Abstract
p53-binding protein 1 (53BP1) is known to be an important mediator of the DNA-damage response1, with di-methylation of histone H4 lysine 20 (H4K20me2) critical to the recruitment of 53BP1 to double strand breaks (DSBs)2,3. However, it is not clear how 53BP1 is specifically targeted to the sites of DNA damage, since the overall level of H4K20me2 does not seem to increase following DNA damage. It has been proposed that DNA breaks may cause exposure of methylated H4K20 previously buried within the chromosome, however experimental evidence for such a model is lacking. Here we found that H4K20 methylation actually increases locally upon the induction of DSBs and that methylation of H4K20 at DSBs is mediated by the histone methyltransferase (HMT) MMSET (also known as NSD2 or WHSC1). Downregulation of MMSET significantly decreases H4K20 methylation at DSBs and the subsequent accumulation of 53BP1. We further found that the recruitment of MMSET to DSBs requires the γH2AX-MDC1 pathway, specifically the interaction between the MDC1 BRCT domain and phosphorylated Ser102 of MMSET. Thus, we propose that a pathway involving γH2AX-MDC1-MMSET regulates the induction of H4K20 methylation on histones around DSBs which, in turn, facilitates 53BP1 recruitment.
In response to DNA damage, 53BP1 rapidly relocalizes to the sites of DNA lesions in a phospho-H2AX (γH2AX)- and MDC1-dependent manner4–7. 53BP1 is also recruited to the sites of DNA damage through a second mechanism that involves the binding of 53BP1’s tandem tudor domains to methylated histones, with di-methylated H4K20 (H4K20me2) being the known physiological binding site for both mammalian 53BP1 and its yeast homolog Crb22,3. However, unlike H2AX phosphorylation, no increase of the total levels of H4K20me2 was observed following DNA damage2,3. It is also not clear whether 53BP1 damage recruitment regulated by H2AX phosphorylation and H4K20 methylation are separate pathways or if they are interconnected. Studies from S. pombe showed that disruption of both H4K20 methylation and H2AX phosphorylation does not cause synergistic or additive effects on the DNA damage response, suggesting that they might function in the same pathway8.
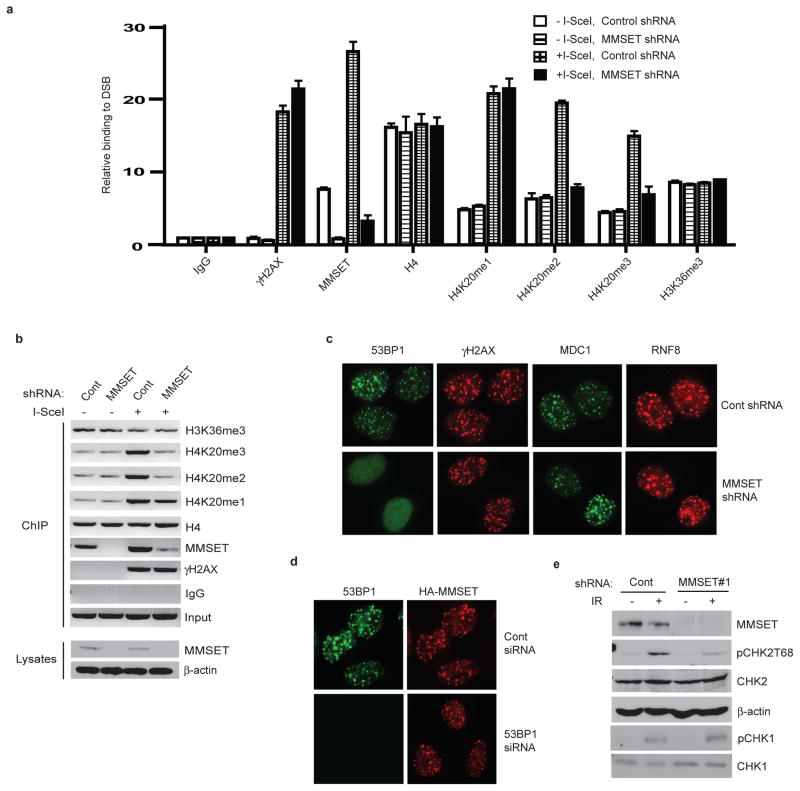
We examined H4K20 methylation at the sites of DNA damage using the cellular system (HeLa DR-GFP), in which expression of exogenous I-SceI introduces a single DSB in the cell’s genome9. After I-SceI induction of DSBs, chromatin was immunoprecipitated from the cells using antibodies directed against mono-, di- or tri-methylated H4K20 (H4K20me1/2/3), and quantitative PCR was used to determine the relative abundance of H4K20me1/2/3 at the induced break sites, while standard PCR gave visual representation of the relative accumulation of these proteins at the DSB sites. Interestingly, H4K20me1/2/3 at the I-SceI break site all increased after DSB induction (Figure 1a and b), as did H4K20me2 signal at the sites of DNA damage induced by laser irradiation (Figure 1c). Consistent with previous reports, we did not observe apparent increase in total H4K20me2 levels2,3 by Western blot at commonly used IR doses (Figure S1a), but we did observe a notable increase following high doses of ionizing radiation (IR). This suggests that local increase of H4K20me2 at DSBs induced by low doses of IR is masked from detection by Western blotting due to the high basal levels of H4K20me2 occurring throughout the genome.
