Abstract
Telomere function is influenced by chromatin structure and organization, which usually involves epigenetic modifications. We describe here the chromatin structure of Arabidopsis thaliana telomeres. Based on the study of six different epigenetic marks we show that Arabidopsis telomeres exhibit euchromatic features. In contrast, subtelomeric regions and telomeric sequences present at interstitial chromosomal loci are heterochromatic. Histone methyltransferases and the chromatin remodeling protein DDM1 control subtelomeric heterochromatin formation. Whereas histone methyltransferases are required for histone H3K92Me and non-CpG DNA methylation, DDM1 directs CpG methylation but not H3K92Me or non-CpG methylation. These results argue that both kinds of proteins participate in different pathways to reinforce subtelomeric heterochromatin formation.
INTRODUCTION
Telomeres play an essential role in cell biology because they contribute to maintain genome stability (1–3). Two basic models of telomeric chromatin organization have been described for telomeres that are replicated by telomerase. The first one can be found in Saccharomyces cerevisiae. In Saccharomyces, the telomeric repeat binding protein Rap1 binds to telomeres and nucleates the assembly of a nucleoprotein complex called telosome (4). Rap1 also recruits the Sir Silencing complex to subtelomeric regions leading to the formation of subtelomeric heterochromatin, which spreads ∼3 kb inside the chromosome (5,6). The integrity of this silencing complex is required for the proper function of telomeres (7,8). The second model of telomeric chromatin organization can be found in mouse where telomeres and subtelomeric regions are heterochromatic. The loss of heterochromatic marks causes telomere dysfunction in mouse, highlighting again the relevance of heterochromatin in telomeres biology (9).
Heterochromatin is highly condensed in interphase nuclei and exhibit defined molecular features including the methylation of DNA and of histone H3 lysine 9 (H3K9) (10–12). In Arabidopsis, heterochromatin is characterized by cytosine methylation, which can be targeted at CpG, CpNpG or CpNpN residues (where N is any nucleotide), and by H3K91,2Me, H3K271,2Me and H4K201Me. In turn, Arabidopsis euchromatin is characterized by H3K41,2,3Me, H3K361,2,3Me, H4K202,3Me and by histones acetylation (13). The levels of these epigenetic marks are regulated by a complex interplay of enzymatic and structural proteins. One of these proteins is the chromatin remodeling factor DDM1 (decrease in DNA methylation), a SWI2/SNF2 homolog. Mutations in DDM1 cause loss of CpG methylation (14–16). CpG methylation have been found to control H3K92Me at different heterochromatic loci, which, in turn, direct non-CpG methylation (17–19). A high number of putative histone methyltransferases are present in Arabidopsis (10). Proteins that are known to be involved in the dimethylation of histone H3K9 include SUVH1, SUVH2, SUVH4, SUVH5 and SUVH6. Mutants lacking of SUVH4, also called KRYPTONYTE, have reduced levels of H3K92Me and non-CpG methylation at different heterochromatic loci (17,18). This decrease of epigenetic marks is more drastic in a triple mutant affected simultaneously in SUVH4, SUVH5 and SUVH6 (20).
Here, we describe the chromatin structure of Arabidopsis thaliana telomeres. We show that whereas telomeres exhibit euchromatic features, subtelomeric regions and interstitial telomeric sequences (ITSs) are heterochromatic. In addition, we show that histone methyltransferases and the chromatin remodeling protein DDM1 control subtelomeric heterochromatin formation. Finally, we compare our results with recently published data that also focus on the chromatin structure of Arabidopsis telomeres.
MATERIALS AND METHODS
Plant material and growth conditions
Seeds from wild-type Arabidopsis thaliana (Columbia ecotype) and from mutant derivatives were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, USA). The ku70 mutant (SALK_123114) and the suvh4 mutant (SALK_105816) were segregated to obtain homozygous lines that were verified by PCR. The Columbia (Col) suvh4 suvh5 suvh6 mutant strain (suvh4-6) was a gift from Dr Judith Bender (Brown University, USA). This strain was made with a Col suvh4 T-DNA insertion disruption into the 10th exon (SALK_044606) obtained from the Arabidopsis Biological Resource Center, the Col suvh5-2 T-DNA insertion allele (20), and the Col suvh6-1 T-DNA insertion allele (21). Each single suvh mutant was backcrossed two times to wild-type Col before being crossed together to make the triple suvh4-6 mutant. The Columbia ddm1 mutant strain used in this study was a gift from Dr Ingo Hoffmann (22). Plants were grown on soil at 22°C with a relative humidity of 70% during 3–4 weeks.
Methylation sensitivity analyses using restriction endonucleases
Southern blot analyses were performed using DNA isolated from rosette leaves following a CTAB based protocol (23). Essentially, 0.3–2 g of leaves were frozen on liquid nitrogen, ground and incubated at 65°C during 1 h in DNA extraction buffer (150 mM Tris pH 7.0; 1 M NaCl; 15 mM EDTA; 1.5% CTAB; 0.1% β-ME; 0.5% polyvinylpyrrolidine). Then, after chloroform extraction, DNA was precipitated with 0.6 volumes of isopropanol, RNase treated, phenolized and precipitated with ethanol. The purified DNA samples were digested with the corresponding restriction endonucleases, phenolized, precipitated with ethanol and resuspended in water. These DNA samples were resolved on agarose gels, transferred to HybondTM-XL membranes (GE Healthcare) and hybridized with a telomeric probe according to manufacturer instructions. Hybridizations were performed overnight at 65°C in the presence of 5× SSPE, 5× Denhardt’s solution, 0.5% SDS and 40 µg/ml of denatured salmon sperm DNA. After hybridization, the membranes were washed three times at room temperature during 10 min with 2× SSPE plus 0.1% SDS and once at 65°C during 45 min with 1× SSPE and 0.1% SDS. The telomeric template used for hybridization was constructed by annealing and ligation of telomeric sequence oligos containing specific restriction sites at their ends. These sites were used for cloning in pUC18 (24). The sequence of the template is as follows: HindIII site – (TTTAGGG)12 - SphI site – (TTTAGGG)10 – SalI site - (TTTAGGG)15 – EcoRI site. Prior to hybridization, it was excised from pUC18 using the HindIII and EcoRI sites, purified and labeled using a Ready-To-GoTM kit from GE Healthcare.
