Abstract
Toll-like receptors (TLRs) are innate receptors that show high conservation throughout the animal kingdom. Most TLRs can be clustered into phylogenetic groups that respond to similar types of ligands. One exception is avian TLR15. This receptor does not categorize into one of the existing groups of TLRs and its ligand is still unknown. Here we report that TLR15 is a sensor for secreted virulence-associated fungal and bacterial proteases. Activation of TLR15 involves proteolytic cleavage of the receptor ectodomain and stimulation of NF-κB–dependent gene transcription. Receptor activation can be mimicked by the expression of a truncated TLR15 of which the entire ectodomain is removed, suggesting that receptor cleavage alleviates receptor inhibition by the leucine-rich repeat domain. Our results indicate TLR15 as a unique type of innate immune receptor that combines TLR characteristics with an activation mechanism typical for the evolutionary distinct protease-activated receptors.
Keywords: chicken, Pseudomonas, Toll-like receptor 9, leucin-rich repeat
The ability to sense the presence of infectious microbes is crucial for human and animal survival. For this reason, mammalian and nonmammalian species have developed an array of receptor proteins that recognizes specific microbial molecules and initiate defensive immune responses (1, 2). One class of innate immune receptors are the Toll-like receptors (TLRs). TLRs consist of a leucine-rich repeat (LRR) ectodomain, a transmembrane domain, and a cytosolic signaling domain [Toll/IL-1 receptor (TIR)], and operate on the cell surface or in endolysosomal compartments (3). The ligand sensing LRR domain of each individual TLR detects a distinct and conserved molecular structure, enabling the host to selectively respond to a broad variety of bacteria, viruses, and fungi. Ligand-binding induces receptor dimerization, which enables the intracellular TIR domains to recruit adaptor proteins including MyD88, Mal, TRAM, and/or TRIF. Subsequent downstream signaling leads to the activation and translocation of transcription factors NF-κB and/or IRF3 to the nucleus, which ultimately results in the production of cytokines and other immune mediators (4).
Numerous functional and crystallization studies have clarified the basic principles of TLR activation (5). Although structurally diverse, all TLR ligands appear to bridge two TLR ligand-sensing domains. For instance, the acyl chains of one molecule of the lipopeptide Pam3CSK4 bind to both TLR2 and TLR1 (6), two TLR3 receptors bind the same stretch of dsRNA (7), and LPS connects two TLR4/MD2 protein complexes (8). However, this TLR activation scenario may be oversimplified and not conserved for all TLR-related receptors. For instance, activation of the Drosophila melanogaster Toll receptor is proposed to involve conformational changes following binding of a “non-crosslinking” ligand (9). Also, recent studies have shown that TLR9 and TLR7 require proteolytic cleavage in lysosomes in a multistep process of activation, adding an additional layer of complexity to the TLR activation (10–12).
The high evolutionary conservation of TLRs throughout the animal kingdom enables phylogenetic clustering into receptor groups that are predicted to respond to similar ligands (13). For instance, all members of the TLR3, TLR4, TLR5, and TLR7/8/9 groups are predicted to sense viral RNA, bacterial LPS, bacterial flagellin, and various types of DNA and RNA, respectively. Even in the avian species that diverted from mammals approximately 300 million years ago (14), functional homologues of most of human TLRs are present (13, 15–19). One exception is the avian-specific TLR15. Phylogenetically, this TLR does not belong to any of the TLR groups that are conserved within the animal kingdom, and its ligand and activation mechanism remain unknown.
In the present study, we identify TLR15 as a sensor for secreted microbial proteases. TLR15 contains a protease-sensitive LRR domain, and proteolytic cleavage of the receptor by microbial proteases induces NF-κB activation.
Results
TLR15 Is a Surface-Localized Glycoprotein with a Typical TLR Architecture.
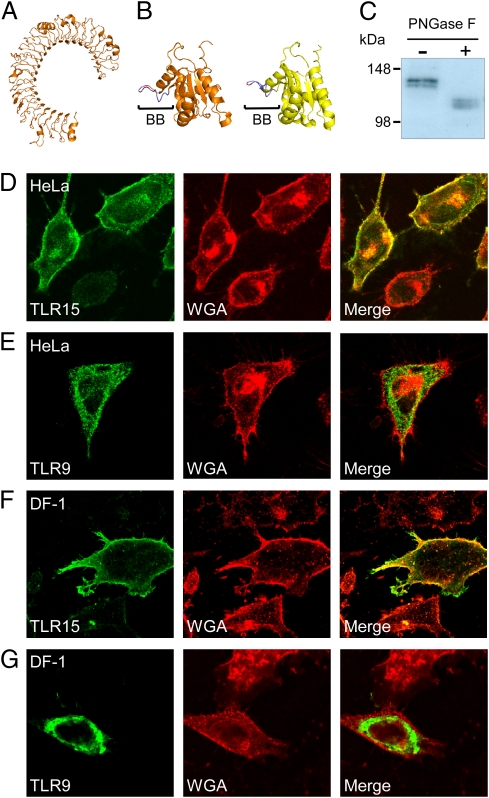
Sequence analysis of TLR15 uncovered a typical TLR makeup, comprising a signal sequence, an N-terminal LRR domain, a transmembrane helix, and a TIR domain (Fig. S1). Comparative modeling of the LRR domain of TLR15 revealed a horseshoe-shaped form typical for TLRs (Fig. 1A). Modeling of the TLR15 TIR domain revealed the presence of a conserved BB-loop structure including a conserved proline residue, known to be required for MyD88-dependent signaling in mammalian TLR family members (Fig. 1B) (20). Expression of flag-tagged TLR15 in COS-7 cells resulted in a TLR15 that migrated as a protein doublet in SDS/PAGE, with apparent molecular masses of approximately 120 kDa and approximately 130 kDa, respectively (Fig. 1C). Deglycosylation of the cell lysates with PNGase F decreased the electrophoretic mobility of both TLR15 forms (Fig. 1C), indicating that TLR15 is highly decorated with N-linked glycans. As the subcellular localization may provide clues on the function of the receptor, we investigated whether TLR15 was situated on the cell surface or in intracellular compartments. In HeLa 57A cells, transfected Flag-tagged TLR15 showed abundant colocalization with the cell surface marker wheat germ agglutinin (WGA; Fig. 1D). Similar TLR15 surface localization was found in COS-7 cells, chicken DF-1 cells (Fig. 1F), and human HEK293 cells. The surface localization of TLR15 was in clear contrast to the control human TLR9, which was detected only intracellularly in these cells (Fig. 1 E and G), consistent with its well documented intracellular location (21).
