Abstract
It has been widely acknowledged that non-coding RNAs are master-regulators of genomic functions. However, the significance of the presence of ncRNA within introns has not received proper attention. ncRNA within introns are commonly produced through the post-splicing process and are specific signals of gene transcription events, impacting many other genes and modulating their expression. This study, along with the following discussion, details the association of thousands of ncRNAs—snoRNA, miRNA, siRNA, piRNA and long ncRNA—within human introns. We propose that such an association between human introns and ncRNAs has a pronounced synergistic effect with important implications for fine-tuning gene expression patterns across the entire genome.
INTRODUCTION
Spliceosomal introns are ubiquitous elements of nuclear genomes. Their evolutionary rise is associated with the origin of eukaryotes (1,2). Recently, a new conception of the co-evolution of introns and nucleus-cytosol compartmentalization has been detailed (3). The existence of introns allows for the alternative splicing of pre-mRNA molecules, thus serving to increase both protein diversity and specialization within the proteome (4,5). Additional intron functions have been reviewed (6). However, the use of introns is a double-edged sword for organisms enriched with these elements, since they require complex processing that can lead to serious problems when splicing goes awry. Particularly, large intron sizes in vertebrate and other complex organisms incur several drawbacks including waste of energy, delay in protein production and increased vulnerability to splicing errors (7). Having acknowledged intron roles, we will focus solely on the non-random presence of non-protein-coding RNAs (ncRNAs) inside these gene elements. At the dawn of small ncRNA discovery, John Mattick first proposed the hypothesis that introns contain information valuable to gene regulation and called it ‘informational RNA’ (8). Since that time a whole new field of RNomics has emerged for the investigation of ncRNAs in genetic regulation. A positive correlation between the number of ncRNAs and the complexity of an organism is evident, while the number of protein-coding genes is relatively constant from worms to humans (9). Non-coding RNAs consist of a diverse group of short molecules including miRNAs, siRNAs, snoRNAs and piRNAs as well as various long ncRNAs. They are involved in a spectrum of regulatory processes within the nucleus and cytoplasm indispensable for the proper organization and functioning of every eukaryotic cell [see reviews (10,11)]. The present study demonstrates how intimately ncRNAs are associated with introns.
MATERIALS AND METHODS
Databases
For the localization of small RNAs within the human genome we used our human Exon–Intron Database (EID), release 36.1 (12) and the NCBI human genome sequence, build 36.1.
Statistics on snoRNA were obtained from snoRNA-LBME-db database, version 3 (13). This is a manually curated database with stringent requirements for experimental verification of each deposited sequence.
A comprehensive set of 462 pre-miRNA was obtained from miRBase(14). Pre-miRNA sequences contained within this database all represent miRNA sequences that have been published in peer reviewed journals. ‘Each sequence represents a predicted hairpin portion of the transcript (14)’.
A comprehensive set of 33 051 human piRNA sequences was obtained from RNAdb (15). This set of human piRNA were obtained from one laboratory using a pyro-sequencing technique (16). The authors provided experimental validation that their sequences are significantly enriched with PIWI-associated small RNA molecules (piRNA).
Complete sets of functional non-coding RNAs for human (124 591 entries) and mouse (110 495 entries) were obtained from functional ncRNA database (fRNAdb) (17).
Sequence processing
Sequences of small ncRNA were matched with the human genome using PERL regular expressions. piRNAs that had perfect matches to multiple locations within the genome were called ‘multi-match’ and were not counted in the distributions for exons, introns, or intergenic regions. The remaining ‘single-match’ ncRNA sequences that had only one perfect match to an exon or intron (transcribed strand) in the human EID were considered to be either exonic or intronic. Those single-match ncRNA sequences that were perfectly matched to complementary sequences of exons or introns from EID were designated as being ‘complementary to’ exons or introns, respectively. All other small ncRNA locations (i.e. outside of exons and introns as well as their complementary strands) were considered to be ‘intergenic’.
miRNA
Distances between miRNA sequences were determined using the chromosomal positions given in the miRNA annotations (14).
siRNA
In order to computationally assess the ability of human introns to produce endogenous siRNA the siRNA.pl Perl program—a modified version of the snoTARGET program (http://bpg.utoledo.edu/∼dbs/snotarget/)—was used. The siRNA.pl program scans the entire set of human introns, searching for stem–loop hairpin structures with perfect stems spanning at least 21 nt and with short (0–80 nt) loops. In order to understand the association of these hairpin structures with repetitive elements, we scanned the introns using siRNA.pl after masking them by Repeatmasker (18) followed by the trf (‘tandem repeats finder’) program for masking tandem repeats (19). In order to evaluate the statistical association of hairpins with introns, a search for hairpin structures was undertaken within three control sets. The control sets were generated using our web application ‘SRI-generator’ (20) and consisted of randomized nucleotide sequences that maintained the oligonucleotide frequency composition and length of the natural set of introns. Statistical significance for the comparison of hairpin distribution between introns and control sets was established using the Fisher exact test. Similar analysis was performed within exons and intergenic regions, and the frequencies of occurrence of perfect stems were compared to those found in introns, using the chi-square test. Evolutionary conservation of the hairpins was examined by performing a BLAST search against cow, mouse and rat orthologous introns (21).
piRNA
A comprehensive set of 33 051 human piRNA sequences was processed by first removing sequences with ambiguous nucleotides (e.g. ‘n’), yielding 32 439 remaining sequences. From this set, 5274 sequences had zero matches to the human genome and were removed from consideration; 22 835 sequences had exactly one match and were named ‘single-match’; while each of the remaining 4330 sequences had multiple exact matches to different genomic locations and were named ‘multi-match.’ Furthermore, we showed that among the 22 835 single-match piRNAs, 3138 sequences were redundant, i.e. were mapped exactly within the same site of the human genome as at least one other piRNA from this group. We removed all redundant sequencing creating the final set of 19 697 single-match piRNAs, which was used for the calculation of distributions within exons, introns and intergenic regions.
