Abstract
Hepatitis C virus infection, a major cause of liver disease worldwide, is curable, but currently approved therapies have suboptimal efficacy. Supplementing these therapies with direct-acting antiviral agents has the potential to considerably improve treatment prospects for hepatitis C virus-infected patients. The critical role played by the viral NS3 protease makes it an attractive target, and despite its shallow, solvent-exposed active site, several potent NS3 protease inhibitors are currently in the clinic. BI 201335, which is progressing through Phase IIb trials, contains a unique C-terminal carboxylic acid that binds noncovalently to the active site and a bromo-quinoline substitution on its proline residue that provides significant potency. In this work we have used stopped flow kinetics, x-ray crystallography, and NMR to characterize these distinctive features. Key findings include: slow association and dissociation rates within a single-step binding mechanism; the critical involvement of water molecules in acid binding; and protein side chain rearrangements, a bromine–oxygen halogen bond, and profound pKa changes within the catalytic triad associated with binding of the bromo-quinoline moiety.
Keywords: Enzyme Kinetics, Hepatitis Virus, NMR, Protein-Drug Interactions, X-ray Crystallography, BI 201335, Halogen Bonding, Mechanism of Action, NS3 Protease, Noncanonical Interactions
Introduction
Worldwide, 130–170 million people are chronically infected with hepatitis C virus (HCV)2 (1). Infection can progress to cirrhosis, which can lead to hepatic decompensation or hepatocellular carcinoma. HCV infection can be cured by administering a combination of pegylated interferon and the nonspecific antiviral drug ribavirin for 24–48 weeks. Unfortunately, this treatment is only 40–50% successful in clearing genotype 1 infections, the most common type in most regions of the world (2). Clearly there is considerable need for more efficacious therapies.
HCV is a small, positive-strand RNA virus with a 9600-nucleotide genome encoding a 3000-amino acid polyprotein that is subsequently processed into individual proteins by cellular and viral proteases. Four of the five cleavage sites between the NS (nonstructural) proteins are cleaved by the viral NS3 serine protease (EC 3.4.21.98). HCV NS3 is a bifunctional protein comprised of an N-terminal 180-amino acid serine protease domain and a C-terminal 420-amino acid helicase domain (3). The central portion of the 54-amino acid NS4A protein is structurally integrated into the NS3 protease domain (4) and is necessary for full protease activity (5).
The search for specific, direct-acting, small molecule inhibitors of HCV replication began shortly after the virus was discovered in 1989 (6); the fact that no such drug has yet made it to the market attests to the challenging nature of this research. Throughout this period, the NS3-NS4A protease has remained a major focus of HCV drug discovery efforts. Beginning with the protease inhibitor BILN 2061 in 2002 (7, 8), a number of specific antivirals have been tested in human trials and have shown very promising results (9). The most advanced compounds in the clinic are NS3-NS4A protease inhibitors, with two, telaprevir and boceprevir, currently completing Phase III trials (10, 11).
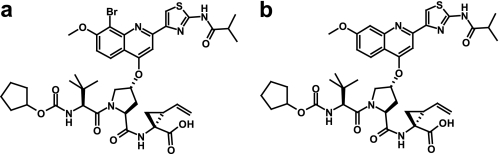
Presently progressing through Phase IIb clinical trials, BI 201335 (Fig. 1a) is a promising drug candidate that potently and selectively targets the viral NS3-NS4A protease (12). Similar to other protease inhibitors, BI 201335 features a peptidic backbone such that it can form extended, substrate-like interactions with an exposed β-strand of the enzyme substrate-binding site. However, distinct from other compounds presently in the clinic, BI 201335 possesses a C-terminal carboxylic acid that contributes significantly to the specificity of the compound for the viral protease (13). Furthermore, the amino acid side chains and N-terminal capping group of BI 201335 have been optimized for potency in both cellular and biochemical assays and to provide good absorption, distribution, metabolism, elimination, and pharmacokinetics properties allowing efficient uptake and distribution of the drug to the target organ, the liver (12). During this optimization, multi-ring aromatic systems extending from the P2 proline were found to be particularly beneficial to C-terminal carboxylate-containing molecules (standardized terminology for protease substrate amino acids (P1–P4) and substrate side chain binding sites (S1–S4) is used throughout (14)).
FIGURE 1.
Chemical structures of the NS3 protease inhibitors used in this study. a, BI 201335. b, analog 1.
Here we present details of BI 201335 binding to NS3-NS4A protease as determined via x-ray crystallography, NMR, and enzyme kinetics. We report the 1.9-Å crystal structure of NS3-NS4A protease in complex with BI 201335. This high resolution structure describes many intricate interactions of the inhibitor with its target, including a noncanonical bromine-oxygen interaction known as a “halogen bond.” To better understand the effects of this halogen bond interaction, we also carried out studies on analog 1 (Fig. 1b), which is identical to BI 201335 except for the replacement of the bromine by a hydrogen atom. We also present NMR evidence that shielding of the catalytic triad by the multi aromatic ring P2 substituent of BI 201335 perturbs the protonation states of its Asp-His pair, which may in turn favor the binding of the carboxylate to the active site. Finally, we elaborate on previously reported steady-state kinetic analyses that indicated BI 201335 is a competitive inhibitor of NS3-NS4A protease activity (12) by reporting the individual association and dissociation rate constants determined by stopped flow methods.
EXPERIMENTAL PROCEDURES
Inhibitors, Peptides, and Proteins
BI 201335 was synthesized as described (15), and analog 1 was synthesized by similar procedures. The HCV genotype 1b Con1 strain NS3-NS4A protein (FL-protease), the HCV genotype 1b J strain NS3 protease domain, and the same domain with an NS4A peptide fused to its N terminus (sc-protease) were expressed and purified as described previously (16). The NS4A peptide used for crystallography (KKGSVVIVGRIILSGRK) has also been described previously (16), as well as the depsipeptide fluorogenic substrate anthranilyl-DDIVP-Abu[C(O)-O]AMY(3-NO2)-TW-OH, which is derived from the NS5A-NS5B cleavage site. FL-protease and sc-protease recognize and process this substrate with a similar efficiency (16).
Note that the efficiency with which the sc-protease processes a different depsipeptide substrate, derived from the NS4A-NS4B cleavage site, is significantly lower under the experimental conditions described here (17) (and data not shown). Efficient cleavage of the NS4A-NS4B substrate by sc-protease can be observed after modification of the reaction buffer, e.g. by the addition of glycerol. No explanation has yet been found for this buffer dependence of NS4A-NS4B substrate cleavage, which is specific to the sc-protease.
