Abstract
Exocytosis involves membrane fusion between granules and the plasma membrane. Nitric oxide (NO) inhibits exocytosis by chemically modifying N-ethylmaleimide-sensitive factor (NSF), a key component of the exocytic machinery. However, cells recover the ability to release messenger molecules within hours of exposure to NO through unknown mechanisms. We now identify thioredoxin (TRX1) as a denitrosylase that reverses NO inhibition of exocytosis. Endogenously synthesized NO increases S-nitrosylated NSF levels, but S-nitrosylated NSF levels decrease within 3 h after exposure to NO. We found that NO increases the interaction between TRX1 and NSF, and endogenous TRX1 removes NO from S-nitrosylated NSF. Knockdown of TRX1 increases the level of S-nitrosylated NSF, prolongs the inhibition of exocytosis, and suppresses leukocyte adhesion. Taken together, these data show that TRX1 promotes exocytosis by denitrosylating NSF. Our findings suggest that TRX1 might regulate exocytosis in a variety of physiological settings, such as vascular inflammation, thrombosis, and insulin release.
Keywords: Endothelium, Exocytosis, Membrane Fusion, Nitric Oxide, von Willebrand Factor
Introduction
NO is a potent anti-inflammatory mediator. NO synthase (NOS)2 inhibitors increase leukocyte trafficking from the blood into inflamed tissues. Conversely, endogenous NO and exogenous NO donors decrease leukocyte extravasation (1–5). Leukocyte trafficking involves five discrete stages (1): rolling (2), activation (3), firm adhesion (4), transmigration (5), and migration (6). NO inhibits leukocyte rolling in part by decreasing expression of P-selectin on the endothelial surface. NO can also inhibit other stages of leukocyte trafficking by decreasing superoxide, integrin expression, and transcellular pore formation (7). Thus NO inhibits vascular inflammation through a variety of mechanisms.
NO inhibits the first stage of leukocyte trafficking by blocking endothelial exocytosis (8). Endothelial granules contain proinflammatory factors such as P-selectin and prothrombotic substances like von Willebrand Factor (VWF) (9). Secretion of the granule contents is driven by a set of proteins inside endothelial cells, including N-ethylmaleimide-sensitive factor (NSF), soluble NSF receptor attachment proteins such as syntaxin 4, and small G-proteins (10–15). NSF is a member of the ATPases associated with diverse cellular activities superfamily that hydrolyzes ATP, disassembles the soluble NSF receptor attachment protein complex, and forms homohexamers (16). NSF is a necessary component of vesicle trafficking.
We previously showed that NO inhibits exocytosis by targeting NSF (17, 18). NO forms a chemical bond with NSF on cysteine residues 21, 91, and 264 (17). This S-nitrosylation of NSF diminishes its ability to disassemble the soluble NSF receptor attachment protein complex, a critical step in exocytosis and the cycle of vesicle trafficking. As a consequence, NO inhibits exocytosis both in vitro and in vivo. However, this study raised several questions. Does NO permanently inhibit NSF and vesicle trafficking? If not, how is NO removed from NSF to restore vesicle trafficking? Which intracellular proteins are capable of reducing NSF?
One denitrosylase candidate is thioredoxin (19). Thioredoxin is a reductase that regulates cell proliferation, apoptosis, and signaling (20, 21). Thioredoxin has three isoforms, of which thioredoxin 1 (TRX1, also referred to as TXN or TRX) is expressed ubiquitously in the cytosol. TRX1 reduces oxidized cysteine residues on target proteins through an interaction with the TRX1 active site CXXC. Several intriguing studies have shown that TRX1 can also remove NO from nitrosothiol groups, both from purified proteins and from intracellular proteins (22–24). We hypothesized that TRX1 regulates exocytosis by removing NO from NSF. We now show that TRX1 interacts with NSF, TRX1 removes NO from NSF, and TRX restores the capacity of endothelial cells to undergo exocytosis.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Histamine and streptavidin-agarose were purchased from Sigma. S-nitroso-N-acetyl-d,l-penicillamine (SNAP), l-nitroarginine methyl ester (l-NAME), and the S-nitrosylation protein detection assay kit were purchased from Cayman Chemical (Ann Arbor, MI). The VWF ELISA kit was purchased from American Diagnostica (Stamford, CT). Lipofectin, Lipofectamine RNAiMAX, Stealth Select RNAi (TXN-HSS144377), Stealth RNAi negative control duplexes, Calcein-AM, and Opti-MEM I media were purchased from Invitrogen. A mammalian c-Myc tag IP/Co-IP kit was purchased from Pierce. Complete Mini EDTA-free protease inhibitor mixture was purchased from Roche. Anti-NSF mouse IgG was purchased from BD Biosciences. Anti-TRX1 goat IgG was from R&D Systems, Inc. (Minneapolis, MN). Anti-GAPDH mouse IgG, anti-c-Myc mouse IgG, protein A/G PLUS-agarose, and HRP-conjugated secondary antibodies to mouse IgG and to goat IgG were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Pooled human umbilical vein endothelial cells (HUVEC) and endothelial growth medium-2 medium were obtained from Lonza-Clonetics (Walkersville, MD).