Chromatin immunoprecipitation
Wild-type or mutant leaves were used to perform ChIP assays as previously described (25). The antibody against 5-methylcytosine was provided by Merck, the antibodies against H3K42Me, H3K92Me and H3K27Me were provided by Upstate Biotechnology and the antibodies against H3K9Ac and H4K16Ac were provided by Abcam. ChIP assays were analyzed by multiplex PCR or by hybridization. When the ChIP assays were analyzed by multiplex PCR, two sequential PCR reactions were performed. In the first one, the DNA was subjected to 22 cycles of amplification. In the second one, 1/100 of the first reaction was amplified. For each input and immunoprecipitated DNA sample, 4 second PCR reactions were performed at increasing number of cycles. Reactions in which the amount of PCR products increased exponentially were quantified and used for final figures. The oligos used for amplification are listed in Supplementary Table S1. Oligos used to display the euchromatic CYP5 were included in all PCR reactions. To calculate enrichment values, the intensities of the PCR bands were quantified using the computer program Quantity One (BioRad). The intensity of the bands corresponding to the loci of interest were made relative to the intensity of the CYP5 bands and normalized against the input samples. When the ChIP assays were analyzed by hybridization, the input and immunoprecipitated DNA samples were amplified following a whole genome amplification protocol to increase hybridization sensitivity (26). For each input and immunoprecipitated DNA sample, equal amounts of amplified DNA were either digested with Tru9I or undigested, resolved on agarose gels and hybridized with the telomeric probe as indicated in the previous section. Enrichment values were calculated as indicated in the legend of Figure 1.
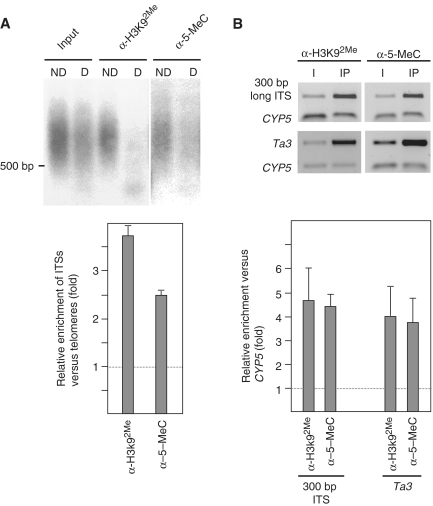
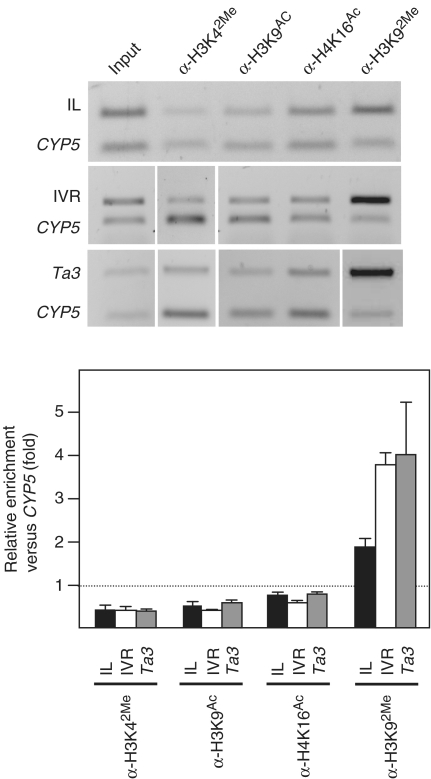
Figure 1.
Arabidopsis telomeres exhibit euchromatic features. (A) Analysis of H3K92Me and 5-methylcytosine levels at telomeres and ITSs. ChIP experiments were performed using antibodies against H3K92Me (α-H3K92Me) and 5-methylcytosine (α-5-MeC). Input and immunoprecipitated DNA samples were amplified as indicated in the ‘Materials and Methods’ section. Then, equal amounts of DNA were digested with Tru9I or undigested, resolved on agarose gels and hybridized with the telomeric probe. D and ND indicate digested or not digested with Tru9I, respectively. Relative enrichment values of ITSs versus telomeres were calculated as follows: the intensity of the hybridization signals above 500 bp was quantified for all lanes. Whereas the signals of undigested samples corresponded to both, telomeres and ITSs, the signals of digested samples corresponded only to telomeres because telomeres are not digested by Tru9I. The relative amounts of ITSs versus telomeres present in the input and in the immunoprecipitated DNA samples were determined by comparing digested and undigested lanes. Relative enrichment values were calculated by dividing the relative amounts of ITSs versus telomeres in the immunoprecipitated DNA samples between the relative amounts found in the input samples. (B) Analysis of H3K92Me and 5-methylcytosine levels at a specific ITS and at the Ta3 retrotransposon. The specific ITS studied was a 300-bp long telomeric repeats array present in the pericentromeric region of chromosome III (inside At3g33072). ChIP experiments were analyzed by multiplex PCR reactions including the CYP5 gene as euchromatic reference. The CYP5 gene corresponds always to the bottom band of the multiplex PCRs. I, input; IP, immunoprecipitated DNA. Graphic representations of enrichment values, calculated as indicated in the ‘Materials and Methods’ section, are shown at the bottom. Mean values of different experiments are represented together with the standard deviation.
RESULTS
Arabidopsis telomeres exhibit euchromatic features
Arabidopsis telomeres are composed of telomeric repeat arrays (of the TTTAGGG type) that are also abundant at interstitial chromosomal loci (27–32). Therefore, it is important to differentiate between telomeres and ITSs when telomeric studies are based on hybridizations with a telomeric probe. This problem has been previously addressed using frequently cutting restriction enzymes like Tru9I or MboI (33,34). Since Arabidopsis telomeres are composed of perfect telomeric repeat arrays, they remain uncut after digestion with the restriction enzymes. In contrast, ITSs are frequently cut because they are composed of very short arrays of perfect telomeric repeats interspersed with degenerated repeats (30,31,35,36). When Arabidopsis genomic DNA is digested with Tru9I and hybridized with a telomeric probe, most of the signals corresponding to ITSs disappear. Only three ITSs bands smaller than 500 bp remain (35). Therefore, the signals detected above 500 bp after Tru9I digestion correspond only to telomeres. In turn, the signals detected above 500 bp when the DNA is undigested correspond to both, telomeres and ITSs (35). This observation has allowed us to study the chromatin structure of Arabidopsis telomeres and ITSs independently. After performing ChIP experiments, we amplified the input and the immunoprecipitated DNA samples following a whole genome amplification protocol, resolved the DNA samples undigested or digested with Tru9I in agarose gels and hybridized them with a telomeric probe (see ‘Materials and Methods’ section). Then, we calculated the relative enrichment of ITSs versus telomeres as indicated in the legend of Figure 1.