Fig. 1.
TLR15 is a surface-localized glycoprotein. (A) Computational modeling of the extracellular domain of TLR15 predicts a typical horseshoe-shape structure consisting of LRRs. (B) Predicted structure of the TLR15 TIR domain (orange), modeled on the human TLR1 TIR domain (yellow). The conserved signaling BB loop (BB) and proline residue are indicated in blue and red, respectively. (C) Flag-tagged TLR15 was expressed in COS-7 cells, deglycosylated by using PNGase F, and detected by immunoblotting with M2-α-FLAG, revealing that TLR15 is expressed as two high molecular mass glycoproteins. (D–G) Confocal microscopy of HeLa 57A and chicken DF-1 cells transfected with Flag-tagged TLR15 or TLR9. In HeLa 57A cells (D) and DF-1 cells (E), TLR15 (green) colocalized with the cell surface marker WGA (red). No colocalization of TLR9 (green) with WGA was observed in HeLa 57A cells (E) or DF-1 cells (G).
TLR15 Is Activated by Secreted Microbial Proteases.
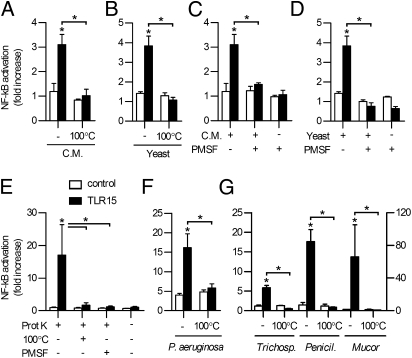
Although no sequence homology between TLR15 and other TLRs was apparent, various types of TLR ligands (LPS, di- and triacylated lipopeptides, zymosan, flagellin, TLR7/8 ligand, CpG DNA, and profilin-like protein) were tested for their ability to activate TLR15 expressed in HeLa 57A cells, with or without the presence of putative heterodimerizing chicken surface TLRs (TLR2t1, TLR2t2, TLR16, TLR4, or TLR5). These experiments did not result in TLR15-mediated NF-κB activation, suggesting that the ligand for TLR15 differs from the known TLR activators. Previous reports indicated that TLR15 is expressed in the chicken cecum (22, 23). In search for an activating ligand of TLR15 we therefore tested freshly isolated chicken cecal content. Stimulation of TLR15-transfected HeLa 57A cells with diluted cecal content resulted in TLR15-dependent NF-κB activation (Fig. 2A), which was abolished after heat treatment (100 °C) of the active material. These results identify a heat-sensitive ligand present in cecal content as ligand for TLR15.
Fig. 2.
TLR15 is activated by microbial proteases. (A and B) NF-κB activation of TLR15- or control-transfected HeLa 57A stimulated for 5 h with (A) cecal matter (C.M.) or (B) C. guilliermondii (yeast) supernatant. Heat inactivation (100 °C, 10 min) of cecal matter and yeast supernatant abolished TLR15-mediated NF-κB activation. (C and D) Cecal matter-induced (C) or yeast-induced (D) TLR15-mediated NF-κB activation in HeLa 57A cells is inhibited by PMSF (1 mM) treatment. (E) Proteinase K (10 ng mL−1, 5 h) specifically activated TLR15- but not control-transfected HeLa 57A cells in a heat- and PMSF treatment-dependent manner. (F and G) TLR15-mediated NF-κB activation in HeLa 57A cells induced by culture supernatant of (F) P. aeruginosa or (G) Trichosporon spp., Aspergillus spp. (left y axis), and Mucor spp. (right y axis), which is abolished by heat treatment (100 °C, 10 min). Data are presented as mean ± SEM of stimulated compared with unstimulated cells from three independent experiments (*P < 0.05).
One of the organisms recovered and isolated from the cecum was typed as the yeast Candida guilliermondii. Sterile supernatant of the pure yeast culture, grown overnight in DMEM at 37 °C, activated TLR15 in a similar manner as the cecal content. Again, this activity was destroyed by heat treatment (Fig. 2B). Fungi are notorious for secreting high levels of (heat-labile) proteases as virulence factors into the environment. To test the involvement of secreted fungal proteases in the activation of TLR15, the protease inhibitor PMSF was added to the cecal content and the fungal supernatant (Fig. 2C and D). In both cases, TLR15 activation was reduced to background levels, indicating a crucial role for proteases in the activation of the receptor. NF-κB activation of other human and chicken TLRs was not affected by the addition of PMSF.
