SUMMARY
A hallmark of Parkinson’s disease (PD) is the preferential loss of substantia nigra dopamine neurons. Here we identify a new Parkin Interacting Substrate, PARIS (ZNF746), whose levels are regulated by the ubiquitin proteasome system via binding to and ubiquitination by the E3 ubiquitin ligase, parkin. PARIS is a novel KRAB and zinc finger protein that accumulates in models of parkin inactivation and in human PD brain. PARIS represses the expression of the transcriptional co-activator, PGC-1α and the PGC-1α target gene, NRF-1 by binding to insulin response sequences in the PGC-1α promoter. Conditional knockout of parkin in adult animals leads to progressive loss of dopamine (DA) neurons that is PARIS dependent. Moreover overexpression of PARIS leads to the selective loss of DA neurons in the substantia nigra, which is reversed by either parkin or PGC-1α co-expression. The identification of PARIS provides a molecular mechanism for neurodegeneration due to parkin inactivation.
INTRODUCTION
Parkinson’s disease (PD) is a progressive neurodegenerative disorder that is characterized phenotypically by bradykinesia, rigidity, tremor, and neuropsychiatric disturbances (Savitt et al., 2006). Although the cause of PD in the majority of cases is unknown, there are rare familial cases for which the genes have been identified. There are at least sixteen PD associated loci (Gasser, 2007). Mutations in α-synuclein and leucine rich repeat kinase 2 (LRRK2) cause autosomal dominant PD. Four genes have been linked to autosomal recessive PD (AR-PD) and include mutations in parkin, DJ-1, PINK1, and ATP13A2. Investigating the biology of these genes and their mutant protein has provided tremendous insight into the pathogenesis of both familial and sporadic PD (Gasser, 2007; Savitt et al., 2006).
Parkin is an ubiquitin E3 ligase (Shimura et al., 2000; Zhang et al., 2000). In general, PD-associated mutations in parkin lead to loss of its E3 ligase function (Tanaka et al., 2004). Moreover, oxidative, nitrosative, and dopaminergic stress, which play important pathogenic roles in PD inactivate parkin, suggesting that parkin inactivation may play a role in sporadic PD (Chung et al., 2004; LaVoie et al., 2005; Winklhofer et al., 2003). Thus, substrates of parkin that are subject to proteasomal degradation should accumulate in animal and cellular models of parkin inactivation and AR-PD due to parkin mutations, and also in sporadic PD. There are a diverse array of parkin substrates that has hindered the generation of a consensus in the field on parkin’s physiologic function and pathologic role in PD. Moreover, parkin’s ability to mono- and polyubiquitinate as well as ubiquitinate proteins with both lysine-48 and lysine-63 chains has made it difficult to reconcile a common biochemical pathway for parkin’s role in PD (Dawson and Dawson, 2010).
Here we identify a new Parkin Interacting Substrate, PARIS, which provides a molecular mechanism for neurodegeneration due to parkin inactivation in PD. Our results show that parkin regulates the levels of PARIS via the ubiquitin proteasome system (UPS). We show that PARIS is a major transcriptional repressor of peroxisome proliferator-activated receptor gamma (PPARγ) coactivator-1α (PGC-1α) expression and that conditional knockout (KO) of parkin in adult mice leads to progressive loss of dopamine (DA) neurons through PARIS overexpression and transcriptional repression of PGC-1α.
RESULTS
PARIS is a KRAB and C2H2 zinc finger protein
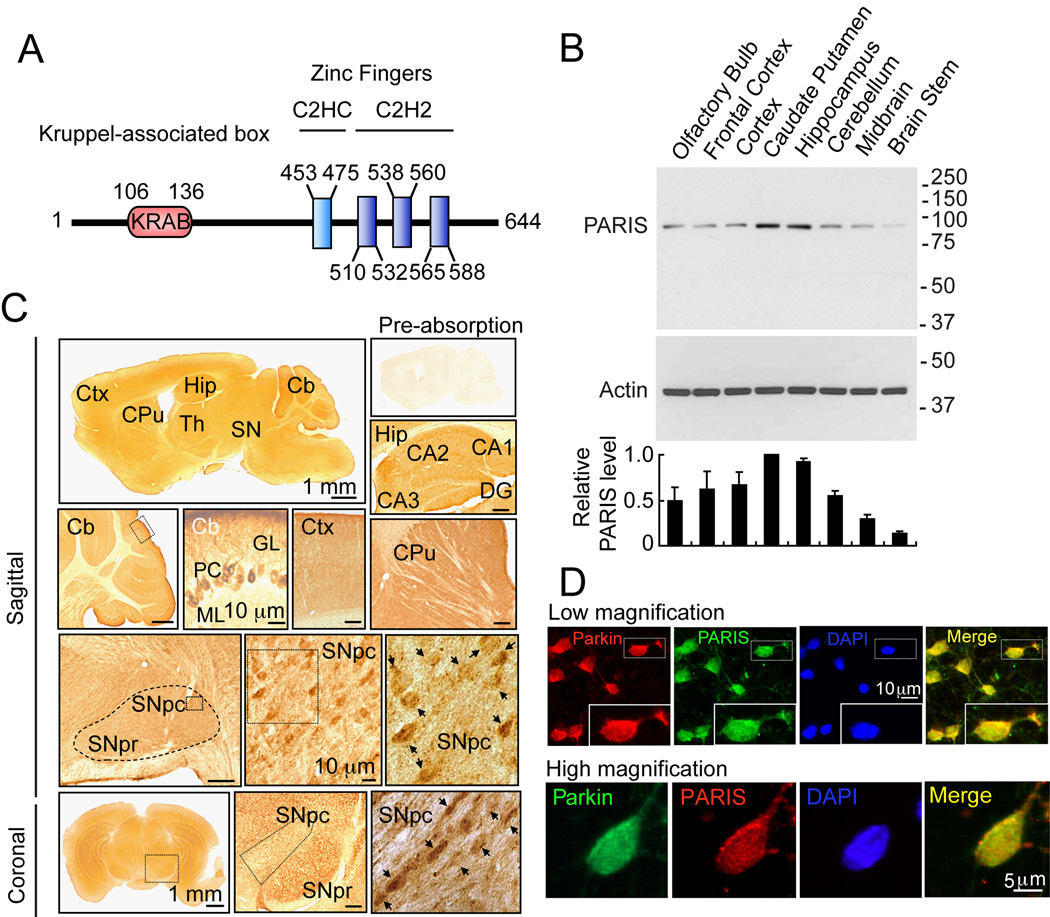
PARIS was identified by yeast two-hybrid screening using parkin as bait (Zhang et al., 2000). Human PARIS (ZNF746) is a 644 amino acid protein that contains a Kruppel-associated box (KRAB) at its N-terminus and a C2HC/C2H2 type zinc finger at its C-terminus (Figure 1A). There is a high degree of homology among human, mouse and rat PARIS proteins (Figure S1A). Northern blot and immunoblot analysis reveals that PARIS is expressed in all organs examined (Figure S1B and S1C). PARIS is differentially expressed in the brain with low levels in cerebellum and midbrain (Figure 1B). Immunohistochemisty reveals that PARIS is heterogeneously distributed throughout the brain and that it is localized to neurons, including substantia nigra (SN) pars compacta DA containing neurons (Figure 1C). Confocal imaging indicates that PARIS is co-localized with parkin in primary cortical neuron cultures (Figure 1D) and in SH-SY5Y dopaminergic-like cells (Figure S1D).
Figure 1. Identification of a novel parkin interacting substrate, PARIS.
(A) Schematic representation of PARIS. The conserved Kruppel associated Box (KRAB) and Zinc Finger motifs and their location are indicated.
(B) Regional analysis and levels of PARIS protein expression via immunoblot in various brain regions. Data = mean ± S.E.M., n = 3.
(C) Immunohistochemical distribution of PARIS in a sagittal and coronal sections of six-week-old male C57BL mouse brain. Right upper panel shows an antigen preabsorption control. Ctx, cerebral cortex; Hip, hippocampus; CPu, caudate putamen; SNpr, SN pars reticulata; SNpc, SN pars compacta; Th, thalamus; Cb, cerebellum; (Cb); PC, Purkinje cells; ML, molecular cell layer; GL, granule cell layer; DG, dentate gyrus. Dashed (−) line outlines SN. High power view of SNpc (rectangles in third row) is shown in the third row middle and right panels and lower middle and right panel. Scale bars, 200 µm unless indicated, n = 3.
(D) Confocal microscopy demonstrates that endogenous PARIS and parkin are co-localized mainly in the cytoplasm of rat cortical neurons. Top Panel, Parkin-red; PARIS-green. Inset – high power view of an individual neuron. Bottom panel, Parkin-green; PARIS-red, Nucleus-DAPI-blue, merge-yellow, n = 4.
See also Figure S1.