Since telomeres are composed of repetitive DNA sequences, which are usually organized as heterochromatin, we decided to study whether Arabidopsis telomeres exhibit heterochromatic features. As mentioned above, H3K92Me and cytosine methylation are diagnostic hallmarks that label Arabidopsis heterochromatin (18,19). We analyzed these heterochromatic modifications at Arabidopsis telomeres and ITSs and found that ITSs had higher levels than telomeres (Figure 1A). After immunoprecipitation with the antibody against H3K92Me, ITSs were enriched 3.7 times versus telomeres. Similarly, ITSs were enriched 2.5 times after 5-methylcytosine immunoprecipitation.
To further characterize the enhanced levels of heterochromatic marks present at ITSs, we studied a specific 300-bp long ITS present in the pericentromeric region of Arabidopsis chromosome III. We analyzed the levels of H3K92Me and 5-methylcytosine at this locus by multiplex PCR including the euchromatic CYP5 gene as a reference. As expected, the 300-bp long ITS showed higher levels of H3K92Me and 5-methylcytosine than CYP5, 4.7 and 4.5 times higher, respectively (Figure 1B). Similar levels of heterochromatic marks were found at the Ta3 retrotransposon (Figure 1B), which has been previously shown to be heterochromatic. Since these levels of enrichment were similar to the levels of ITSs enrichment versus telomeres, we concluded that, whereas ITSs are heterochromatic, telomeres exhibit euchromatic features.
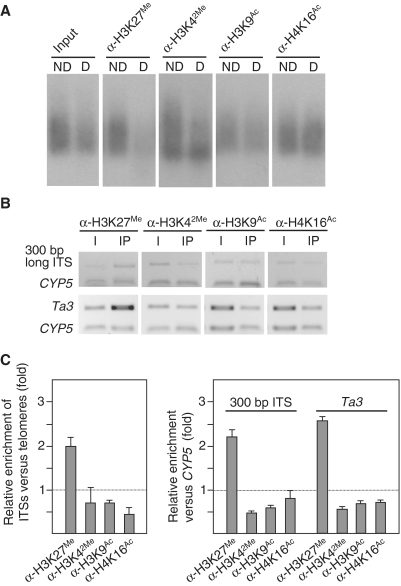
The results mentioned above were very surprising because telomeres have been traditionally considered as part of the heterochromatin present in the cell nucleus. Therefore, we decided to corroborate the euchromatic nature of Arabidopsis telomeres by studying four additional histone modifications: histone H3K42Me, histone H3K27Me and histones H3K9 and H4K16 acetylation (H3K9Ac, H4K16Ac). It is known that Arabidopsis heterochromatin have lower levels of H3K42Me, H3K9Ac and H4K16Ac than euchromatin. In contrast, heterochromatin in Arabidopsis has higher levels of H3K27Me (13). We determined the levels of ITSs enrichment versus telomeres for all these epigenetic modifications. The results obtained for each chromatin marker were consistent with a euchromatic structure for telomeres and an heterochromatic structure for ITSs (Figure 2).
Figure 2.
Additional euchromatic features present at Arabidopsis telomeres. (A) Analysis of H3K27Me, H3K42Me, H3K9Ac and H4K16Ac levels at telomeres and ITSs. ChIP experiments were performed using antibodies against H3K27Me (α-H3K27Me), H3K42Me (α-H3K42Me), H3K9Ac (α-H3K9Ac) and H4K16Ac (α-H4K16Ac). Input and immunoprecipitated DNA samples were amplified as indicated in the ‘Materials and Methods’ section. Then, equal amounts of DNA were digested with Tru9I (D) or undigested (ND), resolved on agarose gels and hybridized with the telomeric probe. (B) Analysis of H3K27Me, H3K42Me, H3K9Ac and H4K16Ac levels at the 300-bp long ITS and at the Ta3 retrotransposon. ChIP experiments were analyzed by multiplex PCR reactions including the CYP5 gene as euchromatic reference. The CYP5 gene corresponds always to the bottom band of the multiplex PCR. I, input; IP, immunoprecipitated DNA. (C) Graphic representation of relative enrichment values of ITSs versus telomeres, of Ta3 versus CYP5 and of the 300-bp long ITS versus CYP5.
As mentioned above, we amplified the input and the immunoprecipitated DNA samples before hybridization. During this process we noticed that telomeres were amplified more efficiently than ITSs. Before amplification, telomeres generated ∼30–40% of the signal displayed when the input samples were hybridized with the telomeric probe. After amplification, telomeres generated 80% of the input signal. We assumed that this amplification bias did not affect the calculation of relative enrichment values because they were calculated by comparing input and immunoprecipitated DNA samples. To support this notion, we analyzed unamplified input and immunoprecipitated DNA samples obtained with α-H3K27Me and α-H3K92Me (Supplementary Figure S1). We estimated the relative amounts of CYP5 in these samples and calculated the relative enrichment of ITSs versus telomeres and of telomeres versus CYP5. We found that ITSs had higher levels of H3K27Me and α-H3K92Me than telomeres, which were similar to the levels determined with the amplified DNA samples (Figures 1 and 2). In addition, we found that telomeres had levels of H3K27Me and α-H3K92Me similar to the euchromatic CYP5 gene (Supplementary Figure S1). These results further supported the euchromatic nature of Arabidopsis telomeres.