Crystallography
HCV NS3 protease domain (J-strain) was prepared at 3.4 mg/ml in 50 mm sodium phosphate, pH 6.0, 0.1% CHAPS, 5% glycerol, 1 mm Tris (2-carboxyethyl) phosphine hydrochloride, 300 mm NaCl, and a 2-fold molar excess of NS4A peptide. Crystallization experiments were performed using the hanging drop vapor diffusion method with a crystallization solution of 3 m NaCl, 0.1 m sodium citrate, pH 6.0, 2.5% t-butanol, 20 mm n-octyl-β-d-glucoside, 5 mm DTT, and 3 mm sodium azide. Crystallization was achieved by suspending drops containing equal volumes of protein and crystallization solutions over a 500-μl reservoir of the same crystallization solution. Rice-shaped crystals appeared within 1 week of incubation at 18 °C and continued to grow for up to 1 month. The crystal soaking solution was comprised of 4 m NaCl, 0.1 m Tris, pH 8.0, 2.5% t-butanol, 20 mm n-octyl-β-d-glucoside, 5 mm DTT, 3 mm sodium azide, and 1% Me2SO (v/v). The crystals were transferred to a drop containing soaking solution and 1 mm of either BI 201335 or analog 1. The drops were suspended over a reservoir of the same soaking solution and allowed to incubate at 18 °C for 4–8 weeks. The crystals were then harvested in rayon loops and directly flash frozen in liquid nitrogen. Diffraction data were collected at 100 K on an FR-E x-ray generator equipped with Osmic HiRes2 optics and a MAR345dtb image plate detector. Data reduction and scaling were performed using HKL2000 (18).
Preliminary models for the BI 201335 and analog 1 structures were obtained via rigid body refinement in CNX (19, 20) using a model originally based on the Protein Data Bank structure of 1DY9 (21). Further iterations of refinement followed by manual model building using COOT (22) yielded final models containing residues 1–182 with C-terminal residues Arg-180 and Ser-182 being artificially truncated to alanine. Despite the presence of reducing agent, long soaking times led to the oxidation of several cysteine residues, including those that bind the structural zinc atom. In the context of the crystal, cysteine oxidation did not structurally impact the NS3 active site as evidenced by comparison of unoxidized, zinc-containing crystal structures with oxidized structures that have partially or completely lost their zinc (data not shown). The data processing statistics and model refinement statistics are listed in Table 1. Stereochemical quality was assessed with the program PROCHECK (v 3.4.4) (23). All of the residues of the final models lie in the “most favored” or “additional allowed” regions. The asymmetric unit of the P61 crystal form is comprised of two independently refined NS3 protease monomers, both of which are capable of binding active site inhibitors. The high degree of agreement between the monomers of these structures reinforces their validity.
TABLE 1.
Data collection and refinement statistics
Each data set was obtained from a single crystal. The values in parentheses are for the highest resolution shell.
| BI 201335 | Analog 1 | |
|---|---|---|
| Data collection | ||
| Space group | P61 | P61 |
| Cell dimensions | ||
| a, b, c (Å) | 94.9, 94.9, 81.9 | 95.0, 95.0, 81.8 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 40.0-1.9 (1.97-1.90) | 40.0-2.3 (2.38-2.30) |
| Rmerge | 7.3 (61.2) | 4.8 (38.3) |
| I/σI | 12.6 (2.3) | 23.2 (4.9) |
| Completeness (%) | 98.0 (85.4) | 98.6 (88.6) |
| Redundancy | 10.6 (5.8) | 10.6 (7.7) |
| Refinement | ||
| Resolution (Å) | 1.9 | 2.3 |
| No. reflections | 30638 | 17940 |
| Rwork/Rfree | 20.2/24.2 | 21.7/25.5 |
| No. atoms | ||
| Protein | 2834 | 2834 |
| Ligand/ion | 116 | 114 |
| Water | 176 | 66 |
| B-factors | ||
| Protein | 37.9 | 48.5 |
| Ligand/ion | 32.5 | 46.7 |
| Water | 45.8 | 44.9 |
| Root mean square deviations | ||
| Bond lengths (Å) | 0.010 | 0.009 |
| Bond angles (°) | 1.13 | 0.98 |
Calculation of the Electrostatic Surface Potential of the P2 Group of BI 201335
The geometry of the isolated P2 substituent of BI 201335 was optimized with the DFT method at the B3LYP/LACV3P** level using the program Jaguar, version 7.5 (Schrödinger LLC, New York, NY). The optimized geometry shows only minor changes with regard to the conformation observed by crystallography. The electrostatic surface potential was calculated using the settings iplotesp = 1 and iplotden = 1 and mapped onto the molecular surface as calculated with a probe of radius 1.4 Å.
NMR Sample Preparation and Spectroscopy
Uniformly 15N-labeled samples of the sc-protease were expressed and purified as described previously (16). The final buffer consisted of 25 mm NaPO4, 150 mm NaCl, 1 mm deuterated Tris (2-carboxyethyl) phosphine hydrochloride at pH 6.0 in 90% H2O, 10% D2O. The concentration of the sc-protease or its complex with BI 201335 was 0.2–0.4 mm. All of the NMR experiments were acquired on a 600-MHz Bruker AVANCE III spectrometer equipped with a 5-mm z-gradient triple resonance cryoprobe at 300 K. One-dimensional jump-return, two-dimensional 1H-15N heteronuclear single quantum coherence, 1H-15N long range heteronuclear multiple quantum coherence (22-ms delay), and two-dimensional NOESY (200-ms mixing) experiments were acquired using standard pulse sequences. All of the NMR data were processed and analyzed using Topspin 2.1 (Bruker BioSpin).
Kinetic Analysis of NS3 Inhibitor Binding
Kinetic experiments were performed using the KinTek stopped flow instrument (SF-2005; excitation, 325 nm; and emission, 410 nm) with a programmable shutter to minimize photobleaching. Enzyme solution (1–100 nm) and substrate-inhibitor solution (20 μm substrate and 2–2000 nm inhibitor) were prepared in 50 mm Tris-HCl, pH 8.0, 0.25 m sodium citrate, 0.01% n-dodecyl-β-d-maltoside, 1 mm Tris (2-carboxyethyl) phosphine hydrochloride, supplemented with 5% Me2SO (v/v). These two solutions were mixed at 1:1 ratio in the instrument. Mixing time was ∼2 ms, and the first data point was collected at 5–50 ms. Upon mixing, the fluorescence data were collected continuously (at least 20 data points/s) for 50–3000 s. Three to four independent data sets were collected for each enzyme-inhibitor combination. The values for kcat and Km were determined using 0.5–12.5 μm substrate.
The data sets were analyzed using the Global Kinetic Explorer (KinTek) nonlinear regression global fit, and the limits of variation for each parameter were verified by confidence contour analysis using the program FitSpace (24). It was assumed that product formation is irreversible. Inhibitor association (kon) and dissociation (koff) rate constants obtained from this fitting procedure are not dependent on the specific values for substrate association and dissociation rate constants or the rate constant for conversion to product. To carry out the data fitting procedure, it was assumed that k1 = 100 μm−1 s−1 (close to the diffusion limited rate). The values for k−1 and k2 were then calculated from the values of Km and kcat as described in Fig. 6. Dissociation half-lives were calculated from values of koff according to the equation t½ = ln(2)/koff. Each progress curve was also fitted directly to the burst exponential equation (Equation 1) to obtain the apparent rate constant at each inhibitor concentration,
where Y is the amount of product present at time t; kobs is the apparent rate constant; A1 is the burst amplitude term corresponding to (vi − vf)/kobs, where vi and vf are initial and steady-state velocity, respectively; C is a correction factor to account for the background signal. Apparent rate constants were plotted against inhibitor concentrations to provide an alternative assessment of the binding mechanism for each enzyme-inhibitor pair.
FIGURE 6.
