Abstract
The history of the genus Pan is a topic of enduring interest. Chimpanzees (Pan troglodytes) are often divided into subspecies, but the population structure and genetic history of chimpanzees across Africa remain unclear. Some population genetics studies have led to speculation that, until recently, this species constituted a single population with ongoing gene flow across its range, which resulted in a continuous gradient of allele frequencies. Chimpanzees, designated here as P. t. ellioti, occupy the Gulf of Guinea region that spans southern Nigeria and western Cameroon at the center of the distribution of this species. Remarkably, few studies have included individuals from this region, hindering the examination of chimpanzee population structure across Africa. Here, we analyzed microsatellite genotypes of 94 chimpanzees, including 32 designated as P. t. ellioti. We find that chimpanzees fall into three major populations: (i) Upper Guinea in western Africa (P. t. verus); (ii) the Gulf of Guinea region (P. t. ellioti); and (iii) equatorial Africa (P. t. troglodytes and P. t. schweinfurthii). Importantly, the Gulf of Guinea population is significantly different genetically from the others, sharing a last common ancestor with the populations in Upper Guinea ~0.46 million years ago (mya) and equatorial Africa ~0.32 mya. Equatorial chimpanzees are subdivided into up to three populations occupying southern Cameroon, central Africa, and eastern Africa, which may have constituted a single population until ~0.10–0.11 mya. Finally, occasional hybridization may be occurring between the Gulf of Guinea and southern Cameroon populations.
Keywords: chimpanzees, phylogenetics, population biology, microsatellites
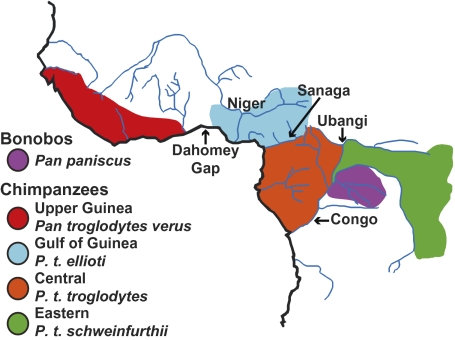
Revealing the histories of chimpanzees (Pan troglodytes) and bonobos (Pan paniscus) is of widespread and enduring interest due to their close relationship with humans. A recent proliferation of research shows that we are just beginning to unravel their complex histories (1), and even now, we still debate the number of species and subspecies belonging to the genus Pan. Most authorities agree that chimpanzees and bonobos should be classified as different species, with their ranges separated by the Congo River (Fig. 1; ref. 2). Chimpanzees are frequently further divided into subspecies across tropical Africa (2, 3), but this classification is disputed (4). P. t. verus occupies the Upper Guinea region of western Africa (2, 3). The range of P. t. troglodytes extends through central Africa. P. t. schweinfurthii occupies eastern Africa (2, 3) and has been proposed to be split into two subspecies, P. t. schweinfurthii and P. t. marungensis, based on craniometric variation (5).
Fig. 1.
Distribution and current taxonomy of Pan.
The subspecies name P. t. ellioti (6), formerly P. t. vellerosus (7, 8), has been proposed for chimpanzees from the Gulf of Guinea region, which spans southern Nigeria and western Cameroon (9). However, relatively little is known about the genetic history of chimpanzees from this region, and some question their distinctiveness from adjacent populations (4). Analyses of mtDNA sequences (8, 10, 11) suggest the following: (i) P. t. ellioti shared a last common ancestor with P. t. verus between 0.4 and 0.6 mya; (ii) the ranges of P. t. ellioti and P. t. troglodytes converge in central Cameroon at the Sanaga River; and (iii) a small hybrid zone between these subspecies may exist in central Cameroon around the confluence of the upper Sanaga River and its main tributary, the Mbam River. Thus, this evidence suggests that chimpanzees found in the Gulf of Guinea region may have a unique population history.
Moreover, Cameroon appears to be important in understanding the natural history of some infectious diseases that crossed from apes to humans, notably HIV–AIDS (12). Chimpanzee simian immunodeficiency virus (SIVcpz) found in southern Cameroon is the progenitor of HIV-1 groups M and N in humans (12). SIVcpz is found at rates exceeding 30% at some localities in the ranges of P. t. troglodytes and P. t. schweinfurthii (12). Natural SIVcpz infection is associated with a lethal AIDS-like syndrome in P. t. schweinfurthii (13), which contributed to a rapid decline of the Kalande community at Gombe National Park in Tanzania (14). SIVcpz does not appear to occur naturally in P. t. ellioti (12) or in P. t. verus (15, 16). The reasons for this difference in the distribution of SIVcpz between chimpanzee subspecies remain unclear, but might be explained by a lack of migration (17), extinctions of infected communities (14), resistance to SIVcpz infection in P. t. ellioti and P. t. verus, or a combination of these factors. A clearer understanding of chimpanzee population structure is key to understanding the natural history of SIVcpz.