Cell Culture and Analysis of VWF Release
HUVEC were grown in EGM-2 medium supplemented with 2% serum and growth factors in a kit. The endothelial exocytosis assay was performed with an ELISA as described previously (17). Briefly, HUVEC were placed in Opti-MEM I medium, pretreated with SNAP (0–20 μm for 6 h) or l-NAME (0–500 μm for 4 h), washed, and stimulated with 20 μm histamine. In some experiments, HUVEC pretreated with SNAP were placed in fresh Opti-MEM I medium without NO donors, incubated for up to 3 h, washed, and stimulated with 20 μm histamine. Then the amount of VWF released into the medium was measured by an ELISA.
Leukocyte Adhesion to HUVEC
HL-60 cells were loaded with 5 μm Calcein-AM, washed with Opti-MEM I medium, and then added to HUVEC. The cells were co-cultured at 4 °C for 15 min and washed with Opti-MEM I medium. The number of adherent leukocytes was evaluated with a fluorescence microscope and a fluorescence microplate reader.
Biotin Switch Assay for the Detection of S-Nitrosylated Proteins
S-nitrosylated proteins were analyzed by the biotin switch method as described previously (25). Briefly, protein-free thiols were blocked, and then S-nitrosothiols were reduced to yield free thiols, which were then covalently labeled with maleimide-biotin (Cayman Chemical). The biotinylated proteins were resuspended with radioimmune precipitation assay buffer (137 mm NaCl, 25 mm Tris, 3 mm KCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.2% sodium dodecyl sulfate), pulled down by streptavidin-agarose, and detected by immunoblotting. Densitometric analysis was performed using the public-domain Java image processing program ImageJ.
Knockdown of TRX1 in HUVEC and in HeLa Cells
HUVEC and HeLa cells were transfected with either 5 nm Stealth RNAi TXN-HSS144377 or 5 nm Stealth RNAi negative control according to the guidelines for transfection with Lipofectamine RNAiMAX. The transfected cells were incubated for 48 h and then treated with SNAP (20–500 μm) or l-NAME (500 μm).
Co-immunoprecipitation of Myc-tagged TRX1 and Its Interacting Proteins
HeLa cells were transfected with either Myc-tagged TRX1 (wild-type) or Myc-tagged TRX1 (C35S mutant) according to the guidelines for transfection with Lipofectin. The transfected cells were incubated for 24 h and then treated with SNAP (5–500 μm). The cells were washed with ice-cold Tris buffered saline and lysed with radioimmune precipitation assay buffer (137 mm NaCl, 25 mm Tris, 3 mm KCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, protease inhibitor mixture). An immunoprecipitation (IP) assay was performed according to the guidelines for IP with a mammalian c-Myc tag IP/Co-IP kit.
Co-immunoprecipitation of NSF and TRX1
HUVEC were treated with SNAP (500 μm) or l-NAME (500 μm) for 4 h. The cells were washed with ice-cold Tris-buffered saline and lysed with radioimmune precipitation assay buffer. The IP assay was performed with anti-NSF mouse IgG and protein A/G PLUS-agarose.
Statistical Analyses
Data were analyzed by Student's t test for comparison of two groups. Statistical significance is defined as p < 0.05.
RESULTS
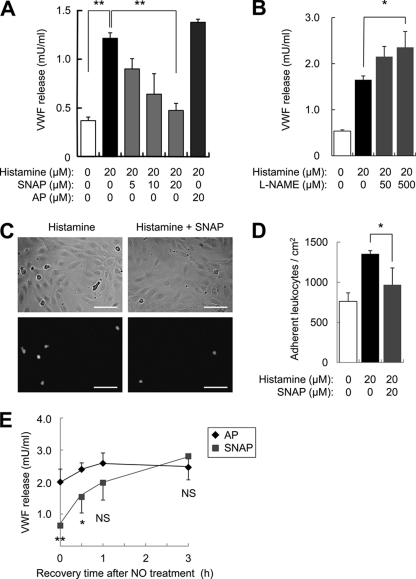
We first explored the effect of NO upon exocytosis. We pretreated HUVEC with the NO donor SNAP or its control acetyl-penicillamine (AP), washed the cells, and then added histamine to trigger exocytosis. Histamine activates exocytosis, and SNAP inhibits exocytosis, as measured by endothelial release of VWF (Fig. 1A). Endogenous NO also inhibits exocytosis because the NOS inhibitor l-NAME increases histamine-induced VWF release (Fig. 1B). SNAP also inhibits endothelial exocytosis as measured by the leukocyte adhesion assay, which depends upon endothelial display of P-selectin (Fig. 1, C and D). Next, we determined how long NO inhibition persists. We pretreated HUVEC with SNAP, washed the cells, and allowed the cells to recover from NO for various times before stimulating exocytosis. SNAP inhibits exocytosis, and this inhibition disappears over 1 h (Fig. 1E). Taken together, these data show that NO inhibits exocytosis and that NO inhibition of exocytosis is reversible.