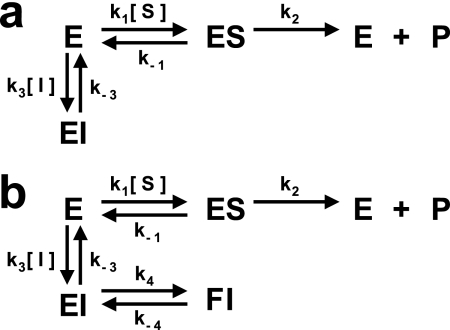
Mechanisms for competitive inhibition. In both cases, kcat is equal to k2, and Km is defined by (k−1 + k2)/k1. a, one-step inhibitor binding in which k3 is the association rate constant (kon) and k−3 is the dissociation rate constant (koff). b, two-step inhibitor binding involving a loosely bound complex (EI) that isomerizes slowly to form a more tightly bound complex (FI).
RESULTS
The Structure of BI 201335 Bound to NS3-NS4A Protease
High quality electron density clearly describes bound BI 201335 and the protein residues in direct contact with it (supplemental Fig. S1). Importantly, the electron density also clearly indicates the presence of numerous water molecules at the active site and throughout the structure. Only the isopropyl moiety of the inhibitor and the side chain of Lys-136 show any noteworthy disorder. The NS3–4A protease adopts the canonical chymotrypsin-like fold, as described previously (4). Compared with other chymotrypsin-like serine proteases, the NS3 protease domain lacks several surface loops near the S2 and S4 subsites, resulting in a shallower and more solvent-exposed binding cleft (25).
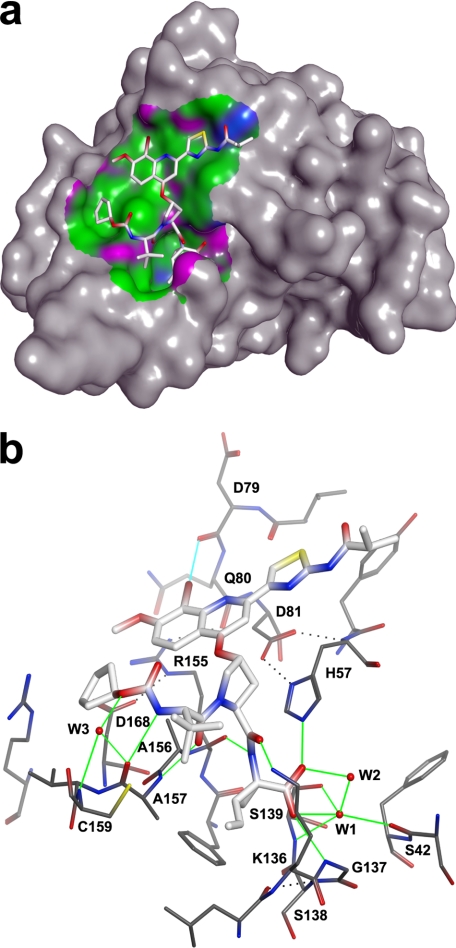
Bound BI 201335 adopts an extended conformation generating over 450 Å2 of interaction surface spanning from S1–S4 (Fig. 2a). The binding site is primarily hydrophobic with discrete polar patches, the pattern of which is precisely complemented by BI 201335. The extended peptidic inhibitor is paired with an exposed strand of the C-terminal β-barrel, forming three substrate-like intermolecular hydrogen bonds: the P1-P2 amide N with Arg-155:O, the P2-P3 amide O with Ala-157:N, and the carbamate nitrogen of the N-terminal capping group with Ala-157:O (Fig. 2b). The carbamate oxygen makes a water-mediated interaction with Ala-157:O and Cys-159:N, and a weaker, likely transient H-bond is also observed between the amine nitrogen of solvent-exposed Lys-136 and the P1-P2 amide oxygen.
FIGURE 2.
Hydrophobic and hydrogen bond interactions of BI 201335 with NS3-NS4A protease. a, 3.6-Å interaction surface colored by hydrophobic (green), mildly polar (blue), and H-bonding (purple). b, intermolecular hydrogen and halogen bond interactions are indicated by green and cyan lines, respectively. Select intramolecular hydrogen bonds are indicated by black dotted lines.
The C-terminal acid of BI 201335 forms good geometry hydrogen bonds to catalytic His-57:NE2 and Gly-137:NH, as seen in previous NS3 protease structures featuring acids bound at the active site (26, 27). The improved data quality of our structure further reveals a bound water molecule that is central to acid binding (labeled W1 in Fig. 2b). This water is positioned within H-bonding distance of the inhibitor acid, the catalytic hydroxyl Ser-139:OG, Ser-139:NH of the oxyanion hole, Ser-42:O, and a second ordered water molecule (labeled W2) that in turn hydrogen bonds back to the inhibitor acid. Although this results in five potential hydrogen bonding partners for W1, the geometries suggest that hydrogen bonding with Ser-139:NH and with that of the second ordered water molecule are mutually exclusive. Although previous structures suggested that catalytic Ser-139:OH hydrogen-bonds with the bound carboxylate, this structure clearly shows long distances and poor angles for such an interaction.
As we and others have previously reported, the binding of large P2 substituents can require the side chain of Arg-155 to adopt a conformation distinct from that observed in unliganded NS3 structures (27, 28). This effect is also observed in the structure of bound BI 201335, in which Arg-155 is largely desolvated and has gained new interactions with residues Gln-80 and Asp-168 (Fig. 2b). The side chain conformations required to form the salt bridge between Arg-155 and Asp-168 result in a relatively flat, hydrophobic surface that spans ∼15 Å from Asp-168 through to catalytic His-57 and serves to bind the aromatic P2 substituent of BI 201335.
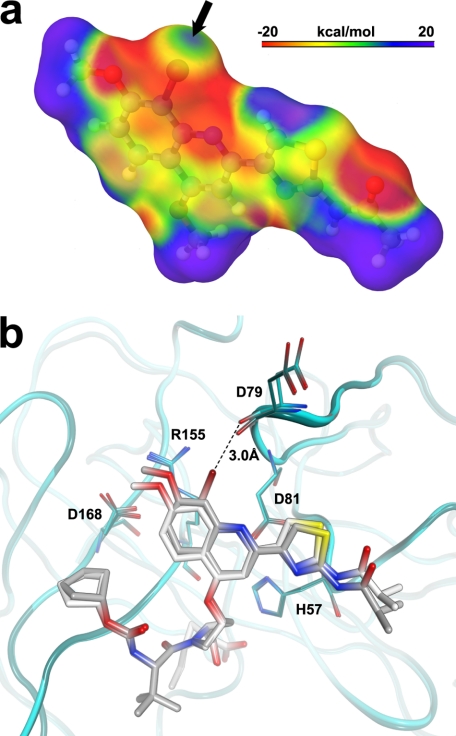
Despite comprising approximately half of the inhibitor, no intermolecular hydrogen bonds are observed for the P2 side chain moiety, thus underscoring the importance of hydrophobic interactions in BI 201335 binding. The structure does, however, indicate the presence of a halogen bond between the bromine and the backbone carbonyl oxygen of Asp-79 (Fig. 2b). A halogen bond is a noncovalent interaction occurring between a polarizable halogen atom (which acts as a Lewis acid) and a Lewis base. In our structure, a lone pair of electrons on the oxygen points toward the tip of the bromine in such a way that the relatively strict geometrical constraints of a halogen bond (29) are satisfied (∠ carbon-bromine-oxygen = 169°; ∠ carbon-oxygen-bromine = 133°). In addition, the quantum-mechanics derived electrostatic surface potential of the BI 201335 P2 proline substituent (Fig. 3a) shows a positive electrostatic potential for the portion of the bromine facing the carbonyl oxygen of Asp-79. Taken together, the geometry and electrostatics clearly support the presence of a halogen bond, thus rationalizing the observed short bromine–oxygen distance of only 3.0 Å.
