Abstract
High mobility group A1 proteins are architectural transcriptional factors that are over-expressed in a wide range of human malignancies. Recently published evidence suggests High mobility group A1 is a promising candidate biomarker and therapeutic target in pancreatic cancer. This review summarises data implicating High mobility group A1 as an important mediator of progression in human cancer and in pancreatic cancer.
Keywords: HMGA1, Akt, ERK, growth, survival, invasion, chemoresistance, pancreatic adenocarcinoma
Pancreatic adenocarcinoma: a leading cause of cancer-related death
Pancreatic cancer is the sixth and fourth most common cause of cancer death in the UK and in the USA, respectively.1,2 In 2006, pancreatic cancer resulted in 3610 male and 3705 female deaths in the UK.3 In USA, the American Cancer Society estimates that there will be 37,680 new cases of pancreatic cancer (18,770 men and 18,910 women) with 34,290 deaths from this disease in 2008.4 Patients diagnosed with pancreatic cancer have a median survival of less than six months and a five-year survival rate of 0.4–5%.5,6 Only a small minority (10–20%) have surgically resectable disease at the time of diagnosis. Even among patients who undergo ‘curative’ resection, most succumb to local recurrence or metastatic disease.7 Adjuvant chemotherapy can improve outcomes, but the magnitude of survival prolongation associated with currently available agents has been modest.2
We believe the strategy most likely to yield substantive improvements in outcomes for patients with pancreatic cancer is the rational development of early detection methods and of targeted therapies, based on an understanding of the biology underlying the aggressive phenotype of pancreatic cancer. This review focuses on High mobility group A1 (HMGA1), a molecular determinant of the malignant phenotype in pancreatic cancer, for which this gene has emerged as a candidate biomarker and therapeutic target.
HMGA1: Identification in gene expression profiling of pancreatic cancers
Nearly all pancreatic ductal adenocarcinomas (PDACs) have an activating mutation in K-ras (95%) and inactivation of p161NK4a (80–95%). Other high frequency genetic alterations in pancreatic cancer include those of p53 (50–75%) and of DPC4/Smad4 (55%).8 Cumulative aberrations in these and other genes have been hypothesised to drive the malignant phenotypic features of pancreatic cancer cells.9 Recent work has begun to comprehensively profile genes over- or under-expressed in PDACs relative to normal noncancerous pancreatic tissues.10–12
Among genes consistently found to be over-expressed in PDACs is HMGA1. Using cDNA microarrays, Iacobuzio-Donahue et al identified HMGA1 transcripts as highly over-expressed in both human pancreatic cancer tissues and cell lines relative to non-cancerous counterparts.13 Similarly, Han et al showed that HMGA1 is significantly over-expressed (between 4 to 14-fold higher than normal pancreas) in PDAC cell lines.14 Further, Tarbe et al found that a degree of HMGA1 over-expression is correlated with metastatic potential in PDAC cells.15 Using immunohistochemistry, Abe et al confirmed that HMGA1 is over-expressed in PDAC tissues at the protein level.16
HMGA1 proteins: nomenclature, molecular structure and mechanisms of action
The high mobility group (HMG) proteins were first isolated from mammalian cells 30 years ago.17 They were named according to their high electrophoretic mobility in polyacrylamide gels.18 The high mobility group A1 proteins consist of HMGA1a and HMGA1b, which differentially spliced products of the HMGA1 gene (located on the short arm of chromosome [6p21] in humans).19 They are identical except for internal deletion of 11 amino acids in HMGA1b. Known as ‘architectural transcription factors’, the HMGA1 proteins are small non-histone chromatin-associated proteins that bind DNA through a mechanism distinct from that of classical transcription factors.20 Each HMGA1 protein has three DNA-binding motifs called ‘AT-hooks’ that preferentially bind to adenine and thymine (AT)-rich sequences in the minor groove of DNA.21 The AT-hook domain consists of a highly conserved palindromic pentapeptide sequence (Pro-Arg-Gly-Arg-Pro [PRGRP]).21
HMGA1-DNA interactions are hypothesised to promote regulation of gene expression through several distinct (but possibly over-lapping) mechanisms. Firstly, HMGA1 is capable of binding to AT-rich promoter regions of genes. When bound to AT-rich promoter elements, HMGA1 proteins induce conformational changes in DNA that promote binding of transcriptional factors. Secondly, they form and stabilise multiprotein complexes together with other sequence-specific transcriptional factors through direct protein interactions. These complexes are known as ‘enhanceosomes’ which act as transcription activating complexes. HMGA1 proteins are known to physically interact with transcriptional factors such as Oct 2, ATF-2/c-Jun, AP-1, NFκB, Elf-1, SRF, PU-1, RAR, NFAT, Sp1 and CAAT/enhancer binding protein-β to modulate gene activation.22 Their multiple protein partners provide HMGA1 proteins with considerable flexibility in modulating transcriptional activity of a large number of genes. Thirdly, HMGA1 proteins can act as ‘anti-repressor’ molecules by competitively inhibiting histones (known transcriptional repressor) from binding to scaffold attachment regions (SAR) of specific DNA segments and thus induce an open chromatin conformation that promotes binding of transcriptional factors and transcriptional activation.23 Finally, post-translational modifications such as phosphorylation, acetylation and methylation can also influence the functions of HMGA1 proteins.24 Interestingly, upon inspection of the peptide sequence of the AT-hook region of HMGA1, it matches the consensus sequence for phosphorylation by cdc2 kinase. With this, it was further demonstrated that HMGA1 is phosphorylated in a cell cycle-dependent manner by cdc2 kinase in vitro, and this modification reduces the binding affinity of HMGA1 proteins for DNA.25,26
HMGA1 as a bona fide oncogene
HMGA1 proteins are present at high levels during embryogenesis while their expression is low or completely absent in differentiated tissues in adult rodents and humans.27 In quiescent, non-transformed cells, HMGA1 expression is rapidly induced following treatment with phorbol esters or growth factors such as EGF and PDGF.19,28 Moreover, transformation with viral oncogenes such as v-ras, v-mos and polyoma middle T antigen results in increased expression of HMGA1 proteins.29 These observations raise the question if HMGA1 expression is merely associated with proliferation, or if it truly has an oncogenic function.
