conservative for duplicate loci undergoing concerted evolution (Innan 2003; Thornton 2007). In four combinations of subspecies and loci, FRP-A (D. m. mojavensis), FRP-C (D. m. sonorensis), FRP-D (D. m. baja), and FRP-E (D. m. wrigleyi), these statistics indicated a significant excess of intermediate frequency or old polymorphisms (Table 3), suggesting that selection acts to retain genetic variation relative to a neutral model. Although no tests remain significant after a conservative Bonferroni correction for multiple measures, this frequency of rejections of the null hypothesis is unlikely to occur by chance (cumulative binomial P = 0.048). All values remained significant when sites undergoing gene conversion were excluded from the alignment (Table 3), further confirming that the observed deviations from neutrality are not the result of gene conversion.
TABLE 3.
Site frequency spectra
| Tajima's D | Fu and Li's F | Fay and Wu's H | |
|---|---|---|---|
| FRP-A | |||
| D. m. baja | −0.28 | 0.21 | −6.93 |
| D. m. wrigleyi | 0.69 | 1.49 | 0.19 |
| D. m. sonorensis | 0.42 | 0.52 | 0.14 |
| D. m. mojavensis | 1.30 | 1.625* | −3.71 |
| FRP-B | |||
| D. m. baja | −0.76 (−0.88) | −0.63 (−0.67) | −5.71 (4.95) |
| D. m. wrigleyi | 0.47 (0.61) | 0.87 (0.88) | −2.61 (−2.48) |
| D. m. sonorensis | 0.5 (0.72) | 0.88 (0.93) | 1.52 (1.33) |
| D. m. mojavensis | 0.46 (0.618) | 1.05 (0.59) | −5.14 (−1.86) |
| FRP-C | |||
| D. m. baja | −1.3 (−1.07) | −1.7 (−1.05) | −2.48 (−3.49) |
| D. m. wrigleyi | 0.21 | −0.18 | 0.38 |
| D. m. sonorensis | 1.57* (1.65*) | 1.66 (1.41) | −0.91 (−0.05) |
| D. m. mojavensis | −1.36 | 0.02 | 0.01 |
| FRP-D | |||
| D. m. baja | 1.60* (1.58*) | 1.67 (1.63*) | −1.10 (−1.24) |
| D. m. wrigleyi | −1.01 | −1.34 | 0.24 |
| D. m. sonorensis | 0.42 (−1.11*) | 0.61 (−0.98) | −0.24 (−4.19) |
| D. m. mojavensis | 0.42 | 0.47 | −1.5 |
| FRP-E | |||
| D. m. baja | 1.19 (1.04) | 1.55 (1.51) | 1.62 (−1.048) |
| D. m. wrigleyi | 1.52* | 1.84* | 2.52 |
| D. m. sonorensis | −0.99 | −0.51 | −5.71* |
| D. m. mojavensis | 0.82 | 0.75 | 3.24 |
Tajima's D (Tajima 1989) and Fu and Li's F (Fu and Li 1993) were calculated for all combinations of races and loci in DnaSP and statistical significance was assessed by coalescent simulations under the conservative assumption of no recombination (Rozas et al. 2003). Values in parentheses are generated from samples in which haplotypes with evidence for gene conversion are excluded. *P < 0.05.
We furthermore examined the relationship between polymorphism within subspecies of D. mojavensis to divergence from D.arizonae using the MK test, (McDonald and Kreitman 1991). Positive directional selection is predicted to result in significant excess of replacement divergence (McDonald and Kreitman 1991). In contrast, an excess of replacement polymorphism may indicate balancing selection, segregation of mildly deleterious variation, or recently relaxed constraint (Nachman 1998). FRP-A exhibited an excess of replacement polymorphism in D. m. mojavensis in a standard MK test, a deviation from neutrality that is consistent with balancing selection (Table 4). While this value must be interpreted with caution, as it does not remain significant after correction for multiple testing, it is consistent with the observed excess of old polymorphisms of this same locus (Table 3). Interestingly, lineage-specific tests at this locus also exhibited a high frequency of nonsynonymous polymorphisms, but lacked sufficient fixed differences on the D. mojavensis branch to reject neutrality (Table 3).
