Abstract
Serological expression cloning of antigens eliciting a humoral immune response to a syngeneic mouse sarcoma identified pem (mouse placenta and embryonic expression gene) as a new member of the cancer/testis family. To identify the human homologue of pem, mouse pem sequences and pem-related expressed sequence tags from human testis were used as PCR primers for amplification using human testis cDNA. However, rather than pem, another gene, designated OY-TES-1, was isolated and found to be the human homologue of proacrosin binding protein sp32 precursor originally identified in mouse, guinea pig, and pig. OY-TES-1 maps to chromosome 12p12-p13 and contains 10 exons. Southern blot analysis suggests the presence of two OY-TES-1-related genes in the human genome. In normal tissues, OY-TES-1 mRNA was expressed only in testis, whereas in malignant tissues, a variable proportion of a wide array of cancers, including bladder, breast, lung, liver, and colon cancers, expressed OY-TES-1. Serological survey of 362 cancer patients with a range of different cancers showed antibody to OY-TES-1 in 25 patients. No OY-TES-1 sera reactivity was found in 20 normal individuals. These findings indicate that OY-TES-1 is an additional member of the cancer/testis family of antigens and that OY-TES-1 is immunogenic in humans.
Keywords: testis cDNA, PCR cloning, antibody response
The list of human tumor antigens recognized by human CD8 T cells (1–4), CD4 T cells (5–8), and antibodies (9, 10) is growing rapidly. They can be categorized into one or more of the following groups: differentiation antigens, e.g., Melan A/MART-1 (11, 12); tyrosinase (13, 14); mutational antigens, e.g., p53 (15); amplified/overexpressed antigens, e.g., HER-2/neu (16); splice variant antigens (9, 17); viral antigens (18, 19); and cancer/testis (CT) antigens, e.g., MAGE (1), NY-ESO-1 (20). CT antigens have received particular attention because of their unique expression pattern and their potential as targets for cancer vaccines. The defining characteristics of CT antigens are high levels of expression in male germ cells, lack of expression in other normal tissue, and aberrant expression in a wide range of different tumor types. To date, more than 10 genes or gene families coding for CT antigens have been defined: MAGE (1), GAGE (21), BAGE (22), SSX (23, 24), NY-ESO-1/LAGE (20, 25), SCP-1 (26), CT7/MAGE-C1 (27, 28), CT9 (29), CT10/MAGE-C2 (30, 31), and CTp11 (32). Of these, NY-ESO-1, initially defined by SEREX (serological analysis of cDNA expression libraries) in esophageal cancer (20), is particularly immunogenic, eliciting both cellular and humoral immune responses in a high proportion of patients with advanced NY-ESO-1-expressing tumors (33–35). In contrast, cellular and humoral immune responses to other CT antigens appear to be less frequent.
In a recent SEREX analysis of BALB/c methylcholanthrene-induced sarcoma Meth A, we identified pem (placenta and embryonic expression gene) (36) as an additional member of the CT family of antigens (37). In attempting to clone the human homologue of pem, we isolated another gene, which turned out to be the human homologue of proacrosin binding protein sp32 precursor (38). Expression profiling of the gene showed that it has the characteristics of a CT antigen and that it elicits a strong immune response in a subset of patients with cancer.
Materials and Methods
RNA Isolation and cDNA Synthesis.
Total RNA was isolated from human testis by using the RNeasy kit (Qiagen, Hilden, Germany). Poly(A)+ RNA was isolated from tumor tissue by using a QuickPrep Micro mRNA Purification kit (Amersham Pharmacia). The mRNA was reverse-transcribed into single-strand cDNA by using Moloney murine leukemia virus reverse transcriptase and oligo(dT)15 as a primer (Amersham Pharmacia).
PCR Amplification and OY-TES-1 Cloning.
Human testis cDNA was amplified by PCR using primers pem5 (5′-GTGGACAAGAGGAAGCACAA-3′) corresponding to nucleotides 65–84 of mouse OY-MS-4 (37) and EST-2 (5′-TCTCCCCATCTCACTCCAC-3′) derived from human testis expressed sequence tag (EST) clone AA397852, matched to nucleotides 700–682 of OY-MS-4. PCR conditions were 1 min at 94°C, 1 min at 54°C, and 1.5 min at 72°C for 35 cycles. These cycles were followed by a 10-min elongation step at 72°C. A PCR product of 1.1 kb was obtained and sequenced by using an ABI PRISM automated sequencer (Perkin–Elmer). This 1.1-kb product showed a high degree of homology with many human ESTs in the EST database (http://www.ncbi.nlm.nih.gov/dbEST), and most of the homologous sequences were cDNA clones derived from human testis. The 3′ end of the gene was extended by sequence information from 12 of these homologous ESTs (Fig. 1). The actual nucleotide sequence (261–1796) of the 1.5-kb PCR product obtained by using anti-sense primer ht-1 (human testis-1) (5′-ATGTGAGTAGGGGCCGAGTA-3′) was identical to the deduced sequence from the homologous ESTs in the EST database. The 1.5-kb product was designated OY-TES-1.