FIGURE 3.
Halogen bonding of the P2 substituent. a, the electrostatic surface potential of the P2 substituent of BI 201335 as calculated using a quantum mechanics approach that considers electronic effects exerted by the chemical environment of the bromine (see “Experimental Procedures”). The black arrow indicates the area of positive surface potential located at the tip of the C-8 bromine. b, superposition of BI 201335 and its des-bromine analog 1 illustrating the structural shifts incurred upon halogen bond formation.
The x-ray structure of analog 1 was also solved in complex with NS3 to directly compare structural effects of the bromine substituent. Although analog 1 binds similarly to BI 201335, the comparison shows that the bromine causes both the P2 quinoline and the Asp-79:O to shift ∼0.5 Å in opposite directions (Fig. 3b). These shifts are significantly larger than the 0.15 Å root mean square deviation for the remaining residues found within 7 Å of the inhibitor and allow the appropriate geometry for a halogen bond to be achieved.
Determination of the Protonation State of the Catalytic Histidine by NMR
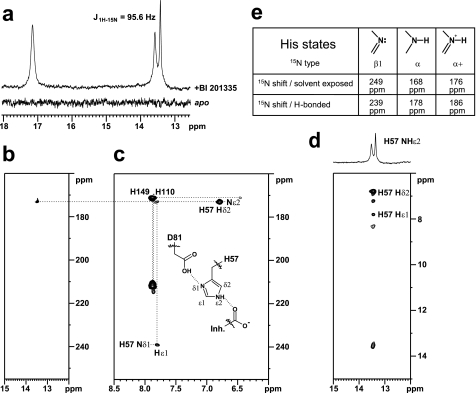
To further refine our understanding of the BI 201335-NS3 interaction, we used NMR to characterize the protonation state and hydrogen bonding of the catalytic histidine (His-57) in the bound state. In serine proteases, the δ1-NH proton of the catalytic histidine is typically involved in a strong hydrogen bond with the carboxylate of the catalytic aspartic acid and is observable in the 1H spectra of most apo enzymes (30, 31). Previous NMR studies with NS3 protease have shown that in the free state His-57 has a pKa of 6.8 and at pH 6.6 protonated and neutral forms exist in an approximate 1:1 ratio (32). However, the solvent-exposed nature of His-57 in apo NS3 protease prevents the NMR detection of its side chain δ1-NH proton, even at −8 °C (25). Similarly, this signal was not detected using an NS4A-NS3 tethered protein, even though the catalytic triad was shown to be properly aligned in the presence of the NS4A peptide (33). Consistent with these results, we were also unable to detect a specific signal for the His-57 imidazole NH protons in a 1H jump-return spectrum recorded in the free state using a uniformly 15N-labeled NS4A peptide-NS3 protease-tethered protein (15N-sc-protease) (Fig. 4a, bottom).
FIGURE 4.
His-57 resonance characterization. a, 1H jump-return spectra of 15N-labeled sc-protease apo (bottom) and in complex with BI 201335 (top). b, selected region of the 1H-15N heteronuclear single quantum coherence spectrum of 15N-labeled sc-protease-BI 201335 complex highlighting the nitrogen correlation (172.9 ppm) of the proton at 13.5 ppm. c, 1H-15N long range heteronuclear multiple quantum coherence spectrum of 15N-labeled sc-protease-BI 201335 complex highlighting a reverse L-shaped pattern for His-57 characteristic of a ϵ-NH tautomer. d, portion of the two-dimensional NOESY spectrum (tm = 200 ms) of the sc-protease-BI 201335 complex highlighting the intermolecular NOEs between the proton at 13.5 ppm and both the His-57 side chain δ2 and ϵ1 protons. e, typical 15N chemical shifts for various types of imidazole ring nitrogen atoms and the effect of H-bonding upon these shifts (31).
In contrast, binding of BI 201335 to 15N-sc-protease led to the observation of two distinct signals in the 1H spectrum, at 17.2 ppm (singlet) and 13.5 ppm (doublet, J = 95.6 Hz) (Fig. 4a, top). Similar signals have been observed in other serine proteases at low pH and assigned to the protonated active site histidine δ1-NH (∼17–18 ppm) and ϵ2-NH (∼13–16 ppm) protons, respectively (30, 31). However, despite the observation of two new proton signals, only one of them appears to have originated from a proton bound to nitrogen, because only one displayed the expected ∼90 Hz 1JN-H coupling. This was also reflected in the two-dimensional 1H-15N heteronuclear single quantum coherence spectrum (Fig. 4b) in which only the resonance at 13.5 ppm correlated with a nitrogen atom (172.9 ppm). Together these data suggest that only one nitrogen atom of the active site His-57 is protonated, i.e. that His-57 is neutral in the BI 201335-bound state at pH 6.0. This was further corroborated by the long range 1H-15N heteronuclear multiple quantum coherence spectrum (Fig. 4c), in which a large difference in the 15N chemical shifts of the ϵ2 and δ1 nitrogens was detected (172.9 and 239.2 ppm), an observation typical of neutral histidines (31, 34).
The long range 1H-15N heteronuclear multiple quantum coherence spectrum also showed an inverted L-shaped peak pattern for His-57 (Fig. 4c), a clear indication that it exists in the ϵ2-NH tautomeric form when bound to BI 201335 (31, 34). Protonation at Nϵ is further supported by specific intraresidue NOEs between the His-57 ϵ2-NH proton (13.5 ppm) and both the ϵ1-H and δ2-H ring protons at 7.82 and 6.83 ppm, respectively (Fig. 4d). Moreover the ϵ2-N chemical shift at 172.9 ppm (∼5 ppm downfield from reference values reported for solvent-exposed α-type imidazole nitrogen; Fig. 4e) and the ϵ2-NH chemical shift at 13.5 ppm are reflective of their involvement in hydrogen bonding, which is in full agreement with the present BI 201335 complexed x-ray structure in which the ϵ2-N of His-57 and the inhibitor P1 carboxylate oxygen are separated by only 2.8 Å. Finally, the chemical shift for the δ1-N nucleus at 239.2 ppm (Fig. 4e) is indicative of its involvement as an acceptor in a strong hydrogen bond most likely with the side chain of Asp-81 as observed in the x-ray complex structure (Asp-81:OG-His-57:Nδ1, d = 2.6 Å).
Taken together, these results have the following interesting consequence: if the δ1-N of His-57 is not protonated but is involved as an acceptor in a strong hydrogen bond, then the proton must be located on the Asp-81 acid side chain (and could correspond to the singlet resonance observed at 17.2 ppm in the 1H spectrum; Fig. 4b). Unfortunately, we were unable to observe any specific NOEs for this proton, thus preventing its unambiguous assignment. Nevertheless, these observations imply both that with BI 201335 bound, the pKa of His-57 is significantly lower than expected and that the pKa of Asp-81 is significantly higher than anticipated. To our knowledge this would be an unprecedented protonation pattern for the active site histidine and aspartic acid pair in serine proteases.