In a study by Ram et al. they followed the expression of HMGA1 during the process of transformation of mouse mammary epithelial cells.30 The expression of HMGA1 was examined in cell lines derived from spontaneously arising mammary epithelial hyperplasia in BALB/C mice. Three cell lines (CL-S1, +SA and −SA) were derived from the same hyperplastic cell line, called the D1 parent line. The CL-S1 cell line is an immortalised cell line that does not form colonies in soft agar or tumour in vivo. The +SA and −SA cell lines were derived from an adenocarcinoma which developed spontaneously from the original D1 parent line when the D1 cells were transplanted in vivo. +SA and −SA cells were differentiated by their ability (+SA) or inability (−SA) to grow in soft agar. Notably, the expression of HMGA1 was highest in the highly tumourigenic +SA line which formed poorly differentiated adenocarcinomas that metastasised early. Intermediate levels of HMGA1 were found in the −SA line which formed well-differentiated adenocarcinomas with low propensity for metastasis. The lowest levels were found in the non-tumourigenic CL-S1 line. One of the important observations from this study was that the elevated expression of HMGA1 was probably related to the stage of neoplastic transformation rather than the rate of cellular proliferation. Additional evidence supporting the roles of HMGA1 in tumourigenesis came from studies of transgenic mice by Xu et al.31 Transgenic mice expressing HMGA1 under control of the murine H-2K promoter and immunoglobulin μ enhancer drove the over-expression of HMGA1 transgene in B and T-cells. The mice spontaneously developed aggressive lymphoma consistent with a mature T-cell phenotype, leading to death between 1 and 8.5 months of age.31 More recently, it was shown that transgenic mice with HMGA1 overexpression targeted to uterine tissues developed adenocarcinomas by nine months of age.32
Although HMGA1 over-expression is a phenomenon observed in several cancer types, the basis for the elevated expression remains mostly unknown. So far, HMGA1 has been shown to be a c-Myc target gene.33 More recently, it was shown that the HMGA1 promoter is strongly regulated by oncogenic Ras.34 This suggests that in tumours with a mutated Ras gene, the Ras GTPase signalling could be a mechanism through which the tumours over-express HMGA1. This could be particularly important in PDAC cells which have a high incidence of K-ras mutations. In Burkitt’s lymphoma cell lines, which are known to over-express c-Myc protein, abrogation of HMGA1 expression through antisense ribozyme approach prevents their transformation.33 Similarly, HMGA1 gene suppression using antisense approach inhibited the transformation of human breast cancer cells.35 In a study by Reeves et al, the human breast epithelial cells harbouring the tetracy cline-regulated HMGA1 transgene developed primary and metastatic tumours in nude mice only when the transgene was actively expressed, implicating the roles of HMGA1 in carcinogenesis.35 Over-expression of HMGA1 proteins induces transformation of Rat-1 fibroblast cells and CB33 human lymphoblastoid cells.33,36 Studies using Rat-1 cells have indicated that HMGA1 is required for c-Jun/Ap-1 induced transformation.37 Antisense-mediated targeting of HMGA1 prevents or inhibits retrovirus-induced transformation of rat thyroid cell line and induces apoptosis in two human thyroid anaplastic carcinoma cell lines.38,39 Of note, the suppression of HMGA1 expression did not have an impact on normal thyroid cells.
HMGA1 has been found to bind to the BRCA1 gene promoter and down regulates its expression in breast cancer cells.40 Given that BRCA1 functions as a tumour suppressor gene involved in DNA damage response, the ability of HMGA1 to suppress its expression will render HMGA1 a pro-oncogenic protein. Moreover, it was speculated that the down regulation of BRCA1 commonly found in aggressive breast tumours could be due to the inhibitory effect of HMGA1 over-expression in these tumours. More recently, HMGA1 has been found to be capable of interfering with p53 tumour suppressor functions, in particular cellular apoptosis.41,42 Pierantoni et al found that HMGA1 binds p53 in vitro and in vivo, and interferes with p53-mediated transcription of p53 effectors (i.e. BCL2-associated X protein and cyclin-dependent kinase inhibitor 1A). Furthermore, HMGA1 is able to cooperate with p53 to activate transcription of p53 inhibitor MDM2. The HMGA1 has also been found to interfere with functions of p53 by another mechanism: HMGA1 promotes HIPK2 (homeodomain-interacting protein kinase 2) relocalisation in cytoplasm which inhibits p53 functions as HIPK2 nuclear localisation is required for p53 activity.42 HMGA1 is able to inhibit p53-mediated apoptosis by modulating p53 target genes and cytoplasmic relocalisation of HIPK2.
Interestingly, Takaha et al demonstrated that HMGA1 over-expression may predispose tumour cells to accumulate unbalanced translocations, leading to cumulative mutations, and potentially more aggressive tumour phenotype.43 Using cDNA array analyses, Reeves et al demonstrated that HMGA1 over-expression in a breast epithelial cell line resulted in a dramatic elevation of integrin expression and their potential signalling pathways.35 Though the array used was only limited in the number of genes investigated, it was hypothesised that HMGA1 may execute its pro-tumourigenic functions by enhancing the integrin and its related downstream pathways. Functional studies demonstrating the dependence of HMGA1-induced tumourigenesis on integrin-related signalling pathways are warranted.
Table 1 shows an extensive body of literature documents HMGA1 over-expression in a wide range of cancer types.
Table 1.
Evidence suggesting a role for HMGA1 in tumourigenesis and metastasis
| Tumour type | Author (Year) | Experimental method | Key results |
|---|---|---|---|
| Breast | Ram et al, 1993 30 | Murine cell lines, HMGA1 mRNA levels | ↑HMGA1 expression correlates with neoplastic transformation |
| Liu et al, 1999 44 | Human cell lines, HMGA1 mRNA and protein levels | ↑HMGA1 expression correlates with metastatic potential HMGA1 expression induced by heregulin |
|
| Nestl et al, 2001 45 | Rat cell lines, SSH | HMGA1 expression correlates with metastatic potential | |