The combined 19 697 single-match and the 4330 multi-match sets were analyzed for their association with repetitive elements from Repbase (22) using the BLAST program without filters. Of the 24 027 piRNA sequences, 1249 demonstrated significant similarity (e < 10−4) to known repeats (5.2%). The corresponding random set was analyzed under identical conditions, yielding 1776 sequences demonstrating significant similarity (e < 10−4) to human repeats (13.2%). Due to the short length of these sequences, a substantial number of false negatives are expected. These results were also confirmed by using RepeatMasker to mask piRNA and random sequences under sensitive conditions using the slow search option.
Among the final set of 19 697 single-match sequences, 15 047 were characterized as intergenic piRNAs and 4650 piRNAs were mapped within the exons and/or introns of protein-coding genes or their complementary strands. From the latter group, 300 piRNA corresponded to loci containing both exons and introns (i.e. overlapping splicing junctions, overlapping genes on opposite strands or alternate transcripts) and were excluded from the calculations regarding the exon/intron distributions.
In order to determine if there were positional preferences for piRNAs within introns, we divided each intron into quintiles (20% portions) based on the entire length of the given intron. Each piRNA sequence was assigned to a quintile based on its position within an intron. The total number of occurrences was calculated for each quintile. The positional preference of piRNAs within mRNAs was determined in a similar manner. The calculation of the standard error of means was determined using the Binominal distribution.
Analyzing the distribution of ‘random’ ncRNAs within genomic regions
We created a PERL program for the selection of 13 500 random positions along the entire human genome. From these positions, 30-bp long sequences were collected and listed as a set of 13 500 ‘random’ ncRNAs. Each of these random sequences was aligned to the entire human genome using the same protocol as for real piRNA (see previous paragraph). Among them, 2068 random sequences matched to several genomic locations and were grouped as ‘multi-match’. Each of the remaining 11 432 sequences had a single match to the genome. Alignment with BLAST demonstrated that 1776 random sequences out of 13 500 [13.2 ± 0.3% standard error (SE)] had a significant similarity to repetitive elements (e < 10−4). The same proportion among the real set of piRNA comprised 5.2 ± 0.14% SE (1249/24027).
SE for each percentage was calculated using the formula SEp = sqrt[p(1 – p) / n], where p is the sample proportion and n is the sample size, using the Binomial distribution. A chi-square test was used to compare the distribution of piRNA sequences classified as exonic, intronic and intergenic to the distribution of 11 432 randomly placed sequences within these genomic regions.
Long intronic ncRNA
A total of 63 077 groups of orthologous introns for five mammalian species (human, mouse, rat, dog, cow) was obtained from the latest release (July 2010) of our Mammalian Orthologous Intron Database (21), available on our website (www.bioinfo.utoledo.edu/domino5). We defined ‘orthologous introns’ as introns from orthologous genes that have the same position and phase relative to the coding sequence.
Each group of orthologous intron sequences from the five species was aligned using MAFFT, a stand-alone program which can align a set of sequences flanking around alignable domains (23) (using the L-INS-I parameters: mafft –localpair –maxiterate 1000 input_file > output_file). A Perl program was developed to process the obtained alignments and investigate the degree of conservation among the different species. The program required that each conserved intronic region (CIR) spanned at least 400 nt in length, so as to exclude small ncRNAs from our results (Explanations in MOID web page). Additionally, CIRs qualified as evolutionarily conserved only if they had at least 50% sequence identity among the five species. This threshold was chosen to be high enough so that regions of identity occurring by chance would be eliminated, and yet low enough to take into consideration the wide degree of divergence among the five species. Various filters were applied to reduce the possibility of the conserved segment being a part of an alternatively spliced exon, as explained in (24).
The corresponding human and mouse sequences of the CIRs with masked repeats (RepeatMasker, version-3.2.8) were compared to the respective Functional non-coding RNA database, fRNAdb, (17) using the stand-alone BLAST program. The results were parsed to enumerate the overlap with ncRNA, in instances where the BLAST score was more than 80 bits (e-value < 2*10−16).
Statistics
Statistical analysis with the chi-square test and Fisher exact test was performed using the R package (v2.7.1).
Programs
The new release of our snoRNA.r3.pl mentioned in the results section is available on our website (http://www.utoledo.edu/med/depts/bioinfo/database.html). All programs used to perform calculations were written in Perl and are available upon request.
RESULTS
snoRNAs, a byproduct of intron splicing in animals
All known snoRNAs in vertebrates (and possibly in Drosophila) are a byproduct of splicing because they are created by the exonucleolytic processing of debranched introns after their excision from the pre-mRNA (25). The vast majority of animal snoRNAs have been found inside the introns of protein-coding genes, while only a few of them have been reported to be inside the introns of ncRNAs transcribed by RNA polymerase II (26,27). The current release of the snoRNA-LBME-db database, version 3, contains 402 experimentally confirmed human snoRNAs (13). The majority of them are involved in the chemical modification of 184 bases of ribosomal 28S, 18S and 5.8S rRNAs and 33 bases of spliceosomal U1, U2, U4, U5, U6 and U12 snRNAs. Moreover, 136 snoRNAs in this database belong to so-called orphan molecules that do not display antisense elements compatible with a modification for rRNA or snRNA. In addition to the described sample of natural snoRNAs, there are many computationally predicted snoRNA-like sequences within human introns whose existence have not been confirmed experimentally and therefore, are not featured in snoRNA-LBME-db. These snoRNA-like sequences have been identified inside genomes using several computational approaches (21,28–31). The computationally predicted sequences possess all the major characteristics of natural snoRNAs such as conserved sequence motifs (boxes) and secondary structures; hence, a portion of them could represent uncharacterized natural snoRNAs. Supplementary Table S1 contains a list of 324 novel C/D-box snoRNA-like sequences within human introns produced by our snoRNA.r3.pl, program (21). We project that the total number of snoRNA-like sequences in the human genome may exceed 1000. The facts testify that the presence of introns in animals is crucial for the biosynthesis of snoRNAs.