Subtelomeric regions organize as well defined heterochromatin domains
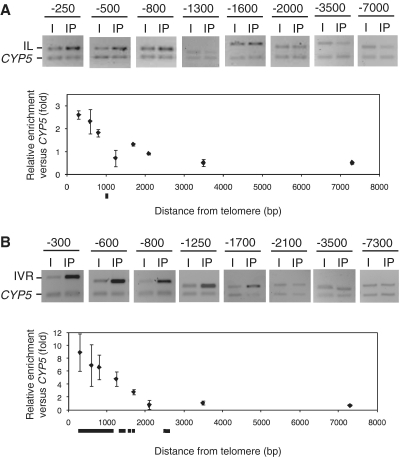
Since subtelomeric regions usually organize as heterochromatin, we decided to analyze the levels of DNA methylation and histone H3K92Me in the subtelomeric regions of Arabidopsis. We studied H3K92Me at different subtelomeric positions in the left arm of chromosome I and in the right arm of chromosome IV (IL and IVR, respectively). H3K92Me enrichment was observed from the telomeric repeats borders up to ∼1 kb in IL and up to around to 2 kb in IVR (Figure 3A and B). However, it was not detected further inside the chromosomes, indicating that these H3K92Me domains are small and well defined. We performed in silico analyses of the subtelomeric regions present in IL and in IVR looking for repetitive DNA sequences and found that whereas the subtelomeric region present in IVR is mostly repetitive up to 2 kb, subtelomeric sequences in IL are essentially non-repetitive (see black rectangles in Figure 3). Therefore, extensive repetitiveness at subtelomeric regions is not required for H3K92Me enrichment although it might influence H3K92Me levels.
Figure 3.
Subtelomeric regions undergo H3K92Me. (A) H3K92Me levels at the left telomeric region of chromosome I (IL). ChIP experiments were analyzed by multiplex PCR using the CYP5 gene as euchromatic reference. The CYP5 gene corresponds always to the bottom band of the multiplex PCRs. I, input; IP, immunoprecipitated DNA. H3K92Me levels were analyzed at different distances from the centromeric border of telomere IL. These distances (in bp) are indicated at the top of each panel. Relative enrichment values are represented at the bottom. Subtelomeric DNA regions longer than 50 bp that share >80% of identity with at least one different region of the genome are indicated by black rectangles. (B) H3K92Me levels at the right telomeric region of chromosome IV (IVR).
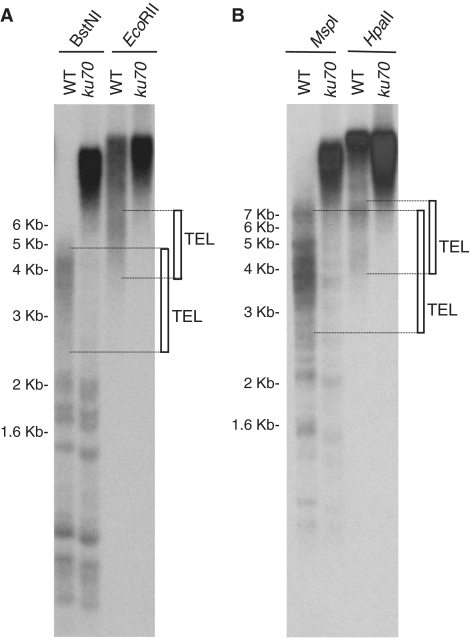
We mapped subtelomeric DNA methylation using an experimental approach that allowed us to analyze the average methylation state of all the subtelomeric regions and of ITSs simultaneously. First, we used two restriction endonucleases, BstNI and EcoRII, which are two isoschizomers that cut a 5-bp DNA sequence motif (CCWGG). Whereas BstNI is insensitive to DNA methylation and digests all their DNA targets, EcoRII is sensitive to CpNpG or CpNpN methylation and does not digest targets that contain methylated cytosines. Therefore, BstNI and EcoRII allow detection of non-CpG methylation. We digested wild-type Arabidopsis DNA samples with these restriction enzymes, resolved them on agarose gels and displayed telomeres and ITSs digestion profiles by hybridization with the telomeric probe. To distinguish telomeres from ITSs, we also analyzed DNA from a ku70 mutant, which has longer telomeres than the wild-type (37). Telomeric bands from ku70 migrate slower in the gel than wild-type telomeric bands. In contrast, wild-type and ku70 bands corresponding to ITSs migrate at the same distance. A smeared band of ∼2.5–5 kb was detected after digesting wild-type genomic DNA with BstNI (indicated by a rectangle in Figure 4A). This band corresponded to telomeres because it shifted upwards in the gel in the ku70 mutant (Figure 4A). This telomeric band was ∼1–2 kb shorter than the telomeric band observed after digesting wild-type DNA with EcoRII (Figure 4A). Therefore, we concluded that subtelomeric regions undergo non-CpG methylation up to ∼1–2 kb from the telomeric repeats border. The lower molecular weight bands observed after digesting wild-type DNA with BstNI corresponded to ITSs because they were also present in the ku70 mutant. These bands were not detected when wild-type and ku70 genomic DNA samples were digested with EcoRII. Instead, higher molecular weight smears were detected at the top of the EcoRII lanes indicating that ITSs undergo non-CpG methylation.
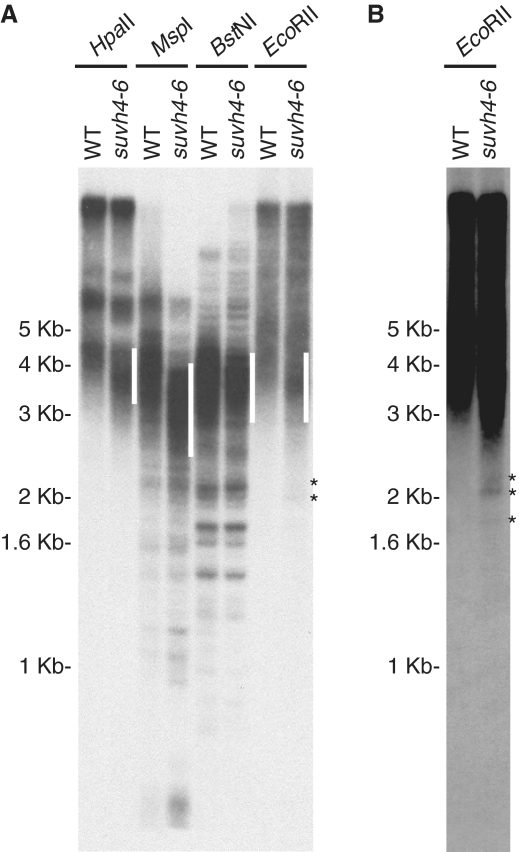
Figure 4.