| Dolde et al, 2002 46 | Human cell lines, HMGA1 protein levels, antisense approach | ↑HMGA1 expression correlates with matastatic potential ↓HMGA1 by antisense -> ↓transformation but no effect on proliferation |
|
| Baldassarre et al, 2003 40 | Human cell lines and tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 expression in highly tumourigenic cell lines and tumour tissues HMGA1 binds and inhibits BRCA1 gene promoter ↑HMGA1 -> ↑growth in vitro |
|
| Flohr et al, 2003 47 | Human tissue samples, HMGA1 protein levels | HMGA1 expression correlates with tumour grade | |
| Chiappetta et al, 2004 48 | Human tissue samples, HMGA1 protein levels | ↑HMGA1 in breast carcinomas with expression correlated with erbB2 | |
| Cervical | Bandiera et al, 1998 49 | Human cell lines and tissue samples, HMGA1 protein levels | ↑HMGA1 expression in pre-invasive and invasive carcinomas |
| Colorectal | Fedele et al, 1996 50 | Human cell lines and tissue samples, HMGA1 protein levels | ↑HMGA1 expression in cell lines and tumour tissues No expression in normal epithelium |
| Abe et al, 1999 51 | Human tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 expression in adenocarcinoma and adenoma with severe atypia | |
| Kim et al, 1999 52 | Human tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 expression in adenocarcinoma No expression in normal epithelium No correlation with clinicopathological variables |
|
| Chiapetta et al, 2001 53 | Human tissue samples, HMGA1 protein levels | ↑HMGA1 expression in adenocarcinoma and adenoma with severe atypia HMGA1 expression associated with early stages of neoplastic transformation |
|
| Gastric | Xiang et al, 1997 54 | Human tissue samples, HMGA1 mRNA levels | ↑HMGA1 correlates with tumour grade |
| Nam et al, 2003, 55 | Human tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 in gastric cancers | |
| Head and neck | Rho et al, 2007 56 | Human tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 in squamous cell carcinomas, with higher expression in recurrent cases |
| Hepatocellular | Abe et al, 2003 57 | Human tissue samples, HMGA1 protein levels | HMGA1 is not overexpressed in hepatocellular carcinoma, but overexpressed in metastatic adenocarcinomas and intrahepatic cholangiocarcinomas |
| Chang et al, 2005 58 | Human tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 correlates with disease recurrence and poor prognosis | |
| Leukemia | Pierantoni et al, 2003 59 | Human cell lines and tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 in most leukemia tissues and all cell lines studied |
| Xu et al, 2004 31 | Transgenic mice, human tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 results in development of aggressive lymphoma in mice ↑HMGA1 found in leukemia tissues |
|
| Neuroblastoma | Giannini et al, 2000 60 | Human tissue samples, HMGA1 protein levels | ↑HMGA1 expression correlates with poor tumour differentiation |
| Ovarian | Masciullo et al, 2003 61 | Human tissue samples, HMGA1 protein levels | ↑HMGA1 in primary ovarian carcinomas |
| Pancreatic | Abe et al, 2000 16 | Human tissue samples, HMGA1 protein levels | ↑HMGA1 is diagnostic of carcinoma ↑HMGA1 is benign tumours with atypia |
| Nestl et al, 2001 45 | Rat cell lines, SSH | HMGA1 expression correlates with metastatic potential | |
| Tarbe et al, 2001 15 | Human cell lines, cDNA microarray analysis | ↑HMGA1 associated with metastasis | |
| Abe et al, 2002 62 | Human tissue samples, HMGA1 protein levels | ↑HMGA1 in IPMT tumors with dysplasia | |
| Abe et al, 2002 57 | Human tissue samples, HMGA1 protein levels | ↑HMGA1 in liver metastases from pancreatic cancer | |
| Prostate | Bussemakers et al, 1991 63 | Rat cell lines, differential hybridization analysis | HMGA1 expression correlates with metastatic potential |
| Tamimi et al, 1996 64 | Human tissue samples, HMGA1 mRNA levels | ↑HMGA1 expression in high grade tumors | |
| Nestl et al, 2001 45 | Rat cell lines, SSH | HMGA1 expression correlates with matastatic potential | |
| Leman et al, 2003 65 | Transgenic mice, HMGA1 protein levels | HMGA1 expression correlates with neoplastic transformation | |
| Thyroid | Kim et al, 1989 66 | Human tissue samples, HMGA1 mRNA levels | ↑HMGA1 correlates with diagnosis of carcinoma |
| Chiappetta et al, 1995 67 | Human cell lines and tissue samples, HMGA1 protein levels | ↑HMGA1 expression in cell lines and tumour tissues | |
| Chiappetta et al, 1998 68 | Human tissue samples, HMGA1 mRNA and protein levels | ↑HMGA1 is highly diagnostic of carcinoma | |
| Czyz et al, 2004 69 | Human tissue, HMGA1 mRNA levels | ↑HMGA1 in follicular carcinomas | |
SSH: suppression subtractive hybridisation
Roles of HMGA1 in pancreatic cancer
HMGA1: a biomarker of clinical relevance and a mediator of the malignant phenotype
Our group recently sought to characterise the biology of HMGA1 in the context of PDAC. The first aim of this work was to address the clinical relevance of HMGA1 in pancreatic cancer. To do this, we determined the prevalence of HMGA1 overexpression in cancers resected from patients with PDAC. Using a tissue microarray constructed with specimens from consecutive patients having undergone pancreatic resection, we found that more than 90% of pancreatic cancers have tumoural HMGA1 overexpression (Figure 1).70 In contrast, HMGA1 expression is absent (or present at very low levels) in normal pancreatic tissues. Importantly, tumoural HMGA1 expression was predictive of poor patient survival and was an independent prognostic indicator on multivariate analysis.
Fig. 1.
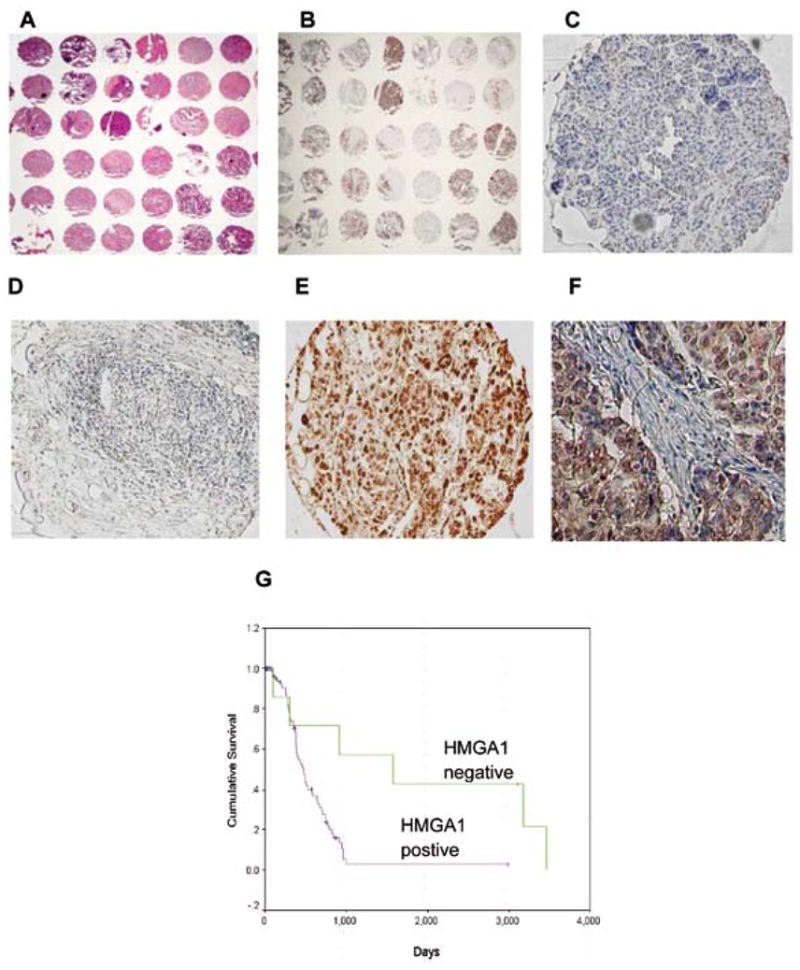
Overview of the pancreatic cancer TMA stained with H&E (A) and HMGA1 immunostaining (B). Magnified view (X100) of HMGA1 immunostaining in: (C) normal pancreas (HMGA1-negative), (D) pancreatic adenocarcinoma (HMGA1-negative), (E) Pancreatic adenocarcinoma (HMGA1-positive). F) A high power magnification (X400) of HMGA1-positive cancer section showing intense nuclear staining for HMGA1 G) Survivals of HMGA1-negative patients were compared with HMGA1-positive patients using Kaplan Meier analysis with log-rank test (P=0.0028). Adapted from Liau et al, Cancer 2008;113(2): 302–314, with permission of Wiley-Liss, Inc., a subsidiary of John Wiley & Sons, Inc. Copyright 2008 American Cancer Society.