FIGURE 1.
Nitric oxide inhibition of exocytosis is reversible. A, NO inhibition of VWF release. HUVEC were pretreated with the NO donor SNAP or its control AP for 6 h, stimulated with histamine, and the amount of VWF released into the media over 45 min was measured by an ELISA (n = 3, mean ± S.D.). The NO donor SNAP decreases endothelial exocytosis. B, endogenous NO inhibits VWF release. HUVEC were pretreated with the NOS inhibitor l-NAME for 4 h, stimulated with histamine, and the VWF released into the media was measured as above (n = 3, mean ± S.D.). The NOS inhibitor l-NAME increases endothelial exocytosis. C, NO inhibition of leukocyte adhesion. HUVEC were pretreated with SNAP for 4 h, stimulated with histamine for 20 min, then co-cultured with calcein-labeled HL-60 cells for 15 min. The upper panels show bright field images of HUVEC and HL-60, and the lower panels show calcein-labeled HL-60 co-cultured with HUVEC. Scale bars = 100 μm. D, quantitation of NO inhibition of leukocyte adhesion in C (n = 3, mean ± S.D.). The NO donor SNAP decreases leukocyte adhesion to endothelial cells. E, NO inhibition of exocytosis is reversible. HUVEC were pretreated with SNAP (20 μm) for 6 h, and then after recovery for 0, 0.5, 1, or 3 h the cells were stimulated with histamine (20 μm). Exocytosis was measured as above (n = 4, mean ± S.D.). The effect of NO upon exocytosis diminishes over time. *, p < 0.05; **, p < 0.01; NS, not significant.
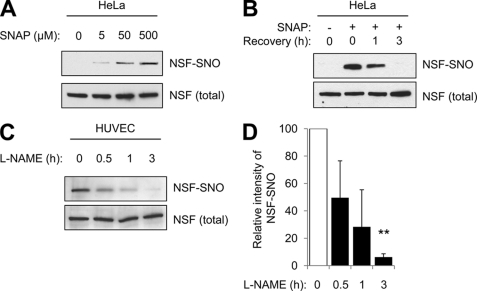
We had previously shown that NO inhibits exocytosis in part by chemically modifying a component of the exocytic machinery, NSF. Because NO inhibition of exocytosis is reversible, we examined whether or not NO modification of NSF is reversible. We treated HeLa cells with SNAP, washed the cells, and permitted them to recover for 0–3 h. We then harvested cell lysates and measured S-nitrosylated NSF (NSF-SNO) by the biotin-switch assay. SNAP increases NSF-SNO (Fig. 2A). After the NO donor is removed, levels of NSF-SNO decrease (Fig. 2B). SNAP also increases S-nitrosylation of other proteins in HeLa cells such as GAPDH, and after the NO donor is removed, GAPDH-SNO levels decrease (data not shown).
FIGURE 2.
S-nitrosylation of NSF is reversible. A, S-nitrosylation of NSF in HeLa cells. HeLa cells were treated with the NO donor SNAP for 6 h. Then, cell lysates were harvested and levels of NSF-SNO were measured by the biotin switch assay. The NO donor SNAP increases NSF-SNO in a dose-dependent manner. B, S-nitrosylation of NSF decreases over time in HeLa cells. HeLa cells were treated with SNAP for 6 h and then washed. After 0–3 h of recovery, cell lysates were harvested, and levels of NSF-SNO were measured by the biotin switch assay. After exposure to an NO donor, the amount of S-nitrosylated NSF decreases over time. C, endogenous S-nitrosylation of NSF decreases over time in endothelial cells. HUVEC were treated with the NOS inhibitor l-NAME for 0–3 h, and then levels of NSF-SNO were measured as above. After NO synthase is inhibited, the amount of endogenous S-nitrosylated NSF decreases over time. D, quantitation of C (mean ± S.D). **, p < 0.01.