Figure 1. Induction of H4K20 methylation and recruitment of MMSET at DSBs.
a and d, Examples of ChIP analysis by PCR of indicated proteins on a DSB induced by I-SceI, where input demonstrates equal amount of DNA for ChIP. b and e Quantitative PCR (Q-PCR) of indicated ChIP samples, where the Y axis represents the relative enrichment of the indicated proteins compared to the IgG control (after normalized with a PCR internal control to a locus other than the DSB; ± s.e.m., n=3). c and f immunofluorescence (IF) staining of 293T cells after indicated treatments, then stained with indicated antibodies.
We next investigated how the increase of H4K20me2/3 was induced at DSBs. It has been proposed that SET8 is mainly responsible for H4K20me110,11, which is required for subsequent di- and tri-methylation of H4K20. SUV4-20h1 and SUV4-20h2 are the major enzymes responsible for H4K20me2 and H4K20me3, respectively12,13. However, despite Suv4-20h1/2 loss and the subsequent lack of most H4K20me2/3, 53BP1 accumulation at DSBs was not abolished and only slightly delayed13. We did not observe substantial accumulation of Suv4-20h1 at the DSBs, while small amounts of SET8 localized to the I-SceI site both prior to and after DNA cleavage (Figure 1d and e). This suggests that other HMTs methylate H4K20 specifically at DSBs. Interestingly, we found that MMSET (also called WHSC1 or NSD2), a newly identified HMT14–16, accumulated at DSBs (Figure 1d, 1e and S1b).
Consistent with the results obtained from ChIP assays, MMSET formed discrete foci following IR, colocalizing with 53BP1 (Figure 1f). MMSET has been shown to methylate H3K36, H3K27 and H4K2014–16, while misregulation of MMSET due to haplo-insufficiency in Wolf-Hirschhorn syndrome17, and by t(4;14) chromosome translocation in multiple myeloma (MM)18,19 suggests it has an important role in the pathogenesis of these diseases. However, the cellular function of MMSET is largely uncharacterized. Our results in Figure 1d–f imply that MMSET plays a role in the DNA damage response. In support of this, downregulation of MMSET resulted in cellular hypersensitivity to IR (Figure S1c).