TABLE 4.
MK tests of FRP-A
| Standard MK test |
Lineage-specific MK test |
||||||
|---|---|---|---|---|---|---|---|
| Poly | Fixed | Test | Poly | Fixed | Test | ||
| S | 19 | 7 | G | 12 | 1 | FET | |
| D. m. baja | N | 26 | 6 | NS | 19 | 1 | NS |
| S | 10 | 9 | G | 6 | 2 | FET | |
| D. m. wrigleyi | N | 8 | 5 | NS | 1 | 4 | NS |
| S | 17 | 7 | G | 10 | 1 | FET | |
| D. m. sonorensis | N | 32 | 5 | NS | 25 | 1 | NS |
| S | 11 | 8 | G | 7 | 2 | FET | |
| D. m. mojavensis | N | 26 | 5 | * | 19 | 1 | NS |
Silent and noncoding (S) or replacement (N) polymorphism (Poly) was compared to fixed divergence (Fixed) from the D. arizonae ortholog. D. mojavensis lineage-specific tests were polarized with D. navojoa outgroup. Significance was assessed by Fisher's exact test (FET) or a G-test (G) when appropriate. *P < 0.05, NS P > 0.05.
In contrast to the FRP-A, as well as the other deviations from neutrality observed in this study, a modified McDonald–Kreitman test comparing polymorphism within populations for FRP-D and FRP-E, to divergence between these two paralogs (see materials and methods), revealed an excess of replacement divergence in FRP-D for D. m. baja and D. m. sonorensis (Table 5). This value remains significant in D. m. baja after correction for multiple testing. This signature of directional selection was inconsistent with the presence of two differentiated haplogroups in both of these subspecies (Figure 4, Table 2), as well as with the excess of both intermediate frequency and old polymorphism in D. m. baja (Table 3). Although these deviations from neutrality may appear contradictory, it is important to remember that they are sensitive to different evolutionary signatures and time scales. While site-frequency spectra may suggest recent selective events at a given locus, MK tests will be more sensitive to the history of the locus since its divergence from the outgroup. Deviations from neutrality in opposite directions, therefore, may indicate that the evolutionary history of these loci has been more complex than simple models of directional or diversifying selection.
TABLE 5.
Modified MK tests of FRP-D and FRP-E
| FRP-D lineage-specific MK test |
FRP-E lineage-specific MK test |
||||||
|---|---|---|---|---|---|---|---|
| Poly | Fixed | Test | Poly | Fixed | Test | ||
| S | 20 | 3 | G | 16 | 12 | G | |
| D. m. baja | N | 8 | 11 | ** | 25 | 9 | NS |
| S | 0 | 13 | FET | 7 | 11 | G | |
| D. m. wrigleyi | N | 1 | 16 | NS | 9 | 12 | NS |
| S | 27 | 3 | G | 3 | 12 | FET | |
| D. m. sonorensis | N | 23 | 13 | **a | 8 | 6 | P = 0.06 |
| S | 0 | 11 | FET | 18 | 10 | G | |
| D. m. mojavensis | S | 1 | 14 | NS | 21 | 9 | NS |
Silent and noncoding (S) or replacement (N) polymorphism (Poly) was compared to fixed divergence (Fixed) from the opposing paralog, as in Thornton and Long (2005). The D. mojavensis lineage-specific tests were polarized with D. navojoa outgroup. Significance was assessed by Fisher's exact test (FET) or a G-test (G) when appropriate. *P < 0.05, **P < 0.01.
Values that remain significant after a conservative Bonferroni correction for multiple measures. NS P > 0.05.