To corroborate our findings, we stimulated TLR15-expressing cells with recombinant fungal proteinase K. The enzyme activated NF-κB in a TLR15-dependent fashion. The effect was dose-dependent (Fig. S2) and again abolished by heat and PMSF treatment of proteinase K (Fig. 2E). Sterile culture supernatant of the Gram-negative opportunistic pathogen Pseudomonas aeruginosa, which is well known to secrete multiple virulence-associated proteases, as well as sterile culture supernatants of three other fungi (Trichosporon spp., Penicillium spp., and Mucor spp.), were similarly able to specifically activate TLR15 in a heat-dependent manner (Fig. 2 F and G). All but one of these activities could be inhibited by PMSF (Fig. S3), further showing that proteolytic activity is required for TLR15 activation. Collectively, these data indicate that TLR15 senses and responds to proteases from fungal and bacterial origin, thereby detecting a conserved factor from distinct microbial species.
Specificity of Protease-Dependent Activation of TLR15.
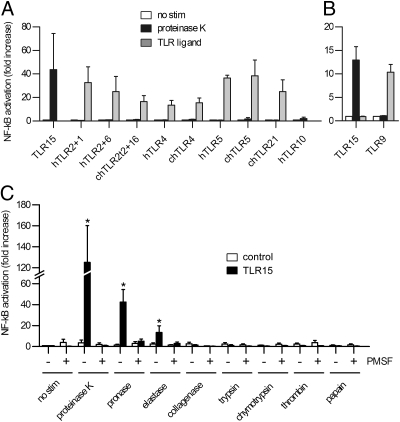
To investigate whether protease-dependent activation is common for TLRs or specific for TLR15, we expressed human TLR2/TLR1, TLR2/TLR6, TLR4, TLR5, and TLR10, and chicken TLR2t2/TLR16, TLR4, TLR5, and TLR21 in HeLa 57A cells and stimulated the cells with proteinase K or the appropriate TLR ligand (Fig. 3A). Whereas all TLRs activated NF-κB after stimulation with their own ligand (with the exception of TLR10, of which the ligand is still unknown), only TLR15 was activated (~40-fold) by the addition of protease. As human TLR9 is not functional in the HeLa 57A cell line, HEK293 cells were transfected with TLR15 and TLR9. As observed for the other TLRs, TLR9 was successfully activated by the CpG DNA ligand ODN 2006 but not by proteinase K (Fig. 3B).
Fig. 3.
Protease-induced TLR activation is specific for TLR15 and restricted to proteases with low substrate specificity. (A and B) NF-κB activation in HeLa 57A cells (A) or HEK293 cells (B) transfected with (combinations of) human or chicken TLRs and stimulated with 10 ng mL−1 proteinase K or with 100 ng mL−1 Pam3CSK4 (hTLR1+2, chTLR2t2+16), 100 ng mL−1 FSL-1 (hTLR2+6), 100 ng mL−1 LPS (hTLR4, chTLR4), 1 μg mL−1 flagellin (hTLR5, chTLR5), or 0.5 μM ODN2006 (chTLR21, hTLR9) for 5 h. hTLR10 was stimulated with 10 ng mL−1 proteinase K only. None of the proteinase K-stimulated TLRs was significantly activated except for TLR15 (P < 0.05), whereas stimulation of all of the TLRs with their own ligand resulted in significant NF-κB activation (P < 0.05). (C) NF-κB activation of TLR15- or control-transfected HeLa 57A cells stimulated (for 3 h) with 25 ng mL−1 proteinase K, 25 ng mL−1 pronase, 25 ng mL−1 bovine elastase, 25 ng mL−1 collagenase, 25 ng mL−1 trypsin, 25 ng mL−1 chymotrypsin, 25 ng mL−1 thrombin, or 25 ng mL−1 papain. Proteases were inhibited by PMSF (1 mM) treatment. Data are presented as mean ± SEM of stimulated versus unstimulated cells from three independent experiments (*P < 0.05).
To determine which type/class of proteases is able to activate TLR15, a panel of seven proteases with variable substrate specificity was selected (Fig. 3C). When HeLa 57A cells transfected with TLR15 were stimulated with 25 ng mL−1 proteinase K or pronase, a strong increase in NF-κB activation was observed compared with unstimulated cells. The stimulatory effect was absent in HeLa 57A cells transfected with control vector (Fig. 3C). When TLR15-expressing cells were stimulated with 25 ng mL−1 porcine elastase, moderate but significant activation was observed (~15-fold increase). Serine proteases with more strict substrate specificity (collagenase, trypsin, chymotrypsin, and thrombin) and the cysteine protease papain did not activate TLR15. Together, these results indicate that protease-induced activation is restricted to TLR15, and that the receptor is activated by proteases with broad substrate specificity.
TLR15 Is Proteolytically Cleaved by Activating Proteases.
