Abstract
Decades of biochemical research have identified most of the enzymes that catalyze metabolic reactions in the yeast Saccharomyces cerevisiae. The adaptation of metabolism to changing nutritional conditions, in contrast, is much less well understood. As an important stepping stone toward such understanding, we exploit the power of proteomics assays based on selected reaction monitoring (SRM) mass spectrometry to quantify abundance changes of the 228 proteins that constitute the central carbon and amino-acid metabolic network in the yeast Saccharomyces cerevisiae, at five different metabolic steady states. Overall, 90% of the targeted proteins, including families of isoenzymes, were consistently detected and quantified in each sample, generating a proteomic data set that represents a nutritionally perturbed biological system at high reproducibility. The data set is near comprehensive because we detect 95–99% of all proteins that are required under a given condition. Interpreted through flux balance modeling, the data indicate that S. cerevisiae retains proteins not necessarily used in a particular environment. Further, the data suggest differential functionality for several metabolic isoenzymes.
Keywords: metabolism, S. cerevisiae, SRM, targeted proteomics
Introduction
Systems biology aims at the comprehensive description of biological systems and ultimately at predicting the behavior of the system from the dynamic and quantitative interactions of its constituting components (Kitano, 2002b; Sauer et al, 2007). Among the cellular systems, metabolism is unique because the topology of the network is almost completely known (Duarte et al, 2007; Feist et al, 2009). Specifically, most of the reactions, the catalyzing enzymes, the enzyme-encoding genes and the converted metabolites are known. While we can also monitor the integrated network operation in the form of metabolite fluxes (Sauer, 2006), we do not yet understand how the behavior of the system emerges from the interaction of the system's components. To establish this link, mathematical models are needed (Kitano, 2002a; Aldridge et al, 2006), whose development in turn requires computational tools (Heinemann and Sauer, 2010) and quantitative, comprehensive data on the response of network components to external and internal stimuli (Sauer et al, 2007).
A genome-scale stoichiometric model of the metabolic reaction network in yeast (Kuepfer et al, 2005) that was recently updated through a community consensus (Herrgård et al, 2008) is arguably one of the topologically most complete biological systems. In this model, 177 stoichiometrically distinct metabolic reactions—catalyzed by 210 enzymes, including isoenzymes—represent central carbon and amino-acid metabolism. A particular feature of this metabolic system is the large number of isoenzymes; i.e., distinct proteins that catalyze identical reactions and often exhibit a high degree of amino-acid sequence identity (e.g., see Wilson, 2003). While the existence of isoenzymes has been known for a long time, the reasons that might explain their preservation in the genome have been much less clear. Suggested reasons include redundancy as a means to buffer against mutations, differential regulation, gene dosage, facilitation of evolutionary innovation and functional diversification (Kuepfer et al, 2005; Ihmels et al, 2007; Wagner, 2008). For the metabolic network considered here, 51 out of 177 reactions can be catalyzed by more than one isoenzyme, with up to seven isoenzymes catalyzing one reaction and up to 99.5% amino-acid sequence identity between them. Furthermore, 31 proteins involved in 11 multi-subunit protein complexes also show up to 95% amino-acid sequence identity. Because proteins with a high degree of sequence similarity generate similar peptides upon tryptic digestion, their distinction by mass spectrometric analysis poses a particular analytical challenge. The quantification of the comprehensive set of enzymes and isoenzymes representing the central carbon and amino-acid metabolism in yeast, under different metabolic states, would provide a unique opportunity to observe the change of the system as a whole and generate key information for the modeling of this system (Kotte and Heinemann, 2009; Oberhardt et al, 2009; Heinemann and Sauer, 2010).
In spite of significant advances in the standard shotgun proteomic technology, the consistent and reproducible detection and quantification of proteins across different complex samples remain challenging (Bell et al, 2009). This is due to the fact that this mass spectrometric method contains stochastic elements, particularly in the selection of precursor ions for collision-activated dissociation, that are difficult to control even if extraordinary precautions are taken to control experimental variability (Tabb et al, 2010). This problem becomes particularly apparent when predetermined sets of proteins of interest need to be measured, and is compounded if they share extensive sequence similarity, such as the isoenzymes in the metabolic sub-proteome.
In this study, we therefore chose to apply a targeted proteomics approach based on selected reaction monitoring (SRM), whereby the proteotypic peptides (PTPs) for each protein on the target list are selectively detected and quantified (Baty and Robinson, 1977; Anderson and Hunter, 2006; Lange et al, 2008b). The pivotal element of this technique is the a priori development of highly specific mass spectrometric assays for each protein and the use of these assays for the detection and quantification of the proteins on a target list in multiple biological samples. We have recently shown that SRM assays can be generated at high throughput using synthetic peptide libraries (Picotti et al, 2010) and that proteins spanning the whole dynamic range of abundance can be detected by the technique in minimally fractionated yeast whole-proteome digests (Picotti et al, 2009). The technique is also highly multiplexed, highly reproducible (Addona et al, 2009) and quantitatively accurate, attributes that are critical for the generation of consistent, comprehensive and quantitative data sets from multiple samples (Stahl-Zeng et al, 2007).
In this study, we demonstrate comprehensive analysis of the 210 enzymes (comprising 228 proteins) that constitute our model-defined sub-proteome and the quantification of this protein set under five metabolic states. We thereby extend previous proteomics investigations of yeast metabolism which focused primarily on the comparison between aerobic and anaerobic growth (Daran-Lapujade et al, 2007; de Groot et al, 2007) or different nutrient limitations (Gutteridge et al, 2010) in chemostat cultures. Our five conditions were chosen to cause major differences in metabolic fluxes, particularly through the here studied metabolic sub-network. In combination with model-derived predictions, we used our comprehensive data set to address two key questions. First, are enzymes absent or only downregulated under conditions where they are not necessary for metabolic operation? And, second, what is the role of the large number of isoenzymes in the sub-network studied?
Results
Selection of the system components from a yeast metabolic network model
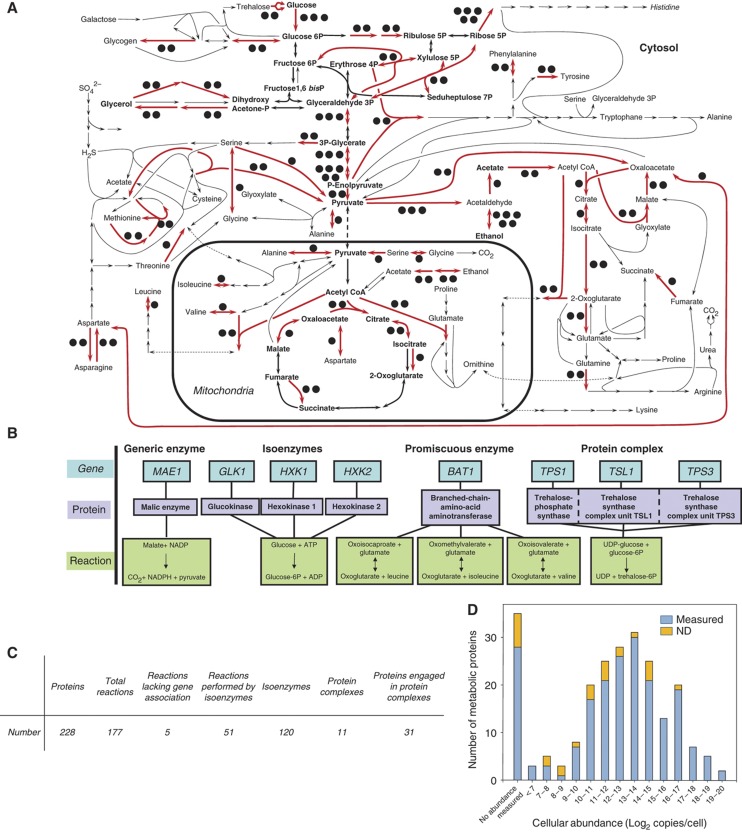
On the basis of a genome-scale stoichiometric model of the yeast metabolic network (Kuepfer et al, 2005; Herrgård et al, 2008), we defined the components of the biological sub-system under study. It consists of the central carbon and amino-acid metabolism in the yeast S. cerevisiae and includes all reactions of intermediary carbon, nitrogen and sulfur metabolism as well as anabolism and catabolism of amino acids, trehalose and glycogen (Figure 1A, Supplementary Table 1). This network is well characterized and there is general agreement on its topology and components (Herrgård et al, 2008). The 230 genes of this network encode 228 different proteins, which assemble into 210 enzymes that catalyze the 177 stoichiometrically distinct biochemical reactions (Figure 1C). The presence of isoenzymes, promiscuous enzymes and protein complexes in the protein set precluded a one-to-one mapping of proteins to reactions due to the more complex relationships schematically shown in Figure 1B. Only five reactions in the chosen network have not yet been associated with an open reading frame. The protein abundance in this network ranges from more than a million to less than a hundred copies per cell (Figure 1D), as determined by an antibody-based quantification approach in a different study (Ghaemmaghami et al, 2003). The system also included several proteins for which the cellular abundance could not be determined in the previous study.