Given these promising clinical correlations, we moved to ‘the bench’ to conduct studies designed to characterise the roles of HMGA1 in mediating the malignant phenotypic features of pancreatic cancer. One important goal was to determine whether HMGA1 over-expression in pancreatic cancer cells is an epiphenomenon related to the rapid cell cycle progression or if HMGA1 over-expression is truly an oncogenic event driving neoplastic progression. Because accumulating evidence suggests that HMGA1-dependent phenomena vary depending on the cellular context, we conducted our studies using a panel of human pancreatic adenocarcinoma cells of varying baseline HMGA1 expression.71 Experiments were performed using the loss-of-function (RNA interference, RNAi) and gain-of-function (ectopic over-expression) approaches to analyse the effects of modulating HMGA1 expression on cancer phenotype. We demonstrated that targeted suppression of HMGA1 expression using RNAi attenuates cellular invasiveness and anchorage-independent proliferation.70,72 In contrast, HMGA1 over-expression promotes resistance to anoikis (apoptosis induced when cells are deprived of attachment to matrix; resistance to anoikis is hypothesised to facilitate metastasis by allowing cancer cells to survive when they are detached from their stroma).73 We also showed that HMGA1 is an important regulator of chemoresistance in PDAC cells.74,75 We translated the in vitro findings to the in vivo setting, as we demonstrated that targeted silencing of HMGA1 significantly attenuated metastasis and tumour growth and promoted chemosensitivity in xenograft mouse models of pancreatic cancer.70,72,74 These findings provide compelling evidence that HMGA1 expression plays a biologically significant role in pancreatic cancer progression rather than being an epiphenomenon without functional significance, as previously speculated.76
Pro-oncogenic downstream signaling pathways
Gene array studies have identified genes for which expression is potentially regulated by HMGA1, however, there has been limited functional data supporting the roles of these genes in mediating the pro-oncogenic properties of HMGA1.22 The best established downstream effector pathway mediating HMGA1-dependent effects has been MAPK/ERK signalling.77 Our goal was to identify novel HMGA1 downstream effectors in the context of pancreatic cancer.70, 72–74 Our results suggest that HMGA1 regulates MMP-9 expression through its effects on the transcriptional activity of the MMP-9 gene promoter.72 Given that the PI3-K/Akt activity is well-known to regulate MMP-9 expression, we hypothesised that HMGA1 would modulate PI3-K/Akt activity.78–80 Further clues on the nature of downstream HMGA1 mediators were gleaned from studies of HMGA1 in non-cancer models. Foti et al reported a genetic flaw in four diabetic patients that markedly reduced their HMGA1 expression, leading to decreased insulin receptor expression in their tissues.81 They subsequently demonstrated that HMGA1-knockout mice have decreased insulin receptor expression and impaired insulin signaling, leading to a phenotype characteristic of type II diabetes. With the knowledge that PI3-K/Akt is a crucial downstream mediator of insulin receptor signalling, they demonstrated that muscle tissues from HMGA1-knockout mice have significantly lower PI3-kinase activities and hence lower levels of Akt activation. Based on these findings, we sought to determine if modulating HMGA1 expression in pancreatic cancer cells would affect pro-oncogenic PI3-K/Akt signaling. We found that HMGA1 modulation directly regulates Akt activation.72 In particular, we described a novel HMGA1/Akt/MMP9 pathway with strong impact on cellular invasiveness in PDAC cells. To verify that the effects of HMGA1 on Akt activation were functional, we further demonstrated that HMGA1 expression also directly regulated the phosphorylation of mammalian target of rapamycin (mTOR) which is a known downstream target of Akt.72 Additional HMGA1-induced malignant cellular characteristics (anoikis resistance and soft agar growth) were each dependent on PI3-K/Akt activation. In addition to Akt, we also found that ERK and caspase pathways are downstream mediators of HMGA1. Our findings on the effects of HMGA1 on ERK activation concur with previous studies by Treff et al, who found that HMGA1 regulates ras/ERK signaling.77
Given that activated Akt provides protection from apoptosis and conventional chemotherapy kills by apoptosis, we postulated that HMGA1 will affect chemoresistance in pancreatic cancer cells.82–86 We found that HMGA1 silencing markedly chemosensitises pancreatic cancer cells to gemcitabine. Given the profound chemoresistance characteristic of pancreatic cancer, this finding highlights the potential role for HMGA1 as a therapeutic target. Recently, another group has described that HMGA1 positively regulates insulin receptor expression in pancreatic cancer cells.87 As such, this provides a possible mechanism through which HMGA1 is able to regulate the PI3-K/Akt pathway - by its regulation of insulin receptor expression. Future studies will need to test this hypothesis. Our findings are summarised in Figure 2. Together, our work has established the clinical relevance of HMGA1 in pancreatic cancer and has characterised the roles of HMGA1 as a novel mediator of the malignant phenotype in this cancer.
Fig. 2.
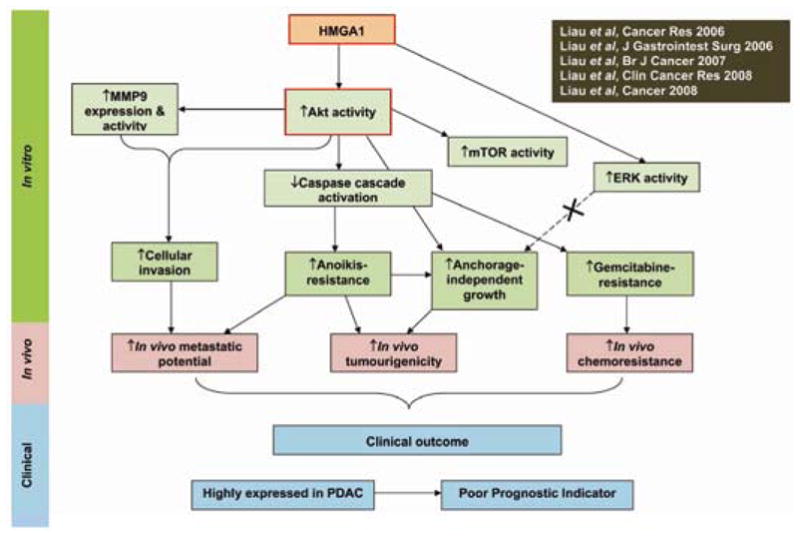
Schematic diagram summarising the roles and mediators of HMGA1 in pancreatic adenocarcinoma
Targeting HMGA1 in pancreatic adenocarcinoma
The failure of conventional therapy in improving outcomes of patients with pancreatic cancer highlights an urgent need for novel therapeutic approaches. Accumulating evidence suggests that targeting HMGA1 in cancer cells may represent such a strategy. Targeting HMGA1 in cancer has several advantages. Given that HMGA1 is over-expressed in a broad range of malignancies in which it regulates a large number of genes involved in cancer progression, its potential as a therapeutic target in oncology is wide ranging. HMGA1 is expressed at high levels during embryogenesis, and its expression reduces to almost absent levels during adulthood.27 As such, targeting HMGA1 would theoretically affect only cancer cells which over-express HMGA1, leaving the normal tissues unaffected or with only minimal collateral damage.27 Furthermore, our findings demonstrated HMGA1 silencing had no effect on anchorage-dependent cellular proliferation (i.e. standard monolayer culture) whilst there was a significant inhibition of anchorage-independent growth. Normal tissue grows under anchorage-dependent conditions. As such, one could speculate that targeted suppression of HMGA1 may have a lesser effect on this mode of growth and hence, would have minimal impact on normal tissues.