We also explored the effect of endogenous NO upon NSF. We treated HUVEC with the NOS inhibitor l-NAME for increasing amounts of time and measured NSF-SNO levels. At baseline, endothelial cells contain NSF-SNO, and after endogenous NO production is limited, NSF-SNO levels gradually decline (Fig. 2, C and D). Taken together, these data show that NO modification of NSF is reversible.
We have previously shown that superoxide or NO can modify specific cysteine residues of NSF. Because others have shown that antioxidant enzymes such as TRX1 can reduce oxidized cysteine residues, we explored the effect of TRX1 upon NSF. We began by searching for an interaction between NSF and TRX1.
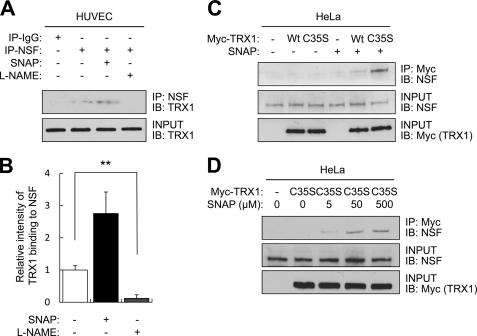
To see if endogenous NSF and TRX1 interact, we immunoprecipitated HUVEC lysates with antibody to NSF and immunoblotted the precipitants with antibody to TRX1. Endogenous NSF and endogenous TRX1 interact (Figs. 3, A and B). We tested the effect of endogenous NO upon this interaction by pretreating endothelial cells with l-NAME and then repeating the immunoprecipitation. NSF and TRX1 fail to interact in the absence of NO (Fig. 3, A and B).
FIGURE 3.
Thioredoxin interacts with NSF in the presence of NO. A, interaction between endogenous NSF and endogenous TRX1. HUVEC were treated with SNAP (500 μm) or l-NAME (500 μm) for 4 h. Cell lysates were immunoprecipitated with antibody to NSF and immunoblotted for TRX1. NSF interacts with TRX1 in an NO-dependent manner. B, quantitation of A (mean ± S.D). **, p < 0.01. C, stable interaction between NSF and TRTX1 depends upon TRX1 residue Cys-35. HeLa cells were transfected with a vector expressing wild-type Myc-TRX1(WT) or mutant Myc-TRX1(C35S) and treated with SNAP for 4 h. Cell lysates were immunoprecipitated with antibody to Myc and immunoblotted for NSF. TRX1 interacts with NSF, through the CXXC disulfide reductase domain of TRX1. D, HeLa cells were transfected with a vector expressing mutant Myc-TRX1(C35S) and treated with increasing amounts of SNAP (5–500 μm) for 4 h. The interaction between NSF and TRX1 was analyzed as above. SNAP increases the interaction in a dose-dependent manner.
We next transfected HeLa cells with a vector expressing myc-tagged wild-type TRX1, designated myc-TRX1 (WT), or with a vector expressing myc-tagged C35S mutant TRX1, designated myc-TRX1 (C35S). We chose to construct this particular mutant, TRX1 (C35S), so that TRX1 would form a more stable mixed disulfide intermediates with potential partners (26, 27). After the cells were transfected with TRX1 expression vectors, we added SNAP, immunoprecipitated with antibody to myc, and immunoblotted for endogenous NSF. TRX1 does not interact with NSF in untreated cells (Fig. 3C, left lanes). However, the NO donor SNAP increases the interaction of TRX1 with NSF (Fig. 3C, right lanes, and D). Furthermore, the mutant TRX1 interacts more with NSF than the wild-type TRX1 (Fig. 3C). Taken together, these data show that TRX1 interacts with NSF, possibly through the CXXC disulfide reductase domain of TRX1 (Cys-32 and Cys-35) and that NO influences this interaction.
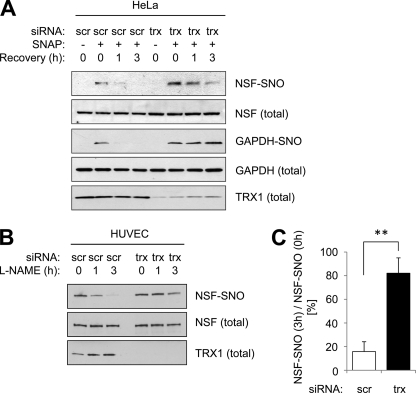
We hypothesized that TRX1 can remove NO from NSF-SNO. This hypothesis is supported by prior work showing that TRX1 can remove NO from purified proteins (22, 28) and from proteins in cancer cell lines (24). To test this hypothesis, we knocked down HeLa expression of endogenous TRX1 by siRNA techniques, then treated the cells with the NO donor SNAP, and then washed the cells and permitted them to recover. The biotin-switch assay reveals that decreasing TRX1 slows the decline in NSF-SNO levels (Fig. 4A). TRX1 also controls levels of GAPDH-SNO (Fig. 4A). We also determined the effect of TRX1 upon endogenous NSF-SNO levels in endothelial cells. Knocking down TRX1 expression prolongs the expression of NSF-NO (Fig. 4, B and C). Taken together, our results suggest that TRX1 directly or indirectly plays a role in the removal of NO from NSF-NO.
