Abstract
Experiments of Pränting and Andersson demonstrate how bacteria adapt to the growth limitation caused by antibiotic resistance mutations. The process of adaptation relies on gene copy number changes that arise at high rates, including duplications (10−4/cell/generation), amplifications (10−2/cell/generation) and mutant copy loss (10−2/cell/division). Reversible increases in copy number improve growth by small steps and provide more targets for rare sequence alterations (10−9/cell/division) that can stably improve growth. After sequence alteration, selection favors loss of the still mutant gene copies that accelerated adaptation. The results strongly support the amplification-reversion model for fast adaptation and argue against the alternative idea of “stress-induced mutagenesis”.
When conditions limit growth, populations of bacterial cells (and higher organisms) adapt genetically at surprisingly high rates. Adaptation is a joy, when your population adapts to a new environment. It is a terror, when pathogens or malignant cells adapt to you. Thus adaptation is important to both the process of evolution and for many aspects of disease. The speed of adaptation under natural conditions is shocking in the light of our experience with stringent lab selections, which prevent growth of the parent population and detect only pre-existing large-effect mutants (Luria & Delbruck, 1943, Lederberg & Lederberg, 1952). By preventing adaptation, lab selections make bacterial genetics possible, but they do not prepare us to appreciate the speed of natural selection during growth. The work of Pränting and Andersson contributes to an understanding of how this occurs.
Over the past 20 years, a controversy has surrounded attempts to explain the fast adaptation seen in several systems that, unlike standard lab procedures, use non-stringent selective conditions. How does selection work so well? Could selective stress be mutagenic? The new results presented here show that selection acts alone and adaptation does not involve “stress-induced mutagenesis”. In explaining fast adaptation without mutagenesis, Pränting and Andersson show how antibiotic-resistant bacteria escape the fitness costs associated with resistance and regain full growth ability. This process is central to understanding microbial populations and how they respond to our use of antibiotics.
The antibiotic used here is protamine, a lethal anti-microbial peptide (AMP) that resembles those produced by metazoans as part of innate immunity (Pränting & Andersson, 2010, Proctor et al., 2006). Mutants resistant to protamine arise at a rate of 10−7/cell/division. This high rate reflects the large size of the mutational target. Resistance is provided by mutations that reduce proton-motive force (PMF). Since PMF depends on many proteins, there are many sites in the genome at which resistance mutations can arise, including heme biosynthetic genes. Impairment of these essential processes provides resistance to the lethal antibiotic, but compromises the general growth rate. Pränting and Andersson show that bacteria rapidly circumvent growth limitation because natural selection can detect small-effect mutations that are common under all circumstances.
A growth-compromised protamine-resistant (hemC) mutant improves its growth ability in small steps due to increases in the copy number of the partially functional mutant gene. Adaptation is initiated by cells with a hemC duplication, which are present in the population at a high steady-state frequency (1/100) before selection (Reams et al., 2010). Cells with extra copies grow slowly and improve further by a series of additional copy number increases, each of which arises at about 10−2/cell/generation. See Figure 1A and B.
Figure 1. The process of amplification-selection.
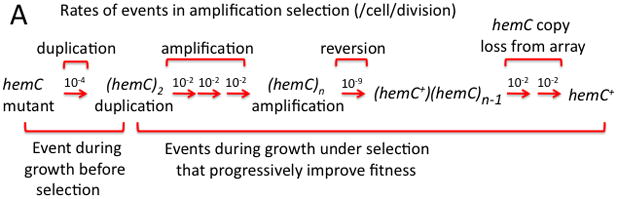
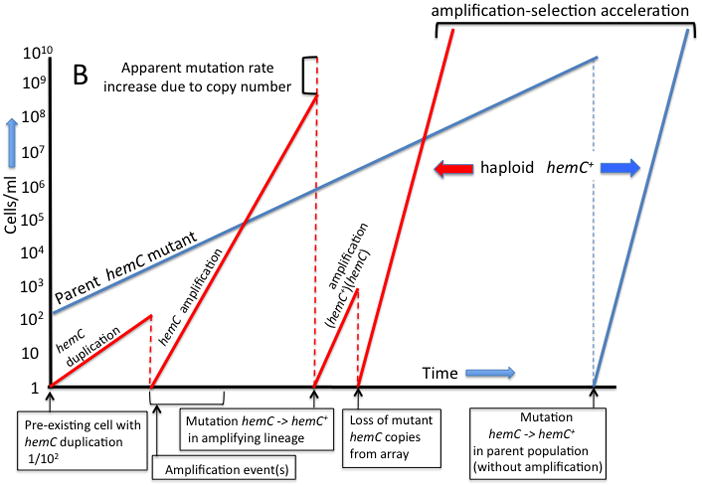
Part A Duplications form at a high rate (10−4/cell/division) and steps in subsequent copy number change occur even more frequently (10−2/cell/division). The frequency of duplications in an unselected population reaches a steady state of about 10−2. Part B When a hemC mutant initiates a culture, 1% of cells have a duplication. Cells with extra copies of hemC grow faster than the parent and acquire amplifications that further enhance fitness. The amplification process is likely to involve multiple steps. A sequence change (hemC -> hemC+) occurs at a standard mutation rate when sufficient targets are present. Amplification causes a modest increase in the rate (/cell) of reversion to hemC+ (top left), but accelerates appearance of full revertants (haploid hemC+) primarily by allowing exponential growth of the improving lineage (top right). The amplifying lineage reaches a high population density and produces stable fitter progeny sooner that the unamplified parent lineage.