Recent studies of chimpanzee genetic diversity have included many regions of the genomes of individuals designated as P. t. verus, P. t. troglodytes, and P. t. schweinfurthii (4, 11, 18–23), but few from P. t. ellioti. These studies suggest that P. t. verus shared a last common ancestor with P. t. troglodytes/P. t. schweinfurthii 0.38–0.55 mya. P. t. troglodytes and P. t. schweinfurthii from central and eastern Africa share more recent ancestry with each other, and the differences separating them appear to be similar in scale to those observed to distinguish some human populations from each other (4). These findings have led some researchers to hypothesize that chimpanzees were, until recently, a single population characterized by a continuous gradient of gene frequencies across their entire range, and thus to propose that chimpanzees should not be classified into subspecies (4). The ways that P. t. ellioti contributes to this picture remain unclear. Becquet et al. (19) have speculated, based on simulations rather than genetic data from multiple individuals, that P. t. ellioti would not differ from the other subspecies by allele frequency differentiation values (FST) of >0.09, leading them to conclude that individuals designated as P. t. ellioti are unlikely to comprise a population that is distinguishable from other chimpanzees (19).
Here, we infer the genetic structure of chimpanzee populations from wild-born chimpanzees (P. troglodytes) and bonobos (P. paniscus) to examine how chimpanzees from Cameroon contribute to continental patterns of chimpanzee population structure. Previously, we inferred the geographic origins of new chimpanzee samples included in this study (n = 45) and concluded that 32 originated in western Cameroon and may belong to P. t. ellioti, 12 originated from southern Cameron and may belong to P. t. troglodytes, and 1 may be a recent hybrid between these subspecies (24). We genotyped these individuals at 27 autosomal microsatellite loci (Table S1) that had been used previously by Becquet et al. (19) to study chimpanzees from other regions of Africa. We analyzed these newly generated data (Table S2) along with genotypes for the same loci for six bonobos and 49 chimpanzees reported (19) to be from P. t. verus (Upper Guinea, n = 31), P. t. troglodytes (central Africa, n = 12), and P. t. schweinfurthii (eastern Africa, n = 6). This approach allowed us to address whether autosomal DNA data can be used to assign chimpanzees from the Gulf of Guinea region into a category that is separate from those in Upper Guinea or central and eastern Africa as well as to compare their population histories.
Results
Population Structure.
Cluster analysis.
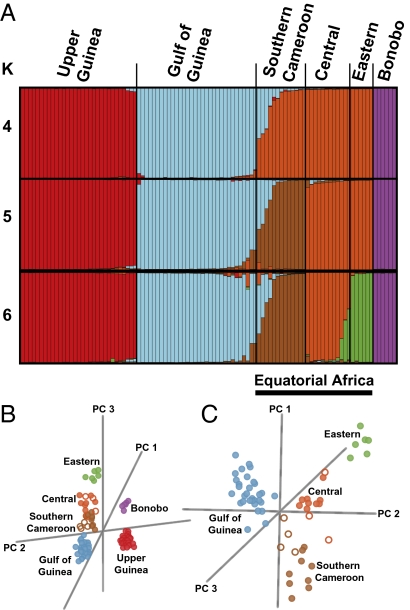
We examined individual ancestry and the number of ancestral chimpanzee population clusters (KMAX) by applying a Bayesian model-based clustering approach (Fig. 2A) implemented in the STRUCTURE software package (25). These analyses were done blinded to a priori population labels. We used several criteria to infer KMAX; this revealed a minimum of four (K = 4, ΔK; ref. 26) and a maximum of six (K = 6, posterior probability distribution, PPD, values; ref. 25) ancestral population clusters on a continental level (Fig. S1 A and B). Chimpanzees from the Gulf of Guinea were separated from chimpanzees in Upper Guinea, chimpanzees in equatorial Africa (including southern Cameroon, other parts of central Africa, and eastern Africa), and bonobos at K = 4. Setting K = 5 distinguished chimpanzees in southern Cameroon from those in central and eastern Africa, whereas setting K = 6 distinguished chimpanzees in central Africa from those originating in eastern Africa. Seven individuals were identified as having >25% ancestry from more than one population. Four of these individuals have inferred origins in southern Cameroon (24) and show shared ancestry with individuals from the Gulf of Guinea and southern Cameroon. The three remaining individuals have inferred origins in central Africa (19) and show shared ancestry with individuals from eastern Africa and central Africa.
Fig. 2.