Figure 1.
The targeted network of central carbon and amino-acid metabolism of S. cerevisiae. (A) Schematic representation of the network. Reactions indicated with red arrows can be catalyzed by isoenzymes and the black dots indicate the number of isoenzymes (Supplementary Table 1). Bold, central carbon metabolism. (B) Four classes of gene–protein-reaction relations in the metabolic network. (C) Characteristic numbers for the considered network. (D) Distribution of cellular abundances for the proteins in central metabolism of S. cerevisiae, as derived from antibody-based expression data (Ghaemmaghami et al, 2003). The left most bar (‘no abundance measured’) represents 35 proteins whose abundance could not be determined in that study. ND, not detected in this study.
Selection of perturbing conditions
To elucidate how this network of proteins responds to environmental challenges, we chose five nutritional conditions resulting in maximal difference in magnitude and direction of metabolic fluxes through this network. The reference condition was aerobic growth on high glucose as the sole carbon and energy source, and ammonia as the sole nitrogen source. The specific nutritional conditions and their phenotypic characteristics are summarized in Table I. The nutritional perturbations concerned the carbon source (galactose or ethanol instead of glucose), oxygen availability (anaerobic instead of aerobic) or the nitrogen source (complex medium consisting of an amino-acid mix lacking carbohydrates, instead of ammonium). Reversal of metabolic flux was expected for (i) glycolysis and gluconeogenesis in glucose-/galactose-grown versus ethanol-grown cells, and (ii) for amino-acid catabolism and anabolism in complex medium-grown versus all other cultures. We observed substrate consumption rates that differed several fold in the selected culture conditions, with the highest overall rate of metabolism on glucose and the lowest on ethanol and in complex medium (Table I). Thus, by the selected conditions the metabolic fluxes through the network must also change, presumably leading to different abundances and presence of proteins.
Table 1. Specific growth, consumption and production rates of substrates and products during exponential growth.
| Condition | Growth rate (h−1) | Glucose consumption rate (mmol (g biomass dry weight h)−1) | Galactose consumption rate (mmol (g biomass dry weight h)−1) | Ethanol production rate (mmol (gram biomass dry weight h)−1) | Acetate production rate (mmol (g biomass dry weight h)−1) | Glycerol production rate (mmol (g biomass dry weight h)−1) |
|---|---|---|---|---|---|---|
| NA, not applicable. | ||||||
| aBecause of evaporation of ethanol from the broth under the experimental conditions, these ethanol production rates for glucose, galactose and anaerobically grown conditions were estimated from mass balance calculations using the measured values as lower boundaries (See Supplementary Table 2). The ethanol consumption rate for the ethanol-grown condition was corrected for evaporation by determining the evaporation rate of ethanol in a separate cell-free experiment. | ||||||
| Glucose grown | 0.30±0.01 | 15.1±1.4 | NA | 29.5a | 0.4±0.1 | 1.0±0.3 |
| Galactose grown | 0.25±0.01 | NA | 7.6±0.4 | 11.0a | 0.6±0.1 | 0.1±0.1 |
| Anaerobically glucose grown | 0.25±0.03 | 16.0±2.2 | NA | 27.1a | 0.1±0.0 | 1.9±0.6 |
| Ethanol grown | 0.07±0.01 | NA | NA | −4.2±0.9a | <0.01 | <0.002 |
| Grown on complex medium | 0.17±0.05 | NA | NA | <3.1 | <0.08 | <0.03 |
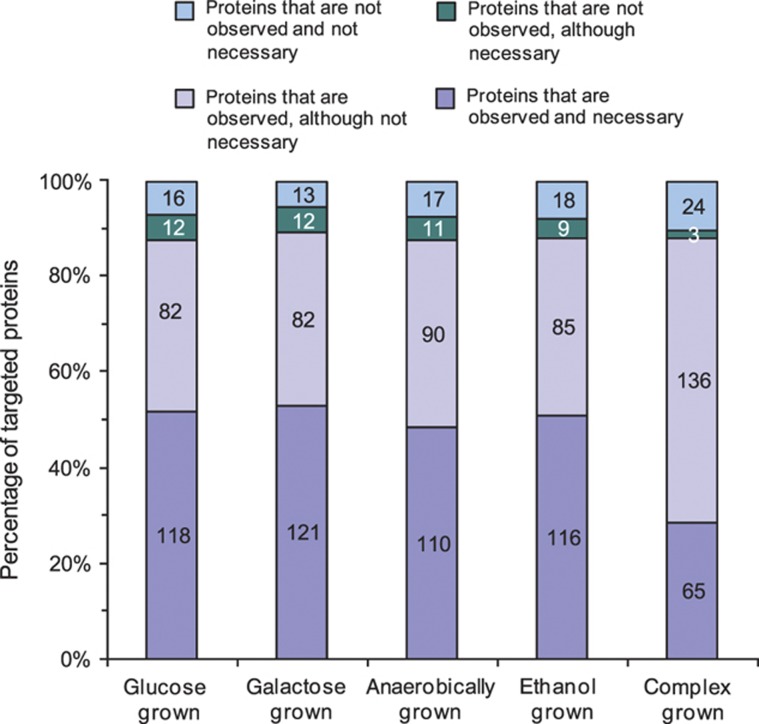
To predict condition-specific enzyme necessity in this system, we used the computational framework of flux balance analysis (FBA; Schellenberger and Palsson, 2009). On the basis of the determined physiological data (Table I), we identified the most efficient, fully connected network of biochemical reactions by minimizing the Euclidean norm of fluxes (Supplementary Table 2). From these FBA solutions, we determined for each of the targeted proteins its condition-dependent necessity to attain the determined physiology, thus obtaining a measure for a protein's expected presence under each condition. Depending on the condition, 121–133 of the targeted proteins were deemed necessary to establish a network that is sufficient to generate biomass (Figure 2, Supplementary Table 3). The notable exception was the complex medium culture, in which only 68 of the targeted proteins were deemed necessary. This model prediction is intuitively logical because biosynthetic pathways for amino acids are not required in the presence of these amino acids. This prediction is consistent with existing genetic evidence, for instance, for the leucine synthesis pathway in S. cerevisiae upon supplementation of leucine (Chin et al, 2008) or for the downregulation of all synthesis pathways upon supplementation of amino acids (Zaslaver et al, 2004).
Figure 2.