Parkin Interacts with PARIS
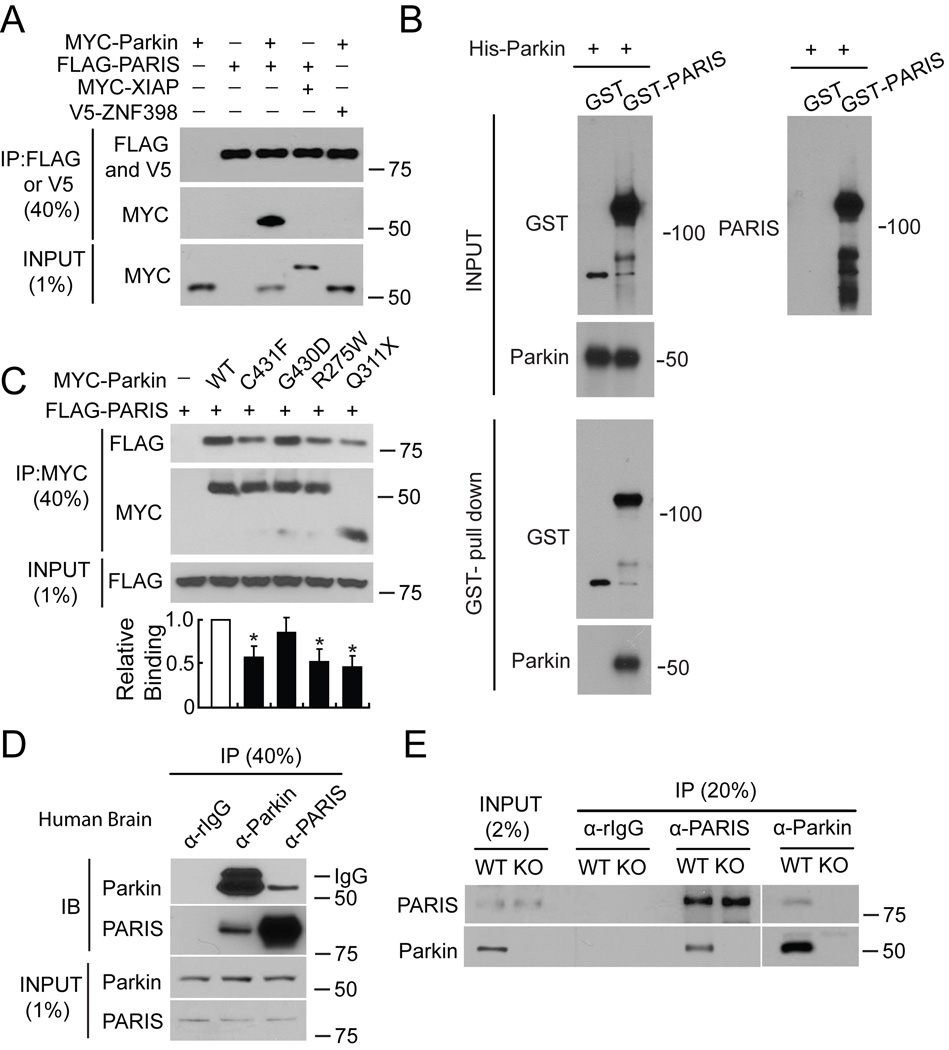
MYC-tagged parkin and FLAG-tagged PARIS co-immunoprecipitate in SH-SY5Y cells, whereas XIAP, a RING finger ubiquitin E3 ligase and ZNF398, a highly conserved homologue of PARIS do not interact with PARIS and parkin, respectively (Figure 2A). Recombinant GST-PARIS pulls down recombinant His-Parkin indicating that PARIS and parkin directly interact (Figure 2B). The familial PD-associated mutations in parkin, C431F, R275W and Q311X bind to PARIS less avidly than WT parkin, whereas G430D mutant parkin binds to PARIS in a manner similar to WT parkin (Figure 2C). PARIS co-immunoprecipitates with parkin and parkin co-immunoprecipitates with PARIS from whole human striatum (Figure 2D) or mouse brain (Figure S2A). This endogenous interaction between parkin and PARIS is not observed in parkin exon 7 KO brain (Figure 2E) confirming the specificity of the interaction.
Figure 2. Parkin interacts with PARIS.
(A) Immunoprecipitated (IP) FLAG-PARIS interacts with MYC-Parkin, but not MYC-XIAP or the PARIS homologue, V5-ZNF398 (lane 6), n = 3.
(B) GST-pull down assay between parkin and PARIS indicates a robust interaction between parkin and PARIS, n = 4.
(C) Immunoprecipitated FLAG-PARIS interacts with WT Parkin and Parkin mutants (C431F, G430D, R275W, Q311X) in SH-SY5Y cells. Lower panel, relative binding, data = mean ± S.E.M., n = 3, *p < 0.05, Student’s t-test.
(D) Immunoblot (IB) shows that Parkin and PARIS co-immunoprecipitate in human striatum, n = 3.
(E) Parkin and PARIS co-immunoprecipitate from WT mouse ventral midbrain, but not parkin KO ventral midbrain, n = 3.
See also Figure S2.
Domain mapping indicates that parkin binds to the C-terminal domain, but not the N-terminal domain of PARIS (Figure S2B). To ascertain which domain of parkin binds to PARIS, various deletion constructs of MYC-tagged parkin were utilized. Co-immunoprecipitation experiments show that both the RING1 or RING2 domains are required for parkin binding to PARIS (Figure S2C and S2D). Taken together these results indicate that parkin interacts with the zinc finger domain of PARIS and PARIS binds to either the RING1 or RING2 domain of parkin.
Parkin ubiquitinates PARIS and regulates PARIS levels
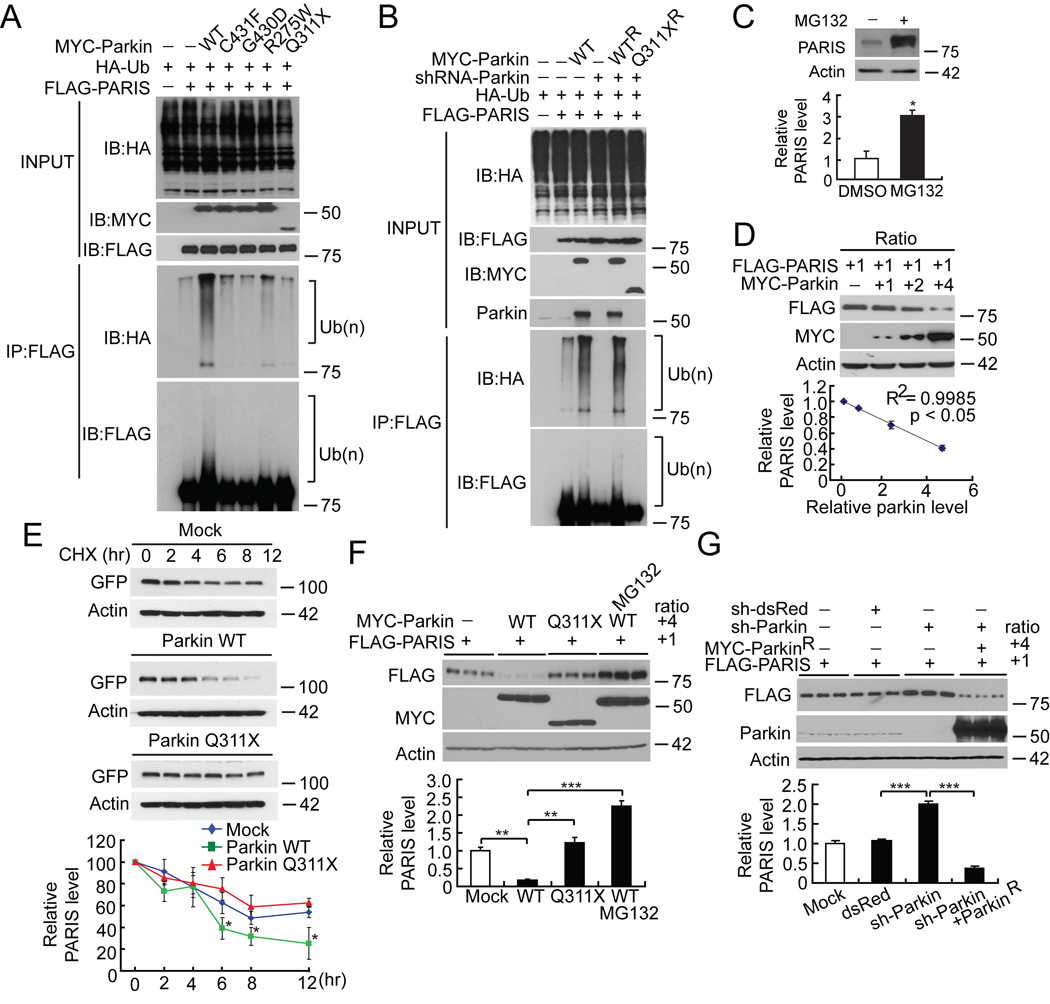
FLAG-tagged PARIS is ubiquitinated by MYC-tagged parkin in SH-SY5Y cells as shown by the substantial HA immunoreactivity in the form of a smear, which is characteristic of polyubiquitinated proteins (lane 3, Figure 3A). Familial linked parkin mutations C431F, G430D and Q311X have substantially reduced ubiquitination activity against PARIS, whereas R275W has modest activity (Figure 3A). The reduction in ubiquitination of PARIS by these mutants is due, in part, to their reduced binding (see Figure 2C) as well as reduced E3 ligase activity of these parkin mutants (Sriram et al., 2005).
Figure 3. Parkin ubiquitinates and regulates the ubiquitin proteasomal degradation of PARIS.
(A) WT MYC-Parkin ubiquitinates FLAG-PARIS (lane 3). Parkin mutants (C431F, G430D, Q311X) are unable to efficiently ubiquitinate FLAG-PARIS (lanes 4–7). Ubiquitination (Ub(n)) is indicated on right with brackets, n = 3.
(B) Endogenous ubiquitination of PARIS (lane 2) is enhanced with exogenous WT Parkin (lane 3) and it is eliminated with shRNA-Parkin (lane 4). In the presence of shRNA-Parkin, robust ubiquitination of PARIS is observed via co-expression of shRNA-resistant WT parkin (WTR) but not shRNA-resistant Q311X mutant parkin (Q311XR), n = 3.
(C) 10 µM MG-132 increases PARIS steady state levels compared to DMSO control. Bottom panel, relative PARIS levels normalized to β-actin, n = 3.
(D) Increasing ratio (1:1 to 4:1) of MYC-Parkin results in decreased steady-state levels of FLAG-PARIS (lanes 1–4). Bottom panel, relative PARIS and parkin levels normalized to β-actin, n = 3; regression analysis, R2=0.9985, p<0.05).
(E) WT Parkin decreases the steady-state levels of PARIS compared to mutant Q311X parkin or GFP transfected control cells in cyclohexamide (CHX)-chase experiments in SH-SY5Y cells transiently expressing FLAG-PARIS. Bottom panel, relative PARIS levels normalized to β-actin, n = 3.
(F) MYC-parkin leads to degradation of FLAG-PARIS at a 4 to 1 ratio, respectively. 10 µM MG-132 prevents the degradation of FLAG-PARIS and MYC-Q311X parkin has no effect, n = 3)
(G) PARIS accumulates after shRNA-Parkin and co-expression of shRNA resistant parkin (MYC-ParkinR) leads to robust degradation of PARIS, n = 3. Data = mean ± S.E.M., *p < 0.05, **p < 0.01 and ***p < 0.001, ANOVA with Student-Newman-Keuls post-hoc analysis (E, F, G) or Student’s t-test (C).
See also Figure S3.
FLAG-PARIS is ubiquitinated in the absence of exogenous parkin (lane 2, Figure 3A and lane 2 Figure 3B). This endogenous ubiquitination is enhanced with co-expression of WT parkin (lane 3, Figure 3A and lane 3 Figure 3B). ShRNA-Parkin eliminates the endogenous ubiquitination (lane 4, Figure 3B). To control for potential off-targets effects of shRNA-parkin, a shRNA-resistant WT parkin, but not a shRNA-resistant Q311X parkin mutant restores the ubiquitination of PARIS in the presence of shRNA-parkin (Figure 3B).