Subtelomeric regions undergo DNA methylation. (A) Southern blot analysis of Arabidopsis genomic DNA digested with BstNI or EcoRII and hybridized with the telomeric probe. Wild-type (WT) and ku70 genomic DNA samples were digested with the restriction enzymes, resolved on an agarose gel and hybridized. The migration distances of molecular weight markers are indicated on the left. Rectangles on the right indicate the position of wild-type telomeric bands (TEL). (B) Southern blot analysis of Arabidopsis genomic DNA digested with MspI or HpaII and hybridized with the telomeric probe.
We also mapped subtelomeric DNA methylation using MspI and HpaII (Figure 4B). These enzymes are two isoschizomers that cut a 4-bp DNA sequence motif (CCGG). Whereas MspI is sensitive to CpNpG methylation, HpaII is sensitive to CpG and CpNpG methylation. Therefore, the comparison of the digestion profiles generated by these enzymes indicates the presence or absence of CpG methylation. The fuzzy telomeric band generated after digesting wild-type genomic DNA with MspI was 1–2 kb shorter than the telomeric band generated after digesting wild-type DNA with HpaII (Figure 4B). Therefore, we concluded that subtelomeric regions undergo CpG methylation. The extent of this DNA methylation is in close agreement with the extent of DNA methylation detected with BstNI and EcoRII and with the extent of H3K92Me enrichment detected in IL and IVR. These results indicated that subtelomeric regions in Arabidopsis organize as small and well-defined heterochromatin domains characterized by DNA and histone H3K9 methylation.
To further characterize subtelomeric heterochromatin, we analyzed the levels of three euchromatic marks in IL and in IVR (Figure 5). These marks were H3K42Me, H3K9Ac and H4K16Ac. We found that both subtelomeric regions had lower levels of all these marks than CYP5. These levels were similar to those present in the Ta3 retrotransposon (Figure 5). Therefore, subtelomeric regions seem to have the typical heterochromatin structure found at other genomic loci in Arabidopsis. These results are supported by high-resolution genomic maps (http://epigenomics.mcdb.ucla.edu) (38–40).
Figure 5.
Subtelomeric regions have similar levels of euchromatic marks than Ta3. Different euchromatic marks (H3K42Me, H3K9Ac and H4K16Ac) were analyzed at 500 bp from the centromeric border of telomere IL, at 600 bp from the centromeric border of telomere IVR and at the Ta3 retrotransposon. A heterochromatic mark (H3K92Me) was included as a control. ChIP experiments were analyzed by multiplex PCR using the CYP5 gene as euchromatic reference. The CYP5 gene corresponds always to the bottom band of the multiplex PCRs. Relative enrichment values are represented at the bottom together with the standard deviation.
Histone methyltransferases control subtelomeric DNA and histone H3K9 methylation
To start deciphering the molecular basis responsible for the formation of subtelomeric heterochromatin, we decided to study histone methyltransferase mutants. Histone methyltransferases are conserved in eukaryotes and control heterochromatin formation (41). There are many putative histone methyltransferase genes in Arabidopsis (10). Some of these genes are involved in H3K92Me. A triple mutant affected in three histone methyltransferases (suvh4, suvh5 and suvh6; herein referred to as suvh4-6) has been previously found to lose H3K92Me and non-CpG methylation at different target loci (20). We analyzed the levels of histone H3K92Me at subtelomeric regions in this triple mutant. We found that suvh4-6 had levels of subtelomeric H3K92Me similar to those present in CYP5 (Figure 6). Therefore, we concluded that subtelomeric heterochromatin formation was altered in this mutant. Interestingly, subtelomeric H3K92Me was decreased but not completely lost in a single suvh4 mutant (also named kyp), indicating that the establishment of subtelomeric H3K92Me involves multiple histone methyltransferases (Supplementary Figure S2).
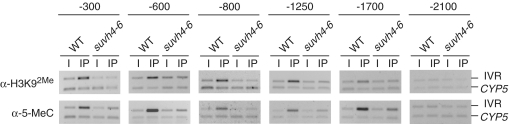
Figure 6.
Histone methyltransferases control subtelomeric H3K92Me and DNA methylation. H3K92Me and DNA methylation levels were analyzed at the right telomeric region of chromosome IV. WT refers to wild-type Arabidopsis whereas suvh4-6 refers to a triple mutant affected simultaneously in SUVH4, SUVH5 and SUVH6. ChIP experiments were analyzed by multiplex PCR using the CYP5 gene as euchromatic reference. The CYP5 gene corresponds always to the bottom band of the multiplex PCRs. I, input; IP, immunoprecipitated DNA. Distances from the centromeric border of telomere IVR (in bp) are indicated at the top of the figure.
To analyze whether the triple suvh4-6 mutant was affected in subtelomeric DNA methylation, we studied the levels of 5-methylcytosine at different subtelomeric positions by ChIP (Figure 6). As previously shown for H3K92Me, we found that cytosine methylation was enriched in IVR up to ∼2 kb in the wild-type strain. However, although cytosine methylation could also be observed in the suvh4-6 mutant, the levels of methylation enrichment were lower in the mutant than in the wild-type. To study whether the lower levels of cytosine methylation found in the mutant involved the loss of non-CpG methylation, we performed methylation sensitivity analyses using HpaII, MspI, BstNI and EcoRII (Figure 7). We digested suvh4-6 DNA with these restriction enzymes and hybridized the resulting DNA samples with the telomeric probe. As a control, we also analyzed wild-type DNA. The fuzzy telomeric band generated after digesting suvh4-6 DNA with HpaII was longer than the telomeric band generated after digesting the same DNA with MspI (see white lines in Figure 7A). Since similar differences were observed with wild-type DNA, we concluded that subtelomeric CpG methylation largely remained in the mutant. Therefore, the lower levels of cytosine methylation detected by ChIP in suvh4-6 should be due to the loss of non-CpG methylation. The restriction profiles obtained with BstNI and EcoRII confirmed this conclusion because the suvh4-6 telomeric bands generated by these enzymes had similar average size (see white lines in Figure 7A). In addition, the restriction profiles generated after digesting wild-type and suvh4-6 DNA with MspI and EcoRII indicated that ITSs methylation was also affected in the mutant (Figure 7A and B). ITSs were digested more extensively by these enzymes in the suvh4-6 mutant than in the wild-type (see low molecular weight bands including those marked with asterisks). These results indicated that part of the non-CpG methylation associated with ITSs was affected in the mutant.
Figure 7.