Comparison of protein necessity and presence. Protein necessity under each of the five tested conditions was determined by flux balance analysis through minimization of the Euclidean norm of intracellular fluxes on the basis of the physiological data (Table I). The numbers on the bars indicate amounts of proteins in each class.
Protein measurements at different metabolic states
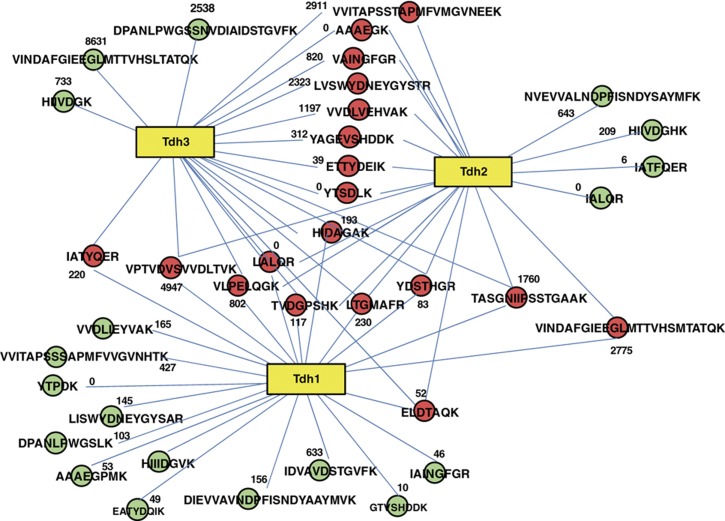
To quantitatively monitor the set of proteins that constitute the selected sub-proteome, we developed protein-specific SRM assays for each of the 228 target proteins. For each protein, we selected a set of representative PTPs and for each PTP we developed an SRM assay consisting of 3–4 validated precursor-to-fragment ion transitions. We measured the protein species in a total proteome digest background and determined their abundance relative to a common, stable isotope-labeled reference sample. The selection of targeted peptides for highly sequence similar isoenzymes is illustrated in Figure 3. Tryptic peptides were selected such that they were detectable by electrospray mass spectrometry, were unique for the proteome and contained sufficient sequence divergence to distinguish between the targeted proteins. The only exception was the isoenzymes Err1, Err2 and Err3, which show 100% nucleic and amino-acid sequence identity between ERR1 and ERR2 and 99% nucleic acid sequence identity (resulting in two differing amino acids) between ERR3 and ERR1/ERR2. As the two peptides distinguishing between ERR3 and ERR1/ERR2 could not be analyzed by liquid chromatography coupled-mass spectrometry, we used non-unique peptides to monitor the cumulative amount of these three proteins. For all other proteins we measured only unique peptides, between two and ten per protein for about two-thirds of the sub-proteome. Several proteins, e.g., isoenzymes with very large sequence overlap (up to 99.5% sequence identity), could be specifically quantified only via one or few distinguishing peptides. The distribution of peptides targeted for each protein is shown in Supplementary Figure 3. The complete set of targeted proteins, the selected PTPs and their respective SRM assays are available in Supplementary Table 4 or via the SRMAtlas interface (www.srmatlas.org; Picotti et al, 2008).
Figure 3.
Example of proteolytic pattern complexity for isoenzymes. Tryptic peptides for three isoenzymes of glyceraldehyde-3-phosphate dehydrogenase are shown as connected circles. Red circles indicate peptides shared by at least two of the three isoenzymes. Green circles indicate peptides unique to one isoenzyme. Numbers indicate the number of observations of the peptide in the PeptideAtlas database (S. cerevisiae build, 2009).
The 228 developed SRM assays were then applied to detect and quantify the proteins in trypsinized extracts of S. cerevisiae cells, harvested during mid-exponential growth under each of the five growth conditions from three independent replicate experiments. A total of ∼35 000 SRM traces were recorded, and 205 of the 228 targeted proteins (90%) were detected under at least one condition. Overall, 199 proteins were detected under all five conditions (Supplementary Table 3). The 23 proteins that were not detected under any condition are listed in Supplementary Table 5, including plausible explanations for the failure of their detection. Mostly, these undetected proteins were not necessary under any of the tested metabolic conditions; hence, not expected to be present. Alternatively, potential technical reasons for missing these proteins include low abundance of the proteins that lack PTPs with good MS properties or whose PTPs are highly modified or membrane localization. Variations of the protocols applied here, e.g., the use of proteases other than trypsin, testing a higher number of PTPs or adapting the protein extraction procedure to detect membrane or cell wall proteins might help covering the missed proteins. A small number of proteins were not detected under four (Gal1, Gal7, Gal10 and Aro9), three (Agx1) or one (Met3) condition(s). The inability to detect these six proteins under the respective condition was always due to a decrease in the signal of the best-responding peptide below the noise level, indicating reduction of the abundance of the respective protein to a very low level or disappearance under that particular condition. For these six proteins, the pattern of the observed abundance reduction was consistent with their reported functions in the Saccharomyces Genome Database (www.yeastgenome.org). The 205 detected proteins covered the full range of previously reported cellular abundances (Ghaemmaghami et al, 2003), without a bias against low-abundant proteins, and included also proteins whose abundance could not be measured in a previous study (Ghaemmaghami et al, 2003). The lack of an abundance bias in this data set is further supported by the even spread of the 23 undetected proteins over all abundance regimes (Figure 1D).
Condition-dependent changes in protein abundance were expressed as log2 of the ratio of a protein's abundance at a given condition relative to the aerobic glucose-grown condition (see complete quantitative data set in Supplementary Table 6 and Supplementary Figure 1, including standard errors and P-values for significance of the abundance change). Globally, the range of protein abundance changes with respect to the glucose-grown culture was drastically broader for the complex medium and ethanol-grown yeast (2−5 to 29- and 2−3 to 28-fold change, respectively) than for the anaerobic and galactose-grown cells (2−3 to 21- and 2−2 to 22-fold change, respectively). For the galactose-grown condition, only the three proteins involved in the galactose-assimilation pathway (Gal1, Gal7 and Gal10) showed large fold changes (up to 212, see Supplementary Table 6), compared with the more moderate abundance changes of the other target proteins. The number of proteins that showed significant abundance changes of at least twofold, either up or downregulated (P-value ⩽0.05, relative to glucose-grown cells), was 145, 96, 46 and 39 for the complex medium, ethanol, galactose and anaerobically grown cultures, respectively (Supplementary Table 6 and Supplementary Figure 1). Overall, a large fraction of proteins shows significant changes across the different experiments, in agreement with our initial choice of conditions that introduce drastically different modes of metabolic operation.
Besides enzymes in the galactose-assimilation pathway, which strongly increase abundance upon growth on galactose, the largest measurable increases in protein abundance (>26-fold) were observed for Acs1, Adh2, Icl1, Idp2, Mdh2, Mls1, Pck1 and Tkl2 on ethanol- or complex medium (Supplementary Table 6 and Supplementary Figure 1). Most of these proteins are gluconeogenic or glyoxylate shunt enzymes, illustrating the importance of these two pathways under those conditions. The largest identified abundance decreases (>24-fold) were observed on complex medium for Arg3, Gdh1, His4, Met10, Met16, Met6 and Ser3, key enzymes of amino-acid biosynthesis and sulfate assimilation, which are not required in complex medium (Supplementary Table 6, and Supplementary Figure 1).
Presence of non-necessary proteins
On the basis of the FBA simulations, we expected to detect only 121–133 of the targeted proteins under the different conditions if only proteins necessary for that condition would be present. In contrast to these expectations, we detected the vast majority of the 228 targeted proteins to be present under all conditions (Figure 2, Supplementary Table 3). Only a small fraction of the non-necessary proteins was indeed not detected. We conclude from these data that in fact many more proteins are present—at least at a basal level—than that are actually necessary for a given condition. One potential explanation for the presence of non-necessary enzymes is to enable the organism to maintain growth and realize immediate basal metabolic fluxes even upon a rapid change to new environmental conditions (Kotte et al, 2010).