Kinetic Analysis of BI 201335 and Analog 1 Binding to NS3-NS4A Protease
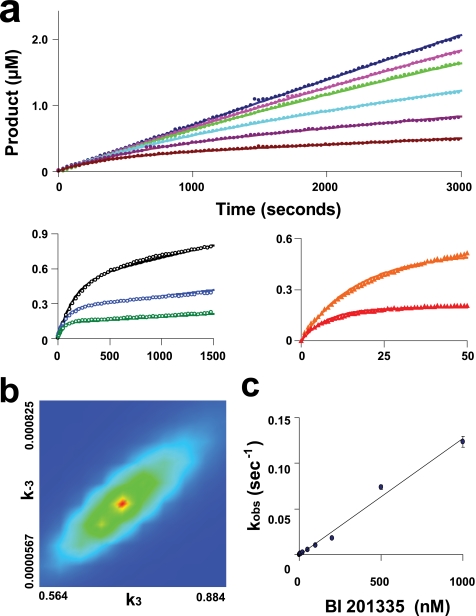
To elucidate the mechanism by which the BI 201335-NS3 complex is formed, kinetic data for both the full-length NS3-NS4A protein (FL-protease) and the sc-protease were collected using a stopped flow spectrophotometer. Kinetic parameters determined by progress curve analysis for FL-protease (kcat = 0.78 s−1 and Km = 2.2 μm) were in agreement with values previously determined by steady-state methods (16). Progress curves for substrate cleavage were obtained in the presence of multiple BI 201335 and FL-protease concentrations (Fig. 5a). At higher concentrations of BI 201335, the enzyme concentration was increased to accurately measure the pre-steady-state product formation. Progress curves clearly showed curvature caused by the slow binding of BI 201335. An excellent fit of the experimental data to the one-step slow-binding mechanism (Fig. 6a) was obtained, giving the inhibitor association and dissociation rate constants shown in Table 2. The value of koff corresponds to a dissociation half-life of 26 min. Confidence contour analysis using the program FitSpace (24) demonstrated that inhibitor binding parameters were well fit within tight confidence intervals (see example in Fig. 5b). A global fit of each data set to the two-step binding mechanism model depicted in Fig. 6b also yielded a good fit (supplemental Fig. S2a). However, confidence contour analysis revealed that in this case some individual parameters were not well constrained (supplemental Fig. S2b). Overall, data analysis of BI 201335 binding to FL-protease suggests that the formation of an initial encounter complex, if it exists, is too weak to be distinguished as a separate step, and binding of BI 201335 to FL-protease is best characterized by a single-step binding mechanism.
FIGURE 5.
Kinetics of binding of BI 201335 to FL-protease. The time dependence of product formation is shown for the hydrolysis of 10 μm substrate by various concentrations of FL-protease and in the presence of different concentrations of BI 201335 (Table 2, data set 3 for the FL-protease-BI 201335 combination). a, activity of 1 nm FL-protease in the presence of 0, 1, 2, 5, 10, and 20 nm BI 201335 (top panel); activity of 5 nm FL-protease in the presence of 50, 100, and 200 nm BI 201335 (bottom left panel); and activity of 50 nm FL-protease in the presence of 500 and 1000 nm BI 201335 (bottom right panel). Axis units for bottom graphs are identical to those for the top graph. The solid lines correspond to the global nonlinear regression fit of the data. Only a portion of the collected data points are graphed to allow visualization of these curves. b, confidence contour showing the limits of k3 and k−3 parameters, and the goodness of constraint by the raw data for these fitted parameters in a one-step binding mechanism fit. c, analytically determined apparent rates of inhibition (kobs) plotted against BI 201335 concentration. Standard deviations were calculated from the values of four individual data sets at each BI 201335 concentration and are indicated by vertical bars on each point in the graph.
TABLE 2.
Summary of kinetic parameters
The error displayed for each individual data set is derived from nonlinear regression fitting of the progress curves. For the average values, the standard deviations are given.
| Enzyme | Inhibitor | Data set | kon | koff | Ki |
|---|---|---|---|---|---|
| m−1s−1 | s−1 | nm | |||
| FL-Protease | BI 201335 | 1 | (9.2 ± 0.04) × 105 | (5.1 ± 1.2) × 10−4 | 0.56 |
| 2 | (8.0 ± 0.02) × 105 | (4.3 ± 0.6) × 10−4 | 0.54 | ||
| 3 | (7.1 ± 0.01) × 105 | (4.0 ± 0.6) × 10−4 | 0.56 | ||
| 4 | (9.6 ± 0.03) × 105 | (4.4 ± 0.9) × 10−4 | 0.46 | ||
| Average | (8.5 ± 1.2) × 105 | (4.4 ± 0.5) × 10−4 | 0.53 ± 0.05 | ||
| FL-Protease | 1 | 1 | (13.9 ± 0.04) × 105 | (26.2 ± 1.0) × 10−4 | 1.88 |
| 2 | (8.3 ± 0.03) × 105 | (15.2 ± 1.0) × 10−4 | 1.83 | ||
| 3 | (12.1 ± 0.04) × 105 | (23.6 ± 1.0) × 10−4 | 1.95 | ||
| Average | (11 ± 3) × 105 | (22 ± 6) × 10−4 | 1.89 ± 0.06 | ||
| sc-Protease | BI 201335 | 1 | (4.9 ± 0.02) × 105 | (5.0 ± 1.0) × 10−4 | 1.03 |
| 2 | (5.8 ± 0.01) × 105 | (5.1 ± 0.6) × 10−4 | 0.88 | ||
| 3 | (5.5 ± 0.01) × 105 | (5.8 ± 0.4) × 10−4 | 0.99 | ||
| Average | (5.4 ± 0.5) × 105 | (5.3 ± 0.4) × 10−4 | 0.97 ± 0.08 |
Further support for single-step binding was provided by fitting data from individual progress curves to the burst exponential equation. The linear relationship between the calculated apparent rate constant kobs and concentrations of BI 201335 up to 1 μm (Fig. 5c) indicates that the dissociation constant for an initial encounter complex (assuming the two-step model of Fig. 6b) is close to or greater than 1 μm, and such a value would be technically very difficult to measure.
Analog 1 differs from BI 201335 only by the absence of the bromine atom on the quinoline moiety (Fig. 1). Progress curves for the binding of analog 1 to FL-protease gave a nearly identical association rate to that for BI 201335 but a 5-fold higher dissociation rate, thus yielding a 3.6-fold higher Ki value (Table 2). The data were again best described by a one-step slow binding mechanism. Dahl et al. (17) reported that binding of low concentrations of NS3 protease inhibitors can activate NS3 protease. We have made observations consistent with this finding under certain conditions and believe that NS3 protease is subject to slow conformational changes under some conditions, for example when buffer compositions are changed, even subtly. However, we saw no evidence for such isomerization under the conditions used in this work.
We also studied the binding of BI 201335 to the sc-protease used in the NMR experiments described above. Kinetic parameters determined by progress curve analysis (kcat = 2.9 s−1 and Km = 4.1 μm) were again in agreement with values previously determined by steady-state methods (16). Both global nonlinear regression and analytical analysis of individual progress curves yielded results very similar to those obtained for FL-protease (Table 2), with a dissociation half-life of 22 min. These data are consistent with previously reported data showing that the sc-protease can be reliably used as a model for the binding of inhibitors to FL-protease.