miRNAs are significantly enriched in the transcribed strands of human introns
Table 1 illustrates the distribution of all known human pre-miRNAs from miRBase within the introns and exons of protein-coding genes as well as the regions between these genes, which we refer to as intergenic regions. The data demonstrates a preference of pre-miRNA to exist inside introns and exons over intergenic regions. The bias of pre-miRNA to favor genic regions while avoiding intergenic regions is statistically significant (X21df = 117.6; P < 2.2 × 10−16). Among the 19 pre-miRNAs found inside exons, 5 occur within the complementary strand of the Retrotransposon-like (RTL1) gene. Their function is associated with the chromosomal methylation and regulation involved in imprinting of the RTL1 locus (32). Only two other exonic pre-miRNAs correspond to coding regions (Supplementary Table S2), while the rest correspond to 5′- or 3′-UTRs. Ten of these are found on the transcribed strand and four are found on the complementary strand.
Table 1.
Distribution of human pre-miRNAs within exons, introns and intergenic regions
| Genomic region | Number of pre-miRNA | Percentage of pre-miRNA | Number of random sequences | Percentage of random sequences |
|---|---|---|---|---|
| Intergenic | 237 | 51.3 ± 2.3 | 8480 | 74.2 ± 0.4 |
| Intron | 206 | 44.6 ± 2.3 | 2779 | 24.3 ± 0.4 |
| Exon | 19 | 4.1 ± 0.9 | 173 | 1.5 ± 0.1 |
| Total | 462 | 100 | 11,432 | 100 |
The distribution of pre-miRNA within specific genomic regions is compared to the estimated probability of localization within these regions, calculated by classifying 11 432 sequences from randomly chosen locations within the human genome. Both transcribed and complementary strands are represented for exons and introns. Percentages are shown ± (SE).
The distribution of pre-miRNAs inside introns is shown in Table 2. The data demonstrates a strong preference of pre-miRNAs to associate with the transcribed strand of introns (87%) while 13% are associated with the complementary strand. Twenty-four percent of intronic pre-miRNAs are found in clusters (two or more pre-miRNAs inside the same intron) while the majority (76%) of these pre-miRNAs are sparsely populated (one pre-miRNA per intron). In intergenic regions, there is a stronger tendency for several pre-miRNAs to be located in close proximity to each other (64% of intergenic pre-miRNAs are separated from each other by <5 kb). The largest cluster of the pre-miRNAs exists in human chromosome 19, where 42 different pre-miRNAs were found within a 150-kb region (Supplementary Table S2). Tables 1 and 2 demonstrate that 39% of all human pre-miRNAs originate from the transcribed strand of introns, while a random distribution would put 12% on the transcribed strand as well as 12% on the complementary strand of introns. A chi-square test confirms that this association of pre-miRNA with the transcribed strand of introns is statistically significant (X21df = 63.2, P = 1.9 × 10−15). The calculated association of pre-miRNAs with introns is likely to be underestimated due to the dearth of information on introns located between 5′- or 3′-untranslated exons. For example, the IC-SNURF-SNRPN gene has 137 introns within the 3′-untranslated portion of the gene, which includes 94 orphan snoRNAs (33). Moreover, the introns in the untranslated part of this gene have not been annotated properly (GenBank NG_002690.1) and thus are not classified as intronic elements. Due to such inaccuracies, we reason that ∼50% of all human miRNA correspond to the transcribed strand of introns and are byproducts of splicing, as is also the case of snoRNAs.
Table 2.
The distribution of pre-miRNAs inside introns
| Orientation and grouping | Number of pre-miRNAs (%) |
|---|---|
| DNA strand | |
| Transcribed | 179 (86.9 ± 2.4) |
| Complementary | 27 (13.1 ± 2.4) |
| Pre-miRNA clustering | |
| One per intron | 157 (76.2 ± 3.0) |
| In clusters (≥2) | 49 (23.8 ± 3.0) |
The data represents intronic pre-miRNA among transcribed and complementary strands as well as the tendency for pre-miRNA to form clusters within introns. A cluster is defined as any intron containing more than one pre-miRNA, irrespective of strand orientation. Percentages are shown ± SE.