Cells with extra mutant hemC copies grow until the population has enough mutant alleles to permit a rare sequence change that improves one copy of the growth-limiting gene. Such sequence changes occur at about 10−9–10−10/cell/generation (Figure 1A and center of Figure 1B). The probability of a sequence change (per cell) is increased slightly by the additional mutational target sites (more copies per cell), but selection accelerates the appearance of revertants primarily by allowing an exponential increase in the number of cells with an amplification (both effects are noted at the top Figure 1B. This process involves no increase in mutation rate (mutation/target).
The added copies of the mutant hemC allele improve fitness sufficiently to offset the basic fitness cost of a gene amplification (Reams et al., 2010). This situation changes after mutation generates an improved hemC allele. At this point, selection holds the improved allele and counter-selects the costly mutant copies. Thus a gene amplification contributes to the formation of a rare mutation by providing more target copies and growth, but is counter-selected once the improved allele is in place. Ultimately haploid revertant cells appear that carry a single improved hemC allele and show no evidence of the gene copy number increase that hastened their formation. (top right of Figure 1B)
The sequence of events demonstrated here was initially suggested (Andersson et al., 1998, Roth et al., 2006) to explain fast adaptation in a system described by Cairns and Foster (Cairns & Foster, 1991). In this system, a partially defective lac mutant is plated on lactose. The parent cells cannot (quite) grow, and the revertant colonies that accumulate over 6 days have been widely attributed to stress-induced mutagenesis of non-growing cells (Foster, 2007, Galhardo et al., 2007). In the alternative amplification-selection model (left side of Figure 2), common pre-existing copy number variants grow under selection and initiate colonies that adapt quickly. The multiple steps of the adaptation process have been hard to resolve, because of idiosyncratic features of Cairns-Foster system that accelerate adaptation. Notably, the lac allele is located on an F’lac plasmid, whose conjugative replication origin produces DNA ends that stimulate amplification. Furthermore, selected lac amplification is modestly mutagenic (Rosche & Foster, 1999), due to co-amplification of the nearby dinB gene (Slechta et al., 2003), which encodes an error-prone polymerase that is mutagenic when over-expressed (Kim et al., 2001, Wagner & Nohmi, 2000). Because of these features, the system allows pre-existing cells with multiple lac copies to initiate colonies that rapidly grow and adapt under selection, producing within stable Lac+ revertant cells within developing colonies by amplification, reversion, segregation. The mutagenesis caused by lac-dinB amplification is not essential to adaptation (Slechta et al., 2003). The rapid adaptation of pre-existing partially-revertant mutants during growth under selection is easily mistaken for stress-induced appearance of single-step mutants in a non-growing population.
Figure 2. Amplification-selection accounts for fast adaptation in multiple systems.
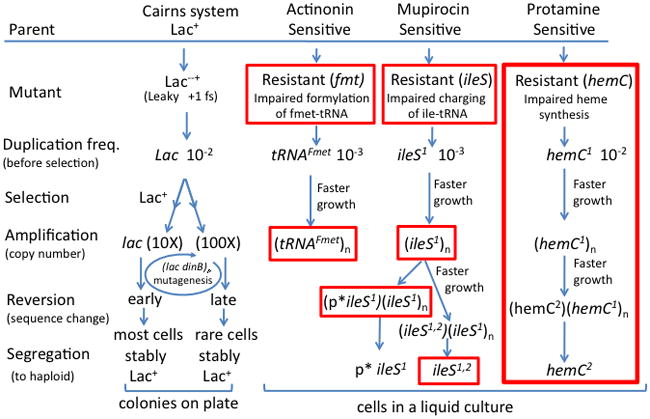
The amplification-selection model was proposed to explain development of Lac+ revertant colonies in the Cairns system (at left). The general applicability of this model is shown by its role in three different antibiotic systems. In these cases, selection favors a series of faster-growing cells in liquid culture. Intermediates in red boxes have been demonstrated experimentally. The work presented here on a protamine-resistant hemC mutant (diagrammed at right) demonstrates all of the intermediates and steps in the amplification selection process.