In vitro ubiquitin assays shows that parkin ubiquitinates PARIS in the presence of various E2 enzymes including UbcH5c (Figure S3A). There is no ubiquitination signal in the absence of PARIS (Figure S3B) and in the absence of parkin (Figure S3C) indicating that the ubiquitination signal is specific for PARIS ubiqutination. CHIP, which acts as a E4 for parkin (Imai et al., 2002), enhances the ubiquitination of PARIS by parkin, but it has no affect in the absence of parkin indicating that CHIP does not ubiquitinate PARIS directly (Figure S3D). OTU1, a K48-linkage specific deubiquitinating enzyme (Messick et al., 2008), successfully hydrolyzed the poly-ubiquitin chain on PARIS (Figure S3E and S3F) and a K48-specific anti-ubiquitin antibody, but not a K63 antibody detects the poly-ubiquitin chain on PARIS (Figure S3E), suggesting that endogenous and exogenous parkin ubiquitinates PARIS via K48 linkages.
The steady state level of PARIS is regulated by the ubiquitin proteasome system since the level of PARIS increases approximately two to three fold when SH-SY5Y cells are treated with the proteasome inhibitor, MG132 (Figure 3C). The levels of PARIS and parkin are tightly and significantly correlated (R2=0.9985) in co-expression experiments in SH-SY5Y cells in which increased levels of parkin lead to decreased levels of PARIS (Figure 3D). Cyclohexamide (100 µg/ml) chase experiments in SH-SY5Y cells transiently expressing GFP-tagged PARIS with or without WT parkin or Q311X parkin demonstrate that the decrease in the steady state levels of PARIS is accelerated in the presence of WT parkin compared with cells transfected with GFP-vector or familial linked parkin Q311X mutant (Figure 3E). Parkin at a 4 to 1 ratio with PARIS leads to a statistically significant reduction in PARIS that is blocked by MG132, whereas the catalytically inactive Q311X parkin fails to reduce PARIS levels (Figure 3F). ShRNA-Parkin leads to a significant increase in the level of PARIS, which is reduced by expression of a shRNA-resistant WT parkin (Figure 3G). These results taken together suggest that parkin controls the levels of PARIS via the ubiquitin proteasome system.
PARIS accumulates in the absence of parkin activity
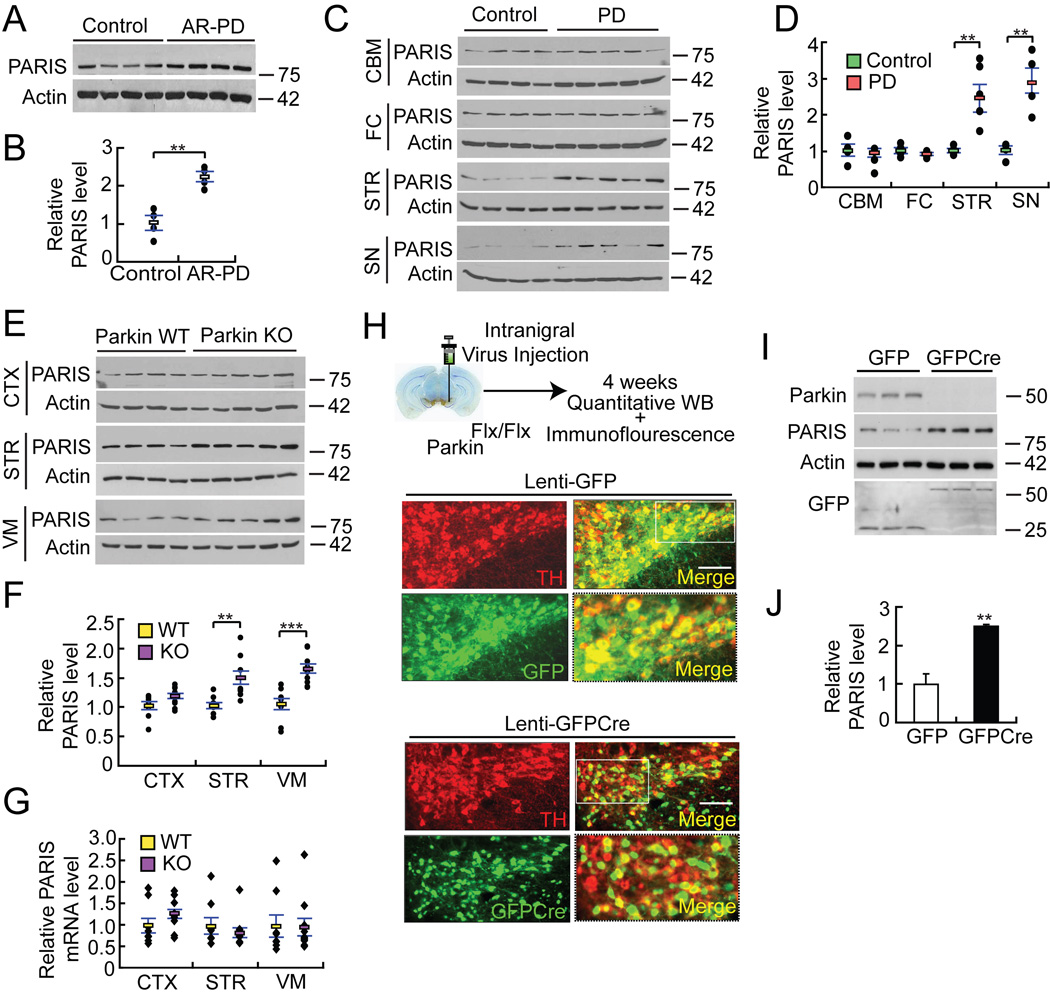
PARIS levels are increased two fold in the cingulate cortex of AR-PD patients, which lack functional parkin, versus age-matched controls (Figure 4A and B). Due to the lack of tissue availability, we were not able to assess the levels of PARIS in other brain regions of AR-PD patients. PARIS levels are also increased by more than two fold in both the striatum and the SN of sporadic PD compared to controls (Figure 4C and 4D). We find no alteration in the levels of PARIS mRNA in PD striatum or PD SN versus control striatum and SN indicating that the increased level of PARIS is not due to a transcriptional effect (see Figure 6A and Figure S5A). PARIS levels are not increased in regions of the brain that are relatively unaffected in PD including the cerebellum and the frontal cortex of sporadic PD patients compared to controls (Figure 4C and 4D) suggesting that the upregulation is primarily within the nigrostriatal pathway.
Figure 4. PARIS accumulates in AR-PD, sporadic PD and in animal models of parkin inactivation.
(A) Immunoblot analysis of PARIS and β-actin in cingulate cortex from age-matched controls and AR-PD patient brains with parkin mutations
(B) Quantitation of the immunoblots in panel A normalized to β-actin, n = 4.
(C) PARIS levels in cerebellum (CBM), frontal cortex (FC), striatum (STR) and SN of sporadic PD patient brains compared to age-matched controls.
(D) Relative PARIS levels normalized to β-actin in panel B, Controls n = 4; PD n = 5.
(E) Immunoblot analysis of PARIS in cortex (CTX), STR and ventral midbrain (VM) from WT and parkin exon 7 KO 18–24 month old mice.
(F) Relative protein levels of PARIS normalized to β-actin for panel E, WT n = 9; parkin KO n = 10.
(G) PARIS mRNA levels in indicated brain regions from WT and parkin exon 7 KO 18–24 month old mice.
(H) Top panel, experimental illustration of stereotaxic intranigral virus injection. Bottom panels, immunofluorescent images of TH (red), GFP (green) and merged (yellow) in exon 7 floxed parkin mice (parkinFlx/Flx) after stereotactic delivery of Lenti-GFP or Lenti-GFPCre into the SNpc. 84.9±1.9% and 78.1±2.6% of TH neurons express GFP and GFPCre, respectively, n = 3 per group. Enlarged images in the right bottom panels were taken from the white rectangle region from the merged images of Lenti-GFPCre and Lenti-GFP, bar = 100 µm.
(I) Immunoblot analysis of parkin, PARIS, actin and GFP 4 weeks after intranigral Lenti-GFPCre or Lenti-GFP injection into parkinFlx/Flx mice.
(J) Relative protein levels of PARIS normalized to β-actin for panel I. Data = mean ± S.E.M., *p < 0.05, **p < 0.01 and ***p < 0.001, unpaired two-tailed Student’s t-test (B, J) and ANOVA test with Student-Newman-Keuls post-hoc analysis (D, F, G).
See also Table S1 and S2.
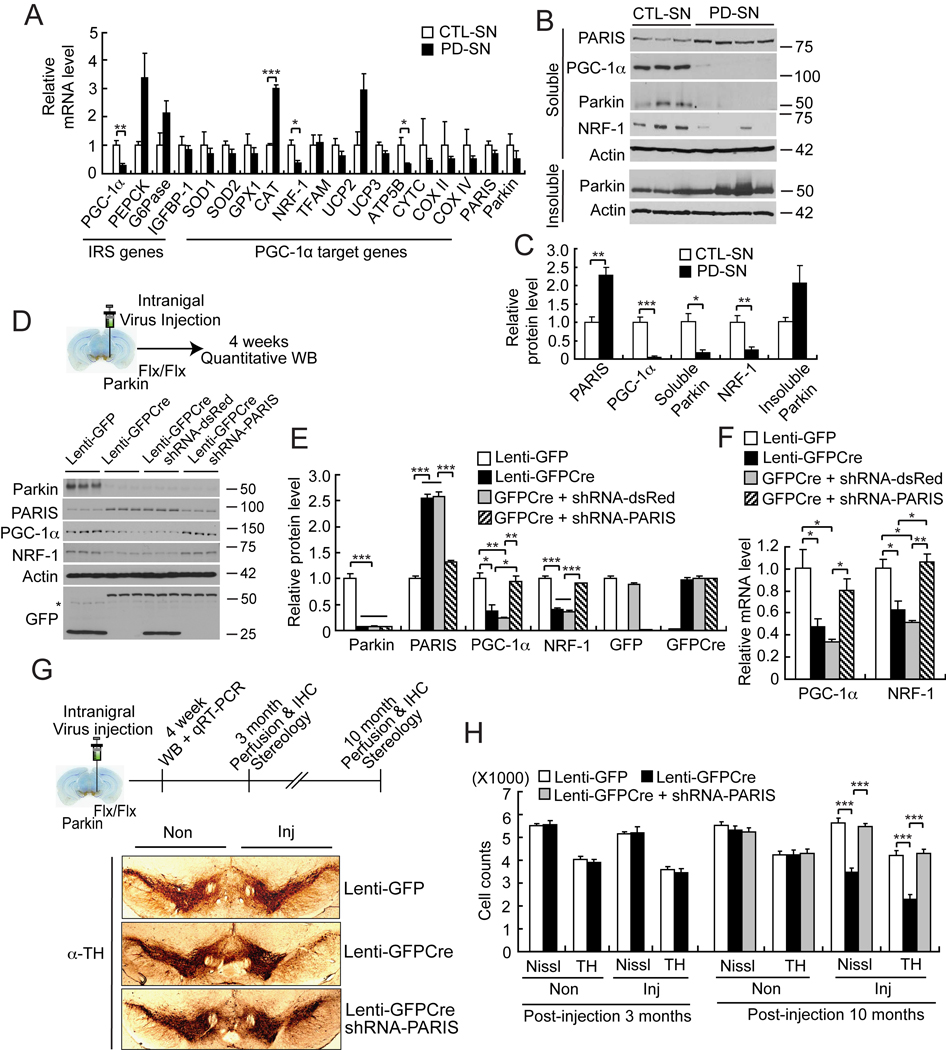
Figure 6. Identification of PGC-1α and NRF-1 as pathological in vivo targets of accumulated PARIS in PD brain and conditional parkin KO mice.