We hypothesized that MMSET regulates DNA damage response through H4K20 methylation at DSBs. As shown in Figure 2a–b and S2a–b, downregulation of MMSET significantly decreased H4K20me2/3 at DSBs, but did not significantly affect H4K20me1 or H3K36 methylation at DSBs. We reasoned that if MMSET regulates H4K20me2 at DSBs, then MMSET should regulate the recruitment 53BP1 to DSBs. Indeed, we found that downregulation of MMSET significantly decreased DNA damage-induced focus formation of 53BP1, but not γH2AX, MDC1 or RNF8 which are upstream regulators of 53BP1 (Figure 2c, Figure S2c–d). Furthermore, 53BP1 focus formation was defective in cells overexpressing a truncated MMSET (H929)15, while in cells expressing full-length MMSET (KMS11), 53BP1 focus formation was unaffected (Figure S2e). Downregulation of 53BP1 did not affect MMSET focus formation (Figure 2d), suggesting that MMSET is an upstream regulator of 53BP1. Importantly, downstream signaling events regulated by 53BP1, such as Chk2 phosphorylation20, were impaired by downregulation of MMSET (Figure 2e, S2f). To determine whether the MMSET methyltransferase activity is required for these processes, we mutated the critical residue (F1117) required for MMSET methyltransferase activity15. We reintroduced shRNA-resistant wild-type (WT) MMSET or MMSET-F1117A to cells stably transfected with MMSET shRNA. As shown in Figure S2g–h, while WT MMSET restored H4K20 methylation and 53BP1 recruitment to DSBs, MMSET-F1117A did not. These data suggest that MMSET methylates H4K20 at DSBs, which facilitates the subsequent accumulation of 53BP1.
Figure 2. MMSET is required for H4K20me2/3 and 53BP1 accumulation at DSBs.
a, Quantitative PCR analysis of ChIP samples from HeLa-DR-GFP cells transfected with control or MMSET shRNA, where the Y axis represents the relative enrichment of the indicated proteins compared to the IgG control (± s.e.m., n=3). b, EtBr staining of ChIP samples from 2a analyzed by PCR, where input demonstrates equal loading of DNA for PCR. c–d, IF of HCT116 cells transfected with the indicated siRNA or shRNA, irradiated (5Gy), and stained with indicated antibodies. e, Phosphorylation of CHK1/2 in the cell lysates from 2c analyzed by immunoblotting.
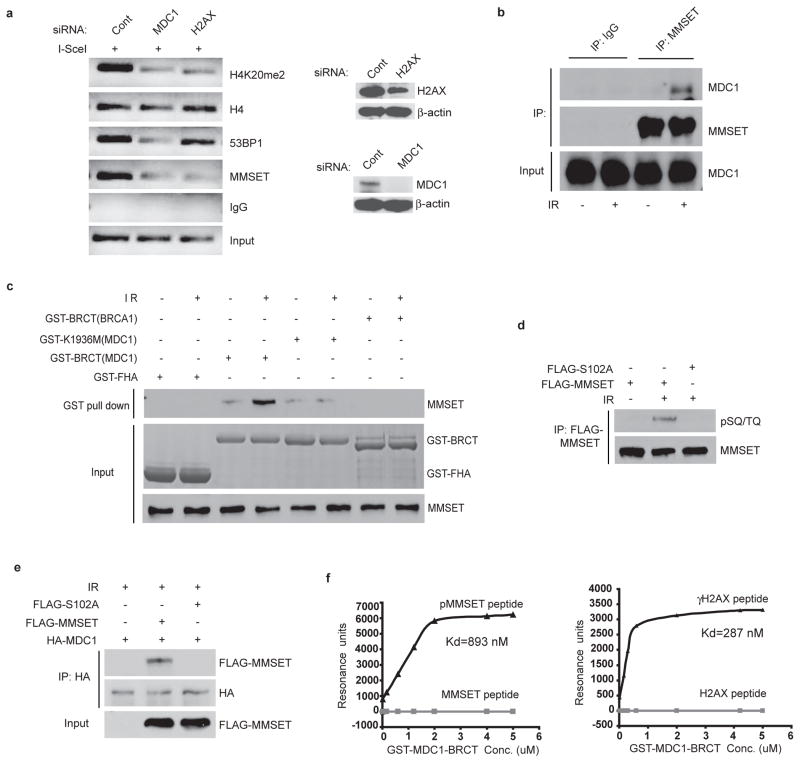
Previous studies indicated that the accumulation of 53BP1 at the sites of DNA damage also requires H2AX and MDC14–7. On investigation of this potential connection we found that MMSET accumulation at DSBs was significantly reduced in cells depleted of H2AX and MDC1 (Figure 3a and S3a), as was H4K20me2 and the accumulation of 53BP1. Furthermore, MDC1 foci appeared earlier than those of MMSET, while MMSET foci formed earlier than those of 53BP1 (Figure S3b). Thus the accumulation of MMSET and the subsequent methylation of H4K20 and 53BP1 recruitment at DSBs appear to require H2AX and MDC1.