Developing HMGA1-specific therapeutics
Molecular approaches such as antisense oligonucleotides and ribozymes have been previously used to knockdown HMGA1 expression. 88 Trapasso et al have shown that adenovirus carrying antisense HMGA1 may result in an apoptotic response in pancreatic cancer cells. Unlike cell surface receptors, HMGA1 is predominantly localised to the nucleus. As such, it will not be amenable to monoclonal antibody targeting due to its inaccessibility in vivo. Development of novel small molecule inhibitors of HMGA1 will move the field closer to clinical therapy. Development of small molecule inhibitors of HMGA1 will need a detailed study of the chemical structure of HMGA1 molecule to identify the potential sites (e.g. AT-hooks) on the HMGA1 molecule that can be inhibited by designed small molecule inhibitors. Distamycin A is an antibiotic that has been used as a small molecule inhibitor of HMGA1 by competing with HMGA1 for binding to AT-rich minor grooves of DNA. Clearly, this mode of action of Distamycin A will not be specific for HMGA1 and will affect other nuclear proteins that bind to DNA minor grooves.89,90 Other means of targeting HMGA1 include cross-linking of HMGA1 molecules to DNA and hence limiting its availability for intranuclear actions such as binding to promoter regions of DNA and forming complexes with other transcriptional factors. This mechanism was described for mitomycin C which has been reported to cross-link HMGA1 molecules to DNA.91
In our previous work, we described a highly effective and specific molecular approach to HMGA1 silencing: the RNA interference. Many pharmaceutical companies are actively pursuing RNAi-based therapeutics with the hope of harnessing the naturally occurring RNAi mechanism as an effective strategy. However, there are several concerns that need to be addressed before RNAi can be a real therapeutic modality. Firstly, the issue of toxicity and off-target effects of RNAi will need to be addressed and warrants intense investigation.92 Secondly, delivery of RNAi to specific tissue sites remains a major stumbling block. Delivery of siRNA is complicated by the serum instability of siRNA, low cellular uptake effi ciency and lack of understanding on its pharmacokinetics and biodistribution.93 Recent developments involving targeted delivery to cancerous tissues is achieved by linking siRNA to ligand, antibody or aptamer are clearly promising.94 Alternatively, short hairpin RNA as transcribed from RNA polymerase promoters from plasmid- and virus-based vectors provide effective strategies for RNA interference. Established gene therapy techniques could be utilised to deliver vectors encoding short hairpin RNA.
Potential adverse effects of therapeutic targeting of HMGA1
Although HMGA1 is absent or present at very low levels in adult tissues, it remains uncertain to what extent HMGA1 expression is required in adult tissues. The loss of HMGA1 function in transgenic mice has led to the development of diabetes through the decreased expression of insulin receptors and impaired insulin signalling pathways. 81 As such, any targeting of HMGA1 can be predicted to lead to glucose intolerance and perhaps, clinical diabetes. Further, in the HMGA1-knockout mouse model, disruption of HMGA1 resulted in cardiac hypertrophy and development of haematological malignancies. 95 Fedele et al found that surprisingly, Hmga1−/− and Hmga1+/− mice developed B-cell expansion, resembling human B-cell lymphomas. This revealed an unsuspected haploinsuffi cient tumour suppressor role of HMGA1. More recently, HMGA1 has been shown to be an essential component of the senescence machinery.96 Given that senescence is a barrier to malignant transformation, this implies that HMGA1 also acts in the tumour suppressor network. The above tumour suppressor roles of HMGA1 are in contrast to its pro-oncogenic properties. It is possible that the dual effect of HMGA1 might depend on cellular context. Clearly, any future clinical trials involving therapeutic targeting of HMGA1 will require close monitoring for possible cardiac, haematological and endocrine complications.
Other potential clinical applications of HMGA1 biology
Previously, we showed that tumoural HMGA1 expression is an indicator of poor prognosis in pancreatic cancer patients.70 Tumoural HMGA1 protein detection could be used to select patients having undergone pancreatic resection for aggressive management of their disease. Development of techniques for detection of HMGA1 proteins or mRNA in readily accessible clinical samples (e.g. blood samples of pancreatic cancer patients) might facilitate monitoring response to chemotherapy and allow early diagnosis of recurrence following surgical resection. Given our findings that HMGA1 expression is low or absent in normal pancreas, there may be a role for HMGA1 as a diagnostic marker for pancreatic cancer. The ability to detect HMGA1 protein or mRNA in pancreatic juice sampled during ERCP could be used as part of a panel of markers to screen for early pancreatic cancer in secondary screening programme such as the EUROPAC programme.97
In addition, the expression of HMGA1 may be an important factor in the choice of therapy. We have shown that HMGA1 expression correlates with resistance to gemcitabine.74 It may be that detection of high HMGA1 expression in tumour samples may indicate a high degree of gemcitabine chemoresistance, and may affect the clinician’s choice of chemotherapeutic agents.
Conclusion
In this review, we have discussed the current evidence for the roles of HMGA1 in cancer, with a particular focus on pancreatic cancer. We have provided evidence to support the potential use of HMGA1 as a prognostic biomarker and therapeutic target in pancreatic cancer.
Acknowledgments
S-S. Liau was in receipt of the IHPBA Kenneth W. Warren Fellowship, British Pancreatic Society Travelling Fellowship, Cancer Research UK Core Skills Bursary, Aid For Cancer Research Grant and Brigham and Women’s Hospital Department of Surgery Research/Equipment Grant. This work was supported by NIH 1 RO1 CA114103 (E.E. Whang) and American Cancer Society RSG-04221-01-CCE (E.E. Whang).
References
- 1.CancerStats. http//:info.cancerresearchuk.org/cancerstats.
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56(2):106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 3.CancerStats. http//:info.cancerresearchuk.org/cancerstats.
- 4.American Cancer Society. Cancer facts and figures 2007. 2007 http://www.cancer.org/docroot/CRI/CRI_2_1x.asp?dt=34.
- 5.Bramhall SR, Allum WH, Jones AG, et al. Treatment and survival in 13,560 patients with pancreatic cancer, and incidence of the disease, in the West Midlands: an epidemiological study. Br J Surg. 1995;82(1):111–15. doi: 10.1002/bjs.1800820137. [DOI] [PubMed] [Google Scholar]
- 6.Hedberg M, Borgstrom A, Genell S, et al. Survival following pancreatic carcinoma: a follow-up study of all cases recorded in Malmo, Sweden, 1977–1991. Br J Surg. 1998;85(12):1641–44. doi: 10.1046/j.1365-2168.1998.00920.x. [DOI] [PubMed] [Google Scholar]