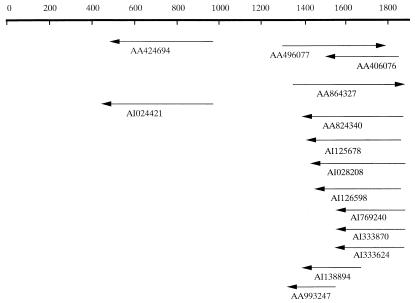
Figure 1.
Top horizontal line represents nucleotide number of OY-TES-1. Arrows represent EST sequences in the EST databank with sequence identity to the 1.1-kb product obtained by PCR using pem-related primers. Twelve of the 14 ESTs cluster to the 3′ end of the product, and 10 of them are derived from testis.
5′ Rapid Amplification of cDNA Ends (RACE).
5′ RACE was performed to identify the 5′ end sequence of OY-TES-1 by using the 5′RACE System for Rapid Amplification kit (GIBCO/BRL). Total RNA from human testis was used as a template, and the first-strand cDNA was synthesized by using specific primer, GSP1-T1 (5′-TTCCTGGGCTGATCGAATGAG-3′) (Fig. 2). dC-tailed cDNA was amplified by using a gene-specific nested primer GSP2-T1 (5′-GCAAAAGAGGAAGGGTTAGAAG-3′) (Fig. 2) and an abridged universal anchor primer (5′-GGCCACGCGTCGACTAGTAC-3′). The RACE product was sequenced with the sequence primer, Tes-N1 (5′-CCGTGGTTTTCATATTGGTC-3′) (Fig. 2).
Figure 2.
Nucleotide and deduced amino acid sequences of OY-TES-1. Primers used for PCR are indicated by arrows. The polyadenylation signal consensus sequence is underlined.
Reverse Transcription–PCR (RT-PCR).
Gene-specific primers ht-5 (human testis-5) (sense: 5′-AAGGACAGGGGACTAAGGAG-3′) and ht-3 (human testis-3) (antisense: 5′-CCGTACAAATCCAGCCCGTA-3′) (Fig. 2) were designed to amplify cDNA segments from normal tissue (MTC panel, CLONTECH) and malignant tissues. mRNA from malignant tissues was purified by using the OuickPrep Micro mRNA Purification Kit (Amersham Pharmacia). mRNA was reverse-transcribed into single-strand cDNA by using Moloney murine leukemia virus reverse transcriptase and oligo(dT)15 as a primer (Amersham Pharmacia), and cDNAs were tested for integrity by amplification of β-actin transcripts in a 30-cycle reaction. RT-PCR was performed by using 30 amplification cycles at an annealing temperature of 62°C, and the products were analyzed by agarose gel electrophoresis.
Radiation Hybrid Mapping.
Chromosomal mapping was performed with the Stanford G3 Radiation Hybrid Panel (purchased from Research Genetics, Huntsville, AL). PCR primers from the 3′ untranslated region of OY-TES-1 were as follows: sense, 5′-CTGGCGTCTATTCTGCCCA-3′; antisense, 5′-TGTAAAGTCATCTTTTAAGGAGG-3′. PCR conditions were 30 sec at 94°C, 30 sec at 57°C, and 30 sec at 72°C for 33 cycles. The PCR products were loaded onto 6% polyacrylamide gel, and the presence of specific PCR products was scored. The screening results then were submitted to the Online RH-server at the Stanford Human Genome Center (Palo Alto, CA).
Isolation of P1-Derived Artificial Chromosome (PAC) Clones Corresponding to OY-TES-1.
To obtain genomic clones for OY-TES-1, PACs were isolated from a PAC library (39) by a PCR screening using the primer set described above.
Fluorescence in Situ Hybridization Analysis.
The PAC DNA probe was prepared by nick-translation with SpectrumGreen-dUTP (Vysis, Dounevs Grove, IL) and hybridized to R-banded metaphase chromosomes (40), with D12Z3 DNA probe (Oncor) as reference. After chromosomes were counterstained with 4′,6-diamidino-2-phenylindole, their fluorescence image was captured by using a monochrome charge-coupled device camera (Zeiss) on an Axioplan fluorescence microscope (Zeiss) with appropriate filters. Multicolor fluorescence signals were merged with counterstaining images by using ISIS2 software (Metasystems, Altlussheim, Germany).