Cluster analysis of Pan. (A) STRUCTURE plots for K = 4–6. Each vertical line represents an individual, and colors represent their inferred ancestry from K ancestral populations. (B) PCA created on the basis of individual genotypes for the Pan dataset. (C) PCA of the Gulf of Guinea/equatorial Africa chimpanzee dataset. Individuals indicated by open circles showed ≥25% ancestry in more than one ancestral cluster (K) by STRUCTURE analysis.
Principal components analysis.
We next applied a principal component analysis (PCA) method that incorporated a significance test of each principal component (PC; ref. 27), again blinded to a priori population labels. The PCA for the full dataset (Fig. 2B) classified the first five eigenvectors as significant (P < 0.05). PC 1 separated bonobos from chimpanzees, and accounted for 33.8% of the extracted variation. PC 2 extracted 28.1% of the variation, and separated the Upper Guinea chimpanzees from the remaining chimpanzees. PC 3 extracted 16.6% of the variation and separated chimpanzees from the Gulf of Guinea from those originating in equatorial Africa (including southern Cameroon, other parts of central Africa, and eastern Africa). PC 1 through PC 3 distinguished the same four populations as the STRUCTURE analysis, but PC 4 and PC 5 did not match the STRUCTURE analysis at K = 5 and K = 6. Instead, PC 4 extracted 11.2% of the variation and distinguished east African chimpanzees from those in central Africa. PC 5 extracted 10.3% of the variation, distinguishing chimpanzees in southern Cameroon from those originating elsewhere in central Africa. We completed a second PCA (Fig. 2C) that excluded bonobos and Upper Guinea chimpanzees to explore whether a gradient might exist across the Gulf of Guinea and equatorial Africa. The Gulf of Guinea/equatorial Africa PCA recovered three significant eigenvectors. PC 1 extracted 66.8% of the variation and separated the Gulf of Guinea population from the equatorial Africa population. PC 2 and PC 3 extracted 16.5% and 16.6% of the variation, respectively, distinguishing three clusters in equatorial Africa: southern Cameroon, central Africa, and eastern Africa.
Population History.
Allele frequency differentiation.
Population pairwise distance matrices were generated for the populations inferred by the cluster analysis using three different methods—D2 (28), RST (29), and (δμ)2 (30)—each sensitive to different underlying models of evolution. RST values are given in Table 1, and all three genetic distance measures were correlated; RST–(δμ)2, r2 = 0.91; (δμ)2–D2, r2 = 0.82; and RST–D2, r2 = 0.92. Each chimpanzee population was differentiated from bonobos. Allele frequency differentiation was highest between bonobos and Upper Guinea chimpanzees compared with the other populations, as reported (19). Among chimpanzees, RST values were highest between the Upper Guinea population and all other regional populations (mean RST = 0.44), followed by comparisons between the Gulf of Guinea population and populations spanning equatorial Africa (mean RST = 0.27). RST values were lowest in comparisons between southern Cameroon, central Africa, and eastern Africa (mean RST = 0.05).
Table 1.
Genetic differentiation among populations
| Bonobos | Upper Guinea | Gulf of Guinea | Southern Cameroon | Central Africa | |
| Upper Guinea | 0.82 | ||||
| Gulf of Guinea | 0.70 | 0.41 | |||
| Southern Cameroon | 0.66 | 0.43 | 0.25 | ||
| Central Africa | 0.70 | 0.46 | 0.27 | 0.07 | |
| Eastern Africa | 0.70 | 0.44 | 0.28 | 0.05 | 0.03 |
Bold values were significant by 10,000 permutations of the data. Values based on an RST (29) model of evolution that assumes that mutations follow the SMM.
We performed an analysis of molecular variance (AMOVA) from this sample of chimpanzees and compared it with AMOVA results from a global sample of humans partitioned into five continental clusters reported in Rosenberg et al. (ref. 31; Table 2). Allele frequency differentiation among the Upper Guinea, Gulf of Guinea, and equatorial Africa populations accounted for 30.1% of the variation among regions and 64.2% of the variation within populations. In the global human sample, 4.3% of the variation among the five regions and 93.2% of the variation within populations was attributable to allele frequency differentiation.
Table 2.
AMOVA in chimpanzees versus humans
| Variance components | |||||
| Sample | Number of regions | Number of populations | Within populations | Among populations within regions | Among regions |
| Chimpanzees | 3* | 5 | 64.2 | 5.7 | 30.1 |
| Humans (global)† | 5 | 52 | 93.2 | 2.5 | 4.3 |
*Three regions were considered in this analysis: Upper Guinea, Gulf of Guinea, and equatorial Africa (including southern Cameroon, other parts of central Africa, and eastern Africa).
†AMOVA results for humans were based on the HDGP–CEPH panel as reported in Rosenberg et al. (31).
Phylogenetic analysis.