FIGURE 4.
Thioredoxin denitrosylates NSF. A, TRX1 denitrosylates NSF in HeLa cells. HeLa cells were transfected with siRNA control or siRNA to TRX1. Cells were treated with the NO donor SNAP (20 μm) for 6 h, washed, and harvested during 0–3 h recovery. Levels of NSF-SNO and GAPDH-SNO were measured as above. Knockdown of TRX1 increases levels of NSF-SNO and GAPDH-SNO. B, TRX1 denitrosylates NSF in endothelial cells. HUVEC were transfected with siRNA control or siRNA to TRX1. Cells were treated with the NOS inhibitor l-NAME for 0–3 h. Levels of NSF-SNO were measured as above. Knockdown of TRX1 increases levels of NSF-SNO. C, quantitation of B (mean ± S.D.). **, p < 0.01.
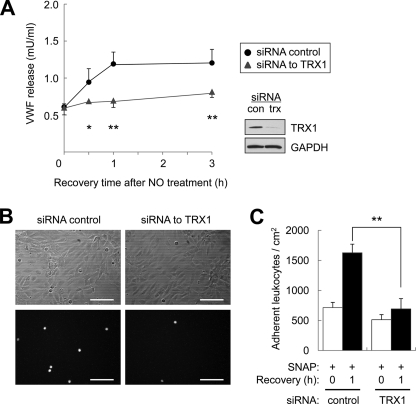
Finally, we determined the biological effect of TRX1 upon cell interactions. NSF is a critical component of the exocytic machinery in endothelial and other secretory cells, and we had previously shown that NO inhibits NSF and its role in exocytosis. We postulated that TRX1 would accelerate the recovery of exocytosis in cells exposed to NO. Accordingly, we knocked down TRX1 in endothelial cells, then treated the cells with the NO donor SNAP, washed the cells, and permitted them to recover. At various times during recovery, we stimulated exocytosis with histamine. Cells recover the ability to undergo exocytosis within 1 h after exposure to NO, but when TRX1 levels are decreased, recovery of exocytosis is prolonged (Fig. 5A). To explore the consequence of TRX1 upon cell interactions, we knocked down TRX1 in endothelial cells, treated them with an NO donor, allowed the cells to recover from NO treatment, and measured leukocyte adhesion. Control cells recover from NO and support leukocyte adhesion (Fig. 5B, left panel and C, left panel). However, after TRX1 silencing, cells recover from NO more slowly and have less leukocyte adhesion (Fig. 5B, right panel and C, right panel). Taken together, these data show that TRX1 speeds the recovery of exocytosis after NO treatment, enhancing leukocyte adhesion.
FIGURE 5.
Thioredoxin restores exocytosis after NO inhibition. A, thioredoxin restores VWF release. HUVEC were transfected with siRNA control or siRNA to TRX1 (insert shows immunoblot of TRX1), pretreated with the NO donor SNAP (20 μm) for 6 h, and washed. After 0–3 h of recovery, HUVEC were stimulated with histamine (20 μm) for 45 min, and exocytosis was measured as above (n = 4, mean ± S.D.). Knockdown of TRX1 delays the recovery of exocytosis from NO inhibition. B, thioredoxin restores leukocyte adhesion after NO inhibition. HUVEC were transfected with siRNA control or siRNA to TRX1, pretreated with SNAP (20 μm) for 4 h, and washed. After 0–1 h of recovery, HUVEC were stimulated with histamine (20 μm) for 20 min, and leukocyte adhesion was evaluated as above. The upper panels show bright field images of HUVEC and HL-60, and the lower panels show fluorescent images of calcein-labeled HL-60. Scale bars = 100 μm. C, quantification of B. Knockdown of TRX1 delays the recovery of leukocyte adhesion from NO inhibition. (n = 3, mean ± S.D.). *, p < 0.05; **, p < 0.01.
DISCUSSION
The major finding of our study is that TRX1 removes NO from NSF, restoring the ability of cells to undergo exocytosis. These data provide an explanation for why NO inhibition of exocytosis is temporary. TRX1 may thus serve as an endogenous modulator of exocytosis.