piRNAs are twice as abundant in the transcribed strand of introns as the complementary strand
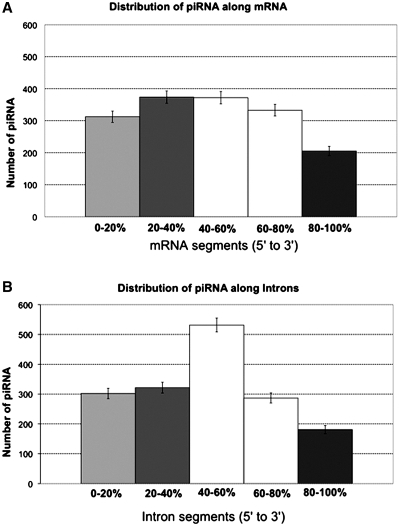
piRNA sequences were mapped to the human genome and classified as multi-match or single-match. Single-match sequences were further filtered and classified as intronic, exonic or intergenic. Table 3 shows these results along with the distribution of 11 432 randomly placed 30-nt long sequences within the human genome (‘Materials and Methods’ section). A comparison of the estimated percentage of piRNA that is repetitive (5.2 ± 0.14% SE) with the estimated percentage of randomly located sequences mapping to repetitive regions (13.2 ± 0.3% SE) suggests that piRNA do not preferentially associate with repetitive elements, in contrast to prior observations (34). The estimated percentage of piRNA that is intergenic (76.4 ± 0.3% SE) does not differ significantly with the estimated percentage of the human genome that is intergenic (74.2 ± 0.4% SE). We observed that 82.1% (16 176/19 697) of human piRNA are produced from intergenic regions (15 047 piRNAs) or from the complementary strands of exons and introns (1129 piRNAs). The rest of the piRNAs are almost equally produced from the transcribed strand of exons (1598/19 697; 8.1 ± 0.2% SE) and introns (1623/19 697; 8.2 ± 0.2% SE). Finally, 300 piRNAs (1.5 ± 0.1% SE) overlap both exons and introns (‘Materials and Methods’ section). Examining the data for intronic and exonic DNA from the human genome, piRNAs are significantly more likely than expected (X21df = 1353.2; P < 2.2 × 10−16) to reside in exons rather than introns, given that introns are on average, approximately 15 times larger than exons. The piRNAs classified as intronic predominantly mapped within the transcribed strand (Table 4). This non-random association (X21df = 177.0, P < 2.2 × 10−16) of piRNAs with the transcribed strand is slightly higher for exons (79.9 ± 1% SE) than for introns (69.1 ± 1% SE). Only 90 piRNAs overlap with the exon–intron splice sites and, intriguingly, are not preferentially associated with the transcribed strand (63 of them are associated with the complementary strand, Supplementary Table S3). Among the entire set of 32 439 human piRNA, we found 36 sequences with a perfect match to regions overlapping the exon–exon splice junctions of mRNA, suggesting that these piRNA are produced from mature mRNA. All 36 piRNAs were found on the transcribed strand (Supplementary Table S3). The observed dearth of piRNAs overlapping exon–exon splice junctions might be explained by the tight binding of splicing proteins at splice sites with mRNA in accordance with the NMD theory (35). This possible protection of mRNA and pre-mRNA by splicing proteins from endonucleolytic cleavage might explain the deficiency of piRNAs corresponding to exon–intron splice sites. Finally, exonic piRNAs tend to be in the internal mRNA regions and notably avoid the 3′-end (Figure 1A), while intronic piRNAs avoid both the 5′- and 3′-termini and prefer to localize within the central regions of introns (Figure 1B).
Table 3.
Distribution of human piRNAs within human genome
| Classification | Number of piRNA (%) | Number of random sequences (%) |
|---|---|---|
| Intergenic | 15 047 (76.4 ± 0.3) | 8480 (74.0 ± 0.4) |
| Intron | 2349 (11.9 ± 0.2) | 2779 (24.3 ± 0.4) |
| Exon | 2001 (10.2 ± 0.2) | 173 (1.5 ± 0.1) |
| Intron/Exon | 300 (1.5 ± 0.1) | 0 (0.0) |
| Total | 19 697 (100) | 11 432 (100) |
The distribution of piRNA within specific genomic regions is compared to the estimated probability of localization within these regions, calculated by classifying 11 432 sequences from randomly chose locations within the human genome. Both transcribed and complementary strands are represented for exons and introns. Percentages are shown ± SE.
Table 4.
The distribution of piRNAs inside introns
| Orientation and grouping | Number of piRNAs (%) |
|---|---|
| DNA strand | |
| Transcribed | 1623 (69.1 ± 1.0) |
| Complementary | 726 (30.9 ± 1.0) |
| piRNA clustering | |
| One per intron | 1043 (44.4 ± 1.0) |
| In clusters (≥2) | 1306 (55.6 ± 1.0) |
The data represents the distribution of intronic piRNA among transcribed and complementary strands as well as the tendency for piRNA to form clusters within introns. A cluster is defined as any intron containing more than one piRNA, irrespective of strand orientation. Percentages are shown ± SE.
Figure 1.
Distribution of piRNA along mRNA and introns. (A) piRNA location along each mRNA was determined by dividing mRNA into five equal segments. The total number of piRNAs within each quintile was determined. (B) The location of piRNA along introns was determined by dividing each intron into quintiles and calculated as in (A). Vertical bars show the standard error of the means.
In summary, we saw a 2.2-fold prevalence of piRNA on the transcribed strand of introns over the complementary strand. A significant enrichment of piRNA within the central regions of introns (Figure 1B) was observed, suggesting that a fraction of piRNAs are likely to be produced from post-spliced introns.
Putative endogenous siRNAs within introns
The number of endogenous siRNA molecules identified so far is quite small (36), therefore any analysis to map their positions within introns and exons at this stage would be uninformative. We instead performed a computational approach in order to assess the ability of human introns to produce endogenous siRNAs. Since hairpin siRNAs are derived from perfect double-stranded segments of RNA, we examined the occurrences of such hairpins within the entire set of human introns which could hypothetically produce siRNAs. This computation resulted in the characterization of 8053 intronic hairpin structures within 6163 introns. These hairpins had perfect stems spanning at least 21 nt in length and a short interlude of 0- to 80-nt long loops. A vast majority of these hairpins are associated with inverted DNA repeats, while only 507 represent unique genomic hairpin sequences unrelated to repetitive DNA (Supplementary Table S4 and Supplementary Figure S1). Similar searches within the three control randomized nucleotide sequence sets derived from naturally occurring introns, yielded no hairpin structures within them. Therefore we infer that there is a statistically significant enrichment of hairpins among natural introns (Fisher exact test, P < 2 × 10−16). No evolutionary conservation of the non-repeat-associated set of hairpins with rodent, dog or cow genomes was found. A similar search for perfect stems within exons (total length: 58 366 965 nt) yielded zero occurrences of perfect stems not associated with DNA repeats. Comparison of hairpin occurrence with introns and exons suggests a significant enrichment of stems within introns compared to exons (X21df = 26.5, P = 2.6 ×10−07). In a representative sample of intergenic regions (total length: 35 374 166 nt) there were 23 stems, which were unassociated with DNA repetitive elements, suggesting that the frequency of perfect stems in intergenic regions is similar to that within introns (X21df = 1.9, P = 0.17). It is unlikely that evolutionarily conserved endogenous stem–loop (cis–trans) siRNAs are produced from introns. Nonetheless, introns might still be a source for endogenous siRNA that are derived from repetitive genomic elements, perhaps inhibiting their propagation.