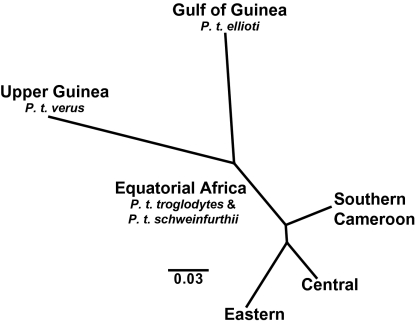
Unrooted neighbor joining trees based on the D2 (28), RST (29), and (δμ)2 (30) genetic distances are shown in Fig. 3 and Fig. S2. The three trees are identical in branching patterns, differing only in relative branch lengths. These trees are consistent with three geographically distinguishable groupings of chimpanzees from (i) Upper Guinea, (ii), the Gulf of Guinea, and (iii) equatorial Africa. Along the equatorial Africa branches of the trees, additional nodes separate the populations from southern Cameroon, central Africa, and eastern Africa.
Fig. 3.
Neighbor-joining phylogram of chimpanzee populations constructed using D2 (28) allele frequency differentiation values.
Population divergence times.
Table 3 lists times to the most recent common genetic ancestor (TMRCA) estimated for various population groupings. TMRCA estimates represent the maximum time of divergence for genetic lineages that predate population split times due to incomplete lineage sorting in the ancestral population before population divergence (32). We estimate that chimpanzees from the Gulf of Guinea shared a last common ancestor with chimpanzees from Upper Guinea 0.46 mya and with chimpanzees from southern Cameroon 0.32 mya. Likewise, we estimate that chimpanzees in southern Cameroon shared a common genetic ancestor with chimpanzees in central and eastern Africa 0.11 mya. Chimpanzees in central and eastern Africa shared a last common ancestor about 0.10 mya. TMRCA dates for chimpanzees from Upper Guinea, central Africa, and eastern Africa from this study are consistent with previous TMRCA estimates from autosomal resequencing data (20–23).
Table 3.
Estimates of population divergence times
| TMRCA* | This study | Wegmann and Excoffier (23) | Hey (22) | Caswell et al. (21) | Becquet and Przeworski (20) |
| Bonobo–Chimpanzee | 1.8 (1.5–2.3) | 1.6 (1.01–2.00) | 0.93 (0.68–1.54) | 1.29 (1.14–1.45) | 0.77 (0.58–1.00) |
| Upper Guinea–Central/Eastern (excluding Cameroon)† | 0.43 (0.34–0.49) | 0.55 (0.34–0.91) | 0.46 (0.35–0.65) | 0.51 (0.43–0.59) | 0.38 (0.27–0.94) |
| Upper Guinea–Gulf of Guinea | 0.46 (0.37–0.53) | — | — | — | — |
| Gulf of Guinea–Equatorial Africa‡ | 0.32 (0.25–0.36) | — | — | — | — |
| Southern Cameroon–Central/Eastern | 0.11 (0.09–0.13) | — | — | — | — |
| Central–Eastern | 0.10 (0.08–0.12) | 0.44 (0.20–0.50) | 0.093 (0.04–0.16) | — | 0.22 (0.14–1.40) |
*Time is expressed in millions of years ago (mya).
†Estimated from data included in the dataset reliability tests performed for this study (SI Text, Table S3).
‡Equatorial Africa includes chimpanzees from southern Cameroon, other parts of central Africa, and eastern Africa.
Dataset Validation.
We applied three methods to examine the reliability of these data, including: (i) analyzing the 27-locus dataset minus chimpanzees from Cameroon compared with the 310-locus dataset analyzed by Becquet et al. (19); (ii) applying a rarefaction procedure of constraining population sample sizes to be equal; and (iii) applying another rarefaction procedure that examined allele richness, unique alleles, and shared private alleles corrected for unequal sample size between populations (33–35). Results of the first test are given in Fig. S3 and Table S3. This test revealed that the number of loci analyzed in this study (n = 27) adequately captured the major population partitions of chimpanzees, as reported by Becquet et al. (19) from a larger set of microsatellite loci; again, note that this previous study did not include samples from Cameroon. Dataset reliability tests based on rarefaction methods are given in Figs. S4–S6. These tests show that unequal sample sizes had a negligible effect on our results. Two important differences, however, were as follows: (i) the distinction between eastern chimpanzees and other populations was detected at lower values of KMAX compared with the full dataset; and (ii) the southern Cameroon sample was not distinguished from those in central Africa by the criteria we used to infer KMAX. The second rarefaction method revealed that the numbers of shared private alleles between population pairs (Fig. S6) were highest for the central-eastern population pair and for the Gulf of Guinea-southern Cameroon population pair. (Details regarding the dataset validation methods and results are discussed in SI Methods and SI Results and Discussion.)