Figure 3. Recruitment of MMSET to DSBs requires the ATM-H2AX-MDC1 pathway.
a, ChIP analysis by PCR of indicated proteins at DSBs in HeLa-DR-GFP cells transfected with the indicated siRNA. Right panels: Western blots of H2AX and MDC1. b, Coimmunoprecipitation of MMSET and MDC1 in HeLa cells before or after IR. c, GST pull down assay of MMSET using indicated GST-fusion proteins. d, 293T cells treated and immunoprecipitated as indicated, then analyzed with anti-pSQ/TQ antibody. e, 293T cells transfected with the indicated constructs were treated as indicated, then immunoprecipitated and immunoblotted with indicated antibodies. f, the interaction between GST-MDC1-BRCT and indicated peptides were measured by BIAcore 3000.
Previous studies also indicate that downstream of MDC1, the E3 ubiquitin ligase RNF8 regulates 53BP1 foci formation through its role in histone ubiquitination21–23. It is unclear whether RNF8 and MMSET regulate 53BP1 accumulation in parallel or in the same pathway. As shown in Figure S4a, downregulation of RNF8 did not affect MMSET recruitment and H4K20 methylation, although 53BP1 recruitment was compromised. In addition, downregulation of MMSET had no effect on the recruitment of RNF8 to DSBs or the ubiquitination of H2A at DSBs (Figures 2c, S2c and S4b) suggesting that RNF8 and MMSET function in distinct pathways. Thus the mechanism through which RNF8 mediated ubiquitination events regulate 53BP1 recruitment remains to be determined. While investigating how the H2AX-MDC1 pathway regulates MMSET accumulation at DSBs, we found that MMSET interacted with MDC1 in a DNA damage-inducible manner (Figure 3b). The interaction appeared specific to the MDC1-BRCT domain, as the BRCA1-BRCT domain and MDC1 BRCT domain mutant K1936M24 was unable to interact (Figure 3c, S4c)).
Since BRCT domains recognize phospho-Ser/Thr motifs25,26, it is likely that MMSET is phosphorylated following DNA damage, thereby facilitating its interaction with MDC1. As shown in Figure S4d, MMSET was phosphorylated at ATM consensus SQ/TQ sites after IR. No phospho-SQ/TQ signal was detected in ATM-deficient MEFs or in samples treated with λ-phosphatase, suggesting that MMSET is phosphorylated in an ATM-dependent manner. A previous large-scale proteomic study demonstrated that Ser102 of MMSET is phosphorylated by ATM following DNA damage27. As shown in Figure 3d, mutation at Ser102 abolished ATM-dependent MMSET phosphorylation following DNA damage, indicating that S102 is the major ATM phosphorylation site of MMSET. Furthermore, mutation at Ser102 abolished the MDC1-MMSET interaction (Figure 3e), verifying that the phosphorylation of Ser102 is required for MDC1 binding. The MDC1-BRCT domain has been shown to bind phospho-139 (pS139QEY) of γH2AX, and a C-terminal Y at the +3 position is critical for the binding specificity, although E at +2 is also positively selected24,28. The MMSET sequence following S102 is QEM, and does not contain Y at the +3 position. To further confirm the specificity of the MDC1-MMSET interaction we used peptides containing either S102 or pS102 to perform several assays. As shown in Figure S4e, phosphopeptides of MMSET preferentially pulled-down endogenous MDC1 from cell lysates. We further determined the binding affinity between MMSET peptides and the MDC1-BRCT domain using surface plasmon resonance. We found that the MDC1-BRCT domain preferentially bound MMSET phosphopeptides (Kd=893 nM), although with a lower affinity than it did γH2AX peptides (Kd=287 nM). No MDC1-binding was found for non-phosphopeptides of MMSET (Figure 3f).