We next asked whether the abundances of non-necessary proteins were lower under conditions where the respective proteins were not necessary. To address this question, we first defined whether a protein needs to change its status from ‘necessary’ to ‘non-necessary’ based on the FBA predictions between the ten possible condition comparisons. In 614 cases, a protein changed its status from ‘necessary’ in one condition to ‘non-necessary’ in another. In about half of these cases (275 cases), the protein's abundance did not change more than twofold. In the other half (303 cases, 49%), a status change from ‘necessary’ to ‘non-necessary’ was accompanied by an at least twofold decrease in abundance. In the remaining 36 cases, protein abundance increased more than twofold, despite a change from ‘necessary’ to ‘non-necessary’, which would be a counterintuitive phenomenon. In the group of proteins that did not need to change from ‘necessary’ to ‘non-necessary’, the protein abundances changed more than twofold in only 27% of a total of 1446 cases. This indicates that changes in necessity status from ‘necessary’ to ‘non-necessary’ were more frequently accompanied by large protein abundance decreases.
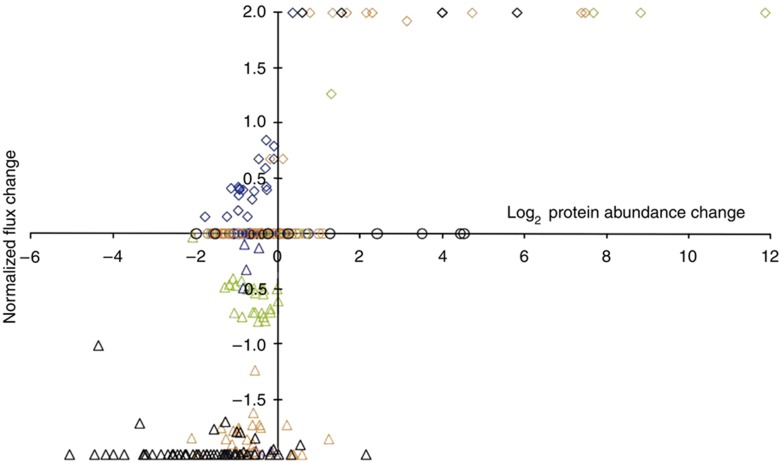
Finally, we asked whether there is a relationship between the abundance of a protein and the predicted metabolic flux through the reaction catalyzed by it. For this purpose, we plotted the normalized changes of the FBA predicted fluxes (Supplementary Table 2) as a function of the change in protein abundance between the condition comparisons (Figure 4). Most flux changes did not require a corresponding change in protein abundance. Changing the flux from zero in one condition to some value in another (normalized flux change of 2 or −2), however, required protein abundance changes in most cases. This on/off flux control by protein abundance was the most pronounced for the comparison of growth on complex versus glucose medium, where essentially all biosynthetic amino-acid fluxes are reduced to zero and most of the catalyzing enzymes are significantly less abundant. This implies that newly required fluxes are predominantly regulated by altered protein abundance while flux modulations are not.
Figure 4.
Correlation of FBA-calculated normalized flux changes with protein abundance changes for the four condition comparisons. Normalized flux changes were calculated using the following formula: (flux A−flux B)/((flux A+flux B)/2), where ‘flux A’ is the flux calculated for condition A, and ‘flux B’ the flux for the same reaction under the reference condition. Data are presented for the comparison of the galactose- (green), anaerobically (blue), ethanol- (red) and complex-grown condition (black) with the aerobic glucose-grown condition. The maximum normalized flux change of 2 (or −2) represents a change from zero in one condition to any magnitude. Fluxes involving promiscuous enzymes, isoenzymes and protein complexes were excluded from the analysis.
Analysis of isoenzyme abundance patterns
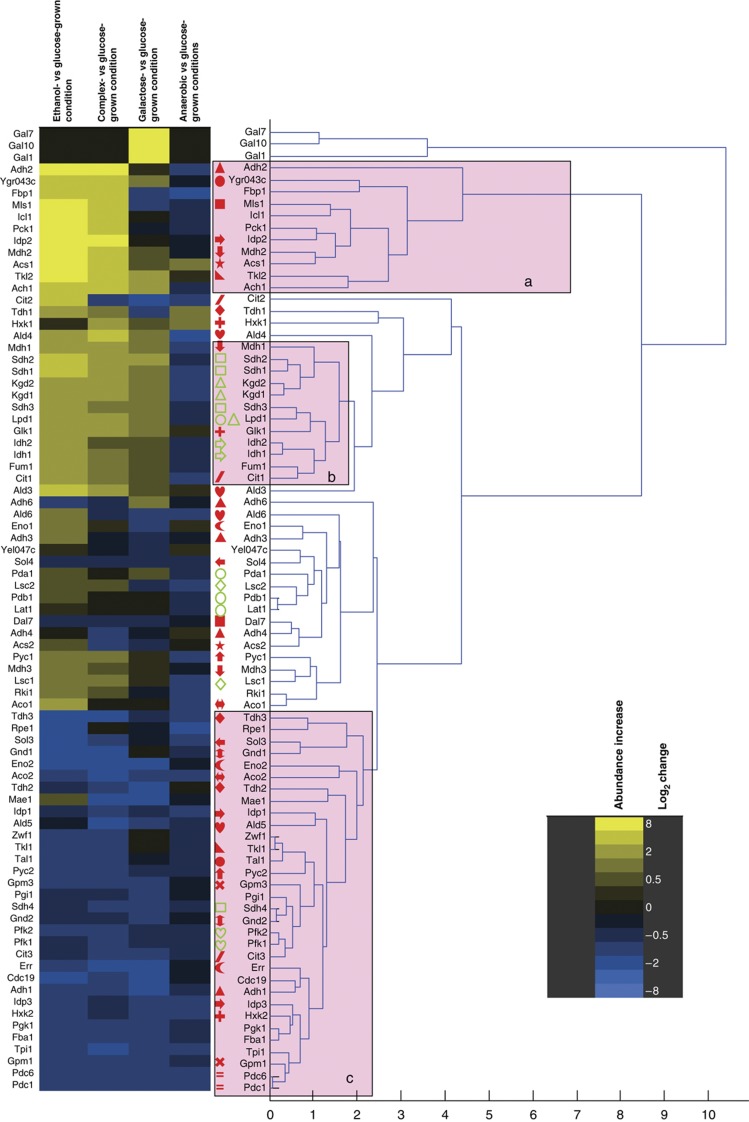
Of the proteins that are not strictly necessary for growth under a given condition, about 40% were isoenzymes of presumably redundant function. Hence, we asked whether the previously proposed functional diversification (Ihmels et al, 2004, 2007) is reflected at the proteome level; i.e., whether isoenzymes exhibit identical or distinct regulation patterns. The data set generated in this study is ideal and unique to answer this question because most isoenzymes in the defined network were completely covered. We used hierarchical clustering (Eisen et al, 1998) to generate patterns of abundance change for all detected isoenzymes over all conditions. Specifically, we relate isoenzyme abundance pattern to the major functional clusters of gluconeogenesis and glyoxylate shunt (Figure 5, block ‘a’), tricarboxylic acid cycle (Figure 5, block ‘b’), and glycolysis and pentose phosphate pathway (Figure 5, block ‘c’).
Figure 5.
Hierarchical clustering analysis of abundance changes in the proteins of central carbon metabolism. Pink boxes indicate branches containing mainly proteins of ‘a’ gluconeogenesis and glyoxylate shunt; ‘b’ tricarboxylic acid cycle and ‘c’ glycolysis, ethanol formation and the pentose phosphate pathway. Isoenzyme families are indicated by identical, closed, red symbols before the protein name; proteins engaged in a protein complex are indicated by identical, open, green symbols. The heat map on the left indicates the direction and magnitude of the abundance changes for each listed protein.