Long ncRNAs inside introns
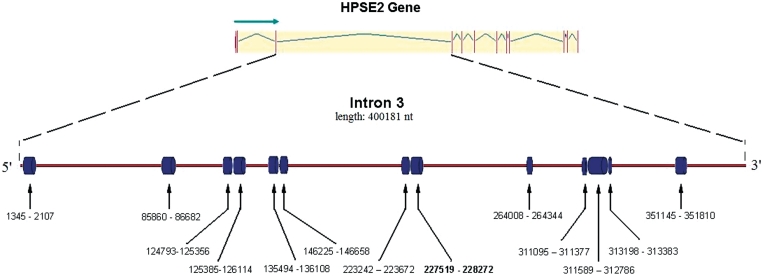
Current estimates suggest that 95% of the human genome is transcribed and produces a vast number of ncRNAs involved in different biological processes (11). Traditionally, ncRNAs are divided into short (<200 nt) and long (>200 nt) categories according to their length (37,38). According to Qureshi, Mattick and Mehler, ‘a major function of long ncRNAs (lncRNAs) appears to modulate the epigenetic status of proximal and distal protein-coding genes through cis- and trans-acting mechanisms’ (39). A considerable proportion of lncRNA exhibit low sequence conservation during evolution (37,39,40). However, in 2009, it was shown that a particular type of lncRNA, known as lincRNA (long intergenic ncRNA, large intervening ncRNA) is highly conserved in mammals (40). Intriguingly, there are also numerous evolutionarily conserved regions in mammalian introns that match the size range of lncRNAs. Recently Louro and co-authors described evolutionary conserved intronic lncRNA sequences from mouse and human (41). Figure 2 demonstrates 13 long conserved regions within one of the largest mammalian introns, intron 3 of Heparanase-2 gene. The average size of these 13 conserved regions is 600 nt, although the size depends on the choice and number of species analyzed. For an example, Supplementary Figure S2 illustrates the alignment of one such conserved region from intron 3 of Heparanase-2. When the introns of a larger selection of vertebrates were aligned, the length of the conserved region became only 100 bp (Supplementary Figure S2A), while in the alignment of a smaller group of closely related species (human–mouse–cow–dog) the evolutionary conservation of the region extended to as much as 750 bp (Supplementary Figure S2B).
Figure 2.
Evolutionarily conserved regions within the third intron of the Heparanase 2 (HPSE2) gene. The intron–exon structure of HPSE2 is shown at the top, with vertical lines depicting exons. In the bottom diagram, the cylinders depict 13 highly conserved regions with the coordinates specified below.
Using the latest release (July 2010) of our Mammalian Orthologous Intron Database (21), we performed a large-scale bioinformatic investigation of the distribution of long evolutionarily conserved regions within the entire set of 63 077 introns from 8161 human genes that have orthologs in each of the four mammalian species: mouse, rat, cow and dog. Only aligned segments >400 nt with at least 50% identity within five mammalian species were taken into account. Furthermore, computational filters removed alignments that could be associated with alternative splicing (‘Materials and Methods’ section). This computation revealed 9833 CIRs with lengths exceeding 400 bp. Since there are several stringent criteria defining orthologous introns, their entire set comprises approximately one-third of the total number of human introns (approximately 180 000). Therefore, the entire number of large CIRs in the human genome may be as large as 30 000. When the threshold for the alignment length was increased to 600 nt, 4848 CIRs were registered. Previous work in our lab showed that distribution of conserved regions within introns is uneven and, in particular, depends on the gene function (24). Such an abundant and uneven distribution of CIRs is in complete accordance with the previously published results by Sironi et al. (42).
Here we present computations in order to check our hypothesis that some CIRs might represent lncRNAs. This hypothesis is strengthened by the resent experimental findings that a fraction of lncRNA is found inside introns (39,41,43–45). BLAST analysis of our 9883 large CIRs (>400 bp and >50% identity) cross-referenced with all known human and mouse ncRNAs from Functional RNA Database (fRNAdb) (17) revealed hundreds of matches between them. Particularly, we found that 415 mouse large non-coding RNA sequences experimentally obtained under the FANTOM3 project and five additional mouse ncRNAs from other sources overlap with the CIRs (Supplementary Table S5). Seventy-seven percent of these 420 mice non-coding RNAs correspond to the transcribed strand of introns, while the remaining 23% correspond to the intronic complementary strand. However, in control calculations with ‘random CIRs’ sequences having the same length and number as natural CIR set, yet placed randomly along the orthologous introns, 438 mouse ncRNA from FANTOM3 dataset matched random CIRs. Moreover, in 86% cases they occur in the transcribed strand of the introns. These results are in accord with the claim of Guttman et al. (40) that ‘current [lincRNA] catalogues may consist largely of transcriptional noise, with a minority of bona fide functional lincRNAs hidden amid this background’.
The human database of experimentally verified large ncRNA is many times smaller than the corresponding mouse set, yet the human ncRNA database contains thousands of putative computer-predicted sequences that have not been predicted for mouse. BLAST analysis of the human ncRNA sequences revealed that 1268 putative ncRNA obtained by RNAz program; 485 putative ncRNA obtained by EvoFold program; and 18 experimentally verified large ncRNA overlap with our entire set of 9833 CIRs (Supplementary Table S6). Not surprisingly, our long intronic conserved regions correspond to 1753 putative ncRNA predicted by RNAz and EvoFold, since the latter algorithms are heavily based on evolution conservation. The EvoFold program considers the evolutionary conservation of RNA secondary structures and, therefore, is capable of predicting the DNA strand which gives rise to the putative ncRNA, since conservation of secondary structure may be strand-specific. However, in many cases it is problematic to infer the orientation of ncRNA when both strands have conserved secondary structures. Among the 485 predicted ncRNA (EvoFold) that overlap with our CIR set, 60.0 ± 2.2% SE correspond to the transcribed intronic strand, while 40.0 ± 2.2% SE to the complementary strand. In control calculations with ‘random CIRs’ they matched only eight EvoFold-predicted sequences and 76 RNAz-predicted sequences from the entire human fRNA database.