DISCUSSION
Although they share common features such as the peptidic backbone, chemical differences among NS3 protease inhibitors have led to a wide variation in binding properties. In this work we have employed an array of biochemical and structural techniques that together provide a detailed understanding of the association between BI 201335 and its target. The stopped flow kinetic experiments presented here show that BI 201335 binds at a rate well below the diffusion limit but also dissociates slowly, yielding an overall dissociation constant in the picomolar range. Binding data were best described by a single-step binding mechanism. Other noncovalent inhibitors have been reported to bind to NS3 by a two-step mechanism (28, 35), and it has recently been suggested that this could be a reflection of an induced conformational change of Arg-155 (28). However, the kinetic data for BI 201335 indicate a single-step mechanism for inhibitor binding despite the requirement for both Arg-155 and Asp-168 to be displaced, as clearly depicted in the x-ray structure. This combination of results suggests either that the Arg-155–Asp-168 conformational change occurs prior to BI 201335 binding (which may explain the observed slow kon) or that the encounter complex, in which Arg-155 and Asp-168 remain in their solvated apo-like conformations, is so weak that it is undetectable over the concentration range we were able to study.
In 2003 we reported the first noncovalently bound NS3 protease/inhibitor crystal structure (27), which described a C-terminal acid inhibitor that bound in a similar manner as the N-terminal cleavage product observed in the structure of the complete NS3 protease-helicase (26). Because of the relatively low resolutions of these structures (3.0 and 2.5 Å, respectively), the details of active site binding were not clear, and a direct hydrogen bond between catalytic Ser-139:OH and the inhibitor acid was suggested (27, 36). The 1.9 Å structure of BI 201335 complexed with the NS3-NS4A protease represents a significant data quality improvement over these previously reported acid-bound structures and clearly shows poor geometry for such an interaction. Instead this structure indicates the importance of a tightly bound water molecule in the binding of carboxylic acid inhibitors. The resulting extensive network of hydrogen bonding interactions now provides a structural basis for product inhibition of NS3 protease and correlates well with mutational analyses indicating that Ser-139 does not play a major role in compound binding (27, 37).
The conformational changes that bring Arg-155 and Asp-168 together create a relatively flat, apolar surface well suited for the binding of multi-cyclic hydrophobic P2 substituents. The P2 substituent of BI 201335 makes excellent use of this surface, with over 190 Å2 of contact, representing more than 40% of the total surface contact of the inhibitor. For similar tripeptide inhibitors, we found that removal of the P2 substituent results in a ∼5-log reduction in IC50 (38–40), thereby demonstrating that this interaction is critical for noncovalent association with the highly exposed binding site of NS3 protease. NMR data indicate that this interaction also greatly influences the pKa values of key catalytic residues, which may in turn further enhance C-terminal carboxylic acid binding (see discussion below). The importance of this surface to BI 201335 binding is further underscored by the fact that the substitutions R155K and D168V are the major causes of clinical resistance to BI 201335, each resulting in a several-hundred-fold loss in antiviral potency (41).
Working with previously reported NS3-NS4A structures, conventional molecular mechanics would predict that the BI 201335 bromine atom should be detrimental because of steric and electrostatic clashes with the backbone carbonyl oxygen of Asp-79. In contrast, BI 201335 demonstrates a 3.6-fold improvement in Ki over its des-bromine analog 1. This discrepancy is resolved with the x-ray structures and quantum mechanical calculations presented herein, which clearly indicate the induced formation of a halogen bond between the bromine and carbonyl oxygen. Within the strict geometric constraints of a halogen bond, these two electronegative atoms participate in an attractive interaction that brings them to 89% of their van der Waals contact distance (42). Halogen bonds have only recently been recognized for their potential in protein-ligand interactions (43, 44), and the magnitude of their contribution is the subject of ongoing research. In a recent report, a combined quantum mechanics/molecular mechanics approach was used to calculate a remarkable interaction energy of 2.13 kcal/mol for an analogous bromine-oxygen interaction (43). In our case, the bromine appears to impart a more modest affinity improvement, possibly because of the shifts in both the ligand and receptor that are required to adopt the correct geometry. Nonetheless, the overall benefit of the bromine to BI 201335 as a drug is even greater because it also imparts desirable antiviral potency and pharmacokinetic properties (12, 15).
NMR experiments have also been conducted to further refine our understanding of BI 201335-active site interactions. Our data clearly indicate that the active site histidine is not protonated in the BI 201335-bound state at pH 6.0, suggesting a significantly lowered pKa for this residue. The lower pKa for His-57 in the bound state is consistent with published NMR results from Merck for two other NS3 protease complexes and may be explained by His-57 desolvation (32, 45, 46). The unusual presence of three basic amino acids (Arg-109, Lys-136, and Arg-155) in the vicinity of the active site may also contribute to the lowered pKa of His-57. Both site-directed mutagenesis and chemical modification have previously been employed to show that increasing the positive surface charge of serine proteases destabilizes the protonated imidazolium group of the active site histidine, thus lowering its pKa (47).
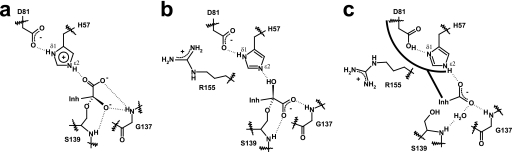
In addition to having a lower pKa in the BI 201335-bound state, our data also indicate that His-57 exists primarily in the ϵ2-NH tautomeric form (ϵ2-NH: 13.5 ppm) and is involved in a hydrogen bond with the inhibitor P1 carboxylate oxygen. Its nonprotonated δ1-N atom is acting as an acceptor in a strong hydrogen bond with the acid proton of Asp-81 (17.2 ppm proposed assignment). One striking difference between our results and those reported by Merck is the tautomeric form that His-57 adopts in the bound state. Indeed, the Merck group clearly demonstrated that His-57 exists in the δ1-NH tautomeric form (δ1-NH: 14.9 ppm) and is involved in a strong hydrogen bond with the carboxylate of Asp-81 in the complexes they studied. Structural differences between the BI 201335 and the Merck inhibitors, particularly at the P1 and P2, are likely responsible for the observed differences in the tautomeric state of His-57. BI 201335 possesses a P1 C-terminal carboxylic acid and a substituted quinoline moiety on the P2 proline, whereas the Merck inhibitors have either an α-ketoacid or an amide moiety at P1 and a much smaller leucine residue at P2 (45, 46). It is interesting to note that optimized NS3-NS4A protease inhibitors containing either a carboxylic acid or an acyl-sulfonamide moiety at P1 generally possess multicyclic aromatic substituents at P2, whereas potent inhibitors containing an activated carbonyl at P1 generally have smaller nonaromatic P2 moieties (48). This difference in the optimized substitution at P2 may be partially explained by differences in how the catalytic histidine interacts with these different inhibitor types. The catalytic serine side chain of a serine protease typically attacks the carbonyl of substrates or covalent inhibitors from the re-side (Fig. 7a) (49), thus orienting carbonyl oxygen toward the oxyanion hole. However, for the NS3 protease in complex with reversible covalent inhibitors, it has been shown that the nucleophilic attack is from the si-side, leading to a tetrahedral intermediate in which the carbonyl oxygen points not toward the oxyanion hole but toward His-57 and is involved in a hydrogen bond with its Nϵ2 atom, which acts as the acceptor (Fig. 7b) (21, 45). For these inhibitors, the histidine is not protonated and exists under its δ1-NH tautomeric form, making a strong hydrogen bond with the carboxylate of Asp-81. The smaller P2 residue of this class of inhibitor allows the side chain of Arg-155 to remain in its apo conformation near the Asp-81 carboxylate, thus leading to an ion pair electrostatic stabilization.