(A) Real-time qRT-PCR of IRS (PEPCK-like motif)-containing genes and PGC-1α dependent genes in PD SN compared to age-matched CTL-SN normalized to GAPDH, n = 3–4 per group.
(B) Immunoblots of PARIS, PGC-1α, parkin and NRF-1 in soluble and insoluble fractions of PD SN compared to age-matched CTL-SN.
(C) Quantitation of the immunoblots in panel B normalized to β-actin, PD, n = 4; Control, n = 3.
(D) Top panel, experimental illustration of stereotaxic intranigral virus injection. Below are immunoblots of parkin, PARIS, PGC-1α, NRF-1, β-actin and GFP, 4 weeks after stereotactic delivery of Lenti-GFP, Lenti-GFPCre, Lenti-GFPCre + shRNA-dsRed, or Lenti-GFPCre + shRNA-PARIS into exon 7 floxed parkin mice (parkinFlx/Flx), n = 3 per group. *nonspecific band.
(E) Quantitation of the immunoblots in panel D normalized to β-actin, n = 3 per group.
(F) The alteration of PGC-1α and NRF1 shown in panel D and E results from transcriptional changes as determined by real-time qRT-PCR, n = 3 per group.
(G) Top panel, experimental illustration of stereotaxic intranigral virus injection. Below is TH immunostaining of representative mice midbrain sections from SN of parkinFlx/Flx mice injected with Lenti-GFP, and Lenti-GFPCre ± Lenti-shRNA-PARIS 10 months post-injection of virus.
(H) Stereological assessment of TH and Nissl positive neurons in the SN of parkinFlx/Flx mice injected with Lenti-GFP, and Lenti-GFPCre ± Lenti-shRNA-PARIS 3 (n = 3 per group) and 10 months (n = 7 per group) after injection of virus. Data = mean ± S.E.M., *p < 0.05, **p < 0.01 and ***p < 0.001, unpaired two-tailed Student’s t-test (panel A and C) and ANOVA with Student-Newman-Keuls post-hoc analysis (panel E, F and H).
See also Figure S5, Table S4 and S5.
We next evaluated whether PARIS is increased in germline parkin exon 7 KO mice (Von Coelln et al., 2004). PARIS is modestly upregulated by approximately 48% in the striatum and by 63% in ventral midbrain of germline parkin exon 7 KO mice compared to age matched WT controls, whereas the levels of PARIS in the cortex is not changed (Figure 4E and 4F). The upregulation of PARIS is not due to a transcriptional effect since we find no difference in the level of PARIS mRNA in germline parkin KO ventral midbrain and striatum versus WT ventral midbrain and striatum(Figure 4G).
Since germline parkin exon 7 KO mice lack parkin from the point of conception, it is possible that compensatory mechanisms account for the lack of a more substantial upregulation of PARIS (for review see (Dawson et al., 2010)). To avoid potential developmental compensation, exon 7 was deleted in 6–8 week old conditional parkin KO mice in which exon 7 was flanked by loxP sites (parkinFlx/Flx) by SN stereotactic injection of a GFP-fused Cre recombinase lentivirus (Lenti-GFPCre) and compared to control SN injections of lentivirus expressing GFP (Lenti-GFP) in parkinFlx/Flx mice (Figure 4H and 4I). Four weeks after injection of the lentiviruses, Lenti-GFP and Lenti-GFPCre effectively transduce neurons in the SN including DA neurons (Figure 4H). Lenti-GFPCre leads to almost a complete loss of parkin from the ventral midbrain of parkinFlx/Flx mice compared to Lenti-GFP mice (4I and 4J). Accompanying the loss of parkin is greater than a two-fold upregulation of PARIS. The upregulation of PARIS in the conditional parkin exon 7 KO model is not due to a transcriptional effect since there is no alteration in PARIS mRNA in the conditional parkin KO ventral midbrain versus WT ventral midbrain (Figure S5E). Thus, the levels of PARIS are increased in conjunction with parkin inactivation and impairment of ubiquitin-mediated proteasomal degradation in sporadic PD, AR-PD and in an animal model of parkin inactivation.
PARIS is a transcriptional repressor of PGC-1α
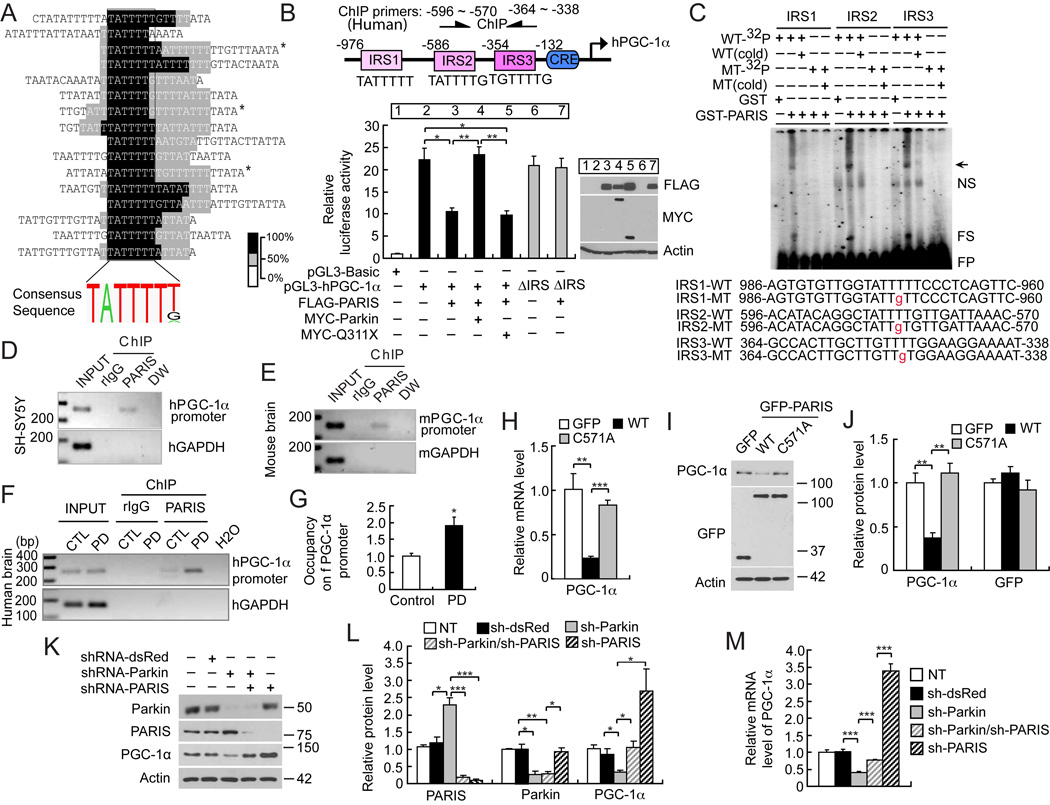
Proteins with KRAB domains can function as transcriptional repressors (Witzgall et al., 1994). GAL4-BD fused PARIS leads to decreased luciferase activity from a 5 × GAL4-luciferase reporter construct, which is restored by co-expression of WT parkin, but not Q311X mutant parkin (Figure S4A). To identify the PARIS DNA binding consensus sequence, a chimeric protein containing the zinc finger domain (ZF) of PARIS (amino acids, 453–589) fused in frame to glutathione-S-tranferase (GST), GST-ZF-PARIS, was used in a Cyclic Amplification and Selection of Targets (CAST) assay (Figure 5A) (Funk and Wright, 1992; Shields and Yang, 1998). Immobilized GST-ZF-PARIS was incubated with a pool of oligonucleotides containing 26 random nucleotides. The final pool of oligonucleotides remaining after four rounds of CASTing followed by three rounds of electrophoretic mobility shift assays (EMSA) were cloned, sequenced and analyzed. Alignment of all the sequences using the program MACAW (National Center for Biotechnology Information) reveals a consensus sequence with a core sequence composed of TATTTT (T/G) (Figure 5A). 19 out of 24 sequences contained the core sequence and 3 out of 19 sequence tags were identified as duplicates indicating this is the primary DNA binding sequence for PARIS. The TATTTT (T/G) consensus sequence is an insulin response sequence (IRS) designated the phosphoenolpyruvate carboxykinase (PEPCK)-like motif (PLM) (Daitoku et al., 2003; O'Brien et al., 2001), which is involved in the regulation of transcripts involved in energy metabolism and insulin responsiveness (Mounier and Posner, 2006).
Figure 5. PARIS acts as transcriptional repressor of PGC-1 α.
(A) Identification and MACAW alignment of the PARIS DNA-binding sequence as determined by CASTing. Darker colors represent a greater degree of overlap of the segment pairs (bottom right - % overlap). *Duplicate sequence tags.
(B) Relative luciferase activity of the 1-kilobase human PGC-1α (−992 to +90) compared to Renilla luciferase ± PARIS or ± parkin or ± familial mutant Q311X parkin, n = 3. Location of IRS, CRE motifs and oligos for human ChIP (arrows) in the PGC-1α promoter construct (top of panel). Immunoblot analysis confirms the expression of FLAG-PARIS, MYC-Parkin and MYC-Q311X parkin (right panel).
(C) EMSA of GST-PARIS of 32P-labeled WT (WT-32P) IRS oligonucleotides (IRS1-WT, IRS2-WT, IRS3-WT) of the human PGC-1α promoter and 32P-labeled mutant (T→ ) (MT-32P) IRS oligonucleotides (IRS1-MT, IRS2-MT, IRS3-MT). Unlabeled WT (WT cold) IRS oligonucleotides disrupt the GST-PARIS-DNA protein complexes with the WT-32P IRS oligonucleotides, n = 3. Unlabeled mutant probes (MT cold) has no effect on mutant (MT-32P). Arrow indicates specific PARIS-shifted probe; NS, nonspecific; FS, fragmented PARIS-shifted probe; FP, free probe.