The strong preference of ncRNA from intronic regions to be associated with the transcribed strand is in accordance to Nakaya et al. (46), who examined 5678 wholly intronic human ‘mRNA clusters’ computed from GenBank entries. They found that 74% of these non-coding ‘mRNA clusters’ correspond to transcribed strand of introns while 26% correspond to the complementary strand.
We conjecture that among large CIRs there may be found thousands of long functional ncRNAs originated through the post-splicing processing.
DISCUSSION
Our calculations demonstrate that human introns may potentially contain thousands of ncRNAs—snoRNAs, miRNAs, piRNAs and, presumably, lincRNA-like molecules. Specifically, introns are enriched with ncRNAs, which mildly regulate gene expression (miRNA and orphan snoRNA). According to Selbach et al. (47) and Baek et al. (48), an individual miRNA modulates (predominantly down-regulates) the expression of hundreds of genes, although modestly (1.5- to 2-fold). Gene array experiments with knockout mice lacking orphan snoRNAs from the IC-SNURF-SNRPN locus revealed that such snoRNAs do not abruptly shut down or turn on genes, but rather, mildly change the expression of dozens of them (49). Lastly, the most abundant group of small ncRNAs in humans (piRNAs), whose functions are restricted to a very specific tissue (spermatocytes), do not show a preference to be either within or outside introns. Recent articles speculate that the role of piRNAs is to defend the genome against transposable elements (50); however, the high percentage of piRNAs not associated with repetitive elements suggests other undefined roles. This idea is supported by a new study demonstrating that piRNAs are also expressed in somatic tissues (51).
Non-coding RNAs regulate gene expression through two major pathways: (i) through transcriptional gene silencing (TGS) occurring within the nucleus, when ncRNAs, after their transcription and processing, are involved in chromatin changes and (ii) through post-transcriptional gene silencing (PTGS) occurring within the cytoplasm, when ncRNAs direct the RISC complex to target mRNAs for either cleavage or translational arrest (52). The TGS pathway is very actively employed in plants and therefore is the most studied pathway in this taxon. Mi et al. (53) characterized more than 300 000 Arabidopsis siRNAs, which are associated with nucleus-localized AGO4 protein and are specifically involved in chromatin changes and methylation. In mammals, the majority of siRNA and miRNA are associated with PTGS, which is the most studied pathway in this group. However, some mammalian miRNA are also involved in chromatin methylation and remodeling. For example, five RTL1-associated miRNAs control imprinting of the RTL1 gene (54). In addition, numerous mammalian piRNA and lincRNA also work through the TGS pathway (55). We see, therefore, that both TGS and PTGS are actively engaged in higher eukaryotes. When intronic ncRNAs (such as miRNA) work via PTGS, they regulate the production of hundreds of different proteins (47,48) some of which could include transcription factors. These transcription factors will in turn modulate the expression of other genes, (although not necessarily the parent gene that initiated this regulation event). However, auto-regulatory feedback loops within the PTGS pathway are not uncommon and have been known since the discovery of miRNAs. One of the first described miRNAs in Caenorhabditis elegans was let-7. Let-7 is regulated by a double-negative feedback loop where the miRNA inhibits the expression of lin-28 and lin-41, while the expression of these target genes inhibits let-7 (56). Another well-known example is an intron of the Arabidopsis Dicer gene containing miRNAs that regulate the expression of its own gene (57). Under the TGS pathway, an intronic ncRNA usually regulates the expression of its host and, potentially, neighboring genes. The regulation of multiple genes via the TGS pathway has not yet been well studied and therefore cannot be ruled out.
How precise should the regulation of genes be in healthy humans? It is well established that within the same cell type and developmental stage there is extensive individual variability in gene expression (58). In many cases the expression levels of genes are heritable and population-specific (58). From the perspective of thermodynamics, gene expression is a fundamentally stochastic process, with randomness in transcription and translation leading to cell-to-cell variations in mRNA and protein levels (59). Raj and Oudenaarden emphasize that the stochastic nature of gene expression has important consequences for cellular function, being beneficial in some contexts and harmful in others (59). In this respect, genetic diseases provide invaluable insight into genomic operation. A majority (87% by our estimate) of the prevalent human genetic autosomal diseases are recessive, which means that one healthy copy of a gene can substitute for two functional copies without much harm. In heterozygous individuals; that is, having one mutant and one normal gene, the expression level of the corresponding protein is often reduced by up to one-half of the average level. Considering the effect of gene overproduction, when the expression level of a large group of genes is even mildly up-regulated, the consequence is usually quite devastating as observed in various cases of human trisomy. One of the most common trisomies is Down syndrome where three copies of chromosome 21 (or a portion of 21) occur in the patient’s karyotype. The phenotype is characterized with some impairment of cognitive ability and physical growth as well as facial abnormalities. A partial trisomy of chromosome 21 can be as small as 2–3 Mb, representing 200 genes with expression levels being elevated 1.5 times on average (60). Perturbed expression of genes on other autosomes as a result of trisomies, such as chromosomes 8, 12, 13 and 18, cause more severe conditions such as Warkany syndrome, chronic lymphocytic leukemia, Patau syndrome and Edward’s syndrome respectively (60). Partial trisomies of these chromosomes produce milder symptoms. The two most frequent autosomal trisomies in humans, 16 and 22, are the most common chromosomal causes of spontaneous first trimester abortions (61). Partial trisomies of the remaining chromosomes are less common and often result in conditions ranging from few phenotypic symptoms, as in the case of Cat eye syndrome (22pter→q11), to lethal birth defects as in the case of chromosome 14 (62). Therefore, even mild up-regulation of a large group of genes is usually deleterious to an organism (60). In 2002 Yan et al. (63) showed that mammals, similar to plants, have allele-specific expression (ASE) of genes also known as allelic imbalance. This heritable allelic variation in gene expression was shown to be a common phenomenon within the human genome (64). De la Chapelle emphasizes the surprising extent of genomic regulation resulting from ASE (65). Many types of ASE dramatically influence susceptibility to disorders such as cancer, autoimmune diseases and diabetes (66,65). It is well documented that ASE is governed by cis-regulatory elements, yet the particular type and location of these elements is yet to be verified and therefore is debatable. De la Chapelle argues that the cis-elements responsible for ASE are likely to be miRNA and lincRNAs (65). From this standpoint, intronic ncRNA are outstanding candidates for the regulation of allelic imbalance via the TGS pathway. The aforementioned example of the five miRNAs that shut down the expression of the maternal RTL1 allele validates the ability of ncRNAs to have allele-specific precision (54).