Southern Blot Hybridization.
Genomic DNA was extracted from testis by using DNeasy (Qiagen). Genomic DNA was digested with 100 units of EcoRI, HindIII, and BamHI at 37°C overnight. The DNA was separated on a 0.8% agarose gel and blotted onto a nylon transfer membrane (Hybond-N+, Amersham Pharmacia). The blot was hybridized to a 603-bp OY-TES-1 cDNA probe (nucleotides 749-1351) directly labeled with alkaline phosphatase (AlkPhos Direct, Amersham Pharmacia), washed, and processed for chemiluminescence according to the manufacturer's instructions.
Recombinant OY-TES-1 Protein.
OY-TES-1 was expressed in Escherichia coli by using the histidine-tag-containing vector pQE32 (Qiagen). cDNA amplification primers were designed to encompass the entire coding sequence of the gene, corresponding to amino acid positions 1–543. Induction of recombinant protein synthesis and subsequent purification by Ni2+-NTA column were performed according to the manufacturer's instructions.
ELISA.
Recombinant OY-TES-1 protein (2 μg/ml) in 0.05 M carbonate buffer (pH 9.6) was absorbed to 96-well plates (Nunc) at 4°C overnight. Plates were washed with PBS/Tween and blocked with 5% FCS/PBS at room temperature for 1 h. After washing, serum dilutions (100 μl) in 5% FCS/PBS were added and incubated at room temperature for 2 h. Plates were washed and incubated with secondary antibody (horseradish peroxidase-conjugated goat-anti human IgG, Medical Biological Laboratory, Tokyo) at 1/2,000 dilution for 1 h at room temperature. Plates were washed and incubated with the substrate solution (1,2-phenylenediamine dihydrochloride) for 20 min at room temperature. After addition of 3 M H2SO4 (100 μl), the absorbance was determined with a microplate reader (Tosoh, Tokyo).
Results
OY-TES-1 cDNA and Predicted Protein Sequence.
Human testis cDNA was amplified by PCR using a sense primer pem5 (5′-GTGGACAAGAGGAAGCACAA-3′) corresponding to nucleotides 65–84 of mouse pem and an antisense primer EST-2 (5′-TCTCCCCATCTCACTCCAC-3′) derived from human testis EST clone AA397852, which matches nucleotides 700–682 of mouse pem (37). A 1.1-kb product was obtained from a putatively negative strand and subjected to EST database analysis. A number of ESTs with sequence homology to the 1.1-kb product were detected, and most of them were derived from human testis. Several of the ESTs cluster at the 3′ end of the clone (Fig. 1). To obtain a full-length cDNA, 5′ RACE was performed. The resulting cDNA, designated OY-TES-1, was found to be 1,895 bp in length. OY-TES-1 contains a single long ORF that extends from base pairs 49 to 1677 and predicts a protein containing 543 aa (Fig. 2). A homology search through the GenBank database revealed that OY-TES-1 is a human homologue of the gene coding for mouse, guinea pig, and porcine proacrosin binding protein sp32 precursor (38). Human proacrosin binding protein sp32 precursor and mouse pem show homology only in the primer sequence: 5′-CCCCATCTCACCCCAC-3′ in EST-2 with 1-nt (underlined) difference corresponding to nucleotides 501–516 in OY-TES-1 and 5′-TTGTGCTTC-3′ in pem5 corresponding to nucleotides 1625–1633 in OY-TES-1. The deduced amino acid sequence of OY-TES-1 showed a high degree of identity with the porcine (81.9%), guinea pig (77.2%), and mouse (75.2%) proacrosin binding protein sp32 precursor sequence (Fig. 3). A glutamic acid and glutamine rich domain (EQ-rich domain) in the amino-terminal half of the precursor molecule and an amino-terminal acidic amino acid region in the mature sp32 characterize the molecule. Twenty cystein residues in the three cystein-rich domains also were totally conserved.
Figure 3.
Sequence alignment of guinea pig (GP), pig, mouse, and human (OY-TES-1) sp32 precursor protein. The conserved residues are indicated by asterisks. Dots indicate identical residues using the guinea pig sp32 precursor sequence as reference. Twenty cysteine residues are totally conserved (highlighted in black). A glutamic acid and glutamine-rich domain (EQ-rich domain) is boxed. Acidic amino acid region is underlined. Cleavage site between 273 arginine and 274 glutamic acid for production of mature sp32 is indicated by vertical arrow. Identity: guinea pig/human, 77.2%; pig/human, 81.9%; mouse/human, 75.2%.