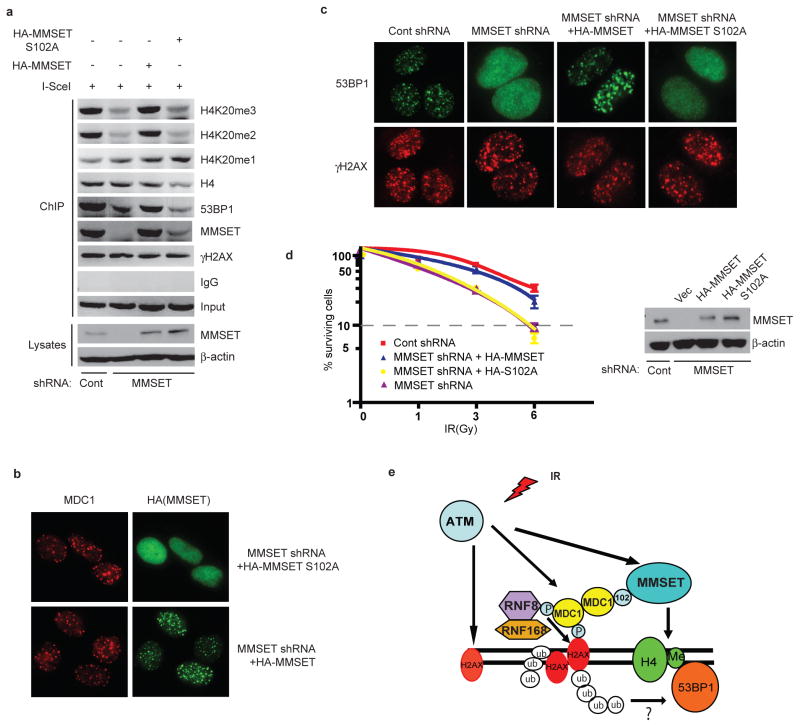
To investigate the functional significance of MMSET phosphorylation, we stably transfected HeLa-DR-GFP cells with MMSET shRNA, and reconstituted these cells with shRNA-resistant MMSET-WT or the MMSET-S102A mutant. As shown in Figure 4a–b and S5a, MMSET-WT was recruited to DSBs, but the recruitment of MMSET-S102A was defective. This suggests that S102 phosphorylation and the MDC1-MMSET interaction are essential for MMSET accumulation at DSBs. Similarly, reconstitution of MMSET-WT, but not MMSET-S102A, rescued H4K20me2/3, 53BP1 accumulation at DSBs and Chk2 phosphorylation (Figure 4a, 4c, S5a–5d). It is possible that S102A mutation affects MMSET methyltransferase activity and subsequent H4K20 methylation. However, we found that the activity of MMSET and MMSET-S102A toward histone H4 is comparable before and after IR, suggesting the above described defects caused by S102A mutation are not due to a decreased methyltransferase activity (Figure S5e–f).
Figure 4. Phosphorylation of MMSET is important for H4K20 methylation, 53BP1 recruitment and DNA damage response.
a, HeLa DR-GFP cells were transfected with the indicated constructs, H4K20 methylation and MMSET recruitment was analyzed by PCR of ChIP samples. b–c, HCT116 cells transfected with indicated constructs were irradiated, and 10 min later, stained with indicated antibodies. d, Radiation sensitivity of cells from Figure 4c was determined by colony formation (±s.e.m., n=3). E, Model demonstrating how the MDC1-MMSET pathway regulates DNA damage-induced histone H4 Lysine 20 methylation and 53BP1 foci formation.
Given that MDC1’s BRCT domain is required for MDC1’s binding to γH2AX at DNA damage sites, how does MDC1 use this same domain to recruit MMSET to DSBs? We found that MDC1 formed oligomers (Figure S5g) and DNA damage increased the oligomerization of MDC1 (Figure S5h). Therefore, it is likely that different molecules in the MDC1 multimers bind γH2AX and MMSET separately at the DNA damage sites. Finally, to examine how MMSET phosphorylation ultimately affects cellular sensitivity to DNA damage, we performed colony formation assays. Depletion of MMSET resulted in a significant increase in IR sensitivity (Figure 4d), and reconstitution with MMSET-WT could reverse this effect, whereas MMSET-S102A could not.