For central carbon metabolism, the only isoenzymes that clustered in the same branch next to each other were Pdc1 and Pdc6. A high degree of functional similarity was seen for the Gpm1/Gpm3 and for the Gnd1/Gnd2 families of isoenzymes because the members of each family clustered in proximate branches. For four families with more than two isoenzymes, always two members clustered close to each other but other members did not (Idp1/Idp3 but not Idp2, Tdh2/Tdh3 but not Tdh1, Eno2/Err, but not Eno1 and Adh3/Adh4, but not Adh1/Adh2/Adh6; Figure 5). For all other families, all members clustered in distant branches indicating functional diversification. Of note, in comparison, subunits of protein complexes preferably clustered in proximate branches with the exception of Sdh4 (Figure 5, green open symbols). Sdh4 is the membrane anchor protein of the succinate dehydrogenase protein complex (Oyedotun and Lemire, 2004) that includes also Sdh1, Sdh2 and Sdh3. The divergent pattern of Sdh4, compared with the other members of the complex, might be a protocol artifact related to sub-optimal extraction of membrane proteins. Although slightly less distinct than for central carbon metabolism, similar observations of distant and proximate clustering were made for isoenzymes and protein complexes in amino-acid metabolism (Supplementary Figure 2). These data, therefore, provide evidence for functional divergence within most isoenzyme families.
Divergent isoenzyme functions
We next attempted to determine the different functions of divergent isoenzymes. This analysis was based on the hypothesis that similar functions should lead to similar protein abundance patterns, therefore resulting in different patterns for functionally divergent isoenzymes. The known pattern of alcohol dehydrogenase isoenzymes Adh1 and Adh2 provide a case supporting this hypothesis. In agreement with their described function in the SGD database (www.yeastgenome.org), we found that the major ethanol-consuming isoenzyme (Adh2) clustered with the glyoxylate shunt and gluconeogenesis proteins, whereas the major ethanol-producing isoenzyme (Adh1) clustered in the glycolytic branch (Figure 5). The extension of this type of analysis to other isoenzyme families indicated new cases of functional diversification.
The first reaction of glucose breakdown can be catalyzed by three hexokinase isoenzymes. While Hxk2 clustered indeed in the glycolytic branch, Glk1 clustered with the tricarboxylic acid cycle proteins and Hxk1 clustered with a protein involved in the synthesis of storage carbohydrates, Glc3 (Supplementary Figure 2), and several other members of the storage pathway can be found in proximate branches (Gsy2, Ugp1, Tsl1, Tps1 and Tps2), supporting the notion that Hxk1 is related to storage carbohydrates (Figure 5). Indeed, both Glk1 and Hxk1 have been speculated to direct glucose toward glycogen storage rather than the regular glycolytic path (Ihmels et al, 2004), as is the case for Hxk2.
The non-oxidative part of the pentose phosphate pathway, i.e., the transketolases Tkl1 and Tkl2 and the transaldolases Tal1 and Ygr043c (recently renamed to Nqm1 (Hua et al, 2008)), where Ygr043c has so far been classified as a transaldolase of unknown function, provide evidence for an entirely novel functional differentiation between isoenzymes. In our data set, Tkl1 and Tal1 cluster, as expected with the pentose phosphate proteins in a glycolytic branch. Their isoenzymes Tkl2 and Ygr043c, however, clustered with the gluconeogenic proteins, indicating that their function is either directly in gluconeogenic metabolism or possibly in the supply of the pentose precursors for amino or nucleic acid synthesis under conditions where their regular synthesis from glucose 6-phosphate is difficult or insufficient.
Of the two isoenzymes for phospoglucomutase (Pgm1 and Pgm2) that catalyze the interconversion of glucose 1-phosphate and glucose 6-phosphate, Pgm2 clustered with the glycogen breakdown to glucose 1-phosphate enzymes Gph1 and Gdb1 (Supplementary Figure 2). The Pgm1 isoenzyme clustered far away with proteins for glycogen and trehalose synthesis (Ugp1, Gsy2, Glc3, Tps1, Tps2 and Tsl1). These data suggest, therefore, that Pgm1 and Pgm2 have alternative functions in synthesis and breakdown of glycogen, respectively.
Finally, the isocitrate dehydrogenase isoforms Idp1 and Idp2 clustered with the glycolytic and gluconeogenic/glyoxylate shunt proteins, respectively (Figure 5). As the cytosolic, NADP-specific isoform Idp2 is not a part of the glyoxylate shunt, our data suggest a gluconeogenic function in NADPH formation, a redox cofactor required for biosynthesis (Minard and McAlister-Henn, 2005). Idp2-based NADPH formation on non-fermentable substrates is further supported by low simulated fluxes for the oxidative pentose-phosphate pathway (the second source for cytosolic NADPH) under gluconeogenesis (Supplementary Table 2) and by clustering of Ald6, the third major source of cytosolic NADPH (Minard and McAlister-Henn, 2005), far away from the gluconeogenic enzymes. This conclusion is consistent with idp2 mutant data during growth on acetate (Minard and McAlister-Henn, 2005). Overall, we thus provide evidence for functional diversification of various isoenzymes and for several cases the data presented here suggest novel functional roles.
Discussion
In this study, we exploited the power of SRM-based targeted proteomics to consistently and reproducibly detect and quantify a target set of yeast metabolic proteins covering a broad range of abundance levels across several samples and experiments. From the 228 target protein set, selected from a consensus stochiometric metabolic model for the central carbon and amino-acid metabolism of S. cerevisiae, ∼90% were successfully identified in minimally one of the samples. This substantially expands the coverage achieved by previous proteomic studies of yeast metabolism. For example, 57, 58 and 55% of the here targeted metabolic proteome was covered by de Groot et al (2007), Gutteridge et al (2010) and Kolkman et al (2006), respectively and only up to 30% of isoenyzme families could be quantitatively resolved (Kolkman et al, 2006; de Groot et al, 2007; Gutteridge et al, 2010). To score the comprehensiveness of our proteomics method, we used a model-based approach to assess which proteins can be expected to be present. On the basis of this analysis, the data reported here are 95–99% comprehensive. Our inability to detect up to 5% remaining proteins may be explained by other factors, such as (i) low abundance and lack of PTPs with good MS properties, (ii) occurrence of post-translational modifications that decrease or eliminate the signal of the corresponding target tryptic peptide or (iii) loss of the protein during the sample preparation steps (e.g., for cell wall or membrane proteins). The detectable proteins were consistently measured in unfractionated yeast proteome tryptic digests. This is in agreement with our earlier demonstration that proteins spanning the whole abundance range of the yeast proteome can be detected by SRM (Picotti et al, 2009) and with the high degree of data reproducibility generated by the SRM technique demonstrated in a previous multi-center study (Addona et al, 2009).
The applied method consistently quantified the target protein set across 15 samples, including biological triplicates, without the problem of missed data points. This number of consistently analyzed samples and replicates constitutes an improvement with respect to the lower reproducibility of data generated by shotgun proteomic studies. In fact, in many proteomic studies to date no replicate data sets were reported and only small numbers of samples were analyzed. This is in large part due to the significant effort and cost associated with generating comprehensive and quantitative proteomic data, especially where approaches based on in-depth fractionation were used. Also, the SRM approach allowed here for the quantitative discrimination of proteins with a high sequence overlap, such as isoenzymes, allowing us to gain insight into their functional diversification. Classical shotgun proteomic measurements based on automated peptide sequencing would be biased against the discrimination of isoenzymes, as their shared peptides are more likely to be high abundant and therefore preferentially detected. Overall, this study has resulted in the largest to date SRM-based proteomic data set (>200 proteins quantified across multiple conditions), with high comprehensiveness for the metabolic network under study, challenged with a set of nutritional conditions that imply radically different modes of metabolic operation. We expect that this will be a useful blueprint for further developing mathematical models of the yeast metabolism and a valuable basis for follow-up studies on the function of target (metabolic) proteins.