Southern Blot Analysis of OY-TES-1.
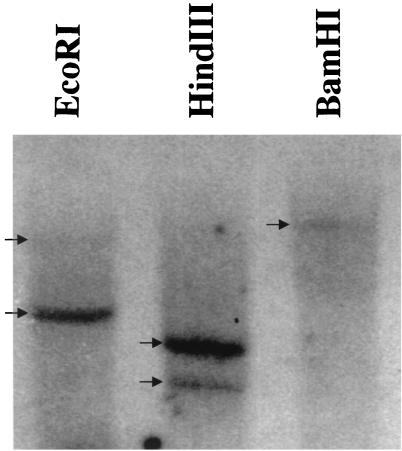
Genomic Southern blot analysis of testis and a renal cancer specimen with the OY-TES-1 probe showed two bands in EcoRI and HindIII digests, suggesting the presence of two OY-TES-1-related genes in the human genome (Fig. 4).
Figure 4.
Southern blot analysis of OY-TES-1 gene. Genomic DNA from normal testis was digested with EcoRI, HindIII, and BamHI, and analyzed with the OY-TES-1 probe. EcoRI and HindIII digests showed a strong band and a weak band, suggesting two OY-TES-1-related genes in the human genome.
Chromosomal Assignment of OY-TES-1.
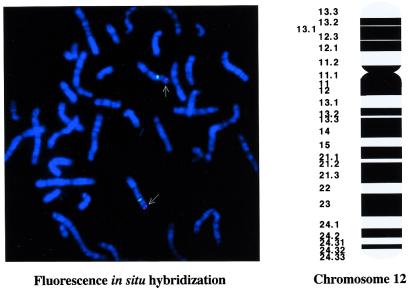
The scores obtained with the radiation hybrid panel were as follows: 00100 - 01001 - 01000 - 01001 -01000 - 10000 - 00000 - 00010 - 00110 - 01100 - 00011 - 00001 - 01000 - 00110 - 00000 - 01000 - 000. According to the online RH-server, this profile indicated mapping to chromosome 12 with an estimated reference interval of 13.9 cM between two markers, D12S99 and D12S358. More precise chromosomal localization was defined by fluorescence in situ hybridization analysis with a PAC probe (145N7). Of the 20 metaphase cells analyzed, 18 showed brightly fluorescent twin signals at 12p12-p13 (Fig. 5).
Figure 5.
Chromosome localization of OY-TES-1 gene by fluorescence in situ hybridization. Metaphases showed twin signals with brightly red fluorescence on 12p12-p13 (arrows). A chromosome 12-specific α satellite probe at the centromere is shown in green fluorescence.
Genomic Structure of OY-TES-1.
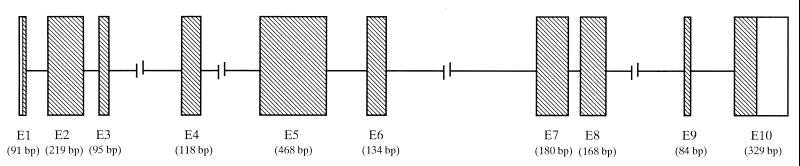
A search of the hgts (http://www.ncbi.nlm.nih.gov/blast/blast.cgi) database was performed to identify the genomic structure of OY-TES-1. The OY-TES-1 cDNA sequence was found in overlapping PAC clone RP4–761J14 and bacterial artificial chromosome clone RP11–433J6 between two markers P941E5/T7 and D12S2320, and confirmed the fluorescence in situ hybridization analysis. As shown in Fig. 6, the gene coding for OY-TES-1 is composed of 10 exons, spanning a distance of 9,339 bp.
Figure 6.
Genomic structure of OY-TES-1. The ORF is shown in shaded boxes and spliced introns as lines. The exon/intron structure is determined based on PAC clone RP4–761J14 and bacterial artificial chromosome clone RP11–433J6.
Expression Pattern of OY-TES-1 in Normal and Malignant Tissues.