Our data extend the findings of other laboratories that show that TRX1 regulates S-nitrosylation of other proteins (29). Nikitovic and Holmgren (22) showed that TRX or the TRX reductase TR cleave S-nitroso-glutathione (GSNO) in solution in vitro, generating GSH and NO. Mitchell and Marletta (28) then demonstrated that TRX can transfer NO from GSNO to caspase 3 in solution in vitro. Next, Stoyanovsky and colleagues (23) found that TRX removes NO from GSNO in solution and also in cells. Stamler and colleagues (24) then showed that TRX1 and TRX2 serve as endogenous denitrosylases. In particular, Stamler found that TRX isoforms can interact with S-nitrosylated caspase 3, denitrosylate caspase 3, and increase apoptosis. Our studies build on these prior works in several ways. First, we show that endogenous TRX1 regulates the NO content of an endogenous protein. Second, we study the role of TRX1 in modulating a signal from endogenous NO, as well as exogenous NO. Third, we show that NO modulates the interaction of TRX1 with a target protein. Finally, we also find that TRX1 regulation of an S-nitrosylated target protein has functional consequences, in this case the modulation of exocytosis.
This study suggests an explanation as to why an inhibitor of vesicle trafficking does not damage target cells: NO inhibition of exocytosis is reversible. We previously found that NO inhibits exocytosis, in part by chemically modifying and inhibiting NSF (17). Although vesicle trafficking is critical to the functioning of all cells, NO treatment in low doses is not cytotoxic. Our current study shows that cells recover from NO inhibition of exocytosis within 1–3 h. This corresponds to a decrease of NSF-SNO within 1–3 h. However, if cellular levels of TRX1 are decreased, NSF-SNO is detected after 3 h of NO treatment (Fig. 4), and exocytosis is still inhibited 3 h after NO exposure (Fig. 4D). Thus TRX1 plays a role in protecting the exocytic machinery from permanent inactivation by NO.
Are other proteins capable of removing NO from protein thiol groups (19)? Glutathione peroxidase can liberate NO from GSNO in vitro (30). In the presence of transition metals, gamma-glutamyl transpeptidase catalyzes the decomposition of GSNO. Xanthine oxidase has been reported to reduce GSNOP and Cys-NO (31). Alcohol dehydrogenase class III isoenzyme uses NAD/NADPH to degrade GSNO in vitro (32, 33) and in vivo (34). Superoxide dismutase has been reported to function as a GSNO denitrosylase, although its activity is weak (35). In addition, enzymes have been reported to remove NO from heme. For example, bacterial flavohemoglobin catalyzes the reaction of NO bound to heme with oxygen to produce nitrate (36). Thus, a distinct set of enzymes are capable of metabolizing nitrosothiols in vitro and in vivo, and some of these enzymes might also be capable of regulating NSF and exocytosis.
Our study implies that TRX1 is capable of promoting vascular inflammation. The conventional view is that TRX1 protects the vasculature from oxidant stress and decreases inflammation (37–39). For example, mice overexpressing TRX1 are resistant to inflammatory conditions such as cerebral ischemia, renal ischemia, adriamycin-induced cardiotoxicity, and pneumonia induced by pathogens or cytokines or toxins (40–45). In addition, TRX1 by binding to ASK1 inhibits vascular inflammation (46). Furthermore, overexpression of TRX1 in the heart limits oxidative stress, decreases myocardial infarct size, and improves cardiac performance (47). In fact, exogenous TRX1 also diminished injury after cardiac ischemia and reperfusion (48). However, some data suggest that TRX1 may play a proinflammatory role under some circumstances. For example, TRX1 increases the ability of NF-κB to bind to DNA by reducing Cys-62, thus increasing proinflammatory gene transcription (49, 50). TRX1 also interacts with Ref-1, promoting AP-1 DNA binding activity (51). Extracellular TRX1 can serve as a chemoattractant (52). Our data predict that TRX1 can play a proinflammatory role in the vasculature by counteracting NO. Although NO inhibits vascular inflammation in part by S-nitrosylation of NSF and inhibition of exocytosis, TRX1 denitrosylates NSF and restores exocytosis, which would be predicted to increase vascular inflammation. Thus the role of TRX1 in inflammation may be more complex than previously thought (53).
Footnotes
- NOS
- nitric-oxide synthase
- VWF
- von Willebrand factor
- NSF
- N-ethylmaleimide-sensitive factor
- TRX1
- thioredoxin 1
- SNAP
- S-nitroso-N-acetyl d,l-penicillamine
- l-NAME
- l-nitroarginine methyl ester
- HUVEC
- human umbilical vein endothelial cell(s)
- IP
- immunoprecipitation
- SNO
- S-nitrosylated
- GSNO
- S-nitroso-glutathione.