Discussion
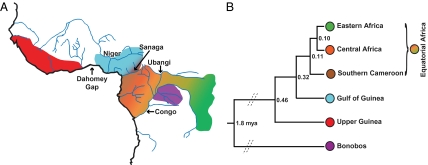
The data presented here suggest that the genus Pan includes a minimum of four (and as many as six) genetically and geographically differentiated populations (Fig. 4A). Bonobos constitute a genetically distinct population from chimpanzees, as expected based on previous studies (4, 11, 18, 19, 21–23). Chimpanzees are divided into three significantly different genetic populations, which inhabit Upper Guinea, the Gulf of Guinea region, and equatorial Africa. The equatorial chimpanzees may be further subdivided into a geographically nested group consisting of up to three populations (southern Cameroon, other parts of central Africa, and eastern Africa) that shared last common ancestors about 0.10–0.11 mya (Fig. 4B and Table 3). We speculate that, until recently, the group of populations spanning equatorial Africa displayed a geographical gradient of allele frequencies with ongoing gene flow between groups (Fig. 4A), as suggested by previous studies (4, 19). However, this closely related series of populations does not include chimpanzees from the Gulf of Guinea region. Instead, chimpanzees from this region constitute a population that is significantly different from all other populations (Fig. 2); this population shared a last common ancestor with the Upper Guinea population about 0.46 mya and with the equatorial Africa population about 0.32 mya. We observed a high number of shared private alleles between individuals from the Gulf of Guinea and the southern Cameroon populations (Fig. S6), and four individuals from Cameroon showed >25% ancestry in both populations. These data, considered jointly with these populations’ 0.32 mya TMRCA (Table 3) and their relatively high allele frequency differentiation values (Table 1), could indicate a pattern of recent introgression between these populations, consistent with previous studies suggesting the existence of a small hybrid zone in central Cameroon (8, 24). Finally, Upper Guinea chimpanzees are differentiated from all other chimpanzee populations and last shared a common ancestor with those further east about 0.46 mya.
Fig. 4.
Proposed population structure of Pan, including chimpanzees from the Gulf of Guinea region. (A) Population distribution map. (B) TMRCA between populations and phylogeny.
The concepts of species and subspecies are fluid and contentious (36), and the classification of chimpanzees is no exception (4, 19). This debate is due in part to limited knowledge about their patterns of genetic differentiation, particularly compared with such patterns in closely related species, including humans. Regional genetic differentiation (Table 2) between chimpanzees from the Upper Guinea, Gulf of Guinea, and equatorial Africa populations is higher (30.1%) than global variation in humans (4.3%). Chimpanzee populations are characterized by TMRCA estimates ranging from 0.32 to 0.55 mya (Table 3), which considerably predate the ~0.20 mya TMRCA for modern humans (37). In addition, humans have experienced a long term Ne of 10,000 and a recent population size expansion (37), whereas chimpanzees have experienced a larger Ne of up to 135,000 with marked variation in Ne between the Upper Guinea, central, and eastern populations (18, 20–23). Thus, these observations do not support the hypothesis that the patterning of genetic differentiation among chimpanzee populations across their entire range is similar to the pattern found among global human populations, as suggested previously (4). However, this hypothesis cannot be rejected if only the populations spanning equatorial Africa are considered. Indeed, evidence from several studies (4, 8, 10, 19, 22, 23) confirms our findings, indicating that the populations which span equatorial Africa exhibit a level of genetic differentiation comparable to that observed among global human populations (4). These observations suggest that there may be little justification for dividing east African chimpanzees (P. t. schweinfurthii and P. t. marungensis) into subspecies separate from P. t. troglodytes, at least on the basis of genetic data.
Based on the genetic data presented here, the simplest subdivision of the genus Pan would include two species: bonobos (P. paniscus) and chimpanzees (P. troglodytes). Chimpanzees would be further subdivided into at least three subspecies (P. t. verus, P. t. ellioti, and P. t. troglodytes), and possibly also P. t. schweinfurthii. Other classification schemes are possible. The majority of the genetic data indicate that P. t verus forms a monophyletic group (8, 11, 38) that is distantly related to other populations (4, 18–22). Consequently, we would be unsurprised to observe the emergence of a consensus supporting an earlier proposal by Morin et al. (38) that P. t. verus be elevated to full species status. If accepted, an alternative subdivision of the genus Pan would include three species: P. paniscus, P. verus, and P. troglodytes. P. troglodytes would be further subdivided into at least two subspecies, P. t. ellioti and P. t. troglodytes. The status of P. t. schweinfurthii and P. t. marungensis remains unclear; although they exhibit morphological differentiation, most of the genetic data suggest that they have not been differentiated from chimpanzees in other parts of equatorial Africa over a long period (4, 8, 10, 18–20, 22).