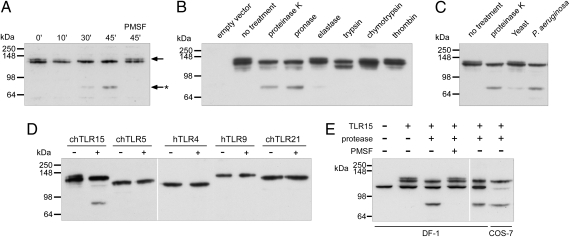
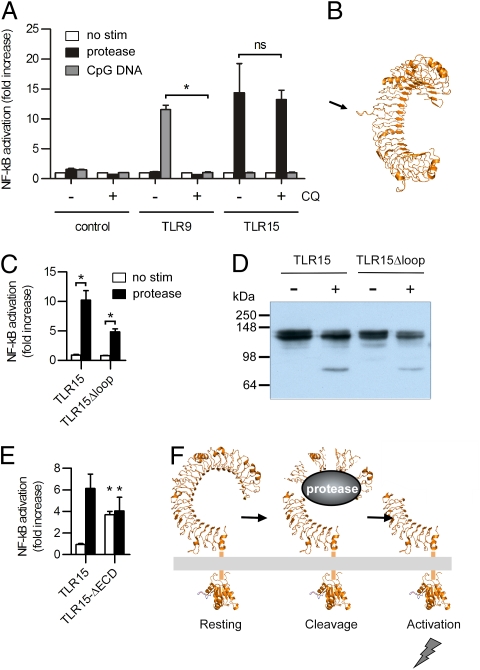
Protease may activate TLR15 indirectly by the release of cellular factors and/or cleavage of the receptor. To investigate whether released factors were involved in the activation of TLR15, we performed transfer experiments in which cell supernatant of protease-treated cells (transfected with TLR15 or control vector), after inhibition of the protease with PMSF, was added to new TLR15-transfected cells. Transfer of protease activity-free supernatants did not stimulate TLR15-dependent NF-κB activation. To test whether proteases directly targeted TLR15, COS-7 cells were transfected with TLR15 containing a C-terminal Flag tag and incubated with proteinase K in the absence and presence of PMSF. Immunoblotting of the cell lysates revealed the TLR15 doublet for the untreated and PMSF-inactivated proteinase K-treated cells. Incubation with active protease, however, resulted in the appearance of a 70-kDa C-terminal TLR15 cleavage fragment, which increased in intensity with prolonged exposure to the protease (Fig. 4A). As the appearance of the TLR15 fragment coincided with disappearance of the upper, 130-kDa band of the TLR15 doublet, we speculated that the upper band represented surface-localized (and thus protease-sensitive) TLR15, whereas the lower 120-kDa band represented intracellular-localized TLR15. This was confirmed by cell surface biotinylation, which labeled only the 130-kDa and cleaved TLR15 protein (Fig. S4). Treatment of chicken DF-1 cells expressing TLR15 yielded similar results (Fig. 4E), and all other (control) TLRs treated with identical amount of proteinase K yielded no cleavage products (Fig. 4D). Furthermore, only proteases that were shown to activate TLR15 (Fig. 3C) gave rise to the specific 70-kDa protein product (Fig. 4B). Finally, when TLR15-expressing COS-7 cells were treated with sterile culture supernatant of C. guilliermondii or P. aeruginosa, a 70-kDa fragment of identical size as the proteinase K-induced fragment was observed (Fig. 4C). Combined, these results show that activation of TLR15 by purified proteases or proteases present in microbial supernatants is accompanied by the formation of a 70-kDa cleavage product and that this effect is unique for TLR15.
Fig. 4.
Activation of TLR15 is accompanied by proteolytic cleavage. (A) Immunoblot analysis of COS-7 cells expressing C-terminal Flag-tagged TLR15 treated with 100 ng mL−1 proteinase K for the indicated times. The upper band of the TLR15 doublet (arrow) is proteolytically cleaved to a C-terminal 70-kDa truncated receptor (arrow with asterisk). Cleavage does not occur after inactivation of the protease by PMSF (1 mM). (B) Immunoblot analysis of TLR15 cleavage in COS-7 cells stimulated with the indicated proteases (100 ng mL−1) for 45 min. (C) Stimulation of TLR15-transfected COS-7 cells with sterile culture supernatants of the yeast C. guilliermondii and P. aeruginosa (45 min) followed by immunoblot analysis of TLR15 cleavage. (D) COS-7 cells expressing C-terminal Flag-tagged chTLR15, chTLR5, hTLR4, hTLR9, or chTLR21 were stimulated with 100 ng mL−1 proteinase K for 45 min and immunoblotted. (E) Proteinase K treatment of TLR15 expressed in chicken DF-1 cells resulted in an identical C-terminal receptor cleavage fragment as seen in COS-7 cells. C-terminal Flag-tagged TLR15 was detected by using M2-α-Flag.
Mechanism of TLR15 Activation.
Proteolytic cleavage has previously been demonstrated for mammalian TLR9. This intracellular TLR receptor for double-stranded CpG DNA undergoes a series of proteolytic cleavage steps by endogenous cysteine endopeptidases to form a functional receptor (10–12). In contrast to TLR9, TLR15 is expressed at the cell surface (Fig. 1 D–G), is not activated by CpG DNA, and is insensitive to chloroquine, which blocks TLR9 activation by inhibiting lysosomal acidification (Fig. 5A). However, comparative modeling of the extracellular LRR domain of TLR15 revealed the presence of an extended loop in LRR9 (Fig. 5B), similar to the loop predicted to be the target of proteases in TLR9 (11, 12). To assess the possible role of the loop in TLR15 cleavage and/or activation, we deleted this proline-rich loop (aa 352–363) in TLR15. Deletion of the loop did not change the proteolytic cleavage of TLR15 (Fig. 5D). Similarly, protease-induced activation of the receptor was not abolished after deletion of the proline-rich loop (Fig. 5C). However, the expression and NF-κB activation of TLR15Δloop were lower compared with WT TLR15, suggesting potential nonoptimal protein stability or transport. Nonetheless, as TLR15Δloop can still be activated and cleaved, we believe the proline-rich loop is not required for TLR15 activation.