Selected sites are structurally associated with determinants of protease inhibitor susceptibility:
The five paralogs examined in this study are serine endoproteases. Serine endoprotease activity in D. arizonae female reproductive tracts is negatively regulated by mating, suggesting susceptibility of female proteases to inhibitors in the male ejaculate (Kelleher and Pennington 2009). If female proteases interact and coevolve with male protease inhibitors, selected sites are predicted to cluster near residues that determine susceptibility to protease inhibitors. Consistent with this hypothesis, selected sites (Kelleher et al. 2007) often are observed to be closely associated with sites important to protease inhibitor susceptibility and resistance (Figure 5, reviewed in Srinivasan et al. 2006). We furthermore observed that 3 of 14 selected sites also are determinants of inhibitor susceptibility, a marginally significant excess (Fisher's exact test, P = 0.058).
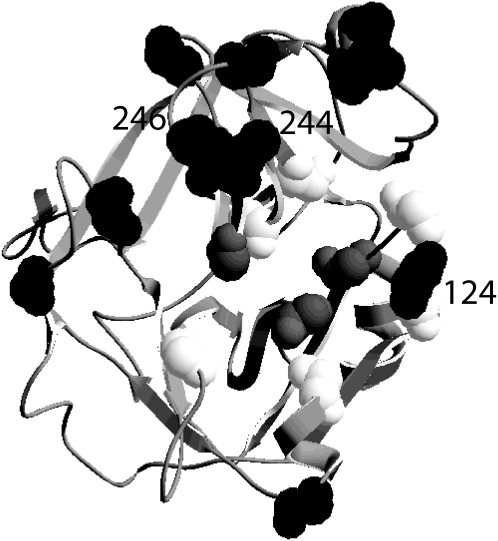
Figure 5.—
Predicted 3D structure of FRP-C. Bayes empirical Bayes selected sites identified under M8 (Yang 1997; Yang et al. 2005) were identified in Kelleher et al. (2007). Sites that are determinants of protease inhibitor susceptibility are open (reviewed in Srinivasan et al. 2006). Shaded sites comprise the catalytic triad (reviewed in Polgar 2005). Selected sites 124, 244, and 246 also are determinants of inhibitor susceptibility.
Examining the distance between selected sites and functional sites in three-dimensional space presents a more robust test for an association between site classes (Clark et al. 2007). We therefore compared the average pairwise distance between each selected site and the closest protease inhibitor interaction site to 106 sets of randomly sampled sites. Selected sites are significantly closer to protease inhibitor interaction sites than expected by chance (P = 0.02220), indicating that these two groups of sites are physically associated within the structure of the protein.
DISCUSSION
Although there is considerable empirical evidence that male reproductive proteins experience both directional and diversifying selective regimes in natural populations, the degree to which these patterns extend to their female interactors, as predicted under models of intersexual coevolution, remains largely unexplored. The exceptional mating system and reproductive biology of D. mojavensis, in conjunction with physiological evidence of intersexual coevolution, presents a particularly compelling system for examining the evolution of female reproductive tract proteins. The female reproductive tract gene family examined here exhibits a history of directional selection, balancing selection, and gene conversion, as well as excess of amino acids substitution at functional sites of protein–protein interaction, results strongly suggestive of molecular coevolution.
Evidence of diversifying selection is particularly intriguing, as the role of selection in maintaining the exceptional genetic variation observed at some reproductive protein loci often remains unresolved.
Female reproductive proteases in D. mojavensis may experience diversity-enhancing selection:
Previous population surveys of D. melanogaster female reproductive tract proteins (Swanson et al. 2004; Panhuis and Swanson 2006; Lawniczak and Begun 2007), as well as a recent study of a second family of female reproductive proteases in D. mojavensis (Kelleher and Markow 2009), report a high frequency of positive directional selection. While an excess of replacement divergence at FRP-D in D. m. baja and D. m. sonorensis suggests a role for directional selection in the evolution of the female reproductive proteins examined here, the majority of deviations from neutrality we observe point to balancing selection as a comparatively more significant force in guiding the evolutionary history of this gene family. All five proteases exhibited nongeographic haplotype structure, and significantly lower FST values than neutral loci, as expected if natural selection acts to maintain genetic variation within isolated subspecies. Four of five loci exhibit significant linkage disequilibrium consistent with positive directional or diversifying selection, and site-frequency spectra tests at four different loci exhibit an excess of intermediate frequency or old polymorphisms, suggesting selective retention of genetic variation relative to a neutral model. McDonald Kreitman tests at one locus, furthermore, exhibit an excess of replacement polymorphisms consistent with balancing selection.