Several lines of evidence now indicate that chimpanzees designated as P. t. ellioti have a unique epidemiological (12), cultural, and ecological heritage (39, 40). However, morphological data for P. t. ellioti are very sparse, and the published studies are confounded by inclusion of specimens (41, 42) collected outside its geographic range (6). The range of P. t. ellioti coincides with the Gulf of Guinea biodiversity hot spot in southern Nigeria and western Cameroon (9), but because our sample of chimpanzees was limited to Cameroon (24), we cannot comment on the uncertain genetic history of chimpanzees west of the Niger River in southwest Nigeria (8). The southern extent of the range of P. t. ellioti appears to extend only to the Sanaga River in central Cameroon, a boundary that coincides with a pronounced ecotone composed of open woodland, savanna, and riparian forest connecting the Guinean and Congolian forest biomes (43). This ecotone, along with the Sanaga River, appears to influence the distribution of many insect (44), avian (45), and mammalian taxa (9, 46), including several pairs of primate species and subspecies (2, 9, 47, 48). Thus, the distribution of chimpanzee subspecies in Cameroon corresponds closely to the distributions of other organisms from the area. Genetic analyses are needed of chimpanzee DNA from known locations covering a dense sampling grid along the length of the Sanaga River and across this ecotone. Such studies will help clarify the complex history of hybridization between the Gulf of Guinea and southern Cameroon chimpanzee populations.
Methods
Dataset Preparation.
This research was carried out with Institutional Animal Care and Use Committee approval from the University at Albany, State University of New York (for M.K.G.) and in full compliance with Convention on International Trade in Endangered Species and Centers for Disease Control export and import regulations. We produced microsatellite genotype profiles for DNA samples isolated from whole blood for 45 chimpanzees housed at the Limbe Wildlife Centre. In a previous study (24), we genotyped these individuals to estimate their geographic origins by a smoothed and continuous assignment test (49) using 10 microsatellite loci that were not included in this study. This analysis revealed that: (i) the 45 chimpanzees are more likely to be from Cameroon than other parts of Africa; and (ii) the estimated geographic origins of these individuals from the assignment tests strongly corresponded to previous population labels given to them based on mtDNA sequence analysis (i.e., P. t. ellioti or P. t. troglodytes). For this study, these 45 chimpanzees were genotyped for 27 autosomal microsatellite loci previously shown to have considerable power for inferring population structure and individual ancestry (19). Information about the markers typed for this study is listed in Table S1. Allele sizes for these individuals (Table S2) were scored between two and four times using a multiplex PCR approach. We integrated newly generated microsatellite genotypes with data from wild-born chimpanzees from Upper Guinea (n = 31), central Africa (n = 12), and eastern Africa (n = 6) previously genotyped for the same loci (19). (SI Methods has detailed laboratory methods and dataset integration procedures.)
Data Analysis.
Population structure.
Cluster analyses were performed using a Bayesian clustering approach (SI Methods) implemented in the STRUCTURE version 2.3 software package (25), assuming admixture and correlated allele frequencies. Fifty iterations of the data at each K = 1–10 with 500,000 Markov Chain Monte Carlo (MCMC) burn-in steps and 1,000,000 MCMC iterations. STRUCTURE output was processed with CLUMPP (50) and plotted with DISTRUCT (51). The EIGENSOFT software package (27) was used for PCA of individual microsatellite genotypes to identify significantly different populations (SI Methods). We used a combination of methods to infer KMAX (SI Methods). Population structure analyses were performed blinded to a priori population labels.
Population history.
Three measures of population genetic differentiation were calculated using the ARLEQUIN 3.5 software package (52): D2 (28), RST (29), and (δμ)2 (ref. 30; SI Methods). The D2 genetic distance is based on a model of genetic drift (28), whereas RST and (δμ)2 assume a stepwise mutation model (SMM) (29, 30). Recent work has shown convincing evidence that the loci typed for this study appear to follow the SMM in Pan (19, 53). Allele sizes were transformed to repeat units before calculation (SI Methods). Individuals with ≥25% ancestry in more than one ancestral cluster (n = 7) from the STRUCTURE analysis (Fig. 2A) were treated as potential hybrids and excluded from population pairwise genetic distance calculations.
We constructed unrooted neighbor joining phylograms with bootstrap values based on the genetic distances for D2, RST, and (δμ)2 using programs distributed with the PHYLIP software package, version 3.5 (ref. 54; SI Methods). We calculated TMRCA dates (SI Methods) using (δμ)2 population pairwise calculations with the method described by Goldstein et al. (30) assuming a mutation rate (μ) of 1.6 × 10−4 and a 20-y generation time.