) (MT-32P) IRS oligonucleotides (IRS1-MT, IRS2-MT, IRS3-MT). Unlabeled WT (WT cold) IRS oligonucleotides disrupt the GST-PARIS-DNA protein complexes with the WT-32P IRS oligonucleotides, n = 3. Unlabeled mutant probes (MT cold) has no effect on mutant (MT-32P). Arrow indicates specific PARIS-shifted probe; NS, nonspecific; FS, fragmented PARIS-shifted probe; FP, free probe.
(D) PARIS occupies the endogenous PGC-1 α promoter as determined by ChIP assay with anti-PARIS polyclonal antibodies in SH-SY5Y cells, n = 3.
(E) PARIS occupies the endogenous mouse PGC-1 α promoter as determined by ChIP in mouse whole brain, n = 3. Mouse specific IRS primers are indicated in Figure S4B.
(F) ChIP assay of endogenous PARIS binding to the IRS region of the human PGC-1a promoter in human PD and aged-matched control (CTL) striatum, control n = 3; PD n = 4.
(G) Quantitation of ChIP in panel F. Human specific IRS primers for D and F are indicated in panel B.
(H) Real-time qRT-PCR of PGC-1α, GFP and β-actin following transient transfection of GFP, GFP-PARIS or GFP-C571A PARIS mutant, n = 4.
(I) Immunoblot analysis of PGC-1α, GFP and β-actin following transient transfection of GFP, GFP-PARIS or GFP-C571A PARIS mutant, n = 4.
(J) Quantitation of the immunoblots in panel I normalized to β-actin, n = 4.
(K) Immunoblot analysis of parkin, PARIS, PGC-1α and β-actin in double knockdown experiments via lentiviral transduction of shRNA-parkin and/or shRNA-PARIS in SH-SY5Y cells, n = 3.
(L) Quantitation of the immunoblots in panel K normalized to β-actin, n = 3, sh = shRNA.
(M) Relative mRNA levels of PGC-1α normalized to GAPDH, n = 3. Quantitative data = mean ± S.E.M., *p < 0.05, **p < 0.01, ***p < 0.001, unpaired two-tailed Student’s t-test (G), ANOVA with Student-Newman-Keuls post-hoc analysis (B, H, J, L, M).
See also Figure S4, Table S2, S3, S5.
A NCBI survey of IRS/PLM responsive transcripts reveals that members of the PPARγ coactivator-1 (PGC-1) family of transcriptional co-activators are regulated by IRS sites. PGC-1α contains three IRS/PLM elements within its 5’-promoter region (Figure 5B). The activity of the 1kb human PGC-1α promoter is decreased approximately 40% in the presence of PARIS, which is rescued by parkin overexpression (Figure 5B). Similar results are observed with the 2 kb mouse PGC-1α promoter (Figure S4B). The familial parkin mutant, Q311X, has minimal effects on the PGC-1α promoter (Figure 5B). PARIS represses PGC-1α promoter activity by binding to the IRS sites, since it fails to inhibit the reporter in which the three IRS sites are deleted in the context of the PGC-1α promoter-reporter construct (Figure 5B).
Individual IRS site mutants (IRS1-M, IRS2-M, IRS3-M) within the PGC-1α reporter that disrupt PARIS binding to the PGC-1α promoter as determined by EMSA (see Figure 5C) and a promoter construct, IRS123-M, containing all three mutations were evaluated in the promoter reporter assay (Figure S4C). IRS1-M and IRS3-M substantially reduce PGC-1α reporter promoter activity, whereas IRS2-M increases the activity (Figure S4C). PARIS overexpression inhibits IRS1-M, IRS2-M, IRS3-M PGC-1α reporter promoter activity suggesting that PARIS binds to and regulates all three sites (Figure S4C) as indicated from the EMSA assays (Figure 5C).
EMSA shows that PARIS binds to the PGC-1α IRS elements since GST-PARIS (full length) elicits a shift of [32P]-labeled oligonucleotides containing the IRS1, IRS2, and IRS3 sequences of the PGC-1α promoter, whereas it fails to cause a shift of IRS1, IRS2 and IRS3 containing a single base substitution within the IRS sequence (Figure 5C). Additionally eight zinc finger mutants of PARIS were assessed for their ability to repress the PGC-1α reporter promoter construct (Figure S4D and S4E). M1, M2, M8 PARIS mutants are able to repress PGC-1α reporter promoter activity, similar to WT PARIS. M3, M4, M5, M6 PARIS mutants partially repress PGC-1α reporter promoter activity, whereas the M7 (C571A) PARIS mutant has no affect on PGC-1α reporter promoter activity. The M7 GST-C571A–PARIS mutant has substantially reduced IRS-binding capacity as determined by EMSA (Figure S4F).
Chromatin immunoprecipitation (ChIP) indicates that PARIS binds to the endogenous PGC-1α promoter in SH-SY5Y cells (Figure 5D) and mouse brain (Figure 5E). PARIS also binds to the endogenous PGC-1α promoter in human brain (Figure 5F) and consistent with its upregulation in PD striatum (see Figure 4C and 4D) there is enhanced PARIS occupancy of endogenous PGC-1α in PD striatum compared to control (Figure 5G). We performed luciferase reporter assay in SH-SY5Y cells and ChIP assays in PD versus control striatum and SH-SY5Y cells with phosphoenolpyruvate carboxykinase (PEPCK) (Figure S4G) and glucose-6-phosphatase (G6Pase) (Figure S4H). The luciferase reporter assay shows that overexpression of PARIS enhances the promoter activity of rat PEPCK, but not mouse G6Pase promoter activity (Figure S4I). The ChIP assay demonstrates that PARIS binds to the endogenous promoter of human PEPCK and G6Pase in SH-SY5Y cells and in control and PD postmortem striatum (Figure S5J). In contrast to PGC-1α, there is not enhanced occupancy of the PEPCK and G6Pase promoter by PARIS. These data suggest that PARIS can bind to the promoter of PEPCK and G6Pase, but in contrast to PGC-1α it positively regulates PEPCK and it has no appreciable effect on G6Pase. Thus, the transcriptional repressive effects of PARIS are relatively specific to PGC-1α.
GFP-WT-PARIS overexpression leads to approximately a 75% reduction in PGC-1α mRNA (Figure 5H) and approximately a 60% reduction in protein levels of PGC-1α (Figure 5I and 5J), whereas the GFP-C571A–PARIS mutant has no effect on PGC1-α protein or message levels (Figure 5H–J). Lentiviral shRNA-parkin leads to a two fold increase in the level of PARIS followed by a 66% reduction of PGC-1α (Figure 5K and 5L). To determine whether the reduction in PGC-1α levels induced by the absence of parkin is dependent on the presence of PARIS, a double knockdown experiment was performed by lentiviral shRNA-parkin and/or shRNA-PARIS in SH-SY5Y cells. Knockdown of PARIS prevents the downregulation of PGC-1α levels induced by parkin knockdown (Figure 5K and 5L). Knockdown of PARIS results in a 3 fold increase in PGC-1α protein levels (Figure 5K and 5L) and a 3.5 fold increase in PGC-1α mRNA (Figure 5M). Knockdown of PARIS in the setting of parkin knockdown also prevents the down regulation of PGC-1α mRNA (Figure 5M). These results taken together indicate that PARIS is a transcriptional repressor that negatively regulates the levels of endogenous PGC-1α and that the downregulation of PGC-1α in the absence of parkin is due to the upregulation of PARIS.
Identification of NRF-1 as a potential in vivo PGC-1α target gene in PD
PGC-1α is a transcriptional coactivator that regulates a variety of genes (Lin et al., 2005; St-Pierre et al., 2006). To determine which PGC-1α target genes are co-regulated by PARIS in PD, real-time quantitative RT-PCR (qRT-PCR) was performed on a variety of PGC-1α target genes in PD SN and striatum. We measured the levels PGC-1α co-regulated genes that play important roles in mitochondrial function and oxidant metabolism including nuclear respiratory factor-1 (NRF-1), copper/zinc superoxide dismutase (SOD1), manganese SOD (SOD2), glutathione peroxidase (GPx1), catalase (CAT), mitochondrial uncoupling proteins (UCP2 and UCP3), mitochondrial transcription factor A (Tfam) and the oxidative phosphorylation regulators, ATP5b, cytochrome C (CytC) and cytochrome C oxidase (COX II and IV) (Figure 6A and Figure S5A). In addition, the levels of other genes containing IRS/PLM in their promoter including PEPCK, G6Pase, insulin-like-growth-factor binding protein 1 (IGFBP-1), tyrosine aminotransferase (TAT), and apolipoprotein C III (APOC3) were monitored along with PGC-1α to assess whether PGC-1α is selectively affected in PD (Figure 6A and Figure S5A). The levels of PARIS and parkin were also assessed as controls. We find that PGC-1α and NRF-1 mRNA are downregulated in PD SN and striatum compared to control SN and striatum (Figure 6A and Figure S5A). In PD SN ATP5B is also significantly downregulated and CAT is significantly upregulated (Figure 6A). All other PGC-1α dependent genes are not significantly altered (Figure 6A and Figure S5A). In addition there is no significant change in the levels of the IRS/PLM responsive transcripts PEPCK, G6Pase and IGFBP-1 (Figure 6A and Figure S5A). TAT and APOC3 are not detectable. No significant alteration in the mRNA level of PARIS and parkin is observed between PD and control SN and striatum (Figure 6A and Figure S5A) indicating that the upregulation in PARIS protein levels (see Figure 4C and 4D) are most likely due to impairment of parkin E3 ubiquitin ligase activity. Moreover, the absence of an alteration in the mRNA levels of PARIS and parkin suggest that the changes in the mRNA levels of PGC-1α and NRF-1 are specific and not due to the degenerative process that occurs in PD.
As shown above (see Figure 4C and 4D) PARIS protein is upregulated almost 3 fold in PD SN (Figure 6B and 6C) and greater than two-fold in PD striatum (Figure S5B and S5C) compared to control SN and striatum respectively. Accompanying the upregulation of PARIS is the down regulation of PGC-1α and NRF-1 in SN (Figure 6B and 6C) and striatum (Figure S5B and S5C). There is a trend towards redistribution of parkin from the soluble to insoluble fraction in SN (Figure 6B and 6C) and parkin shifts from the soluble to insoluble fraction in PD striatum (Figure S5B and S5C). There is a strong negative correlation between the protein levels of PARIS and PGC-1α (R2=0.5195, p < 0.05) and NRF-1 (R2=0.8015, p < 0.01) in the striatum and between PARIS and PGC-1α (R2=0.6955, p < 0.05) and NRF-1 (R2=0.5979, p < 0.05) in the SN and a positive correlation between PGC-1α and NRF-1 striatum (R2=0.6827, p<0.05) and SN (R2=0.6488, p<0.05) (Figure S5D). These results taken together indicate that PARIS accumulates in the nigrostriatal pathway in PD leading to the down regulation of PGC-1α and the PGC-1α dependent gene, NRF-1.