Despite their power in analyzing target proteins across several samples and replicates, SRM approaches are in their infancy and face still considerable technical challenges to their high-throughput application. The first is the need for designing optimal assays for each target protein. Recently, significant advances have been realized to speed up and automate this step and strategies based on unpurified synthetic peptides allow for the fast and low-cost development of SRM assays for essentially any protein or proteome of interest (Picotti et al, 2010). Another challenge is the analysis of SRM data, which involves the detection and assignment of the relevant peaks in the raw MS data. Here, this step was carried out manually, using the most up to date and stringent confidence criteria (Anderson and Hunter, 2006; Lange et al, 2008a; Picotti et al, 2009; MacLean et al, 2010 see details in the Materials and methods). However, manual peak assignment remains tedious and does not allow attributing a false discovery rate based on objective criteria to SRM-based peptide identifications. To this direction, algorithms are currently being developed to automate evaluation of SRM peak matches and their statistical treatment (Reiter L et al, in preparation). The last bottleneck is the number of target proteins that can be concurrently analyzed in a single SRM run. This is at present significantly lower than that of proteins identified in a shotgun proteomics experiment on a high-performance MS (de Godoy et al, 2008) and efforts are currently underway by the MS vendors to improve the multiplexing of this technology. For the specific network under study, the set of ∼200 metabolic proteins can be quantified in ∼4 h of instrument time per sample, measuring multiple SRM transitions per peptide, multiple peptides per protein (where available), with light (endogenous) and heavy (internal standard) signals. The data set presented here, consisting of 15 samples, can be acquired in ∼3 days of mass spectrometric measurements.
Remarkably, the total number of metabolic proteins detectable, and thus expressed, did not change much between the different metabolic states. Although the identity of the necessary proteins varies slightly between conditions (Supplementary Table 3), yeast cells should be able to grow with roughly 120 proteins in the considered network, yet many more are always present. Expression of unneeded proteins has long been known to reduce growth rates and thereby presumably evolutionary fitness (Dekel and Alon, 2005); hence, intuition and genetic evidence (Zaslaver et al, 2004) suggest that enzymes are downregulated under conditions when their reactions are not required. Their here demonstrated unexpected persistence might be explained by the lower than expected costs of unneeded protein synthesis after several generations of exponential growth, at least in Escherichia coli (Shachrai et al, 2010). Alternatively, it can be an adaptive strategy for rapid and flexible responses to environmental changes (Kotte et al, 2010).
About 40% of the apparently ‘superfluous’ proteins are isoenzymes. We showed here that differences in abundance changes among isoenzymes were indicative for different isoenzyme functionality, consistent with earlier studies based on transcriptional data (Ihmels et al, 2004). On the basis of abundance pattern clustering in different functional classes of metabolic pathways, many isoenzymes show evidence for functional diversification in the presented experiments, which might explain their parallel presence in the S. cerevisiae genome (Kuepfer et al, 2005; Ihmels et al, 2007). As we only tested a very limited number of conditions, it is our expectation that also for the isoenzymes that did not show functional diversification so far, such evidence could be found under appropriate metabolic setups.
In conclusion, this study shows that quantitative assays for large sets of biologically related proteins can be developed and deployed to monitor responses of these proteins to a set of different environmental or genetic conditions, providing detailed insights in a cell's physiology. This approach is ideal to explore the dynamics of cellular networks, under physiological or challenged conditions, also for organisms other than yeast, and thus has the potential to find broad applications in systems biology, biomedical and pharmaceutical research.
Materials and methods
Yeast culture conditions and protein extraction
All experiments were performed with the prototrophic strain S. cerevisiae FY4 (Winston et al, 1995). Mineral medium (Verduyn et al, 1990) was supplemented with 10 g l−1 of the carbon sources glucose, galactose or ethanol. Medium for anaerobic cultures was buffered with 10 mM KH-Phthalate buffer. Complex medium contained 20 g l−1 yeast extract and 40 g l−1 peptone. Cultures were grown in 50 ml of respective medium in 500 ml shake flasks at 30°C and shaken at 250 r.p.m. Anaerobic cultures were grown in 50 ml of medium in 150 ml air-tight serum flasks and shaken at 110 r.p.m. Anaerobic medium and flasks were flushed with nitrogen gas (certified <5 p.p.m. O2) upon closure and during sampling, and were sealed with oxygen-impermeable rubber septa.
For each growth condition, triplicate cultures were inoculated from a single pre-culture, pre-grown on identical medium and conditions. Two ml aliquots were withdrawn at regular intervals and treated as described before for biomass content and HPLC analysis (Heer and Sauer, 2008). Aliquots for proteome analysis were withdrawn during the late exponential growth phase at high biomass content when cells still grew exponentially. From each culture, 36 ml of cell broth were harvested on ice and centrifuged for 3 min at 4°C. Pellets were washed once with ice-cold washing buffer (20 mM HEPES, pH 7.5, 2 mM EDTA), frozen in liquid nitrogen and stored at −80°C until extraction.
15N isotopically labeled yeast cells to be used as internal standard for protein abundance quantification were derived from a yeast batch culture that displayed diauxic growth in 2 l minimal medium with 5 g l−1 glucose and 15N-labeled ammonium (99% purity 15N2 (NH4)2SO4, Cambridge Isotope Laboratories) as sole nitrogen source. Cells were grown in a fully aerated bioreactor. To gain high coverage of metabolic proteins, aliquots from the different phases (growth on glucose, transient phase and growth on ethanol) of this experiment were mixed.
Proteins were extracted and precipitated as described previously (Picotti et al, 2009). Before precipitation, the total protein concentration in the lysis buffer was calculated based on the average results of a spectrophotometrical BCA (bicinchoninic acid, Perbio Science, Lausanne, Switzerland) assay, conducted in triplicate for every sample. A 100 μg aliquot of each sample was mixed to an equal amount of 15N-isotopically labeled internal protein standard (see above). Tryptic digestion was conducted as described in Picotti et al (2009). Digested peptide mixtures were cleaned on C18 cartridges (Sep-Pak tC18, Waters) and eluted with 60% (vol/vol) acetonitrile. Samples were dried in a vacuum centrifuge, and solubilized in 0.1% formic acid upon analysis. All samples reported in this work were processed in parallel.
Design of SRM assays
For each protein targeted in this work, up to ten PTPs for detection and quantification via SRM were selected. Preferentially, detectable PTPs were chosen based on their number of observations in the publicly accessible proteomic data repository PeptideAtlas (www.peptideatlas.org, a large proteomic data repository that contains more than 50 000 unique peptides observed in shotgun proteomic experiments (Deutsch et al, 2008)). Peptide identifications deriving from isotope-coded affinity tag experiments were not considered. For proteins with less than five PTPs available from PeptideAtlas, additional PTPs amenable for mass spectrometry analysis were derived by prediction using the publicly available software PeptideSieve (Mallick et al, 2007; tools.proteomecenter.org, Seattle Proteome Centre). For proteins never observed in PeptideAtlas, the five peptides resulting from PeptideSieve prediction were synthesized on a small scale in an unpurified format using the SPOT-synthesis technology (JPT Peptide Technologies) and used as a reference to derive the optimal parameters for the corresponding SRM assays.