Despite the permissible variations in the expression of many individual genes, the entire ensemble of genes must be highly coordinated. Only minor fluctuations in the expression of a number of genes are allowed in healthy humans. Such coordinated regulation of thousands of genes in a cell is unimaginable without numerous feedback loops engaged in the gene expression system. Intronic ncRNAs are perfect elements for such a feedback regulation system. Indeed, intronic ncRNAs are co-produced with the mRNA of their host genes. When a host gene is silent, its pool of ncRNAs is also not produced. However, during transcription, the production of intronic ncRNAs is strictly proportional to the expression level of the host gene. It becomes clear that the fundamental significance of many introns is to provide regulatory ncRNAs for the fine control of genes within complex higher organisms. This view of the subtle yet inextricable value of introns in genomic functioning is what we term the Symbiotic Intron Hypothesis. This hypothesis proposes a ‘non-selfish’ harmony between genes, introns and ncRNAs within higher eukaryotes. Genes provide space for introns inside of them. In turn, introns act as hosts for regulatory ncRNAs. Finally, ncRNAs provide essential regulation for the expression of genes. We conclude, therefore, that there is a natural symbiosis, between genes, introns and ncRNAs—a symbiosis that is only just beginning to be discovered and properly appreciated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation Career award ‘Investigation of intron cellular roles’ (grant number MCB-0643542). Funding for open access charge: National Science Foundation Career award ‘Investigation of intron cellular roles’ (grant number MCB-0643542).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
D.R. and A.M. were responsible for the computational processing and analysis of the miRNA and the piRNA sections of the article. D.R. was responsible for co-referencing CIRs with the mouse database and the analysis of Figure 2. A.M. was responsible for the statistical analysis conducted through the article. A.P. was responsible for the processing and analysis of the siRNA and long ncRNA sections of the article, and was responsible for creating the Mammalian Orthologous Intron Database, their multiple alignments and the websites. S.S.S. was responsible for RepeatMasking of introns for the siRNA and piRNA sections, editing and providing guidance while writing the draft. A.F. and L.F. supervised the project, provided guidance and wrote the draft. All authors have read and approved the article.
REFERENCES
- 1.Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 2.Carmel L, Rogozin IB, Wolf YI, Koonin EV. Patterns of intron gain and conservation in eukaryotic genes. BMC Evol. Biol. 2007;7:192. doi: 10.1186/1471-2148-7-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 4.Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat. Genet. 2008;40:1416–1425. doi: 10.1038/ng.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.House AE, Lynch KW. Regulation of alternative splicing: more than just the ABCs. J. Biol. Chem. 2008;283:1217–1221. doi: 10.1074/jbc.R700031200. [DOI] [PubMed] [Google Scholar]
- 6.Fedorova L, Fedorov A. Introns in gene evolution. Genetica. 2003;118:123–131. [PubMed] [Google Scholar]
- 7.Shepard S, McCreary M, Fedorov A. The peculiarities of large intron splicing in animals. PLoS One. 2009;4:e7853. doi: 10.1371/journal.pone.0007853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattick JS. Introns: evolution and function. Curr. Opin. Genet. Dev. 1994;4:823–831. doi: 10.1016/0959-437x(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 9.Amaral PP, Mattick JS. Noncoding RNA in development. Mamm. Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- 10.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 11.Carninci P, Yasuda J, Hayashizaki Y. Multifaceted mammalian transcriptome. Curr. Opin. Cell Biol. 2008;20:274–280. doi: 10.1016/j.ceb.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Shepelev V, Fedorov A. Advances in the exon-intron database (EID) Brief Bioinform. 2006;7:178–185. doi: 10.1093/bib/bbl003. [DOI] [PubMed] [Google Scholar]
- 13.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang KC, Stephen S, Engstrom PG, Tajul-Arifin K, Chen W, Wahlestedt C, Lenhard B, Hayashizaki Y, Mattick JS. RNAdb–a comprehensive mammalian noncoding RNA database. Nucleic Acids Res. 2005;33:D125–D130. doi: 10.1093/nar/gki089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 17.Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y, Kojima A, Kimura Y, Komori T, Asai K. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35:D145–D148. doi: 10.1093/nar/gkl837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smit AFA, Hubley R, Green P. RepeatMasker Open 3.2.9 < http://www.repeatmasker.org>(26 October 2010, date last accessed) [Google Scholar]
- 19.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bechtel JM, Wittenschlaeger T, Dwyer T, Song J, Arunachalam S, Ramakrishnan SK, Shepard S, Fedorov A. Genomic mid-range inhomogeneity correlates with an abundance of RNA secondary structures. BMC Genomics. 2008;9:284. doi: 10.1186/1471-2164-9-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fedorov A, Stombaugh J, Harr MW, Yu S, Nasalean L, Shepelev V. Computer identification of snoRNA genes using a Mammalian Orthologous Intron Database. Nucleic Acids Res. 2005;33:4578–4583. doi: 10.1093/nar/gki754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 23.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 24.Rais TB, MS thesis . University of Toledo, Toledo; 2009. Conserved signals on non coding RNA across a set of 73 genes associated with autistic spectrum disorders. Biomedical Sciences Program. OH 43614, USA. [Google Scholar]
- 25.Filipowicz W, Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell. Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 26.Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature. 1996;379:464–466. doi: 10.1038/379464a0. [DOI] [PubMed] [Google Scholar]