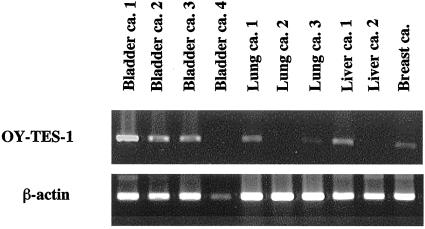
OY-TES-1 mRNA expression was examined by RT-PCR using a panel of normal and malignant tissues. As shown in Fig. 7, the analysis of adult normal tissue showed that OY-TES-1 expression was restricted to testis. In tumors, OY-TES-1 was expressed in a subset of a broad spectrum of tumors of different origins. As shown in Fig. 8 and Table 1, 28% (11/39) of bladder cancer, 40% (2/5) of breast cancer, 40% (2/5) of liver cancer, 20% (1/5) of lung cancer, and 15% (2/13) of colon cancer showed detectable OY-TES-1 mRNA. No expression of OY-TES-1 mRNA was observed in renal or stomach cancers.
Figure 7.
RT-PCR analysis of OY-TES-1 expression in normal tissues. PCR primers were: ht-5, 5′-AAGGACAGGGGACTAAGGAG-3′ and ht-3, 5′-CCGTACAAATCCAGCCCGTA-3′. The same cDNA samples were tested for β-actin as an internal control. OY-TES-1 expression was restricted to testis.
Figure 8.
RT-PCR analysis of OY-TES-1 expression in tumors using primers ht-5 and ht-3. The same cDNA samples were tested for β-actin as an internal control.
Table 1.
OY-TES-1 mRNA expression in human tumors
| Tumor type | mRNA, positive/total |
|---|---|
| Bladder cancer | 11/39 (28%) |
| Breast cancer | 2/5 (40%) |
| Liver cancer | 2/5 (40%) |
| Lung cancer | 1/5 (20%) |
| Colon cancer | 2/13 (15%) |
| Stomach cancer | 0/5 (0%) |
| Renal cancer | 0/10 (0%) |
Seroreactivity Against OY-TES-1 in Cancer Patients.
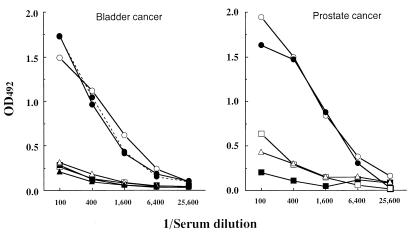
Sera from 362 cancer patients and 20 normal individuals were tested for antibody to OY-TES-1 in ELISA using recombinant OY-TES-1 protein. An unrelated recombinant protein made in the same expression system was used as a control antigen. As shown in Table 2, 5.2% (3/58) of bladder cancer, 3.5% (3/85) of prostate cancer, 10.5% (10/95) of liver cancer, 10.3% (6/58) of colon cancer, and 4.5% (3/66) of lung cancer patients had antibody against OY-TES-1. No OY-TES-1 antibody was detected in the sera of 20 normal individuals. Fig. 9 illustrates titration curves with sera from selected bladder and prostate cancer patients.
Table 2.
ELISA reactivity of sera from normal blood donors and cancer patients: Tests for OY-TES-1 antibody
| Sera | ELISA, positive/total |
|---|---|
| Healthy donors | 0/20 (0%) |
| Bladder cancer | 3/58 (5.2%) |
| Prostate cancer | 3/85 (3.5%) |
| Liver cancer | 10/95 (10.5%) |
| Colon cancer | 6/58 (10.3%) |
| Lung cancer | 3/66 (4.5%) |
| Total | 25/362 (6.9%) |
Figure 9.
Antibody to OY-TES-1 in sera of patients with bladder cancer and prostate cancer. Each line represents a titration curve of a serum sample from a single patient. Test system: ELISA with recombinant OY-TES-1 protein.
In a series of bladder cancer patients, both fresh-frozen specimens and serum samples were available from 13 patients. Tumors were typed for OY-TES-1 mRNA expression by RT-PCR, and sera were assayed for OY-TES-1 antibody by ELISA. One of six patients with an OY-TES-1 mRNA-positive tumor had OY-TES-1 antibody. No OY-TES-1 antibody was detected in sera from seven patients with OY-TES-1 mRNA-negative tumors (Table 3).
Table 3.
Correlation between OY-TES-1 mRNA expression and OY-TES-1 antibody response in bladder cancer patients
| RT-PCR typing | ELISA | Number of cases |
|---|---|---|
| + | + | 1 |
| + | − | 5 |
| − | − | 7 |
Discussion
In this study, we isolated a gene, designated OY-TES-1, from human testis cDNA by direct PCR cloning using primers derived from mouse pem and from pem-related ESTs from human testis. Despite the use of mouse and human pem-related sequences to isolate OY-TES-1, OY-TES-1 shares very little homology with pem. OY-TES-1 expression was restricted to testis in normal adult tissues, whereas it was detected in a range of different human tumor types. Thus, OY-TES-1 has the classical features of a CT antigen. In contrast to most CT antigens that map to the X chromosome, the OY-TES-1 gene was mapped to chromosome 12p12-p13. Two other CT antigens, SCP-1 (26) and CT9 (29), also have a non-X chromosomal localization, in these cases chromosome 1.