- 7.Staley CA, Lee JE, Cleary KR, et al. Preoperative chemoradiation, pancreaticoduodenectomy, and intraoperative radiation therapy for adenocarcinoma of the pancreatic head. Am J Surg. 1996;171(1):118–124. doi: 10.1016/S0002-9610(99)80085-3. discussion 124–15. [DOI] [PubMed] [Google Scholar]
- 8.Jaffee EM, Hruban RH, Canto M, et al. Focus on pancreas cancer. Cancer Cell. 2002;2(1):25–28. doi: 10.1016/s1535-6108(02)00093-4. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 10.Friess H, Ding J, Kleeff J, et al. Microarray-based identification of differentially expressed growth- and metastasis-associated genes in pancreatic cancer. Cell Mol Life Sci. 2003;60(6):1180–99. doi: 10.1007/s00018-003-3036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crnogorac-Jurcevic T, Missiaglia E, et al. Molecular alterations in pancreatic carcinoma: expression profiling shows that dysregulated expression of S100 genes is highly prevalent. J Pathol. 2003;201(1):63–74. doi: 10.1002/path.1418. [DOI] [PubMed] [Google Scholar]
- 12.Logsdon CD, Simeone DM, Binkley C, et al. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63(10):2649–57. [PubMed] [Google Scholar]
- 13.Iacobuzio-Donahue CA, Maitra A, Olsen M, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162(4):1151–62. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han H, Bearss DJ, Browne LW, et al. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002;62(10):2890–96. [PubMed] [Google Scholar]
- 15.Tarbe N, Evtimova V, Burtscher H, et al. Transcriptional profiling of cell lines derived from an orthotopic pancreatic tumor model reveals metastasis-associated genes. Anticancer Res. 2001;21(5):3221–28. [PubMed] [Google Scholar]
- 16.Abe N, Watanabe T, Masaki T, et al. Pancreatic duct cell carcinomas express high levels of high mobility group I(Y) proteins. Cancer Res. 2000;60(12):3117–22. [PubMed] [Google Scholar]
- 17.Goodwin GH, Sanders C, Johns EW. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur J Biochem. 1973;38(1):14–19. doi: 10.1111/j.1432-1033.1973.tb03026.x. [DOI] [PubMed] [Google Scholar]
- 18.Lund T, Holtlund J, Fredriksen M, et al. On the presence of two new high mobility group-like proteins in HeLa S3 cells. FEBS Lett. 1983;152(2):163–67. doi: 10.1016/0014-5793(83)80370-6. [DOI] [PubMed] [Google Scholar]
- 19.Friedmann M, Holth LT, Zoghbi HY, et al. Organization, inducible-expression and chromosome localization of the human HMG-I(Y) nonhistone protein gene. Nucleic Acids Res. 1993;21(18):4259–67. doi: 10.1093/nar/21.18.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bustin M, Lehn DA, Landsman D. Structural features of the HMG chromosomal proteins and their genes. Biochim Biophys Acta. 1990;1049(3):231–43. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- 21.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. Journal of Biological Chemistry. 1990;265(15):8573–82. [PubMed] [Google Scholar]
- 22.Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277(1–2):63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- 23.Zhao K, Kas E, Gonzalez E, et al. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. Embo J. 1993;12(8):3237–47. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sgarra R, Lee J, Tessari MA, et al. The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem. 2006;281(7):3764–72. doi: 10.1074/jbc.M510231200. [DOI] [PubMed] [Google Scholar]
- 25.Reeves R, Langan TA, Nissen MS. Phosphorylation of the DNA-binding domain of nonhistone high-mobility group I protein by cdc2 kinase: reduction of binding affinity. Proc Natl Acad Sci U S A. 1991;88(5):1671–75. doi: 10.1073/pnas.88.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nissen MS, Langan TA, Reeves R. Phosphorylation by cdc2 kinase modulates DNA binding activity of high mobility group I nonhistone chromatin protein. J Biol Chem. 1991;266(30):19945–52. [PubMed] [Google Scholar]
- 27.Chiappetta G, Avantaggiato V, Visconti R, et al. High level expression of the HMGI (Y) gene during embryonic development. Oncogene. 1996;13(11):2439–46. [PubMed] [Google Scholar]
- 28.Ogram SA, Reeves R. Differential regulation of a multipromoter gene. Selective 12-O-tetrade-canoylphorbol-13-acetate induction of a single transcription start site in the HMG-I/Y gene. J Biol Chem. 1995;270(23):14235–42. doi: 10.1074/jbc.270.23.14235. [DOI] [PubMed] [Google Scholar]
- 29.Giancotti V, Pani B, D’Andrea P, et al. Elevated levels of a specific class of nuclear phosphoproteins in cells transformed with v-ras and v-mos oncogenes and by cotransfection with c-myc and polyoma middle T genes. Embo J. 1987;6(7):1981–87. doi: 10.1002/j.1460-2075.1987.tb02461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ram TG, Reeves R, Hosick HL. Elevated high mobility group-I(Y) gene expression is associated with progressive transformation of mouse mammary epithelial cells. Cancer Res. 1993;53(11):2655–60. [PubMed] [Google Scholar]
- 31.Xu Y, Sumter TF, Bhattacharya R, et al. The HMG-I oncogene causes highly penetrant, aggressive lymphoid malignancy in transgenic mice and is overexpressed in human leukemia. Cancer Res. 2004;64(10):3371–75. doi: 10.1158/0008-5472.CAN-04-0044. [DOI] [PubMed] [Google Scholar]
- 32.Tesfaye A, Di Cello F, Hillion J, et al. The high-mobility group A1 gene up-regulates cyclooxygenase 2 expression in uterine tumorigenesis. Cancer Res. 2007;67(9):3998–04. doi: 10.1158/0008-5472.CAN-05-1684. [DOI] [PubMed] [Google Scholar]
- 33.Wood LJ, Mukherjee M, Dolde CE, et al. HMG-I/Y, a new c-Myc target gene and potential oncogene. Mol Cell Biol. 2000;20(15):5490–02. doi: 10.1128/mcb.20.15.5490-5502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cleynen I, Huysmans C, Sasazuki T, et al. Transcriptional control of the human high mobility group A1 gene: basal and oncogenic Ras-regulated expression. Cancer Res. 2007;67(10):4620–29. doi: 10.1158/0008-5472.CAN-06-4325. [DOI] [PubMed] [Google Scholar]
- 35.Reeves R, Edberg DD, Li Y. Architectural transcription factor HMGI(Y) promotes tumor progression and mesenchymal transition of human epithelial cells. Mol Cell Biol. 2001;21(2):575–94. doi: 10.1128/MCB.21.2.575-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wood LJ, Maher JF, Bunton TE, et al. The oncogenic properties of the HMG-I gene family. Cancer Res. 2000;60(15):4256–61. [PubMed] [Google Scholar]
- 37.Hommura F, Katabami M, Leaner VD, et al. HMG-I/Y is a c-Jun/activator protein-1 target gene and is necessary for c-Jun-induced anchorage-independent growth in Rat1a cells. Mol Cancer Res. 2004;2(5):305–14. [PubMed] [Google Scholar]
- 38.Berlingieri MT, Pierantoni GM, Giancotti V, et al. Thyroid cell transformation requires the expression of the HMGA1 proteins. Oncogene. 2002;21(19):2971–80. doi: 10.1038/sj.onc.1205368. [DOI] [PubMed] [Google Scholar]