The evidence of diversifying or balancing selection presented here, is an unexpected evolutionary pattern in light of evidence for ejaculate-female coadaptation within subspecies of D. mojavensis. Crosses between these subspecies exhibit significant differences from within subspecies crosses in egg size (Pitnick et al. 2003), mated female desiccation resistance (Knowles et al. 2005), and the size and duration of the insemination reaction (Knowles and Markow 2001), predicting that the biological molecules that underlie these male by female interactions must also diverge rapidly between populations. While it remains unknown if any of the FRPs examined here play a direct role in ejaculate coadaptation, it is clear that deviations from neutrality often are confined to a single race or group of races, as expected if each race experiences its own unique coevolutionary trajectory. Thus, our data suggest that reproductive divergence between isolated populations of D. mojavensis may not always be underscored by simple directional selection at interacting loci.
Gene conversion, diversifying selection, and adaptive evolution:
Divergence between paralogous members of a multigene family is postulated to result from an antagonistic process between the diversifying force of selection and the homogenizing force of gene conversion. All paralogs examined in this study, except FRP-A, exhibited considerable evidence for interlocus gene conversion, suggesting the process of diversification between paralogs is not yet complete (Figure 2). While the frequency of gene conversion showed no clear relationship with phylogenetic or physical distance (Figure 2), we observed a strong negative association between gene conversion and sites inferred to have undergone adaptive evolution (Figure 3). These results are consistent with a model where gene conversion interferes with the process of adaptive evolution, or may be costly in genetic regions that have experienced positive selection since gene duplication.
In contrast, a different female reproductive protease gene family examined in Kelleher and Markow (2009) revealed no evidence for an antagonistic relationship between gene conversion and adaptive evolution. This same gene family exhibited several indicators of relaxed purifying selection at individual loci, suggesting some paralogs within it may be functionally redundant (Kelleher and Markow 2009). We furthermore found minimal evidence for diversifying selection within individual paralogs (Kelleher and Markow 2009). Thus, the strength of diversifying selection may be an important difference in the selective regimes experienced by these two families.
Balancing selection and gene duplication:
Diversifying selection among D. mojavensis female reproductive proteins presents a stark contrast to the preponderance of directional selection observed among the female reproductive proteins of its congener D. melanogaster (Swanson et al. 2004; Panhuis and Swanson 2006). This selective regime is reminiscent of another emergent pattern of reproductive protein evolution in the D. mojavensis lineage: gene duplication (Kelleher et al. 2007; Wagstaff and Begun 2007; Almeida and Desalle 2008, 2009; Kelleher and Markow 2009). Although there are a few reports of lineage-specific duplications in D. melanogaster seminal fluid proteins (Cirera and Aguadé 1998; Findlay et al. 2008), recent duplicates occur with high frequency among both male seminal proteins (Wagstaff and Begun 2007; Almeida and Desalle 2008) and female reproductive tract proteins (Kelleher et al. 2007; Kelleher and Markow 2009; Kelleher and Pennington 2009) in the repleta species group.
It is exciting to speculate that the selective forces that underlie gene duplication and balancing selection may not be independent. Several models have suggested that if a balanced polymorphism is maintained by overdominant selection, a gene duplication event that unites two functionally diverged alleles on the same chromosome will immediately experience a selective advantage due to heterosis (Spofford 1969; Ohno 1970; Otto and Yong 2002; Walsh 2003; Proulx and Phillips 2006). Balancing selection and gene duplication, therefore, may be iterative steps in the diversification of D. mojavensis FRPs.