FIGURE 7.
Binding modes of various inhibitor types to serine proteases. a, binding mode of a reversible covalent inhibitor to trypsin (49). b, binding mode of an α-ketoacid inhibitor to NS3 protease (21). c, binding mode of an optimized C-terminal carboxylic acid inhibitor to NS3 protease.
Although the Ser-139 side chain is critical for the binding of covalent inhibitors, single amino acid mutations have shown that it does not contribute to binding of C-terminal carboxylic acid inhibitors (27). Instead, the His-57 side chain was found to play a much greater role (27). For these inhibitors it is likely that His-57 becomes temporarily protonated before binding or perhaps immediately upon binding by abstracting a proton from the C-terminal carboxylic acid of the inhibitor (pKa = 6.5, data not shown). A protonated His-57 could then hydrogen bond both with the Asp-81 carboxylate through its δ1-NH and with the inhibitor carboxylate through its ϵ2-NH. However, unlike the case for covalent inhibitors, the nearby guanidinium moiety of Arg-155 in its apo conformation would be unfavorable to the binding of acid inhibitors because such positive charges have been shown to destabilize the histidine protonated imidazolium group (47). The displacement of the Arg-155 side chain away from the His-57/Asp-81 pair and into a salt bridge with Asp-168 (Fig. 7c), as is enforced by the P2 substituent of BI 201335, should therefore be beneficial to His-57 protonation and hence to acid binding. Furthermore, with the aromatic P2 substituent bound directly over it, the His-57/Asp-81 dyad is significantly shielded from solvent, thus providing a rationale as to why the two residues exist in their neutral rather than charged states. Indeed, it has been reported that the desolvation effect arising from the energetically unfavorable process of burying an ionized group in a hydrophobic environment can lead to pronounced shifts (> 2 units) in the pKa values of both acidic and basic residues in favor of their neutral state (50, 51). Despite the energetic costs associated with the conformational and solvation changes described above, the overall rearrangement is ultimately favorable because aromatic substituents on the P2 proline of C-terminal carboxylic acid inhibitors provide a substantial gain in potency (15, 40).
NS3-NS4A protease inhibitors are poised to fill a large unmet medical need in the treatment of HCV infection. For one of these inhibitors, BI 201335, we have employed an array of biochemical and structural techniques to create a detailed understanding of its unique interactions with its target. Such insight is key to understanding the differing properties of the various clinical candidates and to the design of the next generation of NS3-NS4A protease inhibitors.
Supplementary Material
Acknowledgments
We thank Professor Kenneth A. Johnson for discussions on kinetic data fitting and interpretation. We also acknowledge Vida Gorys, who synthesized the compounds used in these experiments.
All of the authors are employees of Boehringer Ingelheim (Canada) Ltd.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
The atomic coordinates and structure factors (codes 3P8N and 3P8O) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- HCV
- hepatitis C virus
- FL-protease
- full-length NS3-NS4A heterodimer
- sc-protease
- NS3 protease domain with NS4A central peptide region covalently fused to the N terminus
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1. Lavanchy D. (2009) Liver Int. 29, 74–81 [DOI] [PubMed] [Google Scholar]
- 2. Ghany M. G., Strader D. B., Thomas D. L., Seeff L. B. American Association for the Study of Liver Diseases (2009) Hepatology 49, 1335–137419330875 [Google Scholar]
- 3. Gallinari P., Brennan D., Nardi C., Brunetti M., Tomei L., Steinkühler C., De Francesco R. (1998) J. Virol. 72, 6758–6769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim J. L., Morgenstern K. A., Lin C., Fox T., Dwyer M. D., Landro J. A., Chambers S. P., Markland W., Lepre C. A., O'Malley E. T., Harbeson S. L., Rice C. M., Murcko M. A., Caron P. R., Thomson J. A. (1996) Cell. 87, 343–355 [DOI] [PubMed] [Google Scholar]
- 5. Bartenschlager R., Lohmann V., Wilkinson T., Koch J. O. (1995) J. Virol. 69, 7519–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choo Q. L., Kuo G., Weiner A. J., Overby L. R., Bradley D. W., Houghton M. (1989) Science 244, 359–362 [DOI] [PubMed] [Google Scholar]
- 7. Lamarre D., Anderson P. C., Bailey M., Beaulieu P., Bolger G., Bonneau P., Bös M., Cameron D. R., Cartier M., Cordingley M. G., Faucher A. M., Goudreau N., Kawai S. H., Kukolj G., Lagacé L., LaPlante S. R., Narjes H., Poupart M. A., Rancourt J., Sentjens R. E., St George R., Simoneau B., Steinmann G., Thibeault D., Tsantrizos Y. S., Weldon S. M., Yong C. L., Llinàs-Brunet M. (2003) Nature 426, 186–189 [DOI] [PubMed] [Google Scholar]
- 8. Hinrichsen H., Benhamou Y., Wedemeyer H., Reiser M., Sentjens R. E., Calleja J. L., Forns X., Erhardt A., Crönlein J., Chaves R. L., Yong C. L., Nehmiz G., Steinmann G. G. (2004) Gastroenterology 127, 1347–1355 [DOI] [PubMed] [Google Scholar]
- 9. Kronenberger B., Zeuzem S. (2009) Ann. Hepatol. 8, 103–112 [PubMed] [Google Scholar]
- 10. Gentile I., Viola C., Borgia F., Castaldo G., Borgia G. (2009) Curr. Med. Chem. 16, 1115–1121 [DOI] [PubMed] [Google Scholar]
- 11. Mederacke I., Wedemeyer H., Manns M. P. (2009) Curr. Opin. Invest. Drugs. 10, 181–189 [PubMed] [Google Scholar]
- 12. White P. W., Llinàs-Brunet M., Amad M., Bethell R. C., Bolger G., Cordingley M. G., Duan J., Garneau M., Lagacé L., Thibeault D., Kukolj G. (2010) Antimicrob. Agents Chemother. 54, 4611–4618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Llinàs-Brunet M., Bailey M., Déziel R., Fazal G., Gorys V., Goulet S., Halmos T., Maurice R., Poirier M., Poupart M. A., Rancourt J., Thibeault D., Wernic D., Lamarre D. (1998) Bioorg. Med. Chem. Lett. 8, 2719–2724 [DOI] [PubMed] [Google Scholar]