- 39.Scala S, Portella G, Fedele M, et al. Adenovirus-mediated suppression of HMGI(Y) protein synthesis as potential therapy of human malignant neoplasias. PNAS. 2000;97(8):4256–61. doi: 10.1073/pnas.070029997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baldassarre G, Battista S, Belletti B, et al. Negative regulation of BRCA1 gene expression by HMGA1 proteins accounts for the reduced BRCA1 protein levels in sporadic breast carcinoma. Mol Cell Biol. 2003;23(7):2225–38. doi: 10.1128/MCB.23.7.2225-2238.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierantoni GM, Rinaldo C, Esposito F, et al. High Mobility Group A1 (HMGA1) proteins interact with p53 and inhibit its apoptotic activity. Cell Death Differ. 2006;13(9):1554–63. doi: 10.1038/sj.cdd.4401839. [DOI] [PubMed] [Google Scholar]
- 42.Pierantoni GM, Rinaldo C, Mottolese M, et al. High-mobility group A1 inhibits p53 by cytoplasmic relocalization of its proapoptotic activator HIPK2. J Clin Invest. 2007 doi: 10.1172/JCI29852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Takaha N, Hawkins AL, Griffin CA, et al. High mobility group protein I(Y): a candidate architectural protein for chromosomal rearrangements in prostate cancer cells. Cancer Res. 2002;62(3):647–51. [PubMed] [Google Scholar]
- 44.Liu WM, Guerra-Vladusic FK, Kurakata S, et al. HMG-I(Y) recognizes base-unpairing regions of matrix attachment sequences and its increased expression is directly linked to metastatic breast cancer phenotype. Cancer Res. 1999;59(22):5695–03. [PubMed] [Google Scholar]
- 45.Nestl A, Von Stein OD, Zatloukal K, et al. Gene expression patterns associated with the metastatic phenotype in rodent and human tumors. Cancer Res. 2001;61(4):1569–77. [PubMed] [Google Scholar]
- 46.Dolde CE, Mukherjee M, Cho C, et al. HMG-I/Y in human breast cancer cell lines. Breast Cancer Res Treat. 2002;71(3):181–91. doi: 10.1023/a:1014444114804. [DOI] [PubMed] [Google Scholar]
- 47.Flohr AM, Rogalla P, Bonk U, et al. High mobility group protein HMGA1 expression in breast cancer reveals a positive correlation with tumour grade. Histol Histopathol. 2003;18(4):999–04. doi: 10.14670/HH-18.999. [DOI] [PubMed] [Google Scholar]
- 48.Chiappetta G, Botti G, Monaco M, et al. HMGA1 protein overexpression in human breast carcinomas: correlation with ErbB2 expression. Clin Cancer Res. 2004;10(22):7637–44. doi: 10.1158/1078-0432.CCR-04-0291. [DOI] [PubMed] [Google Scholar]
- 49.Bandiera A, Bonifacio D, Manfioletti G, et al. Expression of HMGI(Y) proteins in squamous intraepithelial and invasive lesions of the uterine cervix. Cancer Res. 1998;58(3):426–31. [PubMed] [Google Scholar]
- 50.Fedele M, Bandiera A, Chiappetta G, et al. Human colorectal carcinomas express high levels of high mobility group HMGI(Y) proteins. Cancer Res. 1996;56(8):1896–01. [PubMed] [Google Scholar]
- 51.Abe N, Watanabe T, Sugiyama M, et al. Determination of high mobility group I(Y) expression level in colorectal neoplasias: a potential diagnostic marker. Cancer Res. 1999;59(6):1169–74. [PubMed] [Google Scholar]
- 52.Kim DH, Park YS, Park CJ, et al. Expression of the HMGI(Y) gene in human colorectal cancer. Int J Cancer. 1999;84(4):376–80. doi: 10.1002/(sici)1097-0215(19990820)84:4<376::aid-ijc8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 53.Chiappetta G, Manfioletti G, Pentimalli F, et al. High mobility group HMGI(Y) protein expression in human colorectal hyperplastic and neoplastic diseases. Int J Cancer. 2001;91(2):147–51. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1033>3.3.co;2-m. [DOI] [PubMed] [Google Scholar]
- 54.Xiang YY, Wang DY, Tanaka M, et al. Expression of high-mobility group-1 mRNA in human gastrointestinal adenocarcinoma and corresponding non-cancerous mucosa. Int J Cancer. 1997;74(1):1–6. doi: 10.1002/(sici)1097-0215(19970220)74:1<1::aid-ijc1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 55.Nam ES, Kim DH, Cho SJ, et al. Expression of HMGI(Y) associated with malignant phenotype of human gastric tissue. Histopathology. 2003;42(5):466–71. doi: 10.1046/j.1365-2559.2003.01618.x. [DOI] [PubMed] [Google Scholar]
- 56.Rho YS, Lim YC, Park IS, et al. High mobility group HMGI(Y) protein expression in head and neck squamous cell carcinoma. Acta Otolaryngol. 2007;127(1):76–81. doi: 10.1080/00016480600740571. [DOI] [PubMed] [Google Scholar]
- 57.Abe N, Watanabe T, Izumisato Y, et al. High mobility group A1 is expressed in metastatic adenocarcinoma to the liver and intrahepatic cholangiocarcinoma, but not in hepatocellular carcinoma: its potential use in the diagnosis of liver neoplasms. J Gastroenterol. 2003;38(12):1144–49. doi: 10.1007/s00535-003-1221-9. [DOI] [PubMed] [Google Scholar]
- 58.Chang ZG, Yang LY, Wang W, et al. Determination of high mobility group A1 (HMGA1) expression in hepatocellular carcinoma: a potential prognostic marker. Dig Dis Sci. 2005;50(10):1764–70. doi: 10.1007/s10620-005-2934-9. [DOI] [PubMed] [Google Scholar]
- 59.Pierantoni GM, Agosti V, Fedele M, et al. High-mobility group A1 proteins are overexpressed in human leukaemias. Biochem J. 2003;372(Pt 1):145–50. doi: 10.1042/BJ20021493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giannini G, Kim CJ, Di Marcotullio L, et al. Expression of the HMGI(Y) gene products in human neuroblastic tumours correlates with differentiation status. Br J Cancer. 2000;83(11):1503–09. doi: 10.1054/bjoc.2000.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masciullo V, Baldassarre G, Pentimalli F, et al. HMGA1 protein over-expression is a frequent feature of epithelial ovarian carcinomas. Carcinogenesis. 2003;24(7):1191–98. doi: 10.1093/carcin/bgg075. [DOI] [PubMed] [Google Scholar]
- 62.Abe N, Watanabe T, Izumisato Y, et al. Diagnostic significance of high mobility group I(Y) protein expression in intraductal papillary mucinous tumors of the pancreas. Pancreas. 2002;25(2):198–04. doi: 10.1097/00006676-200208000-00015. [DOI] [PubMed] [Google Scholar]
- 63.Bussemakers MJ, van de Ven WJ, Debruyne FM, et al. Identification of high mobility group protein I(Y) as potential progression marker for prostate cancer by differential hybridization analysis. Cancer Res. 1991;51(2):606–11. [PubMed] [Google Scholar]
- 64.Tamimi Y, van der Poel H, Karthaus H, et al. A retrospective study of high mobility group protein I(Y) as progression marker for prostate cancer determined by in situ hybridization. Br J Cancer. 1996;74(4):573–78. doi: 10.1038/bjc.1996.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leman ES, Madigan MC, Brunagel G, et al. Nuclear matrix localization of high mobility group protein I(Y) in a transgenic mouse model for prostate cancer. J Cell Biochem. 2003;88(3):599–08. doi: 10.1002/jcb.10368. [DOI] [PubMed] [Google Scholar]
- 66.Kim YW, Kern HF, Mullins TD, et al. Characterization of clones of a human pancreatic adenocarcinoma cell line representing different stages of differentiation. Pancreas. 1989;4(3):353–62. doi: 10.1097/00006676-198906000-00013. [DOI] [PubMed] [Google Scholar]
- 67.Chiappetta G, Bandiera A, Berlingieri MT, et al. The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene. 1995;10(7):1307–14. [PubMed] [Google Scholar]