Sequence analysis indicates that OY-TES-1 is the human homologue of porcine, guinea pig, and mouse proacrosin binding protein sp32 precursor (38). sp32 is located in the sperm acrosome and appears to function as a binding protein to proacrosin for packaging and condensation of the acrosin zymogen in the acrosomal matrix. The deduced amino acid sequence of OY-TES-1 shows a high degree of homology with the corresponding porcine (81.9%), guinea pig (77.2%), and mouse (75.2%) sp32 precursor product. Twenty cystein residues located in the molecule are totally conserved. The amino terminal region is highly hydrophobic, suggesting that it serves as a signal sequence. The mature sp32 is produced by posttranslational cleavage between 273 arginine and 274 glutamic acid of the precursor molecule. There are abundant acidic amino acids in the amino terminal region of the mature sp32, which is composed of 24 residues in human sp32, 25 residues in porcine sp32, 22 residues in the guinea pig sp32, and 24 residues in mouse sp32.
There is considerable interest in defining the functions of CT antigens in germ cell development, and their role, if any, in cancer cells. Although there is speculation that some CT gene products are transcriptional factors, only two of the previously defined CT antigens, SCP-1 and CT9, have known functions. SCP-1 is a synaptonemal complex protein involved in chromosome reduction during meiosis (41), and CT9, the bromodomain testis-specific gene product (BRDT), appears to be a transcriptional regulator (42). We can now add OY-TES-1 to the list of CT antigens with a known function.
In our survey of 362 cancer patients with various types of cancer, 25 patients produced antibody to OY-TES-1 protein. Antibody production against OY-TES-1 was observed in patients with bladder, prostate, liver, colon, and lung cancer. No reactivity was observed in sera from normal individuals. In an analysis of 13 cases of bladder cancer, where tumor and serum specimens from the same patient were available, one of six bladder cancer patients with OY-TES-1 mRNA-positive tumors had antibody against OY-TES-1: no patients with OY-TES-1 mRNA-negative tumors had OY-TES-1 antibody. If one assumes that OY-TES-1 antibody is found only in patients with OY-TES-1 mRNA-positive tumors (as is the case in the NY-ESO-1 system; ref. 35), the frequency of OY-TES-1 antibody in cancer populations can be estimated by relating the frequency of mRNA expression in a particular tumor type to the frequency of antibodies in patients with that type of tumor. For example, OY-TES-1 mRNA expression was observed in 28% (11/39) of tumors from patients with bladder cancer, and OY-TES-1 antibody production was observed in 5.2% (3/58) of patients with bladder cancer. Thus, we can estimate that approximately 19% of bladder cancer patients with OY-TES-1 mRNA-positive tumors develop OY-TES-1 antibodies. In colon cancer, where the expression of CT antigen is infrequent, OY-TES-1 was expressed in 15% of the tumors. Using the same calculations, it can be estimated that 70% of colon cancer patients with OY-TES-1 mRNA-positive tumors produce OY-TES-1 antibody.
In terms of antibody frequency, OY-TES-1 appears to have high immunogenic potential, and, in this regard, resembles the strong immunogenicity of NY-ESO-1 (33–35, 43) and contrasts with the low frequency of antibody observed against other CT antigens, e.g., MAGE, SSX, and SCP-1. In the case of NY-ESO-1, the presence of a humoral immune response is predictive of a strong CD8 T cell response to NY-ESO-1 (33, 34) as demonstrated by elispot, tetramer analysis, and cytotoxicity. Several HLA-A2 and HLA-A24 binding motifs are present in the OY-TES-1 product, and patients with humoral immunity to OY-TES-1 are good candidates for the analysis of CD8 T cell responses to OY-TES-1.
Acknowledgments
We thank Dr. V. Jongeneel (Swiss Institute of Bioinformatics, Ludwig Institute for Cancer Research) for critical reading of the manuscript, Dr. K. Shimizu (Institute for Cellular and Molecular Biology, Okayama University Medical School) for valuable suggestions, and Mr. Y. Isomoto (DNA Sequencing Facility at Central Laboratory, Okayama University Medical School) for technical assistance. We also thank Ms. M. Isobe for excellent technical assistance and Ms. J. Mizuuchi for preparation of the manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research on Priority Area (C) from the Ministry of Education, Science, Sports and Culture of Japan, and the Cancer Research Institute, New York.