REFERENCES
- 1. Kubes P., McCafferty D. M. (2000) Am. J. Med. 109, 150–158 [DOI] [PubMed] [Google Scholar]
- 2. Lefer A. M., Lefer D. J. (1999) Am. J. Physiol. 276, G572–575 [DOI] [PubMed] [Google Scholar]
- 3. Grisham M. B., Jourd'Heuil D., Wink D. A. (1999) Am. J. Physiol. 276, G315–321 [DOI] [PubMed] [Google Scholar]
- 4. Zamora R., Vodovotz Y., Billiar T. R. (2000) Mol. Med. 6, 347–373 [PMC free article] [PubMed] [Google Scholar]
- 5. Cirino G., Fiorucci S., Sessa W. C. (2003) Trends Pharmacol. Sci. 24, 91–95 [DOI] [PubMed] [Google Scholar]
- 6. Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007) Nat. Rev. Immunol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 7. Davenpeck K. L., Gauthier T. W., Lefer A. M. (1994) Gastroenterology 107, 1050–1058 [DOI] [PubMed] [Google Scholar]
- 8. Lowenstein C. J. (2007) Cardiovasc. Res. 75, 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lowenstein C. J., Morrell C. N., Yamakuchi M. (2005) Trends Cardiovasc. Med. 15, 302–308 [DOI] [PubMed] [Google Scholar]
- 10. Jahn R., Scheller R. H. (2006) Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 11. Mellman I., Warren G. (2000) Cell 100, 99–112 [DOI] [PubMed] [Google Scholar]
- 12. Südhof T. C. (2004) Annu. Rev. Neurosci. 27, 509–547 [DOI] [PubMed] [Google Scholar]
- 13. Südhof T. C., Rothman J. E. (2009) Science 323, 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Whiteheart S. W., Griff I. C., Brunner M., Clary D. O., Mayer T., Buhrow S. A., Rothman J. E. (1993) Nature 362, 353–355 [DOI] [PubMed] [Google Scholar]
- 15. Wickner W., Schekman R. (2008) Nat. Struct. Mol. Biol. 15, 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whiteheart S. W., Brunner M., Wilson D. W., Wiedmann M., Rothman J. E. (1992) J. Biol. Chem. 267, 12239–12243 [PubMed] [Google Scholar]
- 17. Matsushita K., Morrell C. N., Cambien B., Yang S. X., Yamakuchi M., Bao C., Hara M. R., Quick R. A., Cao W., O'Rourke B., Lowenstein J. M., Pevsner J., Wagner D. D., Lowenstein C. J. (2003) Cell 115, 139–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang Y., Man H. Y., Sekine-Aizawa Y., Han Y., Juluri K., Luo H., Cheah J., Lowenstein C., Huganir R. L., Snyder S. H. (2005) Neuron 46, 533–540 [DOI] [PubMed] [Google Scholar]
- 19. Benhar M., Forrester M. T., Stamler J. S. (2009) Nat. Rev. Mol. Cell Biol. 10, 721–732 [DOI] [PubMed] [Google Scholar]
- 20. Hoshino Y., Shioji K., Nakamura H., Masutani H., Yodoi J. (2007) Antioxid. Redox Signal. 9, 689–699 [DOI] [PubMed] [Google Scholar]
- 21. Lillig C. H., Holmgren A. (2007) Antioxid. Redox Signal. 9, 25–47 [DOI] [PubMed] [Google Scholar]
- 22. Nikitovic D., Holmgren A. (1996) J. Biol. Chem. 271, 19180–19185 [DOI] [PubMed] [Google Scholar]
- 23. Sengupta R., Ryter S. W., Zuckerbraun B. S., Tzeng E., Billiar T. R., Stoyanovsky D. A. (2007) Biochemistry 46, 8472–8483 [DOI] [PubMed] [Google Scholar]
- 24. Benhar M., Forrester M. T., Hess D. T., Stamler J. S. (2008) Science 320, 1050–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jaffrey S. R., Snyder S. H. (2001) Sci. STKE 2001, pl1. [DOI] [PubMed] [Google Scholar]
- 26. Verdoucq L., Vignols F., Jacquot J. P., Chartier Y., Meyer Y. (1999) J. Biol. Chem. 274, 19714–19722 [DOI] [PubMed] [Google Scholar]
- 27. Schwertassek U., Balmer Y., Gutscher M., Weingarten L., Preuss M., Engelhard J., Winkler M., Dick T. P. (2007) EMBO J. 26, 3086–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mitchell D. A., Marletta M. A. (2005) Nat. Chem. Biol. 1, 154–158 [DOI] [PubMed] [Google Scholar]
- 29. Lima B., Forrester M. T., Hess D. T., Stamler J. S. Circ. Res. 106, 633–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hou Y., Guo Z., Li J., Wang P. G. (1996) Biochem. Biophys. Res. Commun. 228, 88–93 [DOI] [PubMed] [Google Scholar]