Down regulation of PGC-1α and NRF-1 in conditional parkin KO mice requires PARIS
In adult conditional parkin KO mice (see Figure 4H–J) four weeks after Lenti-GFPCre mediated parkin deletion there is a greater than two fold upregulation of PARIS (Figure 6D, 6E and Figure 4H–J), comparable to that which occurs in sporadic PD SN (see Figure 4C and 4D) and a concomitant down regulation of PGC-1α and NRF-1 (Figure 6D and 6E). The alteration of PGC-1α and NRF1 results from the reduction of their mRNA levels (Figure 6F). Moreover, we analyzed the mRNA levels of PGC-1α target genes in the ventral midbrain of the Lenti-GFPCre-mediated conditional parkin KO and show a significant reduction of PGC-1α, SOD2, and NRF-1 and no significant alteration in other sampled PGC-1α regulated transcripts (Figure S5E) similar to what occurs in sporadic PD brain. In addition, the levels of other genes containing IRS/PLM in their promoter including PEPCK, G6Pase, IGFBP-1, TAT, and APOC were monitored. Of the genes containing IRS/PLM in their promoters, only PGC-1α is significantly down regulated (Figure S5E). The upregulation of PARIS and subsequent downregulation of PGC-1α and NRF-1 occurs prior to the loss of DA neurons since there is no appreciable loss of DA neurons at 3 months after the Lenti-GFPCre injection (see Figure 6H). Moreover, laser capture microdissection (LCM) was performed prior to the loss of DA neurons to obtain mRNA from TH positive neurons transduced with GFP-Cre from conditional parkin KO mice 4 weeks after the Lenti-GFPCre injection to ascertain whether the reduction of PGC-1α is cell autonomous in DA neurons (Figure S5F and S5G). We find a robust reduction of PGC-1α mRNA in TH-positive DA neurons of conditional parkin KO mice, whereas the levels of PARIS mRNA are unchanged (Figure S5F and S5G). PARIS is only modestly upregulated in germline parkin exon 7 KO mice (also see Figure 4E and 4F). PGC-1α and NRF-1 protein levels in germline parkin exon 7 KO mice are comparable to those of WT mice (Figure S5H and S5I). Thus, germline deletion of parkin apparently leads to compensatory changes that prevent substantial alterations in the levels of PGC-1α and NRF-1.
We further developed the Cre-flox conditional parkin exon 7 KO model by introducing lentiviral shRNA-PARIS along with lenti-GFPCre into parkinFlx/Flx mice to address whether the changes in PGC-1α and NRF-1 are due to PARIS (Figure 6D–F). Co-administration of lentiviral shRNA-PARIS along with Lenti-GFPCre prevents the changes in PGC-1α and NRF-1 protein and mRNA as compared to control lentiviral shRNA-dsRed plus Lenti-GFPCre (Figure 6D–F). These results taken together indicate that PARIS accumulates in the nigrostriatal pathway in PD and in models of parkin inactivation leading to the down regulation of PGC-1α and the PGC-1α dependent gene, NRF-1. These changes in PGC-1α and NRF-1 due to the loss of parkin are cell autonomous and precede the loss of DA neurons and are due to the upregulation of PARIS, since knockdown of PARIS prevents these changes. Moreover, since deletion of parkin from adult mice leads to similar events that occur in sporadic PD, it is likely that the absence and/or inactivation of parkin in PD leads to PARIS upregulation and impairment of PGC-1α signaling.
Neurodegeneration in conditional parkin KO mice requires PARIS
Conditional KO of parkin leads to a significant reduction in tyrosine hydroxylase (TH) positive and Nissl stained DA neurons 10 months after stereotaxic injection of Lenti-GPFCre into the SN of 6–8 week old parkinFlx/Flx mice compared to parkinFlx/Flx mice injected with control Lenti-GFP (Figure 6G and 6H). The loss of DA neurons is progressive, since there is no substantial loss of DA neurons after 3 months (Figure 6H). PARIS is required for the loss of DA neurons in conditional parkin KO mice, since co-administration of lentiviral shRNA-PARIS along with Lenti-GFPCre significantly reduces the loss of DA neurons due to conditional KO of parkin (Figure 6G and 6H). Taken together these results indicate that conditional KO of parkin in adult mice leads to degeneration of DA neurons and the upregulation of PARIS is necessary to contribute to the demise of DA neurons.
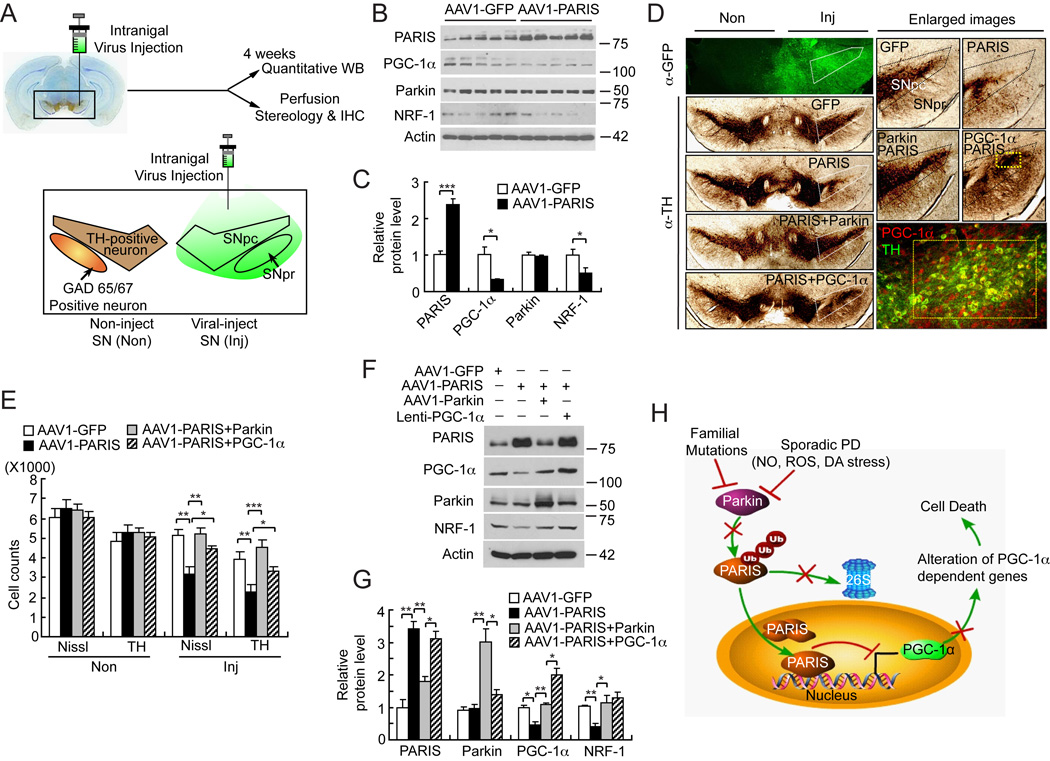
Overexpression of PARIS kills dopamine neurons in vivo: restoration by parkin and PGC-1α
A PARIS overexpression model was developed in which AAV1-PARIS was stereotactically injected into the SN of C57Bl/6 mice and compared to mice injected with control AAV1-GFP virus (Figure 7). Stereotactic intranigral injection of AAV1 effectively transduces the entire SN (Figure 7A). One month after stereotactic injection of the viruses, AAV1 mediated overexpression of PARIS leads to a greater than two-fold upregulation of PARIS levels in the SN of mice (Figure 7B and 7C) and it has no affect on parkin levels (Figure 7B and 7C). Accompanying the increase in PARIS levels is a concomitant downregulation of PGC-1α and NRF-1 protein levels (Figure 7B and 7C).
Figure 7. Introduction of AAV1-Parkin or Lenti-PGC-1α in mice SN protects from AAV1-PARIS-mediated selective dopaminergic neuronal toxicity.
(A) Schematic illustration of intranigral viral injection and transduced brain regions.
(B) Immunoblot analysis of PARIS, PGC-1α, parkin and NRF-1 four-weeks post intranigral injection of AAV1-PARIS, n = 5 per group.
(C) Quantitation of the immunoblots in panel B normalized to β-actin.
(D) TH staining of a representative section of mice injected with AAV1-GFP, AAV1-PARIS ± AAV1-parkin or AAV1-PARIS ± Lenti-PGC-1α. Each panel shows the noninjected side (Non) and contralateral injected side (Inj) and white pentagonal box indicates the SNpc. Enlarged images containing SNpc and SNpr are shown on the right panels. AAV1 encoding GFP was used as transduction control in all injection procedures. Broad regions including SNpc and SNpr were successfully transduced (left top panel). In right bottom panel, yellow rectangle indicates the region that PARIS and lenti- PGC-1α co-transduced. Approximately 30% of the SNpc was transduced with lenti-PGC-1α and this is the region, which is protected from PARIS toxicity, n = 6 per group.
(E) Stereological TH, Nissl-positive neuronal counting, n = 6 per group.
(F) Immunoblot analysis of PARIS, PGC-1α, parkin and NRF-1, n = 3.
(G) Quantitation of the immunoblots in panel F normalized to β-actin.
(H) Parkin-PARIS-PGC-1α pathway as a model in PD. Endogenous PARIS acts to maintain the balance of PGC-1α levels. In PD, parkin is inactivated by diverse insults such as familial mutations, reactive oxygen species (ROS), nitrosative (NO) and dopamine (DA) stress and PARIS accumulates. Accumulated PARIS continuously inhibits PGC-1α transcription leading to reduction in PGC-1α dependent genes. Ultimately this situation results in neurodegeneration in PD. Data = mean ± S.E.M. *p < 0.05, **p < 0.01, ***p < 0.001; ANOVA with the Student-Newman-Keuls post hoc test.
See also Figure S6.