Biochemical basis of molecular coevolution:
The biochemical underpinnings of intersexual coevolution within races of D. mojavensis remain largely unexplored. It is compelling, however, that the female reproductive tracts of D. arizonae, the close sister species to D. mojavensis (MRCA = 0.7 MYA) (Reed et al. 2007; Matzkin 2008), exhibit exceptional levels of proteolytic activity that are negatively regulated by mating (Kelleher and Pennington 2009). While this negative regulation could be transcriptional, it also is possible that female proteases are negatively regulated by known proteases inhibitors in the D. mojavensis male ejaculate (Wagstaff and Begun 2005; Kelleher et al. 2009). Consistent with the hypothesis that female proteases interact and coevolve with protease inhibitors, our structural analysis revealed that previously inferred selected sites are clustered with sites that determine susceptibility to protease inhibitors (Figure 5). A similar result was observed in Kelleher and Markow (2009), suggesting that molecular coevolution with protease inhibitors may be a general property of female reproductive proteases in D. mojavensis.
Conclusion:
Evolutionary dynamics of D. mojavensis female reproductive tracts proteins, both in terms of balancing selection and accelerated gene duplication (Kelleher et al. 2007; Kelleher and Markow 2009; Kelleher and Pennington 2009), present a dramatic contrast to patterns of directional selection and the paucity of gene duplication observed in D. melanogaster (Swanson et al. 2004; Panhuis and Swanson 2006; Lawniczak and Begun 2007). These two congeners also have dramatically different mating systems and levels of female promiscuity, as well as considerably disparate reproductive physiologies. Our data suggest these reproductive traits may leave signatures in the evolutionary histories of the proteins involved and create a framework for comparing the dynamics of reproductive proteins between closely related organisms.
Acknowledgments
The authors acknowledge Michael Nachman for generous use of equipment; Willie Swanson for helpful discussion; and Jeremy Bono, Stephen Schaeffer, and five anonymous reviewers for comments on the manuscript. This research was funded by a National Science Foundation (NSF) doctoral dissertation improvement grant to E.S.K. and the Center for Insect Science and the University of Arizona. E.S.K. was supported by an NSF-Integrative Graduate Education and Research Traineeship in Evolutionary, Functional, and Computational Genomics at the University of Arizona, a dissertation fellowship from the American Association of University Women, a postdoctoral fellowship from the Cornell Center for Comparative and Population Genomics, and a National Institutes of Health National Research Service Award (NIH-NRSA) postdoctoral fellowship (GM090567). N.L.C. was supported by an NIH-NRSA postdoctoral fellowship (GM084592).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.124743/DC1.
References
- Aguadé, M., 1998. Different forces drive the evolution of ACP26Aa and ACP26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics 150 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida, F. C., and R. Desalle, 2008. Evidence of adaptive evolution of accessory gland proteins in closely related species of the Drosophila repleta group. Mol. Biol. Evol. 25 2043–2053. [DOI] [PubMed] [Google Scholar]
- Almeida, F. C., and R. Desalle, 2009. Orthology, function, and evolution of accessory gland proteins in the Drosophila repleta group. Genetics 181 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Pimentel, H., L. P. Tolbert and W. B. Heed, 1994. Ultrastructural examination of the insemination reaction in Drosophila. Cell Tissue Res. 275 467–479. [DOI] [PubMed] [Google Scholar]
- Barnes, A. I., S. Wigby, J. M. Boone, L. Partridge and T. Chapman, 2008. Feeding, fecundity and lifespan in female Drosophila melanogaster. Proc. Biol. Sci. 275 1675–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., and H. A. Lindfors, 2005. Rapid evolution of genomic Acp complement in the melanogaster subgroup of Drosophila. Mol. Biol. Evol. 22 2010–2021. [DOI] [PubMed] [Google Scholar]
- Begun, D. J., H. A. Lindfors, M. E. Thompson and A. K. Holloway, 2006. Recently evolved genes identified from Drosophila yakuba and D. erecta accessory gland expressed sequence tags. Genetics 172 1675–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkhead, T. R., and A. P. Møller (Editors), 1998. Sperm Competition and Sexual Selection. Academic Press, London.