- 68.Chiappetta G, Tallini G, De Biasio MC, et al. Detection of high mobility group I HMGI(Y) protein in the diagnosis of thyroid tumors: HMGI(Y) expression represents a potential diagnostic indicator of carcinoma. Cancer Res. 1998;58(18):4193–98. [PubMed] [Google Scholar]
- 69.Czyz W, Balcerczak E, Jakubiak M, et al. HMGI(Y) gene expression as a potential marker of thyroid follicular carcinoma. Langenbecks Arch Surg. 2004;389(3):193–97. doi: 10.1007/s00423-004-0479-6. [DOI] [PubMed] [Google Scholar]
- 70.Liau SS, Rocha F, Matros E, et al. High mobility group AT-hook 1 (HMGA1) is an independent prognostic factor and novel therapeutic target in pancreatic adenocarcinoma. Cancer. 2008;113(2):302–14. doi: 10.1002/cncr.23560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fedele M, Pierantoni GM, Berlingieri MT, et al. Over-expression of proteins HMGA1 induces cell cycle deregulation and apoptosis in normal rat thyroid cells. Cancer Res. 2001;61(11):4583–90. [PubMed] [Google Scholar]
- 72.Liau SS, Jazag A, Whang EE. HMGA1 is a determinant of cellular invasiveness and in vivo metastatic potential in pancreatic adenocarcinoma. Cancer Res. 2006;66(24):11613–22. doi: 10.1158/0008-5472.CAN-06-1460. [DOI] [PubMed] [Google Scholar]
- 73.Liau SS, Jazag A, Ito K, Whang EE. Overexpression of HMGA1 promotes anoikis resistance and constitutive Akt activation in pancreatic adenocarcinoma cells. Br J Cancer. 2007;96(6):993–1000. doi: 10.1038/sj.bjc.6603654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liau SS, Whang E. HMGA1 is a molecular determinant of chemoresistance to gemcitabine in pancreatic adenocarcinoma. Clin Cancer Res. 2008;14(5):1470–77. doi: 10.1158/1078-0432.CCR-07-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liau SS, Ashley SW, Whang EE. Lentivirus-mediated RNA interference of HMGA1 promotes chemosensitivity to gemcitabine in pancreatic adenocarcinoma. J Gastrointest Surg. 2006;10(9):1254–62. doi: 10.1016/j.gassur.2006.06.011. discussion 1263. [DOI] [PubMed] [Google Scholar]
- 76.Evans A, Lennard TW, Davies BR. High-mobility group protein 1(Y): metastasis-associated or metastasis- inducing? J Surg Oncol. 2004;88(2):86–99. doi: 10.1002/jso.20136. [DOI] [PubMed] [Google Scholar]
- 77.Treff NR, Pouchnik D, Dement GA, et al. High-mobility group A1a protein regulates Ras/ERK signaling in MCF-7 human breast cancer cells. Oncogene. 2004;23(3):777–85. doi: 10.1038/sj.onc.1207167. [DOI] [PubMed] [Google Scholar]
- 78.Charoenrat P, Wongkajornsilp A, Rhys-Evans PH, et al. Signaling pathways required for matrix metalloproteinase-9 induction by betacellulin in head-and-neck squamous carcinoma cells. Int J Cancer. 2004;111(2):174–83. doi: 10.1002/ijc.20228. [DOI] [PubMed] [Google Scholar]
- 79.Ellerbroek SM, Halbleib JM, Benavidez M, et al. Phosphatidylinositol 3-kinase activity in epidermal growth factor-stimulated matrix metalloproteinase-9 production and cell surface association. Cancer Res. 2001;61(5):1855–61. [PubMed] [Google Scholar]
- 80.Lu Y, Wahl LM. Production of matrix metalloproteinase- 9 by activated human monocytes involves a phosphatidylinositol-3 kinase/Akt/IKKalpha/NF-kappaB pathway. J Leukoc Biol. 2005;78(1):259–65. doi: 10.1189/jlb.0904498. [DOI] [PubMed] [Google Scholar]
- 81.Foti D, Chiefari E, Fedele M, et al. Lack of the architectural factor HMGA1 causes insulin resistance and diabetes in humans and mice. Nat Med. 2005;11(7):765–73. doi: 10.1038/nm1254. [DOI] [PubMed] [Google Scholar]
- 82.Peruzzi F, Prisco M, Dews M, et al. Multiple signaling pathways of the insulin-like growth factor 1 receptor in protection from apoptosis. Mol Cell Biol. 1999;19(10):7203–15. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fahy BN, Schlieman M, Virudachalam S, et al. AKT inhibition is associated with chemosensitisation in the pancreatic cancer cell line MIA-PaCa-2. Br J Cancer. 2003;89(2):391–97. doi: 10.1038/sj.bjc.6601037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Arlt A, Gehrz A, Muerkoster S, et al. Role of NF-kappaB and Akt/PI3K in the resistance of pancreatic carcinoma cell lines against gemcitabine-induced cell death. Oncogene. 2003;22(21):3243–51. doi: 10.1038/sj.onc.1206390. [DOI] [PubMed] [Google Scholar]
- 85.Blanco-Aparicio C, Pequeno B, Moneo V, et al. Inhibition of phosphatidylinositol-3-kinase synergizes with gemcitabine in low-passage tumor cell lines correlating with Bax translocation to the mitochondria. Anticancer Drugs. 2005;16(9):977–87. doi: 10.1097/01.cad.0000180117.93535.cf. [DOI] [PubMed] [Google Scholar]
- 86.Ng SSW, Tsao M-S, Chow S, et al. Inhibition of Phosphatidylinositide 3-Kinase Enhances Gemcitabine-induced Apoptosis in Human Pancreatic Cancer Cells. Cancer Res. 2000;60(19):5451–55. [PubMed] [Google Scholar]
- 87.Kolb S, Fritsch R, Saur D, et al. HMGA1 Controls Transcription of Insulin Receptor to Regulate Cyclin D1 Translation in Pancreatic Cancer Cells. Cancer Res. 2007;67(10):4679–86. doi: 10.1158/0008-5472.CAN-06-3308. [DOI] [PubMed] [Google Scholar]
- 88.Trapasso F, Sarti M, Cesari R, et al. Therapy of human pancreatic carcinoma based on suppression of HMGA1 protein synthesis in preclinical models. Cancer Gene Ther. 2004;11(9):633–41. doi: 10.1038/sj.cgt.7700745. [DOI] [PubMed] [Google Scholar]
- 89.Massaad-Massade L, Navarro S, Krummrei U, et al. HMGA1 enhances the transcriptional activity and binding of the estrogen receptor to its responsive element. Biochemistry. 2002;41(8):2760–68. doi: 10.1021/bi011455j. [DOI] [PubMed] [Google Scholar]
- 90.Ghersa P, Whelan J, Cambet Y, et al. Distamycin prolongs E-selectin expression by interacting with a specific NF-kappaB-HMG-I(Y) binding site in the promoter. Nucleic Acids Res. 1997;25(2):339–46. doi: 10.1093/nar/25.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Reeves R, Beckerbauer LM. HMGA proteins as therapeutic drug targets. Prog Cell Cycle Res. 2003;5:279–86. [PubMed] [Google Scholar]
- 92.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–41. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 93.Behlke MA. Progress towards in vivo use of siRNAs. Mol Ther. 2006;13(4):644–70. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pirollo KF, Zon G, Rait A, et al. Tumor-targeting nano-immunoliposome complex for short interfering RNA delivery. Hum Gene Ther. 2006;17(1):117–24. doi: 10.1089/hum.2006.17.117. [DOI] [PubMed] [Google Scholar]
- 95.Fedele M, Fidanza V, Battista S, et al. Haploinsufficiency of the Hmga1 gene causes cardiac hypertrophy and myelo-lymphoproliferative disorders in mice. Cancer Res. 2006;66(5):2536–43. doi: 10.1158/0008-5472.CAN-05-1889. [DOI] [PubMed] [Google Scholar]
- 96.Narita M, Narita M, Krizhanovsky V, et al. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell. 2006;126(3):503–14. doi: 10.1016/j.cell.2006.05.052. [DOI] [PubMed] [Google Scholar]
- 97.EUROPAC Project. Secondary screening for early pancreatic cancer in high risk families. University of Liverpool Department of Surgery; 2007. http://www.liv.ac.uk/surgery/sseuropac.htm. [Google Scholar]