Fig. 5.
TLR15 is activated by a mechanism distinct from TLR9. (A) NF-κB activation of TLR15-, TLR9-, or control-transfected HEK293 cells left untreated (white bars) or stimulated with 20 ng mL−1 proteinase K (black bars) or 0.5 μM ODN 2006 (gray bars) for 5 h. Cells were preincubated with (+) chloroquine (CQ) or control buffer (−) for 30 min before stimulation. (B) Model of TLR15 showing the proline-rich loop from LRR9. (C) HeLa 57A cells were transfected with TLR15 or TLR15Δloop, and stimulated with 20 ng mL−1 proteinase K. (D) Proteinase K (+) or control (−) treatment of TLR15 and TLR15Δloop expressed in COS-7 cells. C-terminal Flag-tagged receptors were detected by using M2-α-Flag. (E) TLR15- or TLR15ΔECD-transfected HeLa 57A cells were stimulated with 20 ng mL−1 proteinase K (black bars) or control buffer (white bars). (F) Model of TLR15 activation. In A, C, and E, data are presented as mean ± SEM of stimulated versus unstimulated cells from three independent experiments (*P < 0.05; ns, not significant).
For protease-activated receptors (PARs), a group of receptors involved in various cellular processes including proinflammatory responses, it has been shown that the presence of an inhibitory receptor domain prevents self-activation of the receptors (24). To investigate the presence of a similar mechanism for TLR15 activation, we constructed a receptor with a deleted extracellular domain (ECD), thereby removing any potential inhibitory protease-sensitive elements. Expression of the truncated TLR15ΔECD at the cell surface (Fig. S5), caused constitutive activation of NF-κB, independent of the presence of proteases (Fig. 5E). These findings are consistent with a model of TLR15 activation in which cleavage of the extracellular LRR domain of TLR15 by microbial proteases results in the release of inhibitory elements causing TLR15 self-activation without the requirement of an external ligand (Fig. 5F).
Discussion
The discovery that chicken TLR15 is activated following cleavage by secreted microbial proteases presents a unique innate immune defense strategy and a unique mechanism of TLR activation. As secreted proteases are widely present as virulence factors of fungal, parasitic, and bacterial pathogens, the detection of proteolytic activity may enable birds to selectively respond to organisms that induce cellular proteolytic damage. Our results demonstrate that chicken TLR15 is exclusively activated through the enzymatic activity of microbe-derived proteases. We excluded contaminating agents like LPS, zymosan, and DNA as potential TLR15 ligands through several methods: (i) proteases from several different microbial and nonmicrobial sources were able to activate the receptor; (ii) TLR15 activation was specifically inhibited after inactivation of the proteases; (iii) a series of highly sensitive TLRs, including TLR2, TLR4, and TLR9, could not be activated by the purified TLR15-activating protease; and (iv) a high dose of purified microbial TLR ligands did not activate TLR15. Recent advances in innate immunity have revealed an immune-stimulating function for damage-associated molecular patterns like HMGB1, heat shock proteins, and uric acid, which are released by stressed or damaged host cells (25, 26). However, as protease-conditioned culture supernatant did not gain TLR15-stimulating ability, damage-associated molecular patterns were eliminated as potential TLR15 ligands.
Immune activation through TLR15 is accompanied by cleavage of the ligand sensing LRR domain, and activation and cleavage can be completely blocked by inactivation of the stimulating protease. Immunoblotting of cleaved TLR15 indicates that only the upper (~130-kDa) band of the TLR15 protein doublet is cleaved, representing the surface localized, protease-accessible TLR15 fraction. Cleavage leads to the generation of a C-terminal TLR15 fragment of 70 kDa, which after deglycosylation is predicted to have a final molecular mass of 62 kDa. This pinpoints the cleavage site to a region of approximately 30 aa (aa ~341–370), which includes a proline-rich insertion within LRR 9 (Fig. S1). Structural modeling revealed that this insertion forms a disordered loop extending outwards from the ligand sensing domain (Fig. 5B), similar to the proteolytic cleavage region in LRR14 of murine TLR9 (10–12). Therefore, TLR15 activation may be evolutionary related to the activation of mammalian TLR9. However, in contrast to deleting the cleavage region in TLR9, removal of the TLR15 insertion (i.e., TLR15Δloop) did not abolish receptor cleavage (Fig. 5D). In addition, TLR15Δloop could still induce protease-dependent NF-κB activation (albeit at lower levels then full-length TLR15; Fig. 5C), suggesting that the deleted region does not contain the protease cleavage site. Several attempts to create a functional, constitutively active TLR15 truncated at the LRR 9 insertion were unsuccessful as a result of the loss of receptor surface expression. This problem was overcome by the expression of TLR15ΔECD, which mimicked the activation of a cleaved full-length TLR15 even without the presence of proteolytic activation. The autoactivation of TLR15ΔECD may indicate a tendency to self-dimerization of the truncated receptor, as has been previously described for TLR4 (27) and Drosophila Toll (28). Thus, the LRR domain of TLRs may not only serve as ligand sensor element but may also act as an inhibitor of self-activation. In the case of TLR15, LRR proteolysis may alleviate the inhibitory function by release of a receptor fragment and/or associated membrane components or via the induction of conformational changes in TLR15 that initiate signaling.