- 31. Trujillo M., Alvarez M. N., Peluffo G., Freeman B. A., Radi R. (1998) J. Biol. Chem. 273, 7828–7834 [DOI] [PubMed] [Google Scholar]
- 32. Kuwada M., Horie S., Ogura Y. (1980) J. Biochem. 88, 859–869 [DOI] [PubMed] [Google Scholar]
- 33. Jensen D. E., Belka G. K., Du Bois G. C. (1998) Biochem. J. 331, 659–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu L., Hausladen A., Zeng M., Que L., Heitman J., Stamler J. S. (2001) Nature 410, 490–494 [DOI] [PubMed] [Google Scholar]
- 35. Jourd'heuil D., Laroux F. S., Miles A. M., Wink D. A., Grisham M. B. (1999) Arch. Biochem. Biophys. 361, 323–330 [DOI] [PubMed] [Google Scholar]
- 36. Hausladen A., Gow A., Stamler J. S. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10108–10112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamawaki H., Haendeler J., Berk B. C. (2003) Circ. Res. 93, 1029–1033 [DOI] [PubMed] [Google Scholar]
- 38. Yoshida T., Nakamura H., Masutani H., Yodoi J. (2005) Ann. N.Y. Acad. Sci. 1055, 1–12 [DOI] [PubMed] [Google Scholar]
- 39. Ebrahimian T., Touyz R. M. (2008) Antioxid. Redox Signal. 10, 1127–1136 [DOI] [PubMed] [Google Scholar]
- 40. Takagi Y., Mitsui A., Nishiyama A., Nozaki K., Sono H., Gon Y., Hashimoto N., Yodoi J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 4131–4136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hattori I., Takagi Y., Nakamura H., Nozaki K., Bai J., Kondo N., Sugino T., Nishimura M., Hashimoto N., Yodoi J. (2004) Antioxid. Redox Signal. 6, 81–87 [DOI] [PubMed] [Google Scholar]
- 42. Kasuno K., Nakamura H., Ono T., Muso E., Yodoi J. (2003) Kidney Int. 64, 1273–1282 [DOI] [PubMed] [Google Scholar]
- 43. Nakamura H., Tamura S., Watanabe I., Iwasaki T., Yodoi J. (2002) Immunol. Lett. 82, 165–170 [DOI] [PubMed] [Google Scholar]
- 44. Hoshino T., Nakamura H., Okamoto M., Kato S., Araya S., Nomiyama K., Oizumi K., Young H. A., Aizawa H., Yodoi J. (2003) Am. J. Respir. Crit. Care Med. 168, 1075–1083 [DOI] [PubMed] [Google Scholar]
- 45. Shioji K., Kishimoto C., Nakamura H., Masutani H., Yuan Z., Oka S., Yodoi J. (2002) Circulation 106, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 46. Yamawaki H., Pan S., Lee R. T., Berk B. C. (2005) J. Clin. Invest. 115, 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Turoczi T., Chang V. W., Engelman R. M., Maulik N., Ho Y. S., Das D. K. (2003) J. Mol. Cell. Cardiol. 35, 695–704 [DOI] [PubMed] [Google Scholar]
- 48. Tao L., Gao E., Bryan N. S., Qu Y., Liu H. R., Hu A., Christopher T. A., Lopez B. L., Yodoi J., Koch W. J., Feelisch M., Ma X. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11471–11476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matthews J. R., Wakasugi N., Virelizier J. L., Yodoi J., Hay R. T. (1992) Nucleic Acids Res. 20, 3821–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sakurai A., Yuasa K., Shoji Y., Himeno S., Tsujimoto M., Kunimoto M., Imura N., Hara S. (2004) J. Cell. Physiol. 198, 22–30 [DOI] [PubMed] [Google Scholar]
- 51. Wei S. J., Botero A., Hirota K., Bradbury C. M., Markovina S., Laszlo A., Spitz D. R., Goswami P. C., Yodoi J., Gius D. (2000) Cancer Res. 60, 6688–6695 [PubMed] [Google Scholar]
- 52. Bertini R., Howard O. M., Dong H. F., Oppenheim J. J., Bizzarri C., Sergi R., Caselli G., Pagliei S., Romines B., Wilshire J. A., Mengozzi M., Nakamura H., Yodoi J., Pekkari K., Gurunath R., Holmgren A., Herzenberg L. A., Herzenberg L. A., Ghezzi P. (1999) J. Exp. Med. 189, 1783–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Burke-Gaffney A., Callister M. E., Nakamura H. (2005) Trends Pharmacol. Sci. 26, 398–404 [DOI] [PubMed] [Google Scholar]