- 14. Schechter I., Berger A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 15. Llinàs-Brunet M., Bailey M. D., Goudreau N., Bhardwaj P. K., Bordeleau J., Bös M., Bousquet Y., Cordingley M. G., Duan J., Forgione P., Garneau M., Ghiro E., Gorys V., Goulet S., Halmos T., Kawai S. H., Naud J., Poupart M. A., White P. W. (2010) J. Med. Chem. 53, 6466–6476 [DOI] [PubMed] [Google Scholar]
- 16. Thibeault D., Massariol M. J., Zhao S., Welchner E., Goudreau N., Gingras R., Llinàs-Brunet M., White P. W. (2009) Biochemistry 48, 744–753 [DOI] [PubMed] [Google Scholar]
- 17. Dahl G., Arenas O. G., Danielson U. H., Dahl G., Arenas O. G., Danielson U. H. (2009) Biochemistry 48, 11592–11602 [DOI] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 20. Brunger A. T. (2007) Nat. Protoc. 2, 2728–2733 [DOI] [PubMed] [Google Scholar]
- 21. Di Marco S., Rizzi M., Volpari C., Walsh M. A., Narjes F., Colarusso S., De Francesco R., Matassa V. G., Sollazzo M. (2000) J. Biol. Chem. 275, 7152–7157 [DOI] [PubMed] [Google Scholar]
- 22. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 23. Laskowski R. A., MacArthur M. W., Hutchinson E. G., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 24. Johnson K. A., Simpson Z. B., Blom T. (2009) Anal. Biochem. 387, 30–41 [DOI] [PubMed] [Google Scholar]
- 25. Barbato G., Cicero D. O., Nardi M. C., Steinkühler C., Cortese R., De Francesco R., Bazzo R. (1999) J. Mol. Biol. 289, 371–384 [DOI] [PubMed] [Google Scholar]
- 26. Yao N., Reichert P., Taremi S. S., Prosise W. W., Weber P. C. (1999) Structure 7, 1353–1363 [DOI] [PubMed] [Google Scholar]
- 27. Tsantrizos Y. S., Bolger G., Bonneau P., Cameron D. R., Goudreau N., Kukolj G., LaPlante S. R., Llinàs-Brunet M., Nar H., Lamarre D. (2003) Angew. Chem. Int. Ed. Engl. 42, 1356–1360 [DOI] [PubMed] [Google Scholar]
- 28. Cummings M. D., Lindberg J., Lin T. I., de Kock H., Lenz O., Lilja E., Felländer S., Baraznenok V., Nyström S., Nilsson M., Vrang L., Edlund M., Rosenquist A., Samuelsson B., Raboisson P., Simmen K. (2010) Angew. Chem. Int. Ed. Engl. 49, 1652–1655 [DOI] [PubMed] [Google Scholar]
- 29. Auffinger P., Hays F. A., Westhof E., Ho P. S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 16789–16794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Markley J. L., Westler W. M. (1996) Biochemistry. 35, 11092–11097 [DOI] [PubMed] [Google Scholar]
- 31. Bachovchin W. W. (2001) Magn. Reson. Chem. 39, S199–S213 [Google Scholar]
- 32. Urbani A., Bazzo R., Nardi M. C., Cicero D. O., De Francesco R., Steinkühler C., Barbato G. (1998) J. Biol. Chem. 273, 18760–18769 [DOI] [PubMed] [Google Scholar]
- 33. McCoy M. A., Senior M. M., Gesell J. J., Ramanathan L., Wyss D. F. (2001) J. Mol. Biol. 305, 1099–1110 [DOI] [PubMed] [Google Scholar]
- 34. Pelton J. G., Torchia D. A., Meadow N. D., Roseman S. (1993) Protein Sci. 2, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rajagopalan R., Misialek S., Stevens S. K., Myszka D. G., Brandhuber B. J., Ballard J. A., Andrews S. W., Seiwert S. D., Kossen K. (2009) Biochemistry 48, 2559–2568 [DOI] [PubMed] [Google Scholar]
- 36. Butkiewicz N., Yao N., Zhong W., Wright-Minogue J., Ingravallo P., Zhang R., Durkin J., Standring D. N., Baroudy B. M., Sangar D. V., Lemon S. M., Lau J. Y., Hong Z. (2000) J. Virol. 74, 4291–4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fattori D., Urbani A., Brunetti M., Ingenito R., Pessi A., Prendergast K., Narjes F., Matassa V. G., De Francesco R., Steinkühler C. (2000) J. Biol. Chem. 275, 15106–15113 [DOI] [PubMed] [Google Scholar]
- 38. Llinàs-Brunet M., Bailey M. D., Ghiro E., Gorys V., Halmos T., Poirier M., Rancourt J., Goudreau N. (2004) J. Med. Chem. 47, 6584–6594 [DOI] [PubMed] [Google Scholar]
- 39. Llinàs-Brunet M., Bailey M., Fazal G., Ghiro E., Gorys V., Goulet S., Halmos T., Maurice R., Poirier M., Poupart M. A., Rancourt J., Thibeault D., Wernic D., Lamarre D. (2000) Bioorg. Med. Chem. Lett. 10, 2267–2270 [DOI] [PubMed] [Google Scholar]
- 40. Goudreau N., Cameron D. R., Bonneau P., Gorys V., Plouffe C., Poirier M., Lamarre D., Llinàs-Brunet M. (2004) J. Med. Chem. 47, 123–132 [DOI] [PubMed] [Google Scholar]
- 41. Manns M. P., Bourlière M., Benhamou Y., Pol S., Bonacini M., Trepo C., Wright D., Berg T., Calleja J. L., White P. W., Stern J. O., Steinmann G., Yong C. L., Kukolj G., Scherer J., Boecher W. O. (2011) J. Hepatol. 10.1016/j.jhep.2010.08.040 [DOI] [PubMed] [Google Scholar]
- 42. Bondi A. (1964) J. Phys. Chem. 68, 441–451 [Google Scholar]
- 43. Lu Y., Shi T., Wang Y., Yang H., Yan X., Luo X., Jiang H., Zhu W. (2009) J. Med. Chem. 52, 2854–2862 [DOI] [PubMed] [Google Scholar]
- 44. Voth A. R., Ho P. S. (2007) Curr. Top. Med. Chem. 7, 1336–1348 [DOI] [PubMed] [Google Scholar]
- 45. Barbato G., Cicero D. O., Cordier F., Narjes F., Gerlach B., Sambucini S., Grzesiek S., Matassa V. G., De Francesco R., Bazzo R. (2000) EMBO J. 19, 1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gallo M., Pennestri M., Bottomley M. J., Barbato G., Eliseo T., Paci M., Narjes F., De Francesco R., Summa V., Koch U., Bazzo R., Cicero D. O. (2009) J. Mol. Biol. 385, 1142–1155 [DOI] [PubMed] [Google Scholar]
- 47. DeSantis G., Jones J. B. (1998) J. Am. Chem. Soc. 120, 8582–8586 [Google Scholar]
- 48. Goudreau N., Llinàs-Brunet M. (2005) Exp. Op. Invest. Drugs 14, 1129–1144 [DOI] [PubMed] [Google Scholar]
- 49. Walter J., Bode W. (1983) Hoppe-Seyler's Z. Physiol. Chem. 364, 949–959 [DOI] [PubMed] [Google Scholar]
- 50. Stranzl G. R., Gruber K., Steinkellner G., Zangger K., Schwab H., Kratky C. (2004) J. Biol. Chem. 279, 3699–3707 [DOI] [PubMed] [Google Scholar]
- 51. Harris T. K., Turner G. J. (2002) IUBMB Life 53, 85–98 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.