Abbreviations
- EST
expressed sequence tag
- CT
cancer/testis
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcription–PCR
- PAC
P1-derived artificial chromosome
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB051833).
References
- 1.van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 2.Van den Eynde B, van der Bruggen P. Curr Opin Immunol. 1997;9:684–693. doi: 10.1016/s0952-7915(97)80050-7. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg S A. Immunity. 1999;10:281–287. doi: 10.1016/s1074-7613(00)80028-x. [DOI] [PubMed] [Google Scholar]
- 4.Jäger E, Chen Y-T, Drijfhout J W, Karbach J, Ringhoffer M, Jäger D, Arand M, Wada H, Noguchi Y, Stockert E, et al. J Exp Med. 1998;187:265–270. doi: 10.1084/jem.187.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang R-F, Wang X, Atwood A C, Topalian S L, Rosenberg S A. Science. 1999;284:1351–1354. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 6.Wang R F, Wang X, Rosenberg S A. J Exp Med. 1999;189:1659–1668. doi: 10.1084/jem.189.10.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaux P, Vantomme V, Stroobant V, Thielemans K, Corthals J, Luiten R, Eggermont A M M, Boon T, van der Bruggen P. J Exp Med. 1999;189:767–777. doi: 10.1084/jem.189.5.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jäger E, Jäger D, Karbach J, Chen Y-T, Ritter G, Nagata Y, Gnjatic S, Stockert E, Arand M, Old L J, Knuth A. J Exp Med. 2000;191:625–630. doi: 10.1084/jem.191.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahin U, Türeci Ö, Schmitt H, Cochlovius B, Johannes T, Schmits R, Stenner F, Luo G, Schobert I, Pfreundschuh M. Proc Natl Acad Sci USA. 1995;92:11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Old L J, Chen Y-T. J Exp Med. 1998;187:1163–1167. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulie P G, Brichard V, Van Pel A, Wölfel T, Schneider J, Traversari C, Mattei S, De Plaen E, Lurquin C, Szikora J-P, et al. J Exp Med. 1994;180:35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawakami Y, Eliyahu S, Delgaldo C H, Robbins P F, Rivoitini L, Topalian S L, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brichard V, Van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethé B, Coulie P, Boon T. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skipper J C A, Hendrickson R C, Gulden P H, Brichard V, Van Pel A, Chen Y, Shabanowitz J, Wolfel T, Slingluff C L, Jr, Boon T, et al. J Exp Med. 1996;183:527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scanlan M J, Chen Y-T, Williamson B, Güre A O, Stockert E, Gordan J D, Türeci Ö, Sahin U, Pfreundshuh M, Old L J. Int J Cancer. 1998;76:652–658. doi: 10.1002/(sici)1097-0215(19980529)76:5<652::aid-ijc7>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 16.Fisk B, Blevins T L, Wharton J T, Ioannides C G. J Exp Med. 1995;181:2109–2117. doi: 10.1084/jem.181.6.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scanlan M J, Williamson B, Jungbluth A, Stockert E, Arden K C, Viars C S, Güre A O, Gordan J D, Chen Y-T, Old L J. Biochim Biophys Acta. 1999;1445:39–52. doi: 10.1016/s0167-4781(99)00033-0. [DOI] [PubMed] [Google Scholar]
- 18.Lennette E T, Winberg G, Yadav M, Enblad G, Klein G. Eur J Cancer. 1995;31:1875–1878. doi: 10.1016/0959-8049(95)00354-l. [DOI] [PubMed] [Google Scholar]
- 19.Türeci Ö, Sahin U, Pfreundshuh M. Mol Med Today. 1997;3:342–349. doi: 10.1016/s1357-4310(97)01081-2. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y-T, Scanlan M J, Sahin U, Türeci Ö, Güre A O, Tsang S, Williamson B, Stockert E, Pfreundschuh M, Old L J. Proc Natl Acad Sci USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. J Exp Med. 1995;182:689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Böel P, Wildmann C, Sensi M-L, Brasseur R, Renauld J-C, Coulie P, Boon T, van der Bruggen P. Immunity. 1995;2:167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 23.Türeci Ö, Sahin U, Schobert I, Koslowski M, Schmitt H, Schild H-J, Stenner F, Seitz G, Rammensee H-G, Pfreundschuh M. Cancer Res. 1996;56:4766–4772. [PubMed] [Google Scholar]