- Calkins, J. D., D. El-Hinn and W. J. Swanson, 2007. Adaptive evolution in an avian reproductive protein: ZP3. J. Mol. Evol. 65 555–563. [DOI] [PubMed] [Google Scholar]
- Cirera, S., and M. Aguadé, 1998. Molecular evolution of a duplication: the sex-peptide (Acp70A) gene region of Drosophila subobscura and Drosophila madeirensis. Mol. Biol. Evol. 15 988–996. [DOI] [PubMed] [Google Scholar]
- Chapman, T., L. F. Liddle, J. M. Kalb, M. F. Wolfner and L. Partridge, 1995. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature 373 241–244. [DOI] [PubMed] [Google Scholar]
- Clark, N. L., J. E. Aagaard and W. J. Swanson, 2006. Evolution of reproductive proteins from animals and plants. Reproduction 131 11–22. [DOI] [PubMed] [Google Scholar]
- Clark, N. L., G. D. Findlay, X. Yi, M. J. MacCoss and W. J. Swanson, 2007. Duplication and selection on abalone sperm lysin in an allopatric population. Mol. Biol. Evol. 24 2081–2090. [DOI] [PubMed] [Google Scholar]
- Clark, N. L., J. Gasper, M. Sekino, S. A. Springer, C. F. Aquadro et al., 2009. Coevolution of interacting fertilization proteins. PLoS Genetics 5 e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopman, E. B., and D. L. Hartl, 2007. A portrait of copy-number polymorphism in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 104 19920–19925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard, W. G., 1996. Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press, Princeton, N J.
- Findlay, G. D., X. Yi, M. J. Maccoss and W. J. Swanson, 2008. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 6 e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraczkiewicz, R., and W. Braun, 1998. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comp. Chem. 19 319. [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasper, J., and W. J. Swanson, 2006. Molecular population genetics of the gene encoding the human fertilization protein zonadhesin reveals rapid adaptive evolution. Am. J. Hum. Genet. 79 820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good, J. M., C. L. Ross and T. A. Markow, 2006. Multiple paternity in wild-caught Drosophila mojavensis. Mol. Ecol. 15 2253–2260. [DOI] [PubMed] [Google Scholar]
- Hamm, D., B. S. Mautz, M. F. Wolfner, C. F. Aquadro and W. J. Swanson, 2007. Evidence of amino acid diversity-enhancing selection within humans and among primates at the candidate sperm-receptor gene PKDREJ. Am. J. Hum. Genet. 81 44–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R. R., M. Kreitman and M. Aguadé, 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116 153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H., 2003. The coalescent and infinite-site model of a small multigene family. Genetics 163 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, E. S., and T. A. Markow, 2007. Reproductive tract interactions contribute to isolation in Drosophila. Fly 1 33–37. [DOI] [PubMed] [Google Scholar]
- Kelleher, E. S., W. J. Swanson and T. A. Markow, 2007. Gene duplication and adaptive evolution of digestive proteases in Drosophila female reproductive tracts. PLoS Genet. 3 1541–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, E. S., and T. A. Markow, 2009. Duplication, selection and gene conversion in a Drosophila mojavensis female reproductive protein family. Genetics 181 1451–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, E. S., and J. E. Pennington, 2009. 2009 Protease gene duplication and proteolytic activity in Drosophila female reproductive tracts. Mol. Biol. Evol. 26 2125–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher, E. S., T. D. Watts, B. A. Laflamme, P. A. Haynes and T. A. Markow, 2009. Proteomic analysis of Drosophila mojavensis male accessory glands suggests novel classes of seminal fluid proteins. Insect Biochem. Mol. Biol. 39 366–371. [DOI] [PubMed] [Google Scholar]