Histone methyltransferases control subtelomeric non-CpG methylation. (A) Southern blot analysis of wild-type (WT) and suvh4-6 genomic DNA digested with HpaII, MspI, BstNI or EcoRII and hybridized with the telomeric probe. The migration distances of molecular weight markers are indicated on the left. White lines indicate the position of the main suvh4-6 telomeric bands. Asterisks indicate ITSs bands generated after digesting suvh4-6 DNA with EcoRII. (B) Longer exposure of the lanes containing wild-type and suvh4-6 DNA digested with EcoRII. Asterisks indicate ITSs bands.
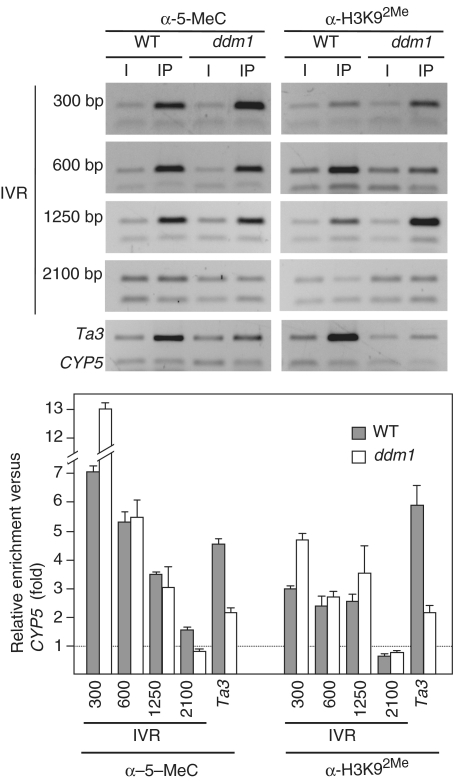
DDM1 controls subtelomeric DNA methylation
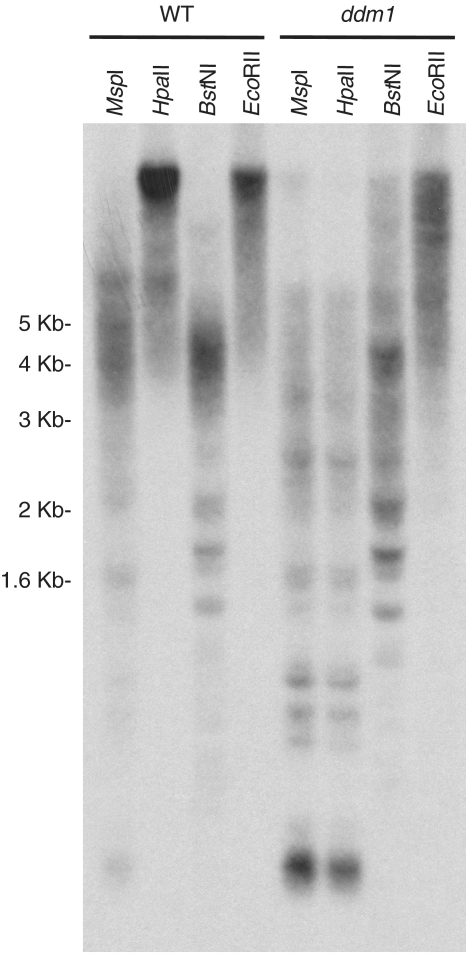
To further characterize subtelomeric heterochromatin we decided to study a ddm1 mutant. DDM1 is a chromatin remodeling protein that is required for heterochromatin formation in Arabidopsis. The ddm1 mutant loses CpG methylation and have reduced non-CpG methylation at different heterochromatic loci (14–16). We analyzed subtelomeric DNA methylation in this mutant by using HpaII, MspI, BstNI and EcoRII (Figure 8). As previously shown, the fuzzy telomeric band generated by MspI after digesting wild-type DNA and hybridizing it with the telomeric probe was shorter than the telomeric band generated after digesting the same DNA with HpaII. However, the telomeric bands generated after digesting ddm1 DNA with both enzymes had similar size. These results indicated that subtelomeric CpG methylation was lost in the ddm1 mutant. By contrast, the restriction profiles generated by BstNI and EcoRII revealed that subtelomeric non-CpG methylation largely remained in the mutant because the telomeric band generated by EcoRII after digesting suvh4-6 DNA was longer than the telomeric band generated by BstNI after digesting the same DNA. In addition, the restriction profiles generated with MspI and HpaII revealed that CpG methylation was also lost at ITSs in the ddm1 mutant (Figure 8). The low molecular weight ITSs bands generated after digesting wild-type DNA with MspI were not detected after HpaII digestion. However, these bands were readily detected after digesting ddm1 DNA with MspI or with HpaII.
Figure 8.
DDM1 controls subtelomeric DNA methylation. A Southern blot analysis of wild-type (WT) and ddm1 genomic DNA digested with HpaII, MspI, BstNI or EcoRII and hybridized with the telomeric probe is shown. The migration distances of molecular weight markers are indicated on the left.
We analyzed subtelomeric H3K92Me and 5-methylcytosine by ChIP in the ddm1 mutant. As a control, we also analyzed the Ta3 retrotransposon (Figure 9). We found that the levels of H3K92Me and 5-methylcytosine were reduced at the Ta3 retrotransposon in the mutant. However, the levels of these epigenetic marks were not reduced at subtelomeric regions. These results argue that most of the subtelomeric DNA methylation is of the non-CpG type and indicate that neither DDM1 nor CpG methylation are required for H3K92Me or for non-CpG methylation at subtelomeric regions.
Figure 9.
CpG methylation is not required for H3K92Me at subtelomeric heterochromatin. H3K92Me and DNA methylation levels were analyzed at the right telomeric region of chromosome IV. WT refers to wild-type Arabidopsis. ChIP experiments were analyzed by multiplex PCR using the CYP5 gene as euchromatic reference. The CYP5 gene corresponds always to the bottom band of the multiplex PCRs. I, input; IP, immunoprecipitated DNA. Distances from the centromeric border of telomere IVR are indicated in the left. Relative enrichment values are represented at the bottom together with the standard deviation.