For each PTP, three to eight SRM transitions for both the double- and the triple-charged state were calculated, corresponding to fragment ions of the y-series. Fragment ions with a mass-to-charge ratio (m/z) greater than the precursor ion m/z were prioritized. Fragments with m/z ratios close to the precursor ion m/z (smaller than 5 Th difference) were discarded. This selection process was automated through in-house written software. The selected transitions were used to detect the peptides by SRM in whole S. cerevisiae protein digests and to trigger acquisition of the full fragment ion spectra of the peptides. For low-abundant PTPs and PTPs predicted by PeptideSieve, full fragment ion spectra were acquired from synthetic peptide preparations. Optimal fragments to be used in the final SRM assays were chosen from the full fragment ion spectra, acquired on the triple quadrupole mass spectrometer (see below).
Mass spectrometry analysis
All peptide samples were analyzed on a hybrid triple quadrupole/ion trap mass spectrometer equipped with a nanoelectrospray ion source (4000QTrap, AB/Sciex). Chromatographic separations of peptides were performed on a nano-LC system (Tempo, AB/Sciex) coupled to a fused silica emitter (length 16 cm, diameter 75 μm) packed with a Magic C18 AQ 5 μm resin (Michrom BioResources). Peptides were loaded on the column from a cooled (4°C) autosampler (Tempo, AB/Sciex) and chromatographically separated using a linear gradient of acetonitrile/water at a flow rate of 300 nl min−1. A gradient from 5 to 30% acetonitrile in 30 or 60 min was used. In the assay validation phase, the mass spectrometer was operated in MRM mode, triggering acquisition of a full MS/MS spectrum upon detection of an SRM trace. The set of SRM transitions generated as described above was split into multiple MS methods and analyzed in several runs. MRM acquisition was performed with Q1 and Q3 operated at unit resolution (0.7 m/z half maximum peak width). MS/MS spectra were acquired in enhanced product ion mode for the two highest MRM transitions, using the following settings: dynamic fill time, Q1 resolution low, scan speed 4000 a.m.u. s−1, m/z range 300–1400. Collision energies (CEs) were calculated according to the formulas: CE=0.044 × m/z+5.5 and CE=0.051 × m/z+0.5 (with m/z being the mass-to-charge ratio of the precursor ion) for doubly and triply charged precursor ions, respectively.
Database search and extraction of optimal SRM transitions
Raw MS/MS .wiff data was converted to .mzXML format with the program mzWiff and searched against the yeast SGD database (version dated 01/26/2006) using Sequest (version 27). A decoy database was generated by randomly reshuffling amino acids in between tryptic cleavage sites, and appended to the target database. Precursor mass tolerance was set at 2.1 Da. Data were searched with full tryptic cleavage and carboxyamidomethylation of cysteine residues as a static modification. The search results were validated and assigned probabilities using the PeptideProphet program implemented in the Trans-Proteomic-Pipeline (Deutsch et al, 2008), with decoy-assisted semiparametric model and retention-time model enabled, as previously described (Picotti et al, 2008) and filtered to a decoy-estimated false discovery rate of 1%.
For each peptide, the 3 or 4 fragment ions resulting in the highest signals were selected from the QQQ MS/MS spectra as previously described (Picotti et al, 2008) as final SRM assay. The corresponding SRM transitions associated with the 15N analog of each peptide were calculated and measured as internal standard. The final SRM assays were used to detect and quantify the proteins in total lysates of yeast cells grown under the set of five conditions mentioned above, using time-scheduled SRM/MRM acquisition (retention time window, 200–300 s; target scan time, 3.5 s, maximally 950 transitions per run). Blank runs (water, 0.1% formic acid, injected) were performed prior to most SRM measurements. In these controls, the same SRM method was used as in the following (sample) run (e.g, a method in which the same set of transitions was measured). The complete set of SRM assays is available in Supplementary Table 4 or via the public SRMAtlas database (www.srmatlas.org; Picotti et al, 2008). The usage of time-scheduled SRM acquisition maximized the throughput and sensitivity of the proteomic analysis (Stahl-Zeng et al, 2007). MS/MS spectra are available at https://db.systemsbiology.net/sbeams/cgi/PeptideAtlas/GetPeptideSpectra.
Quantification of protein abundance changes
Peak heights for the transitions associated with the native and 15N-labelled peptides were quantified using MultiQuant v.1.1 Beta (Applied Biosystems). To accept validation of a set of SRM traces, we checked that the retention time at which the MS/MS spectrum was acquired from the natural or synthetic sources matched that of the SRM peaks defining the peptide of interest. We also confirmed ‘coelution’ and shape similarity of different SRM traces recorded for a given peptide. In some dubious cases, we also checked that relative intensities of SRM traces from different fragments matched those observed in the MS/MS spectrum of the peptide. For relative quantification of each protein across the set of different growth conditions, the ratio between the light and heavy SRM peak height was calculated and normalized to that obtained for the aerobic glucose-grown sample. Protein abundance changes were expressed as the mean log2 ratios out of the different transitions over peptides, protein and the three replicate cultures per condition±the standard error of the mean. Outlier transitions (for instance, shouldered transition traces, noisy transitions or with a signal-to-noise ratio smaller than three) were not considered in the calculations. A Student's one sample t-test was performed on the log2 ratios to determine statistically significant changes in protein abundances (P<0.05). For proteins that were not detected in all five conditions (i.e., the signal of their best-responding PTP decreased to the noise level in one or several of the five growth conditions), we used the noise levels of the SRM traces to estimate a minimal abundance change compared with a condition with an above-noise signal.
Minimal set of necessary proteins
For determination of the minimal set of necessary proteins under each of the five growth conditions, FBA (Fell and Small, 1986) was performed using an updated version of a previously published genome-scale model of the metabolism of S. cerevisiae (Kuepfer et al, 2005; Supplementary Table 7). Measured specific production and consumption rates (Table I) were used as constraints on the metabolic network, and minimization of the Euclidean norm of fluxes (Blank et al, 2005) was used as objective function. It minimizes the total sum of the squared values of all fluxes in the network. Calculations were carried out using MATLAB scripts (The MathWorks) and solved using LINDO numerical solver software package (Lindo Systems). On the basis of the FBA solution, reactions from the targeted sub-network that had a value zero were classified as non-necessary, and reactions with a value were classified as necessary. As necessary reactions do not map one-to-one to necessary proteins, this was corrected for the presence of isoenzymes, protein promiscuity and protein complexes, as explained in Supplementary Table 3.
Hierarchical cluster analysis
The Pearson's linear correlation coefficients of the log2 of the measured abundance changes for each protein were calculated with the log2 of the abundance of this protein under aerobic glucose-grown conditions set to zero. The correlation coefficients were then used to calculate the Euclidean distances between proteins with which hierarchical cluster trees were generated using group average as clustering method. All calculations were performed in MATLAB software. Protein abundance changes of <20% and protein abundance changes with P-values for their significance >0.05 were assumed to be only marginal changes, and their value was set to zero for the cluster analysis. Abundance changes for proteins that were not observed under a particular condition were also set to zero for that condition change. The reported minimal abundance changes for these proteins for conditions where they were observed were then taken as the value to cluster. Proteins for which now five zero values were yielded were excluded from the cluster analysis all together.
Supplementary Material
Supplementary Figures S1–3, Supplementary Tables S1–7
Acknowledgments
We acknowledge SystemsX.ch for funding (YeastX and PhosphoNetX projects). PP is the recipient of an intra-european Marie Curie fellowship, LR was supported by a grant from Forschungskredit of the University of Zurich. RA is supported by the European Research Council (Grant # ERC-2008-AdG 233226).
Author contributions: RC, PP, MH, US and RA conceived and designed the research. RC, PP, MH, RA and US wrote the paper. RC and PP performed experiments, developed and performed analyses and assays and analyzed data. LR devised statistical and computational proteomics tools. RS performed SRM data analysis.