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, L. L., and T. A. Markow, 2001. Sexually antagonistic coevolution of a postmating prezygotic reproductive character in desert Drosophila. Proc. Nat. Acad. Sci. USA 98 8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, L. L., B. B. Hernandez and T. A. Markow, 2005. Non-antagonistic interactions between the sexes revealed by the ecological consequences of reproductive traits. J. Evol. Biol. 18 156–161. [DOI] [PubMed] [Google Scholar]
- Kuijper, B., A. D. Stewart and W. R. Rice, 2006. The cost of mating rises nonlinearly with copulation frequency in a laboratory population of Drosophila melanogaster. J. Evol. Biol. 19 1795–1802. [DOI] [PubMed] [Google Scholar]
- Lawniczak, M. K., and D. J. Begun, 2007. Molecular population genetics of female-expressed mating-induced serine proteases in Drosophila melanogaster. Mol. Biol. Evol. 24 1944–1951. [DOI] [PubMed] [Google Scholar]
- Lee, Y. H., T. Ota and V. D. Vacquier, 1995. Positive selection is a general phenomenon in the evolution of abalone sperm lysin. Mol. Biol. Evol. 12 231–238. [DOI] [PubMed] [Google Scholar]
- Levitan, D. R., and D. L. Ferrell, 2006. Selection on gamete recognition proteins depends on sex, density, and genotype frequency. Science 312 267–269. [DOI] [PubMed] [Google Scholar]
- Machado, C. A., L. M. Matzkin, L. K. Reed and T. A. Markow, 2007. 2007 Multilocus nuclear sequences reveal intra- and interspecific relationships among chromosomally polymorphic species of cactophilic Drosophila. Mol. Ecol. 16 3009–3024. [DOI] [PubMed] [Google Scholar]
- Markow, T. A., 1996. Evolution of Drosophila mating systems. Evol. Biol. 29 73–106. [Google Scholar]
- Markow, T. A., and P. F. Ankney, 1984. Drosophila males contribute to oogenesis in a multiple mating species. Science 224 302–303. [DOI] [PubMed] [Google Scholar]
- Markow, T. A., and P. F. Ankney, 1988. Insemination reaction in Drosophila found in species whose males contribute material to oocytes before fertilization. Evolution 42 1097–1101. [DOI] [PubMed] [Google Scholar]
- Matzkin, L. M., 2008. The molecular basis of host adaptation in cactophilic Drosophila: molecular evolution of a glutathione S-transferase gene (GstD1) in Drosophila mojavensis. Genetics 178 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, J. H., and M. Kreitman, 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351 652–654. [DOI] [PubMed] [Google Scholar]
- Metz, E. C., and S. R. Palumbi, 1996. Positive selection and sequence rearrangements generate extensive polymorphism in the gamete recognition protein bindin. Mol. Biol. Evol. 13 397–406. [DOI] [PubMed] [Google Scholar]
- Moy, G. W., S. A. Springer, S. L. Adams, W. J. Swanson and V. D. Vacquier, 2008. Extraordinary intraspecific diversity in oyster sperm bindin. Proc. Natl. Acad. Sci. USA 105 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, J. L., K. Ravi Ram, L. A. McGraw, M. C. Bloch Qazi, E. D. Siggia et al., 2005. Cross-species comparison of Drosophila male accessory gland protein genes. Genetics 171 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman, M. W., 1998. Deleterious mutations in animal mitochondrial DNA. Genetica 102/103 61–69. [PubMed] [Google Scholar]
- Ohno, S, 1970. Evolution by Gene Duplication. Springer-Verlag, New York.
- Otto, S. P., and P. Yong, 2002. The evolution of gene duplicates. Adv. Genet. 46 451–483. [DOI] [PubMed] [Google Scholar]
- Panhuis, T. M., and W. J. Swanson, 2006. Molecular evolution and population genetic analysis of candidate female reproductive genes in Drosophila. Genetics 173 2039–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panhuis, T. M., N. L. Clark and W. J. Swanson, 2006. Rapid evolution of reproductive proteins in abalone and Drosophila. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, G. A., 1979. Sexual selection and sexual conflict, pp. 123–166 in Sexual Selection and Reproductive Competition in Insects, edited by M. S. Blum and N. A. Blum. Academic Press, London.