To our knowledge, direct proteolytic activation of a full-length TLR has not previously been described, although mammalian TLR4 can be activated indirectly by elastase-cleaved ECM components (29). Mammals appear to lack a TLR that is able to directly sense proteolytic activity. Instead, several alternative systems that initiate immune responses through pathogenic proteases have evolved in the mammalian species. Most studied are the cell surface G protein-coupled PARs (PARs 1–4) (24). PAR2 in particular can be activated by proteases from exogenous sources, like the bacterial pathogens Porphyromonas gingivalis (30) and Helicobacter pylori (31). Dust mite proteases may also activate PAR2 (32) as well as unidentified receptor present on human basophils (33). Although homologues of several PARs and TLR4 are present in the chicken genome, their contribution to sensing microbial proteases is unknown. Our results indicate that, during evolution, the avian species have evolved a unique receptor that combines key properties of PARs and TLRs in a single molecule.
Previous studies showed the expression of TLR15 in the intestinal tract of chickens, similar to where PAR2 is activated by pathogen-induced host protease in mice (34). Although the gut lumen is a rich environment loaded with multiple types of potentially activating proteases, PAR2 and presumably TLR15 are not constantly activated by the normal flora. Under normal conditions, the mucus layer functions as a barrier between the content of the intestinal lumen and the epithelial cell layer, which is further protected from aberrant protease activity by secretory leukocyte peptidase inhibitor (35). In addition, previous studies have shown strong up-regulation of TLR15 in intestinal tissue after Salmonella infection and TLR stimulation (22, 23), suggesting that, under healthy homeostatic conditions, the contact between TLR15 and normal intestinal proteases is limited. This form of regulation may represent an extra safeguard for the chicken for activation of the innate immune system by accidental exposure to proteases.
In conclusion, we show that chicken TLR15 is activated by secreted microbial proteases that cleave the extracellular LRR domain. Identification of a TLR that detects harmful proteolytic activity provides a unique mechanism of immune activation that adds a dimension to the general consensus of TLR activation and function. The activation of the receptor after removal of inhibitory receptor elements may indicate the existence of an additional TLR control mechanism to prevent aberrant activation of the receptor in the absence of ligand.
Materials and Methods
Cell Lines and Chemicals.
HeLa 57A and HEK293 were maintained in DMEM (Invitrogen) supplemented with 10% FCS; COS-7 cells were maintained in Iscove modified Dulbecco media supplemented with 10% FCS; chicken DF-1 and HD11 cells were maintained in DMEM supplemented with minimum essential medium nonessential amino acids and 10% FCS. PMSF, chloroquine, and proteases (pronase, pancreatic porcine elastase, collagenase, trypsin, chymotrypsin, thrombin, and papain) were purchased from Sigma-Aldrich; TLR ligands were purchased from Invivogen; proteinase K was purchased from Roche; and PNGase F was purchased from New England Biolabs.
Homology Modeling.
The amino acid sequence of the TLR15 ectodomain and TIR domain were modeled by using the automative ESyPred3D web server 1.0 on the ectodomain of human TLR3 (Protein Data Bank accession no. 2A0Z) and the TIR domain of human TLR1 (Protein Data Bank accession no. 1FYV), respectively.
Plasmid Constructs.
For the construction of N-terminal Flag-tagged TLR15 (TLR15-Flag-N), the TLR15 ORF was amplified by PCR by using pfu polymerase (Promega) from HD11 chromosomal DNA with primers 5′-CCGAATTCATTCCTAACTCAGAGAACATCTCC-3′ (forward) and 5′- GGTCTAGATTCCATCTCAATTACATCCTC-3′ (reverse), and cloned into p3Xflag-CMV23-c-Myc (Sigma-Aldrich) with the restriction enzymes EcoRI and XbaI. For the construction of C-terminal Flag-tagged TLR15 (TLR15-Flag-C), TLR15 was amplified from HD11 chromosomal DNA using primers 5′-CCGAATTCGCCACCATGGGGATCCTT-ATTGGGAGTC-3′ (forward) and 5′-GGGCGGCCGCCTTCCATCTCAATTACATCCTC-3′ (reverse), and cloned into p3XTracer (18) with restriction enzymes EcoRI and NotI. C-terminal Flag-tagged chTLR5 (chTLR5-Flag-C) was amplified from pTracer-chTLR5 (16) with primers 5′-CCGGATCCGCCACCATGGTACATCAACGGCTAATAATTG-3′ (forward) and 5′-GGGCGGCCGCCTTCCATCTCAATTAC-ATCCTC-3′ (reverse), and cloned into p3XTracer with restriction enzymes BamHI and NotI. chTLR21, hTLR9, hTLR1, hTLR6, hTLR2, chTLR2t2, chTLR16, chTLR4, chTLR5, and hTLR5 were constructed previously (15, 16, 18, 19). C-terminal Flag-tagged human TLR4 was created by digestion of pcDNA3-TLR4-YFP (21) with the restriction enzyme XhoI, subsequent treatment with S1 Nuclease (Promega), and digestion with BamHI. The resulting TLR4 fragment was ligated into p3XTracer, which was first digested with NotI, treated with S1 nuclease, and digested with BamHI to yield hTLR4-Flag-C. TLR15Δloop was created by PCR amplification of the N-terminal fragment of TLR15 by pfu polymerase with primers 5′-CCGAATTCGCCACCATGGGGATCCTTATTGGGAGTC-3′ (forward) and 5′-ATTTGTA-CAGTTCCTGGACGTACGGTATATAT-3′ (reverse), subsequent digestion with restriction enzymes EcoRI and BsrGI, and ligation into EcoRI and BsrGI-digested TLR15-Flag-C, thereby replacing the WT N-terminal fragment of TLR15. TLR15ΔECD was created by PCR amplification of the transmembrane and TIR domain fragment of TLR15 by pfu polymerase with primers 5′-CCGAATTCAGGCATTCAGATGGCCATTACAG-3′ (forward) and 5′-GGTCTAGATTCCATCTCAATTACATCCTC-3′ (reverse), subsequent digestion with restriction enzymes EcoRI and XbaI, and ligation into p3XFlag-CMV23-c-Myc. This truncated TLR15 consists of an N-terminal 3× FLAG, TLR15 aa 654 to 869 (i.e., the entire transmembrane domain and TIR domain; Fig. S1), and a C-terminal c-Myc. All constructs were confirmed by sequencing.