DISCUSSION
Arabidopsis telomeres represent a novel type of telomeric chromatin organization
Two basic types of telomeric chromatin organization have been previously described for telomeres that are replicated by telomerase. The first one can be found in Saccharomyces cerevisiae and consist of short telomeres that associate with a multiprotein complex and subtelomeric regions that are heterochromatic (4,5). The second model can be found in mouse where telomeres and subtelomeric regions associate with heterochromatic marks (9). We have found that Arabidopsis telomeres exhibit euchromatic features whereas subtelomeric regions are heterochromatic. Therefore, Arabidopsis represents a novel type of telomeric chromatin organization.
Arabidopsis heterochromatin share more similarities with mouse than with Saccharomyces heterochromatin. Actually, Saccharomyces heterochromatin is rather different from the heterochromatin found in most eukaryotes. It is characterized by low levels of histones acetylation and by the absence of DNA methylation (42). In turn, although heterochromatin in many eukaryotes is also characterized by histones hypoacetylation, it usually contains methylation marks that are not present in Saccharomyces heterochromatin (13,42). Mouse telomeric and/or subtelomeric heterochromatin have been found to contain low levels of histones H3 and H4 acetylation and high levels of histone H3K93Me, histone H4K203Me and DNA methylation (9). Similarly, we have found that Arabidopsis subtelomeric heterochromatin has low levels of histones H3K9 and H4K16 acetylation, low levels of histone H3K42Me and high levels of histone H3K92Me and DNA methylation. Our results are in agreement with data published in a recent report by Vrbsky and colleagues (43). In this report, they described high levels of subtelomeric histone H3K92Me, histone H3K27Me and DNA methylation in Arabidopsis.
We have found that histone methyltransferases and the chromatin remodeling protein DDM1 control subtelomeric heterochromatin formation in Arabidopsis. These proteins contribute to label subtelomeric heterochromatin with different epigenetic marks. Whereas histone methyltransferases are required for H3K92Me and non-CpG methylation, DDM1 is required for CpG methylation. Neither DDM1 nor CpG methylation are needed for H3K92Me or for non-CpG methylation at subtelomeric regions. However, DDM1 and CpG methylation influence H3K92Me and non-CpG methylation at other heterochromatic loci, including the Ta3 retrotransposon (Figure 9) (14–19). These results reflect that some of the molecular mechanisms involved in the formation of subtelomeric heterochromatin might differ from those found at other heterochromatic loci. In addition, these results argue that both kinds of proteins, histone methyltransferases and DDM1, participate in different pathways to reinforce subtelomeric heterochromatin formation. Vrbsky and colleagues have shown that the RNA dependent DNA Methylation pathway (RdDM) is involved in the formation of subtelomeric heterochromatin in Arabidopsis (43). An rdr2 mutant, which is affected in this pathway, was shown to have lower levels of DNA methylation and histone H3K92Me than the wild-type at subtelomeric regions (43). Here, we have identified the SUVH4-6 proteins as members of this pathway. In a complementary way, DDM1 could participate in the maintenance of subtelomeric CpG methylation associated with the replication of the DNA (44,45).
Vrbsky and colleagues analyzed the chromatin structure of Arabidopsis telomeres and proposed that they exhibit features of intermediate heterochromatin that extend to subtelomeric regions (43). This intermediate heterochromatin was proposed to contain both repressive and active epigenetic marks. The repressive marks were DNA methylation, H3K27Me and H3K92Me. The active mark was H3K43Me. Here, we report that Arabidopsis telomeres exhibit euchromatic features whereas subtelomeric regions are heterochromatic. Our conclusions are based on the analysis of six different epigenetic marks including DNA methylation, H3K27Me and H3K92Me. We understand that the discrepancies between the data published by Vrbsky and colleagues and our results rely on the different methodological approaches that we have followed to analyze epigenetic marks.
To analyze histone H3 epigenetic modifications at telomeres, Vrbsky and colleagues performed ChIP experiments with different antibodies, dot-blotted the input and the immunoprecipitated DNA samples and hybridized them with a telomeric probe. They also hybridized bysulfite treated DNA with the telomeric probe to analyze DNA methylation at telomeres. Although they were aware of the presence of ITSs in the Arabidopsis genome, they assumed that only telomeres were detected in those studies. This assumption relied on some experimental data that they showed (43). They digested Arabidopsis genomic DNA with TruI1, resolved the resulting DNA fragments in a gel and hybridized them at high stringency (65°C) with the telomeric probe. After hybridization, they found that most of the signal corresponded to telomeres. Therefore, they concluded that only telomeres were detected in the chromatin structure analyses. We have published essentially the same experiment mentioned above but using Tru9I, an isoschizomer of TruI1 (35). We also found that most of the hybridization signal corresponded to telomeres after Tru9I digestion. However, if the genomic DNA was not digested with Tru9I before hybridization, we found that most of the signal corresponded to ITSs (35). As mentioned above, ITSs are barely detected after Tru9I digestion because most of them are sliced to a size that do not hybridize with the telomeric probe. In contrast, telomeres are readily detected after Tru9I digestion because they are essentially composed of perfect telomeric repeats, which are not cut by the enzyme (35). Since Vrbsky and colleagues analyzed the chromatin structure of Arabidopsis telomeres by hybridizing DNA samples undigested with TruI1, we believe that they did not analyze the chromatin structure of true telomeres. We think that they analyzed a mix of euchromatic telomeres and heterochromatic ITSs.
Vrbsky and colleagues proposed that subtelomeric regions have an intermediate heterochromatin structure because they found subtelomeric enrichment of a euchromatic mark (H3K43Me). This result does not agree with high-resolution genomic maps (http://epigenomics.mcdb.ucla.edu) (40). Therefore, we believe that the levels of H3K43Me at subtelomeric regions should be further analyzed by quantitative methods.
Arabidopsis telomeres might impair heterochromatin formation
We have shown that subtelomeric heterochromatin occupy small and well-defined domains in Arabidopsis. This heterochromatin might be recruited by telomeric proteins and, then, spread inside the chromosome following a sequential mechanism. However, subtelomeric heterochromatin does not spread through telomeres towards the chromosome end, which might reflect that Arabidopsis telomeres impair heterochromatin formation.