Pränting and Andersson demonstrate all the intermediates of the amplification-selection model. Of particular interest are cells whose array of hemC copies includes both the original mutant allele and a mutationally improved allele. This intermediate has not been seen previously, but is revealed here because the adaptation process moves more slowly for the chromosomal hemC system (> 100 generations), than for the Cairns-Foster system with lac on an F’lac plasmid (9 generations). These systems are compared in Figure 2 (left and right sides).
The amplification-selection model has now been show to operate in four different genetic systems. In Figure 2, three antibiotic systems analyzed by Andersson and coworkers are compared to the Cairns system. In each case, cells overcome growth defects by amplifying a growth-limiting gene. In one case, the drug actinonin inhibits the essential enzyme (Fmt), which formylates methionyl-tRNA and forms the initiator f-Met-tRNA. Resistant mutants (fmt) reduce formyl transferase and cause a general fitness loss. Growth is improved by amplifying the tRNAFMet gene (Nilsson et al., 2006). In another case, the antibiotic mupirocin inhibits isoleucyl-tRNA synthetase (ileS) and resistant mutations have an altered ileS coding sequence that severely limits growth. Growth improves first by amplification of the ileS gene and later by secondary mutations that either improve the ileS promotor or improve the IleS protein (Paulander et al., 2010). Either change enhances fitness and allows rapid loss of mutant alleles from the amplified array. The earlier systems show amplification-selection at work and demonstrate some steps in the process (Indicated in red in the Figure 2). However, the protamine work described here demonstrates all of the intermediates and the progression from one stage to the next (right side of Figure 2).
These results have enormous relevance for understanding effects of antibiotics on real populations. While protamine is not a clinical antibiotic, it resembles the antimicrobial-peptides (AMP), produced by many metazoans. Some chronic and recurring infections are associated with growth-impaired small-colony variants (SCV), which are likely to be positively selected in the host. These resistant bacteria may persist for long periods, only to expand their populations at a later time. Recurring infections can be dire, even when increased fitness restores antibiotic sensitivity.
Pränting and Andersson demonstrate a general mechanism for fast adaptation (whether a joy or a terror). Natural selection (unlike stringent lab selection) detects small growth improvements provided by mutations that are frequent under all growth conditions. These frequent small-effect mutations are not detected in stringent lab selections, so have been largely missed by bacterial genetics. The work presented here shows that such mutations are important to the high rate of genetic adaptation when selection is imposed on growing cells.
References
- Andersson DI, Slechta ES, Roth JR. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol. 2007;42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SR, Matsui K, Yamada M, Gruz P, Nohmi T. Roles of chromosomal and episomal dinB genes encoding DNA pol IV in targeted and untargeted mutagenesis in Escherichia coli. Mol Genet Genomics. 2001;266:207–215. doi: 10.1007/s004380100541. [DOI] [PubMed] [Google Scholar]
- Lederberg J, Lederberg EM. Replica plating and indirect selection of bacterial mutants. J Bacteriol. 1952;63:399–406. doi: 10.1128/jb.63.3.399-406.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Delbruck M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson AI, Zorzet A, Kanth A, Dahlstrom S, Berg OG, Andersson DI. Reducing the fitness cost of antibiotic resistance by amplification of initiator tRNA genes. Proc Natl Acad Sci U S A. 2006;103:6976–6981. doi: 10.1073/pnas.0602171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulander W, Andersson DI, Maisnier-Patin S. Amplification of the gene for isoleucyl-tRNA synthetase facilitates adaptation to the fitness cost of mupirocin resistance in Salmonella enterica. Genetics. 2010;185:305–312. doi: 10.1534/genetics.109.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pränting M, Andersson DI. Mechanisms and physiological effects of protamine resistance in Salmonella enterica serovar Typhimurium LT2. J Antimicrob Chemother. 2010;65:876–887. doi: 10.1093/jac/dkq059. [DOI] [PubMed] [Google Scholar]
- Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- Reams AB, Kofoid E, Savageau M, Roth JR. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics. 2010;184:1077–1094. doi: 10.1534/genetics.109.111963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche WA, Foster PL. The role of transient hypermutators in adaptive mutation in Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:6862–6867. doi: 10.1073/pnas.96.12.6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- Slechta ES, Bunny KL, Kugelberg E, Kofoid E, Andersson DI, Roth JR. Adaptive mutation: general mutagenesis is not a programmed response to stress but results from rare coamplification of dinB with lac. Proc Natl Acad Sci U S A. 2003;100:12847–12852. doi: 10.1073/pnas.1735464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J Bacteriol. 2000;182:4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]