Deglycosylation of TLR15.
N-terminal Flag-tagged TLR15 plasmid DNA (500 ng) was transfected into COS-7 cells in 12-well plates by using FuGENE 6 at a DNA:lipid ratio of 1:2. After 48 h, cells were washed three times with Dulbecco PBS (DPBS) solution and lysed with Reporter Lysis Buffer (RLB; Promega) at −80 °C. Protein lysates were deglycosylated using PNGase F (New England Biolabs) according to the manufacturer's protocol and analyzed by immunoblotting using M2-α-Flag (Sigma-Aldrich).
Confocal Laser Microscopy.
Cells were grown in 24-well plates on glass coverslips and transfected with the appropriate C-terminal Flag-tagged TLR. After 48 h, cells were prepared for confocal laser microscopy as described previously (15). Briefly, cells were washed with DPBS solution and incubated with WGA-biotin for 10 min at 37 °C. After washing with DPBS solution, cells were fixed with DPBS/2% paraformaldehyde for 30 min and permeabilized with 0.2% Triton X-100 for 15 min. Following blocking with DPBS/2% BSA (60 min), cells were incubated with M2-α-Flag antibody (60 min), washed with DPBS solution, and incubated with Alexa Fluor-488 goat anti-mouse IgG (Invitrogen) and Alexa Fluor-568 streptavidin (Invitrogen) for 60 min. Cells were subsequently embedded in FluorSave (Calbiochem) and viewed on a Leica TCA SP confocal laser-scanning microscope.
Stimulation Assays.
For stimulation assays, HeLa 57A and HEK293 cells were transfected in 48-well plates with 125 ng receptor plasmid DNA and 125 ng LacZ plasmid DNA (HeLa 57A), or 125 ng receptor plasmid DNA, 62.5 ng LacZ plasmid DNA, and 62.5 ng NF-κB plasmid DNA (HEK293) by using FuGENE 6 (Roche), at a DNA:lipid ratio of 1:3. After 48 h, cells were washed three times with DMEM and stimulated with the appropriate amount of ligand. After 5 h of stimulation, cells were washed twice with DPBS solution and lysed using RLB at −80 °C. Luciferase activity in the lysates was measured in a luminometer (TD-20/20; Turner Designs) with luciferase reagent (Promega). Experiments were performed at least three times independently. Cecal content was collected from free-ranging chickens, diluted 1:10 in DPBS solution, and filtered; fungal supernatants were prepared by overnight incubation of maltose-cultivated fungi in DMEM at 37 °C followed by filter sterilization; P. aeruginosa supernatants were prepared from 48 h cultures in heart infusion broth (BioTrading) at 37 °C and filter-sterilized.
Detection of TLR15 Cleavage.
For detection of TLR15 cleavage, COS-7 and DF-1 cells were transfected in 12-well plates with 2 μg of receptor plasmid DNA using FuGENE 6 at DNA:lipid ratios of 1:2 (COS-7) and 1:3 (DF-1). After 42 h, cells were washed three times with Iscove modified Dulbecco medium (COS-7) or DMEM (DF-1) and incubated for 45 min with proteases at 37 °C. Cells were subsequently lysed in RLB and subjected to immunoblot analysis. TLRs were detected by using M2-α-Flag antibody.
Statistical Analysis.
Statistical significance was determined by the paired two-tailed Student t test at P values lower than 0.05.
Supplementary Material
Acknowledgments
We thank Barend Blankenstein for fungal typing, Martijn van Herwijnen for technical assistance, and Douglas Golenbock for providing plasmid pcDNA3-TLR4-YFP.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1018135108/-/DCSupplemental.