Supplementary Material
Acknowledgments
We thank the government of Cameroon for permission to conduct this research. We thank the Limbe Wildlife Centre, the Pandrillus Foundation, and the Wildlife Conservation Society for their support in Cameroon. We thank Celine Becquet and John Oates for helpful comments on early drafts of this manuscript. This research was supported by National Science Foundation Physical Anthropology Senior Research Award 0755823 (to M.K.G.), funding provided by the University at Albany — State University of New York (to M.K.G.), a David and Lucile Packard Career Award (to S.A.T.), and National Institutes of Health Pioneer Award DP1-OD-006445 (to S.A.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015422108/-/DCSupplemental.
References
- 1.Cohen J. In the shadow of Jane Goodall. Science. 2010;328:30–35. doi: 10.1126/science.328.5974.30. [DOI] [PubMed] [Google Scholar]
- 2.Groves CP. Primate Taxonomy. Washington, D.C.: Smithsonian Institution; 2001. [Google Scholar]
- 3.Oates JF, et al. Pan troglodytes. Switzerland: IUCN, Gland; 2010. IUCN Red List of Threatened Species. Version 2010.4. Available at www.iucnredlist.org. Accessed October 18, 2010. [Google Scholar]
- 4.Fischer A, Pollack J, Thalmann O, Nickel B, Pääbo S. Demographic history and genetic differentiation in apes. Curr Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 5.Groves CP. Geographic variation within eastern chimpanzees. Australas Primatol. 2005;17:19–46. [Google Scholar]
- 6.Oates JF, Groves CP, Jenkins PD. The type locality of Pan troglodytes vellerosus (Gray, 1862), and implications for the nomenclature of West African chimpanzees. Primates. 2009;50:78–80. doi: 10.1007/s10329-008-0116-z. [DOI] [PubMed] [Google Scholar]
- 7.Gonder MK, et al. A new west African chimpanzee subspecies? Nature. 1997;388:337. doi: 10.1038/41005. [DOI] [PubMed] [Google Scholar]
- 8.Gonder MK, Disotell TR, Oates JF. New genetic evidence on the evolution of chimpanzee populations, and implications for taxonomy. Int J Primatol. 2006;27:1103–1127. [Google Scholar]
- 9.Oates JF, Bergl R, Linder J. Africa's Gulf of Guinea forests: Biodiversity patterns and conservation priorities. Advances in Applied Biodiversity Science. 2004;6:1–95. [Google Scholar]
- 10.Gagneux P, Gonder MK, Goldberg TL, Morin PA. Gene flow in wild chimpanzee populations: what genetic data tell us about chimpanzee movement over space and time. Philos Trans R Soc Lond B Biol Sci. 2001;356:889–897. doi: 10.1098/rstb.2001.0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone AC, et al. More reliable estimates of divergence times in Pan using complete mtDNA sequences and accounting for population structure. Philos Trans R Soc Lond B Biol Sci. 2010;365:3277–3288. doi: 10.1098/rstb.2010.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keele BF, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keele BF, et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature. 2009;460:515–519. doi: 10.1038/nature08200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudicell RS, et al. Impact of simian immunodeficiency virus infection on chimpanzee population dynamics. PLoS Pathog. 2010;6:e1001116. doi: 10.1371/journal.ppat.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santiago ML, et al. SIVcpz in wild chimpanzees. Science. 2002;295:465. doi: 10.1126/science.295.5554.465. [DOI] [PubMed] [Google Scholar]
- 16.Switzer WM, et al. The epidemiology of simian immunodeficiency virus infection in a large number of wild- and captive-born chimpanzees: evidence for a recent introduction following chimpanzee divergence. AIDS Res Hum Retroviruses. 2005;21:335–342. doi: 10.1089/aid.2005.21.335. [DOI] [PubMed] [Google Scholar]
- 17.Sharp PM, Shaw GM, Hahn BH. Simian immunodeficiency virus infection of chimpanzees. J Virol. 2005;79:3891–3902. doi: 10.1128/JVI.79.7.3891-3902.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Won YJ, Hey J. Divergence population genetics of chimpanzees. Mol Biol Evol. 2005;22:297–307. doi: 10.1093/molbev/msi017. [DOI] [PubMed] [Google Scholar]
- 19.Becquet C, Patterson N, Stone AC, Przeworski M, Reich D. Genetic structure of chimpanzee populations. PLoS Genet. 2007;3:e66. doi: 10.1371/journal.pgen.0030066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becquet C, Przeworski M. A new approach to estimate parameters of speciation models with application to apes. Genome Res. 2007;17:1505–1519. doi: 10.1101/gr.6409707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caswell JL, et al. Analysis of chimpanzee history based on genome sequence alignments. PLoS Genet. 2008;4:e1000057. doi: 10.1371/journal.pgen.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hey J. The divergence of chimpanzee species and subspecies as revealed in multipopulation isolation-with-migration analyses. Mol Biol Evol. 2010;27:921–933. doi: 10.1093/molbev/msp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wegmann D, Excoffier L. Bayesian inference of the demographic history of chimpanzees. Mol Biol Evol. 2010;27:1425–1435. doi: 10.1093/molbev/msq028. [DOI] [PubMed] [Google Scholar]