In summary, our studies reveal a critical role of the methyltransferase MMSET in regulating the assembly of 53BP1 foci at DNA lesions (Figure 4e). We show that H4K20 methylation, unlike the previously-held view, is induced at DSBs. We also establish a previously unrecognized link between the H2AX-MDC1 pathway and H4K20 methylation, and show that MMSET connects these two pathways. These results suggest that multiple myeloma tumors with t(4;14) translocation and MMSET dysregulation may have aberrant responses to DNA damage, which may be related to the poor prognosis observed in this subgroup of patients that are treated with DNA alkylating agents.
Methods Summary
Hela DR-GFP and MDA-MB-231 Ros8 cell lines were used for the ChIP assays which were subsequently analyzed by PCR or Q-PCR. Co-immunoprecipitation was used to detect a protein–protein interactions in vivo and SPR were used to detect the affinity for the protein and peptide interaction in vitro. Transient transfection of siRNA or stable downregulation by shRNA was used to decrease the level of specific proteins. Immunofluorescence staining was used to visualize protein accumulation and localization after DNA damage.
Supplementary Material
Acknowledgments
We thank Dr. Marin Jasin (Memorial Sloan-Kettering Cancer Center) for providing the HeLa-DR-GFP cells. We thank Dr. Stepehn Baylin (Johns Hopkins University) for providing MDA-MB-231 cells with inducible I-SceI expression. We thank Dr. Michal Goldbeg (Hebrew Universitu, Isreal), Xiaochun Yu (University of Michigan), Manuel Stucki (University of Zurich) for providing MDC1 and BRCA1 constructs. We thank Dr. Michael Huen (University of Hong Kong) for providing 53BP1 expression constructs. We thank Dr. Jonathan D. Licht (Northwestern University, Chicago, IL) for providing MMSET antibodies and Dr. Ben Ho Park (Johns Hopkins University) for providing MMSET shRNAs. This work was supported by Richard Schulze Family Foundation, and NIH (CA130996 and CA151329, ZL), and a grant from the American Cancer Society (IRG-58-010-52, ZY) and a Siteman Career Award in Breast Cancer Research (ZY).
Footnotes
Author Contributions
H.P. designed and performed the experiments; L.Z., K.L., Y.Q., and F.F. performed some experiments; M.C., P.L.B. provided essential tools; L.W., Z.Y and Z.L designed the experiments and supervised the project.
Author information
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to lou.zhenkun@mayo.edu.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full methods accompany this paper.
References
- 1.FitzGerald JE, Grenon M, Lowndes NF. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- 2.Botuyan MV, et al. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanders SL, et al. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Celeste A, et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol. 2003;5:675–679. doi: 10.1038/ncb1004. [DOI] [PubMed] [Google Scholar]
- 5.Bekker-Jensen S, Lukas C, Melander F, Bartek J, Lukas J. Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J Cell Biol. 2005;170:201–211. doi: 10.1083/jcb.200503043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lou Z, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 8.Du LL, Nakamura TM, Russell P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006;20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 10.Fang J, et al. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- 11.Nishioka K, et al. PR-Set7 is a nucleosome-specific methyltransferase that modifies lysine 20 of histone H4 and is associated with silent chromatin. Mol Cell. 2002;9:1201–1213. doi: 10.1016/s1097-2765(02)00548-8. [DOI] [PubMed] [Google Scholar]
- 12.Schotta G, et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schotta G, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JY, et al. Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol Cell Biol. 2008;28:2023–2034. doi: 10.1128/MCB.02130-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marango J, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–3154. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nimura K, et al. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature. 2009;460:287–291. doi: 10.1038/nature08086. [DOI] [PubMed] [Google Scholar]
- 17.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf-Hirschhorn syndrome. Trends Genet. 2005;21:188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 18.Chesi M, et al. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–3034. [PubMed] [Google Scholar]
- 19.Stec I, et al. WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet. 1998;7:1071–1082. doi: 10.1093/hmg/7.7.1071. [DOI] [PubMed] [Google Scholar]
- 20.Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- 21.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 24.Stucki M, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 26.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 27.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 28.Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J Biol Chem. 2005;280:32053–32056. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- 29.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 30.You Z, et al. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.