References
- 1.Litman GW, Cannon JP, Dishaw LJ. Reconstructing immune phylogeny: New perspectives. Nat Rev Immunol. 2005;5:866–879. doi: 10.1038/nri1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 3.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: Regulation through compartmentalization. Nat Rev Immunol. 2009;9:535–542. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 5.Jin MS, Lee JO. Structures of the Toll-like receptor family and its ligand complexes. Immunity. 2008;29:182–191. doi: 10.1016/j.immuni.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 6.Jin MS, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park BS, et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 9.Gangloff M, et al. Structural insight into the mechanism of activation of the Toll receptor by the dimeric ligand Spätzle. J Biol Chem. 2008;283:14629–14635. doi: 10.1074/jbc.M800112200. [DOI] [PubMed] [Google Scholar]
- 10.Ewald SE, et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park B, et al. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sepulveda FE, et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Roach JC, et al. The evolution of vertebrate Toll-like receptors. Proc Natl Acad Sci USA. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anonymous; International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 15.Keestra AM, de Zoete MR, van Aubel RA, van Putten JPM. The central leucine-rich repeat region of chicken TLR16 dictates unique ligand specificity and species-specific interaction with TLR2. J Immunol. 2007;178:7110–7119. doi: 10.4049/jimmunol.178.11.7110. [DOI] [PubMed] [Google Scholar]
- 16.Keestra AM, de Zoete MR, van Aubel RA, van Putten JPM. Functional characterization of chicken TLR5 reveals species-specific recognition of flagellin. Mol Immunol. 2008;45:1298–1307. doi: 10.1016/j.molimm.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Temperley ND, Berlin S, Paton IR, Griffin DK, Burt DW. Evolution of the chicken Toll-like receptor gene family: A story of gene gain and gene loss. BMC Genomics. 2008;9:62. doi: 10.1186/1471-2164-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keestra AM, de Zoete MR, Bouwman LI, van Putten JP. Chicken TLR21 is an innate CpG DNA receptor distinct from mammalian TLR9. J Immunol. 2010;185:460–467. doi: 10.4049/jimmunol.0901921. [DOI] [PubMed] [Google Scholar]
- 19.Keestra AM, van Putten JPM. Unique properties of the chicken TLR4/MD-2 complex: selective lipopolysaccharide activation of the MyD88-dependent pathway. J Immunol. 2008;181:4354–4362. doi: 10.4049/jimmunol.181.6.4354. [DOI] [PubMed] [Google Scholar]
- 20.O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 21.Latz E, et al. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat Immunol. 2004;5:190–198. doi: 10.1038/ni1028. [DOI] [PubMed] [Google Scholar]
- 22.Higgs R, et al. Induction of a novel chicken Toll-like receptor following Salmonella enterica serovar Typhimurium infection. Infect Immun. 2006;74:1692–1698. doi: 10.1128/IAI.74.3.1692-1698.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaughnessy RG, et al. Innate immune gene expression differentiates the early avian intestinal response between Salmonella and Campylobacter. Vet Immunol Immunopathol. 2009;132:191–198. doi: 10.1016/j.vetimm.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 24.Ossovskaya VS, Bunnett NW. Protease-activated receptors: Contribution to physiology and disease. Physiol Rev. 2004;84:579–621. doi: 10.1152/physrev.00028.2003. [DOI] [PubMed] [Google Scholar]
- 25.Bianchi ME. DAMPs, PAMPs and alarmins: All we need to know about danger. J Leukoc Biol. 2007;81:1–5. doi: 10.1189/jlb.0306164. [DOI] [PubMed] [Google Scholar]
- 26.Kaczorowski DJ, Mollen KP, Edmonds R, Billiar TR. Early events in the recognition of danger signals after tissue injury. J Leukoc Biol. 2008;83:546–552. doi: 10.1189/jlb.0607374. [DOI] [PubMed] [Google Scholar]
- 27.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 28.Schneider DS, Hudson KL, Lin TY, Anderson KV. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 1991;5:797–807. doi: 10.1101/gad.5.5.797. [DOI] [PubMed] [Google Scholar]
- 29.Brunn GJ, Bungum MK, Johnson GB, Platt JL. Conditional signaling by Toll-like receptor 4. FASEB J. 2005;19:872–874. doi: 10.1096/fj.04-3211fje. [DOI] [PubMed] [Google Scholar]
- 30.Holzhausen M, et al. Protease-activated receptor-2 activation: A major role in the pathogenesis of Porphyromonas gingivalis infection. Am J Pathol. 2006;168:1189–1199. doi: 10.2353/ajpath.2006.050658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kajikawa H, et al. Helicobacter pylori activates gastric epithelial cells to produce interleukin-8 via protease-activated receptor 2. Digestion. 2007;76:248–255. doi: 10.1159/000113041. [DOI] [PubMed] [Google Scholar]
- 32.Chignard M, Pidard D. Neutrophil and pathogen proteinases versus proteinase-activated receptor-2 lung epithelial cells: More terminators than activators. Am J Respir Cell Mol Biol. 2006;34:394–398. doi: 10.1165/rcmb.2005-0250TR. [DOI] [PubMed] [Google Scholar]
- 33.Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol. 2008;9:310–318. doi: 10.1038/ni1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hansen KK, et al. A major role for proteolytic activity and proteinase-activated receptor-2 in the pathogenesis of infectious colitis. Proc Natl Acad Sci USA. 2005;102:8363–8368. doi: 10.1073/pnas.0409535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergenfeldt M, et al. Localization of immunoreactive secretory leukocyte protease inhibitor (SLPI) in intestinal mucosa. J Gastroenterol. 1996;31:18–23. doi: 10.1007/BF01211182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.