- Patterson, J. T., 1946. A new type of isolating mechanism in Drosophila. Proc. Nat. Acad. Sci. USA 32 202–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiler, E., S. Castrezana, L. K. Reed and T. A. Markow, 2009. Genetic, ecological and morphological differences among populations of the cactophilic Drosophila mojavensis from southwestern USA and northwestern Mexico, with descriptions of two new subspecies. J. Nat. Hist. 43 923–938. [Google Scholar]
- Pitnick, S., G. S. Spicer and T. A. Markow, 1997. Phylogenetic examination of female incorporation of ejaculate in Drosophila. Evolution 51 833–845. [DOI] [PubMed] [Google Scholar]
- Pitnick, S., and F. García-González, 2002. Harm to females increases with male body size in Drosophila melanogaster. Proc. Biol. Sci. 269 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick, S., G. T. Miller, K. Schneider and T. A. Markow, 2003. Ejaculate-female coevolution in Drosophila mojavensis. Proc. Biol. Sci. 270 507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgar, L., 2005. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 62 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proulx, S. R., and R. C. Phillips, 2006. Allelic divergence precedes and promotes gene duplication. Evolution 60 881–892. [PubMed] [Google Scholar]
- Reed, L. K., M. Nyboer and T. A. Markow, 2007. Evolutionary relationships of Drosophila mojavensis geographic host races and their sister species Drosophila arizonae. Mol. Ecol. 16 1007–1022. [DOI] [PubMed] [Google Scholar]
- Rozas, J., and R. Rozas, 1995. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput. Appl. Biosci. 11 621–625. [DOI] [PubMed] [Google Scholar]
- Rozas, J., M. Gullaud, G. Blandin and M. Aguadé, 2001. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics 158 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sánchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. [DOI] [PubMed] [Google Scholar]
- Sawyer, S., 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6 526–538. [DOI] [PubMed] [Google Scholar]
- Schwede, T., Kopp, J., Guex, N., and M. C. Peitsch, 2003. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spofford, J. B., 1969. Heterosis and evolution of duplications. Am. Nat. 103 407–432. [Google Scholar]
- Springer, S. A., G. W. Moy, D. S. Friend, W. J. Swanson and V. D. Vacquier, 2008. Oyster sperm bindin is a combinatorial fucose lectin with remarkable intra-species diversity. Int. J. Dev. Biol. 52 759–768. [DOI] [PubMed] [Google Scholar]
- Srinivasan, A., A. P. Giri and V. S. Gupta, 2006. Structural and functional diversities in lepidopteran serine proteases. Cell. Mol. Biol. Lett. 11 132–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. a The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. b Reproductive protein evolution. Annu. Rev. Ecol. Syst. 33 161–179. [Google Scholar]
- Swanson, W. J., A. Wong, M. F. Wolfner and C. F. Aquadro, 2004. Evolutionary expressed sequence tag analysis of Drosophila female reproductive tracts identifies genes subjected to positive selection. Genetics 168 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D. L., 2000. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4.0. Sinauer Associates, Sunderland, MA.
- Tajima, F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, K., and M. Long, 2005. Excess of amino acid substitutions relative to polymorphism between X-linked duplications in Drosophila melanogaster. Mol. Biol. Evol. 22 273–284. [DOI] [PubMed] [Google Scholar]
- Thornton, K. R., 2007. The neutral coalescent process for recent gene duplications and copy-number variants. Genetics 177 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2005. Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics 171 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2007. Adaptive evolution of recently duplicated accessory gland protein genes in desert Drosophila. Genetics 177 1023–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, B., 2003. Population-genetic models of the fates of duplicate genes. Genetica 118 279–294. [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., W. S. Wong and R. Nielsen, 2005. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22 1107–1118. [DOI] [PubMed] [Google Scholar]