PARIS overexpression leads to a greater than 40% reduction in TH positive and Nissl stained DA neurons positive neurons (Figure 7D and Figure 7E). We do not observe any substantial decrement in GABAergic neurons as assessed by GAD65/67 immunoreactivity via immunohistochemistry (Figure S6A) or via immunoblot (Figure S6B and S6C) in AAV1-PARIS versus AAV1-GFP transduced SN, indicating that PARIS overexpression is selectively detrimental to DA neurons. Co-administration of AAV1-Parkin with AAV1-PARIS prevents the loss of dopamine neurons induced by PARIS overexpression (Figure 7D and 7E). Lentiviral PGC-1α also prevents the loss of DA neurons induced by PARIS overexpression (Figure 7D and 7E). AAV1-mediated overexpression of PARIS was confirmed by immunoblot analysis and reduces PGC-1α and NRF-1 levels by 52% and 60%, respectively. These reductions are restored by co-overexpression of parkin or PGC-1α (Figure 7F and 7G). These results indicate that PARIS overexpression is sufficient to downregulate PGC-1α, NRF-1 and selectively kill DA kills neurons through a PGC-1α dependent mechanism.
DISCUSSION
PARIS (ZNF746) is a novel parkin substrate
PARIS fulfills several criteria for a parkin substrate and in the absence of parkin activity in PD it accumulates making it an attractive pathogenic substrate. In sporadic PD, PARIS only accumulates in the striatum and SN. The selective inactivation of parkin in the striatum and SN in sporadic PD through nitrosative and dopaminergic stress (Chung et al., 2004; LaVoie et al., 2005) and c-Abl phosphorylation (Ko et al., 2010) most likely accounts for the accumulation of PARIS. Similarly, in parkin exon 7 KO mice, PARIS is only upregulated in the striatum and SN. However, PARIS accumulates in the cingulate cortex of AR-PD brains, suggesting that parkin can regulate the expression of PARIS outside the striatum and SN. Whether there is a differential regional or temporal upregulation of PARIS in AR-PD brains is not known as tissue from these areas was unavailable for analysis. The lack of an upregulation in the cortex of parkin exon 7 mice, suggests that there may differential and selective regulatory mechanisms in the cortex versus the striatum and SN.
PARIS is a novel transcriptional repressor of PGC-1α
PARIS is a member of the family of KRAB zinc-finger proteins (KRAB-ZFPs) transcriptional repressors (Looman et al., 2002). Homologues of PARIS exist in simpler organisms suggesting that PARIS function may be evolutionarily conserved. PARIS seems to bind exclusively to IRS/PLM motifs, which provide an important site of regulation of a variety of IRS/PLM responsive proteins (Daitoku et al., 2003; Mounier and Posner, 2006; O'Brien et al., 2001). Our studies reveal that PGC-1α levels are potently and selectively regulated by PARIS, consistent with the observation that PGC-1α transcription is controlled by IRS/PLM motifs (Finck and Kelly, 2006). In PD SN and striatum and conditional parkin KO midbrain, PGC-1α is the only IRS/PLM regulated gene that is downregulated suggesting that PARIS selectively represses PGC-1α expression in PD.
Notably PARIS is a physiological transcriptional repressor of PGC-1α, which directly and endogenously occupies the cis-regulatory elements of PGC-1α. PGC-1α is a transcriptional co-activator that controls the transcription of many genes involved in cellular metabolism including mitochondrial biogenesis and respiration and ROS metabolism (Finck and Kelly, 2006; St-Pierre et al., 2006). The levels of PGC-1α dependent genes are controlled in large part by the nature and composition of the PGC-1α transcriptional co-activator complex (Finck and Kelly, 2006). The PGC-1α dependent gene, NRF-1, appears to be particularly susceptible to the inhibitory effects of PARIS.
It is likely that there is not only a PARIS transcriptional repressor protein complex that plays important regulatory roles in PARIS-mediated transcriptional repression and specificity but the repression of PARIS relies on the genomic context rather than simple IRS motif. In addition, under certain contexts it appears as though PARIS can act a transcriptional activator. Consistent with this notion are our observations that PARIS can bind the PEPCK or G6Pase endogenous promoters via ChIP and it has no effect on G6Pase promoter-reporter activity, but enhances PEPCK promoter-reporter activity. DNA response elements can function as allosteric effectors that determine the transcriptional activity of regulators, explaining that regulators may activate transcription in the context of one gene, yet repress transcription in another (Lefstin and Yamamoto, 1998). Indeed, the IRS motifs in the PGC-1α promoter are organized differently then the IRS motifs of the PEPCK and G6Pase promoters, which may, in part account for the ability of PARIS to act as both as a repressor and activator. Other IRS/PLM responsive genes are likely to be regulated by PARIS and the identification of these genes, their potential regulation by PARIS and their potential role in PD requires additional study.
Dopaminergic Degeneration in Conditional Parkin KO Mice
Germline deletion of parkin using a variety of approaches created parkin KO mice with minimal phenotypes and no loss of DA neurons (Goldberg et al., 2003; Von Coelln et al., 2004). A number of reasons were recently put forward regarding the lack of overt degeneration of DA neurons in genetic animal models of PD including compensatory mechanisms (Dawson et al., 2010). Similar compensatory mechanisms are likely to occur in PD (Bezard et al., 2003; Palop et al., 2006), and compensation accounts, in part, for the age-dependence of PD. The ultimate failure of these compensatory mechanisms contributes to neurodegeneration.
Since, our germline parkin exon 7 KO mice (Von Coelln et al., 2004) do not have degeneration of DA neurons, we generated conditional parkin exon 7 KO mice and deleted parkin from adult mice to avoid developmental compensation. Similar to germline deletion of parkin, embryonic deletion of glial-cell-line-derived neurotrophic factor (GDNF) had no deleterious effects on DA neurons (Pascual et al., 2008). However, conditional KO of GDNF in adult animals using a tamoxifen sensitive Cre-Lox recombination unmasked the “true physiologic action of GDNF” and led to mice with profound degeneration of catecholaminergic neurons (Pascual et al., 2008), indicating that developmental compensation can be overcome by deleting genes in the DA system in adulthood (for review see (Dawson et al., 2010)). Similar to the adult GDNF deleted mice, we observed progressive loss of DA neurons when parkin is deleted in adult mice. PARIS levels increase in parkin conditional KO mice similar to levels in AR-PD brain and in sporadic PD SN. Accompanying the upregulation of PARIS is downregulation of PGC-1α and NRF in conditional KO mice similar to the downregulation in sporadic PD striatum and SN. Consistent with the notion that compensation occurs in the germline parkin KO mice is the observation that PARIS is only modestly elevated and there is no alteration in the levels of PGC-1α and NRF and there is no loss of DA neurons.
Parkin, PARIS, PGC-1α and neurodegeneration
We cannot be certain that PARIS is the sole substrate or mechanism that contributes to DA neuron degeneration following parkin inactivation. Recent studies suggest that PINK1 in a mitochondrial membrane potential-dependent manner signals and recruits parkin through as of yet unclear mechanisms from the cytoplasm to the mitochondria to initiate degradation of damaged mitochondria through autophagy (mitophagy) (Vives-Bauza and Przedborski, 2010). Since PGC-1α and NRF-1 are major transcriptional regulators of mitochondrial biogenesis (Finck and Kelly, 2006; St-Pierre et al., 2006; Ventura-Clapier et al., 2008; Wu et al., 1999), it is conceivable that when parkin decreases the number of mitochondria through mitophagy in response to mitochondrial damage, it is counter balanced by downregulation of PARIS levels as a homeostatic mechanism to increase mitochondrial size and number through regulation of PGC-1α and NRF-1 levels. The contribution of other parkin substrates that are regulated by the UPS and accumulate in AR-PD, sporadic PD and models of parkin inactivation, such as the aminoacyl-tRNA synthetase interacting multifunctional protein type 2 (AIMP2) also known as p38/JTV-1 (Corti et al., 2003; Ko et al., 2005) and far upstream element-binding protein 1 (FBP-1) (Ko et al., 2006), as well as others are not known. Additional substrates and mechanisms will need to be evaluated in future studies to determine their relative contributions to DA neuron degeneration due to parkin inactivation. However, we can be confident that PARIS upregulation is necessary and sufficient to cause DA neuron degeneration in models of parkin inactivation and that maintaining PGC-1α function is beneficial. Indeed PGC-1α has been implicated in other models of PD (St-Pierre et al., 2006). Moreover, PGC-1α-responsive genes are underexpressed in microdissected dopaminergic neurons of PD suggesting that the alteration of PGC-1α might be a cause of PD pathogenesis, not the consequence (Zheng et al., 2010).
Herein we hypothesize a model that accumulated PARIS in the setting of parkin’s inactivation represses PGC-1α expression leading to neurodegeneration. Consistent with this hypothesis are the observations that the reduction in PGC-1α levels and neurodegeneration are substantially reduced by knocking down PARIS levels in the setting of parkin inactivation. Since PARIS is likely to regulate other genes, the Parkin-PARIS- PGC-1α is one potential contributory mechanism to PD pathogenesis. This parkin-PARIS-PGC-1α neurodegenerative pathway ultimately results in the selective vulnerability of dopamine neurons and accounts, in part, for the neurodegeneration in PD (Figure 7H). Further study is required to determine the full implications of the parkin-PARIS-PGC-1α neurodegenerative pathway in PD. Nonetheless, our results suggest that parkin inactivation acting through PARIS and downregulation of PGC-1α contributes to the pathogenesis of PD.
EXPERIMENTAL PROCEDURES
Yeast Two-Hybrid Screening
Saccharomyces cerevisiae MaV203 was transformed with pDBLeu-R1-parkin, and 3 × 106 stable transformants were further transformed with 15 µg of pPC86 human brain cDNA library (Life Tech/Gibco). Transformants were selected and confirmed according to the manufacturer's instructions as previously described (Zhang et al., 2000).
Antibodies
Polyclonal PARIS antibodies were generated as described in supplemental information. Primary antibodies used include the following: goat anti-PGC-1α (K-15, Santa Cruz Biotechnology), mouse anti-PGC-1α (4C1.3, Calbiochem), goat anti-NRF1 (A-19, Santa Cruz Biotechnology), rabbit anti-NRF1 (ab34682, Abcam), mouse anti-parkin (Park8, Cell Signaling), rabbit anti-TH (Novus Biologicals), rabbit anti-glutamate decarboxylase (GAD) 65&67 (Millipore), rabbit anti-GFP (ab661, Abcam), mouse anti-GFP (ab1218, Abcam); Secondary antibodies used include Biotin-SP-conjugated goat anti-rabbit (Jackson ImmunoResearch lab), donkey anti-goat-Cy3, donkey anti-rabbit-Cy2/Cy3, donkey anti-mouse-Cy2 for immunostaining.