- 24.Güre A O, Türeci Ö, Sahin U, Tsang S, Scanlan M J, Jäger E, Knuth A, Pfreundschuh M, Old L J, Chen Y-T. Int J Cancer. 1997;72:965–971. doi: 10.1002/(sici)1097-0215(19970917)72:6<965::aid-ijc8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 25.Lethé B, Lucas S, Michaux L, De Smet C, Godelaine D, Serrano A, De Plaen E, Boon T. Int J Cancer. 1998;76:903–908. doi: 10.1002/(sici)1097-0215(19980610)76:6<903::aid-ijc22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 26.Türeci Ö, Sahin U, Zwick C, Koslowski M, Seitz G, Pfreundschuh M. Proc Natl Acad Sci USA. 1998;95:5211–5216. doi: 10.1073/pnas.95.9.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y-T, Güre A O, Tsang S, Stockert E, Jäger E, Knuth A, Old L J. Proc Natl Acad Sci USA. 1998;95:6919–6923. doi: 10.1073/pnas.95.12.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lucas S, De Smet C, Arden K C, Viars C S, Lethé B, Lurquin C, Boon T. Cancer Res. 1998;58:743–752. [PubMed] [Google Scholar]
- 29.Scanlan M J, Altorki N K, Güre A O, Williamson B, Jungbluth A, Chen Y-T, Old L J. Cancer Lett. 2000;150:155–164. doi: 10.1016/s0304-3835(99)00385-7. [DOI] [PubMed] [Google Scholar]
- 30.Güre A O, Stockert E, Arden K C, Boyer A D, Viars C S, Scanlan M J, Old L J, Chen Y-T. Int J Cancer. 2000;85:726–732. doi: 10.1002/(sici)1097-0215(20000301)85:5<726::aid-ijc21>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 31.Lucas S, De Plaen E, Boon T. Int J Cancer. 2000;87:55–60. [PubMed] [Google Scholar]
- 32.Zendman A J W, Cornelissen I M H A, Weidle U H, Ruiter D J, van Muijen G N P. Cancer Res. 1999;59:6223–6229. [PubMed] [Google Scholar]
- 33.Jäger E, Nagata Y, Gnjatic S, Wada H, Stockert E, Karbach J, Dunbar P R, Lee S Y, Jungbluth A, Jäger D, et al. Proc Natl Acad Sci USA. 2000;97:4760–4765. doi: 10.1073/pnas.97.9.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gnjatic S, Nagata Y, Jäger E, Stockert E, Shankara S, Roberts B L, Mazzara G P, Lee S Y, Dunbar P R, Dupont B, et al. Proc Natl Acad Sci USA. 2000;97:10917–10922. doi: 10.1073/pnas.97.20.10917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stockert E, Jäger E, Chen Y-T, Scanlan M J, Gout I, Karbach J, Arand M, Knuth A, Old L J. J Exp Med. 1998;187:1349–1354. doi: 10.1084/jem.187.8.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilkinson M F, Kleeman J, Richards J, MacLeod C L. Dev Biol. 1990;141:451–455. doi: 10.1016/0012-1606(90)90400-d. [DOI] [PubMed] [Google Scholar]
- 37.Ono T, Sato S, Kimura N, Tanaka M, Shibuya A, Old L J, Nakayama E. Int J Cancer. 2000;88:845–851. doi: 10.1002/1097-0215(20001215)88:6<845::aid-ijc1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 38.Baba T, Niida Y, Michikawa Y, Kashiwabara S, Kodaira K, Takenaka M, Kohno N, Gerton G L, Arai Y. J Biochem. 1994;269:10133–10140. [PubMed] [Google Scholar]
- 39.Matsumoto N, Soeda E, Ohashi H, Fujimoto M, Kato R, Tsujita T, Tomita H, Kondo S, Fukushima Y, Niikawa N. Genomics. 1997;45:11–16. doi: 10.1006/geno.1997.4897. [DOI] [PubMed] [Google Scholar]
- 40.Takahashi E, Hori T, Oconnell P, Leppert M, White R. Hum Genet. 1990;86:14–16. doi: 10.1007/BF00205165. [DOI] [PubMed] [Google Scholar]
- 41.Meuwissen R J L, Offenberg H H, Dietrich A J, Riesewijk A, van Iersel M, Heyting C. EMBO J. 1992;11:5091–5100. doi: 10.1002/j.1460-2075.1992.tb05616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones M H, Numata M, Shimane M. Genomics. 1997;45:529–534. doi: 10.1006/geno.1997.5000. [DOI] [PubMed] [Google Scholar]
- 43.Valmori D, Dutoit V, Líenard D, Rimoldi D, Pittet M J, Champagne P, Ellefsen K, Sahin U, Speiser D, Lejeune F, et al. Cancer Res. 2000;60:4499–4506. [PubMed] [Google Scholar]