- 27.Makarova JA, Kramerov DA. Analysis of C/D box snoRNA genes in vertebrates: the number of copies decreases in placental mammals. Genomics. 2009;94:11–19. doi: 10.1016/j.ygeno.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. doi: 10.1038/nbt1144. [DOI] [PubMed] [Google Scholar]
- 29.Weber MJ. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2006;2:e205. doi: 10.1371/journal.pgen.0020205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 2006;34:5112–5123. doi: 10.1093/nar/gkl672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo Y, Li S. Genome-wide analyses of retrogenes derived from the human box H/ACA snoRNAs. Nucleic Acids Res. 2007;35:559–571. doi: 10.1093/nar/gkl1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis E, Caiment F, Tordoir X, Cavaille J, Ferguson-Smith A, Cockett N, Georges M, Charlier C. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 2005;15:743–749. doi: 10.1016/j.cub.2005.02.060. [DOI] [PubMed] [Google Scholar]
- 33.Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 34.Ro S, Song R, Park C, Zheng H, Sanders KM, Yan W. Cloning and expression profiling of small RNAs expressed in the mouse ovary. RNA. 2007;13:2366–2380. doi: 10.1261/rna.754207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva AL, Romao L. The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS Lett. 2009;583:499–505. doi: 10.1016/j.febslet.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 37.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 38.Marques AC, Ponting CP. Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol. 2009;10:R124. doi: 10.1186/gb-2009-10-11-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louro R, El-Jundi T, Nakaya HI, Reis EM, Verjovski-Almeida S. Conserved tissue expression signatures of intronic noncoding RNAs transcribed from human and mouse loci. Genomics. 2008;92:18–25. doi: 10.1016/j.ygeno.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Sironi M, Menozzi G, Comi GP, Cagliani R, Bresolin N, Pozzoli U. Analysis of intronic conserved elements indicates that functional complexity might represent a major source of negative selection on non-coding sequences. Hum. Mol. Genet. 2005;14:2533–2546. doi: 10.1093/hmg/ddi257. [DOI] [PubMed] [Google Scholar]
- 43.Hill AE, Hong JS, Wen H, Teng L, McPherson DT, McPherson SA, Levasseur DN, Sorscher EJ. Micro-RNA-like effects of complete intronic sequences. Front Biosci. 2006;11:1998–2006. doi: 10.2741/1941. [DOI] [PubMed] [Google Scholar]
- 44.Louro R, Smirnova AS, Verjovski-Almeida S. Long intronic noncoding RNA transcription: expression noise or expression choice? Genomics. 2009;93:291–298. doi: 10.1016/j.ygeno.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 45.Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakaya HI, Amaral PP, Louro R, Lopes A, Fachel AA, Moreira YB, El-Jundi TA, da Silva AM, Reis EM, Verjovski-Almeida S. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 48.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding F, Li HH, Zhang S, Solomon NM, Camper SA, Cohen P, Francke U. SnoRNA Snord116 (Pwcr1/MBII-85) deletion causes growth deficiency and hyperphagia in mice. PLoS One. 2008;3:e1709. doi: 10.1371/journal.pone.0001709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Halic M, Moazed D. Transposon silencing by piRNAs. Cell. 2009;138:1058–1060. doi: 10.1016/j.cell.2009.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reynolds SH, Ruohola-Baker H. PIWI goes solo in the soma. Dev. Cell. 2009;16:627–628. doi: 10.1016/j.devcel.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–358. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 53.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Youngson NA, Kocialkowski S, Peel N, Ferguson-Smith AC. A small family of sushi-class retrotransposon-derived genes in mammals and their relation to genomic imprinting. J. Mol. Evol. 2005;61:481–490. doi: 10.1007/s00239-004-0332-0. [DOI] [PubMed] [Google Scholar]
- 55.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 56.Nimmo RA, Slack FJ. An elegant miRror: microRNAs in stem cells, developmental timing and cancer. Chromosoma. 2009;118:405–418. doi: 10.1007/s00412-009-0210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie Z, Kasschau KD, Carrington JC. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 2003;13:784–789. doi: 10.1016/s0960-9822(03)00281-1. [DOI] [PubMed] [Google Scholar]
- 58.Cheung VG, Spielman RS. Genetics of human gene expression: mapping DNA variants that influence gene expression. Nat. Rev. Genet. 2009;10:595–604. doi: 10.1038/nrg2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Altug-Teber O, Bonin M, Walter M, Mau-Holzmann UA, Dufke A, Stappert H, Tekesin I, Heilbronner H, Nieselt K, Riess O. Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet. Genome Res. 2007;119:171–184. doi: 10.1159/000112058. [DOI] [PubMed] [Google Scholar]
- 61.Nagaishi M, Yamamoto T, Iinuma K, Shimomura K, Berend SA, Knops J. Chromosome abnormalities identified in 347 spontaneous abortions collected in Japan. J. Obstet. Gynaecol. Res. 2004;30:237–241. doi: 10.1111/j.1447-0756.2004.00191.x. [DOI] [PubMed] [Google Scholar]
- 62.Chen CP, Chern SR, Tsai EJ, Lee CC, Chen LF, Wang W. Prenatal diagnosis of partial trisomy 14q (14q31.1–>qter) and partial monosomy 5p (5p13.2–>pter) associated with polyhydramnios, short limbs, micropenis and brain malformations. Genet. Couns. 2009;20:281–288. [PubMed] [Google Scholar]
- 63.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 64.Lo HS, Wang Z, Hu Y, Yang HH, Gere S, Buetow KH, Lee MP. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de la Chapelle A. Genetic predisposition to human disease: allele-specific expression and low-penetrance regulatory loci. Oncogene. 2009;28:3345–3348. doi: 10.1038/onc.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Knight JC. Allele-specific gene expression uncovered. Trends Genet. 2004;20:113–116. doi: 10.1016/j.tig.2004.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.