Footnotes
The authors declare that they have no conflict of interest.
References
- Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM et al. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol 27: 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldridge BB, Burke JM, Lauffenburger DA, Sorger PK (2006) Physicochemical modelling of cell signalling pathways. Nat Cell Biol 8: 1195–1203 [DOI] [PubMed] [Google Scholar]
- Anderson L, Hunter CL (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics 5: 573–588 [DOI] [PubMed] [Google Scholar]
- Baty JD, Robinson PR (1977) Single and multiple ion recording techniques for the analysis of diphenylhydantoin and its major metabolite in plasma. Biomed Mass Spectrom 4: 36–41 [DOI] [PubMed] [Google Scholar]
- Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJ (2009) A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat Methods 6: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank L, Kuepfer L, Sauer U (2005) Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol 6: R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Chubukov V, Jolly ER, DeRisi J, Li H (2008) Dynamics and design principles of a basic regulatory architecture controlling metabolic pathways. PLoS Biol 6: e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daran-Lapujade P, Rossell S, van Gulik WM, Luttik MA, de Groot MJ, Slijper M, Heck AJ, Daran JM, de Winde JH, Westerhoff HV, Pronk JT, Bakker BM (2007) The fluxes through glycolytic enzymes in Saccharomyces cerevisiae are predominantly regulated at posttranscriptional levels. Proc Natl Acad Sci USA 104: 15753–15758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455: 1251–1254 [DOI] [PubMed] [Google Scholar]
- de Groot MJ, Daran-Lapujade P, van Breukelen B, Knijnenburg TA, de Hulster EA, Reinders MJ, Pronk JT, Heck AJ, Slijper M (2007) Quantitative proteomics and transcriptomics of anaerobic and aerobic yeast cultures reveals post-transcriptional regulation of key cellular processes. Microbiology 153: 3864–3878 [DOI] [PubMed] [Google Scholar]
- Dekel E, Alon U (2005) Optimality and evolutionary tuning of the expression level of a protein. Nature 436: 588–592 [DOI] [PubMed] [Google Scholar]
- Deutsch EW, Lam H, Aebersold R (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep 9: 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte NC, Becker SA, Jamshidi N, Thiele I, Mo ML, Vo TD, Srivas R, Palsson B (2007) Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci USA 104: 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist AM, Herrgård MJ, Thiele I, Reed JL, Palsson B (2009) Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol 7: 129–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell DA, Small JR (1986) Fat synthesis in adipose tissue. An examination of stoichiometric constraints. Biochem J 238: 781–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Gutteridge A, Pir P, Castrillo JI, Charles PD, Lilley KS, Oliver SG (2010) Nutrient control of eukaryote cell growth: a systems biology study in yeast. BMC Biol 8: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heer D, Sauer U (2008) Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb Biotechnol 1: 497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann M, Sauer U (2010) Systems biology of microbial metabolism. Curr Opin Microbiol 13: 337–343 [DOI] [PubMed] [Google Scholar]
- Herrgård MJ, Swainston N, Dobson P, Dunn WB, Arga KY, Arvas M, Büthgen N, Borger S, Costenoble R, Heinemann M, Hucka M, Le Novère N, Li P, Liebermeister W, Mo ML, Oliveira AP, Petranovic D, Pettifer S, Simeonidis E, Smallbone K et al. (2008) A consensus yeast metabolic network reconstruction obtained from a community approach to systems biology. Nat Biotechnol 26: 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua H, Hui R, Xu L, Shuilong T, Zhiqiang Z, Liwen N, Maikun T (2008) The crystal structure and identification of NQM1/YGR043C, a transaldolase from Saccharomyces cerevisiae. Proteins 73: 1076–1081 [DOI] [PubMed] [Google Scholar]
- Ihmels J, Collins SR, Schuldiner M, Krogan NJ, Weissman JS (2007) Backup without redundancy: genetic interactions reveal the cost of duplicate gene loss. Mol Syst Biol 3: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Levy R, Barkai N (2004) Principles of transcriptional control in the metabolic network of Saccharomyces cerevisiae. Nat Biotechnol 22: 86–92 [DOI] [PubMed] [Google Scholar]
- Kitano H (2002a) Computational systems biology. Nature 420: 206–210 [DOI] [PubMed] [Google Scholar]
- Kitano H (2002b) Systems biology: a brief overview. Science 295: 1662–1664 [DOI] [PubMed] [Google Scholar]
- Kolkman A, Daran-Lapujade P, Fullaondo A, Olsthoorn MM, Pronk JT, Slijper M, Heck AJ (2006) Proteome analysis of yeast response to various nutrient limitations. Mol Syst Biol 2: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotte O, Heinemann M (2009) A divide-and-conquer approach to analyze underdetermined biochemical models. Bioinformatics 25: 519–525 [DOI] [PubMed] [Google Scholar]
- Kotte O, Zaugg JB, Heinemann M (2010) Bacterial adaptation through distributed sensing of metabolic fluxes. Mol Syst Biol 6: 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuepfer L, Sauer U, Blank LM (2005) Metabolic functions of duplicate genes in Saccharomyces cerevisiae. Genome Res 15: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Malmstrom JA, Didion J, King NL, Johansson BP, Schafer J, Rameseder J, Wong CH, Deutsch EW, Brusniak MY, Buhlmann P, Bjorck L, Domon B, Aebersold R (2008a) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol Cell Proteomics 7: 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R (2008b) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R (2007) Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol 25: 125–131 [DOI] [PubMed] [Google Scholar]
- Minard KI, McAlister-Henn L (2005) Sources of NADPH in yeast vary with carbon source. J Biol Chem 280: 39890–39896 [DOI] [PubMed] [Google Scholar]
- Oberhardt MA, Palsson BO, Papin JA (2009) Applications of genome-scale metabolic reconstructions. Mol Syst Biol 5: 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyedotun KS, Lemire BD (2004) The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase: homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem 279: 9424–9431 [DOI] [PubMed] [Google Scholar]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Lam H, Campbell D, Deutsch EW, Mirzaei H, Ranish J, Domon B, Aebersold R (2008) A database of mass spectrometric assays for the yeast proteome. Nat Methods 5: 913–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods 7: 43–46 [DOI] [PubMed] [Google Scholar]
- Sauer U (2006) Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol 2: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer U, Heinemann M, Zamboni N (2007) Genetics: getting closer to the whole picture. Science 316: 550–551 [DOI] [PubMed] [Google Scholar]
- Schellenberger J, Palsson B (2009) Use of randomized sampling for analysis of metabolic networks. J Biol Chem 284: 5457–5461 [DOI] [PubMed] [Google Scholar]
- Shachrai I, Zaslaver A, Alon U, Dekel E (2010) Cost of unneeded proteins in E. coli is reduced after several generations in exponential growth. Mol Cell 38: 758–767 [DOI] [PubMed] [Google Scholar]
- Stahl-Zeng J, Lange V, Ossola R, Eckhardt K, Krek W, Aebersold R, Domon B (2007) High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol Cell Proteomics 6: 1809–1817 [DOI] [PubMed] [Google Scholar]
- Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P et al. (2010) Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res 9: 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA, van Dijken JP (1990) Physiology of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J Gen Microbiol 136: 395–403 [DOI] [PubMed] [Google Scholar]
- Wagner A (2008) Gene duplications, robustness and evolutionary innovations. Bioessays 30: 367–373 [DOI] [PubMed] [Google Scholar]
- Wilson JE (2003) Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol 206: 2049–2057 [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55 [DOI] [PubMed] [Google Scholar]
- Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U (2004) Just-in-time transcription program in metabolic pathways. Nat Genet 36: 486–491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1–3, Supplementary Tables S1–7