- 24.Ghobrial L, et al. Tracing the origins of rescued chimpanzees reveals widespread chimpanzee hunting in Cameroon. BMC Ecol. 2010;10:2. doi: 10.1186/1472-6785-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 27.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slatkin M. A measure of population subdivision based on microsatellite allele frequencies. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldstein DB, Ruiz Linares A, Cavalli-Sforza LL, Feldman MW. Genetic absolute dating based on microsatellites and the origin of modern humans. Proc Natl Acad Sci USA. 1995;92:6723–6727. doi: 10.1073/pnas.92.15.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberg NA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 32.Edwards SV, Beerli P. Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution. 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- 33.Petit RJ, Mousadik AEL, Pons O. Identifying populations for conservation on the basis of genetic markers. Conserv Biol. 1998;12:844–855. [Google Scholar]
- 34.Kalinowski ST. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv Genet. 2004;5:539–543. [Google Scholar]
- 35.Szpiech ZA, Jakobsson M, Rosenberg NA. ADZE: a rarefaction approach for counting alleles private to combinations of populations. Bioinformatics. 2008;24:2498–2504. doi: 10.1093/bioinformatics/btn478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hey J. Genes, categories, and species: The evolutionary and cognitive causes of the species problem. Oxford, New York: Oxford University Press; 2001. [Google Scholar]
- 37.Campbell MC, Tishkoff SA. The evolution of human genetic and phenotypic variation in Africa. Curr Biol. 2010;20:R166–R173. doi: 10.1016/j.cub.2009.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morin PA, et al. Kin selection, social structure, gene flow, and the evolution of chimpanzees. Science. 1994;265:1193–1201. doi: 10.1126/science.7915048. [DOI] [PubMed] [Google Scholar]
- 39.Morgan BJ, Abwe EE. Chimpanzees use stone hammers in Cameroon. Curr Biol. 2006;16:R632–R633. doi: 10.1016/j.cub.2006.07.045. [DOI] [PubMed] [Google Scholar]
- 40.Fowler A, Sommer V. Subsistence technology of Nigerian chimpanzees. Int J Primatol. 2007;28:997–1023. [Google Scholar]
- 41.Taylor AB, Groves CP. Patterns of mandibular variation in Pan and Gorilla and implications for African ape taxonomy. J Hum Evol. 2003;44:529–561. doi: 10.1016/s0047-2484(03)00027-7. [DOI] [PubMed] [Google Scholar]
- 42.Pilbrow V. Population systematics of chimpanzees using molar morphometrics. J Hum Evol. 2006;51:646–662. doi: 10.1016/j.jhevol.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 43.White F. The Vegetation of Africa. Paris: UNESCO; 1983. [Google Scholar]
- 44.Simard F, et al. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smith TB, Wayne RK, Girman DJ, Bruford MW. A role for ecotones in generating rainforest biodiversity. Science. 1997;276:1855–1857. [Google Scholar]
- 46.Kingdon J. The Kingdon field guide to African mammals. Princeton: Princeton University Press; 2003. 476 pp. [Google Scholar]
- 47.Grubb P, et al. Assessment of the diversity of African primates. Int J Primatol. 2003;24:1301–1357. [Google Scholar]
- 48.Anthony NM, et al. The role of Pleistocene refugia and rivers in shaping gorilla genetic diversity in central Africa. Proc Natl Acad Sci USA. 2007;104:20432–20436. doi: 10.1073/pnas.0704816105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wasser SK, et al. Assigning African elephant DNA to geographic region of origin: applications to the ivory trade. Proc Natl Acad Sci USA. 2004;101:14847–14852. doi: 10.1073/pnas.0403170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- 51.Rosenberg NA. DISTRUCT: A program for the graphical display of population structure. Mol Ecol Notes. 2004;4:137–138. [Google Scholar]
- 52.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 53.Sun JX, Mullikin JC, Patterson N, Reich DE. Microsatellites are molecular clocks that support accurate inferences about history. Mol Biol Evol. 2009;26:1017–1027. doi: 10.1093/molbev/msp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Felsenstein J. PHYLIP–Phylogeny Inference Package (Version 3.2) Cladistic. 1989;5:164–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.