Plasmid Constructions
Full-length parkin, and deletion mutants, Q311X, R42P, R275W, G430D and C431F parkin, HA-ubiquitin, PARIS, PARIS deletion and point mutations, GST-PARIS, ZNF398 vector were constructed as described in supplemental information. Construct integrity was verified by sequencing. Lentiviral pLV-PGC-1α plasmid was kindly provided by Dr. Dimitri Krainc (Massachusetts General Hospital, Harvard Medical School, Charlestown, USA), MYC-tagged XIAP was generously given from Dr. Kenny K. K. Chung (Hong Kong University of Science and Technology, Clear Water Bay, Hong Kong),
Cell culture and Transfection
Human neuroblastoma SH-SY5Y cells (ATCC, Manassas, VA) were grown in DMEM containing 10% FBS and antibiotics in a humidified 5% CO2/95% air atmosphere at 37°C. For transient transfection, cells were transfected with indicated amounts of target vector using Lipofectamine Plus (Invitrogen), according to manufacturer’s instructions. For co-immunoprecipitation from cell cultures, SH-SY5Y cells were transfected with 2 µg of each plasmid, unless otherwise indicated in supplemental information. For the ubiquitination assay, SH-SY5Y cells were transiently transfected with 2 µg of pRK5-Myc-tagged parkin, Myc-tagged parkin (C431F, G430D, R275W, Q311X) pCMV-FLAG-PARIS, and 2 µg of pMT123-HA-ubiquitin plasmids for 48 h. For the luciferase assay SH-SY5Y cells were transiently transfected with pCMV-empty vector or pCMV-FLAG-PARIS with either wild type or Q311X parkin, or pGL3-Basic, pGL3-PGC-1α promoter-Luciferase, or pGL3-PGC-1α promoter deletion mutant (a gift from Akyoshi Fukamizu, University of Tsukuba, Japan) (Daitoku et al., 2003) for firefly Luciferase assay and 0.1 µg pRL-TK vector (Promega) for Renilla luciferase control.
Immunocytochemistry and Immunblot Analysis
Immunocytochemistry and immunoblot analysis was performed as described in the supplemental information.
In vitro Interaction and Ubiquitination Assays
GST-PARIS and His-Parkin were used in in vitro interaction and ubiquitination assays as described in the supplemental information.
CAST, EMSA, ChIP, qRT-PCR Assays
We followed a previous published protocol with modification for CAST ((Cyclic Amplification and Selection of Targets) (Voz et al., 2000) as described in the supplemental information. GST, GST-PARIS, GST-C571A–PARIS were used for electrophoretic mobility shift assays (EMSA) as described in the supplemental information. Chromatin immunoprecipitation was carried out according to the manufacturer’s instruction as described in the supplemental information. Primers used for real-time pRT-PCR are listed in Table S4.
Conditional parkin knockout
To generate Cre-flox conditional model of parkin knock out, a lentiviral vector expressing GFP fused Cre recombinase (Lenti-GFPCre) was stereotaxically introduced into exon 7 floxed parkin mice (parkinFlx/Flx) using the coordinates in supplemental information. Furthermore lentiviral shRNA-PARIS was co-administrated along with Lenti-GFPCre to demonstrate whether the changes in PGC-1α and NRF-1 are due to PARIS.
Statistics
Quantitative data is presented as the mean ± S.E.M. Statistical significance was either assessed via an unpaired two-tailed Student’s t-test or an ANOVA test with Student-Newman-Keuls post-hoc analysis. Assessments were considered significant with a p < 0.05,.
Supplementary Material
Acknowledgements
This work was supported by NS38377, NS048206, NS051764 and the Bachmann Strauss Dystonia and Parkinson’s Disease Foundation. Y.L. is supported by Samsung Scholarship Foundation. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases. The authors thank Drs. Xin Guo, Sathya Siram, members of the Dawson laboratory for assistance and contributions to the manuscript. We also thank Drs. Alex Kolodkin, David Ginty and Solomon H. Snyder for helpful suggestions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental data includes supplemental experiment procedures, 5 tables and 6 figures.
The authors declare that they have no competing financial interests.
REFERENCES
- Bezard E, Gross CE, Brotchie JM. Presymptomatic compensation in Parkinson's disease is not dopamine-mediated. Trends Neurosci. 2003;26:215–221. doi: 10.1016/S0166-2236(03)00038-9. [DOI] [PubMed] [Google Scholar]
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Corti O, Hampe C, Koutnikova H, Darios F, Jacquier S, Prigent A, Robinson JC, Pradier L, Ruberg M, Mirande M, et al. The p38 subunit of the aminoacyl-tRNA synthetase complex is a Parkin substrate: linking protein biosynthesis and neurodegeneration. Hum Mol Genet. 2003;12:1427–1437. doi: 10.1093/hmg/ddg159. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–649. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- Dawson TM, Dawson VL. The role of parkin in familial and sporadic Parkinson's disease. Mov Disord. 2010;25 Suppl 1:S32–S39. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TM, Ko HS, Dawson VL. Genetic animal models of Parkinson's disease. Neuron. 2010;66:646–661. doi: 10.1016/j.neuron.2010.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WD, Wright WE. Cyclic amplification and selection of targets for multicomponent complexes: myogenin interacts with factors recognizing binding sites for basic helix-loop-helix, nuclear factor 1, myocyte-specific enhancer-binding factor 2, and COMP1 factor. Proc Natl Acad Sci U S A. 1992;89:9484–9488. doi: 10.1073/pnas.89.20.9484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser T. Update on the genetics of Parkinson's disease. Mov Disord. 2007;22:S343–S350. doi: 10.1002/mds.21676. [DOI] [PubMed] [Google Scholar]
- Goldberg MS, Fleming SM, Palacino JJ, Cepeda C, Lam HA, Bhatnagar A, Meloni EG, Wu N, Ackerson LC, Klapstein GJ, et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J Biol Chem. 2003;278:43628–43635. doi: 10.1074/jbc.M308947200. [DOI] [PubMed] [Google Scholar]
- Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, Takahashi R. CHIP is associated with Parkin, a gene responsible for familial Parkinson's disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/s1097-2765(02)00583-x. [DOI] [PubMed] [Google Scholar]
- Ko HS, Kim SW, Sriram SR, Dawson VL, Dawson TM. Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. J Biol Chem. 2006;281:16193–16196. doi: 10.1074/jbc.C600041200. [DOI] [PubMed] [Google Scholar]
- Ko HS, Lee Y, Shin JH, Karuppagounder SS, Gadad BS, Koleske AJ, Pletnikova O, Troncoso JC, Dawson VL, Dawson TM. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin's ubiquitination and protective function. Proc Natl Acad Sci U S A. 2010;107:16691–16696. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HS, von Coelln R, Sriram SR, Kim SW, Chung KK, Pletnikova O, Troncoso J, Johnson B, Saffary R, Goh EL, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25:7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11:1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Looman C, Abrink M, Mark C, Hellman L. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol Biol Evol. 2002;19:2118–2130. doi: 10.1093/oxfordjournals.molbev.a004037. [DOI] [PubMed] [Google Scholar]
- Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, Shanks JR, Reyes-Turcu FE, Wilkinson KD, Marmorstein R. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–11049. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier C, Posner BI. Transcriptional regulation by insulin: from the receptor to the gene. Can J Physiol Pharmacol. 2006;84:713–724. doi: 10.1139/y05-152. [DOI] [PubMed] [Google Scholar]
- O'Brien RM, Streeper RS, Ayala JE, Stadelmaier BT, Hornbuckle LA. Insulin-regulated gene expression. Biochem Soc Trans. 2001;29:552–558. doi: 10.1042/bst0290552. [DOI] [PubMed] [Google Scholar]
- Palop JJ, Chin J, Mucke L. A network dysfunction perspective on neurodegenerative diseases. Nature. 2006;443:768–773. doi: 10.1038/nature05289. [DOI] [PubMed] [Google Scholar]
- Pascual A, Hidalgo-Figueroa M, Piruat JI, Pintado CO, Gomez-Diaz R, Lopez-Barneo J. Absolute requirement of GDNF for adult catecholaminergic neuron survival. Nat Neurosci. 2008;11:755–761. doi: 10.1038/nn.2136. [DOI] [PubMed] [Google Scholar]
- Savitt JM, Dawson VL, Dawson TM. Diagnosis and treatment of Parkinson disease: molecules to medicine. J Clin Invest. 2006;116:1744–1754. doi: 10.1172/JCI29178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields JM, Yang VW. Identification of the DNA sequence that interacts with the gut-enriched Kruppel-like factor. Nucleic Acids Res. 1998;26:796–802. doi: 10.1093/nar/26.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, Shimizu N, Iwai K, Chiba T, Tanaka K, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- Sriram SR, Li X, Ko HS, Chung KK, Wong E, Lim KL, Dawson VL, Dawson TM. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum Mol Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Suzuki T, Hattori N, Mizuno Y. Ubiquitin, proteasome and parkin. Biochim Biophys Acta. 2004;1695:235–247. doi: 10.1016/j.bbamcr.2004.09.026. [DOI] [PubMed] [Google Scholar]
- Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79:208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- Vives-Bauza C, Przedborski S. PINK1 points Parkin to mitochondria. Autophagy. 2010;6 doi: 10.4161/auto.6.5.12068. [DOI] [PubMed] [Google Scholar]
- Von Coelln R, Thomas B, Savitt JM, Lim KL, Sasaki M, Hess EJ, Dawson VL, Dawson TM. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc Natl Acad Sci U S A. 2004;101:10744–10749. doi: 10.1073/pnas.0401297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voz ML, Agten NS, Van de Ven WJ, Kas K. PLAG1, the main translocation target in pleomorphic adenoma of the salivary glands, is a positive regulator of IGF-II. Cancer Res. 2000;60:106–113. [PubMed] [Google Scholar]
- Winklhofer KF, Henn IH, Kay-Jackson PC, Heller U, Tatzelt J. Inactivation of parkin by oxidative stress and C-terminal truncations: a protective role of molecular chaperones. J Biol Chem. 2003;278:47199–47208. doi: 10.1074/jbc.M306769200. [DOI] [PubMed] [Google Scholar]
- Witzgall R, O'Leary E, Leaf A, Onaldi D, Bonventre JV. The Kruppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci U S A. 1994;91:4514–4518. doi: 10.1073/pnas.91.10.4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97:13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Liao Z, Locascio JJ, Lesniak KA, Roderick SS, Watt ML, Eklund AC, Zhang-James Y, Kim PD, Hauser MA, et al. PGC-1alpha, a potential therapeutic target for early intervention in Parkinson's disease. Sci Transl Med. 2010;2 doi: 10.1126/scitranslmed.3001059. 52ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.