In Arabidopsis, like in other eukaryotes, the RNAi pathway participates in the heterochromatinization of repetitive sequences. This pathway involves the action of siRNAs which, in principle, should be able to target any nuclear compartment (45,46). There are siRNAs containing telomeric sequences in Arabidopsis (http://asrp.cgrb.oregonstate.edu) (43,47). These telomeric siRNAs, together with other components of the RNAi pathway, should be involved in the heterochromatinization of Arabidopsis ITSs. However, telomeric siRNAs do not induce telomeres heterochromatinization. We believe that some molecular or architectural features of Arabidopsis telomeres might impair siRNAs induced heterochromatin formation. Some boundary elements might localize at the telomeric/subtelomeric transition regions and contribute to maintain the euchromatic state of telomeres and/or to prevent subtelomeric heterochromatin from spreading into telomeres. The subtelomeric regions that abutt Arabidopsis telomeres usually contain degenerated telomeric repeats, which could be bound by many of the putative telomeric repeat binding factors present in the plant (48). It will be interesting to investigate the influence of these degenerated repeats and of the D-Loop formation on the chromatin organization of Arabidopsis telomeric and subtelomeric regions.
The heterochromatic nature of Arabidopsis ITSs might contribute to confer genomic stability
As mentioned above, whereas telomeres localize at the end of eukaryotic chromosomes and are essentially composed of perfect telomeric repeats, Arabidopsis ITSs are mainly pericentromeric and contain few perfect telomeric repeats interspersed with degenerated repeats (27–32,35,36). We have shown that Arabidopsis ITSs are heterochromatic, which might be influenced by their pericentromeric localization. However, how the differential environment and the primary sequence of Arabidopsis telomeres and ITSs influence their chromatin organization should be the subject of future studies. In general, ITSs have been related to chromosomal aberrations, fragile sites, hot spots for recombination and diseases caused by genomic instability (49). In Arabidopsis, the lack of telomerase and of the XPF-ERCC1 endonuclease leads to developmental problems that are accompanied by genomic instability. This instability involves high levels of recombination events between telomeric regions and ITSs (36), which in the wild-type strain probably occur with low frequency. Since heterochromatin is known to inhibit repeated DNA recombination (50), we believe that the heterochromatic nature of Arabidopsis ITSs might contribute to prevent genomic instability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Education and Science grants (BFU2004-01560/BMC, BFU2008-02497/BMC and GEN2003-20859-C03-03/INTER); FEDER funds (European Regional Development Funds). Funding for open access charge: Instituto de Bioquícmica Vegetal y Fotosíntesis.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Judith Bender and Ingo Hoffmann for their kind gift of suvh4-6 and ddm1 seeds. The authors also thank Carmen López for help with the analysis of subtelomeric heterochromatin and Felix Prado and Andrés Aguilera for critical reading of the manuscript.
REFERENCES
- 1.Cech T. Beginning to understand the end of the chromosome. Cell. 2004;43:405–413. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E. Telomeres and telomerase: their mechanisms of action and the effects of altering their functions. FEBS Lett. 2005;579:859–862. doi: 10.1016/j.febslet.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 3.Greider C. Telomerase RNA levels limit the telomere length equilibrium. Cold Spring Harb. Symp. Quant. Biol. 2006;71:225–229. doi: 10.1101/sqb.2006.71.063. [DOI] [PubMed] [Google Scholar]
- 4.Wright J, Gottschling D, Zakian V. Saccharomyces telomeres assume a non-nucleosomal chromatin structure. Genes Dev. 1992;6:197–210. doi: 10.1101/gad.6.2.197. [DOI] [PubMed] [Google Scholar]
- 5.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 6.Luo K, Vega-Palas M, Grunstein M. Rap1-Sir4 binding independent of other Sir, yKu, or histone interactions initiates the assembly of telomeric heterochromatin in yeast. Genes Dev. 2002;16:1528–1539. doi: 10.1101/gad.988802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lustig A, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 8.Palladino F, Laroche T, Gilson E, Axelrod A, Pillus L, Gasser S. SIR3 and SIR4 proteins are required for the positioning and integrity of yeast telomeres. Cell. 1993;75:543–555. doi: 10.1016/0092-8674(93)90388-7. [DOI] [PubMed] [Google Scholar]
- 9.Blasco M. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 10.Fischer A, Hofmann I, Naumann K, Reuter G. Heterochromatin proteins and the control of heterochromatic gene silencing in Arabidopsis. J. Plant Phys. 2006;163:358–368. doi: 10.1016/j.jplph.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- 12.Grewal S, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs J, Demidov D, Houben A, Schubert I. Chromosomal histone modification patterns-from conservation to diversity. Trends Plant Sci. 2006;11:199–208. doi: 10.1016/j.tplants.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Vongs A, Kakutani T, Martienssen R, Richards E. Arabidopsis thaliana DNA methylation mutants. Science. 1993;260:1926–1928. doi: 10.1126/science.8316832. [DOI] [PubMed] [Google Scholar]
- 15.Ronemus M, Galbiati M, Ticknor C, Chen J, Dellaporta S. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- 16.Jeddeloh J, Stokes T, Richards E. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 1999;22:94–97. doi: 10.1038/8803. [DOI] [PubMed] [Google Scholar]
- 17.Jackson J, Lindroth A, Cao X, Jacobsen S. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 18.Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks: DNA methylation and histone H3 lysine 9 methylation. Curr. Biol. 2002;12:1360–1367. doi: 10.1016/s0960-9822(02)00976-4. [DOI] [PubMed] [Google Scholar]
- 19.Soppe W, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang M, Jacobsen S, Schubert I, Fransz P. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebbs M, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ebbs M, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell. Biol. 2005;25:10507–10515. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, Hause G, Aurich A, Dorn R, Jenuwein T, et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 2005;24:1418–1429. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murray M, Thompson F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4325. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 25.Gendrel A, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- 26.Lippman Z, Gendrel A, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat. Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 27.Richards E, Ausubel F. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53:127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 28.Richards E, Goodman H, Ausubel F. The centromere region of Arabidopsis thaliana chromosome 1 contains telomere-similar sequences. Nucleic Acids Res. 1991;19:3351–3357. doi: 10.1093/nar/19.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards E, Chao S, Vongs A, Yang J. Characterization of Arabidopsis thaliana telomeres isolated in yeast. Nucleic Acids Res. 1992;20:4039–4046. doi: 10.1093/nar/20.15.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regad F, Lebas M, Lescure B. ITSs within the Arabidopsis thaliana genome. J. Mol. Biol. 1994;239:163–169. doi: 10.1006/jmbi.1994.1360. [DOI] [PubMed] [Google Scholar]
- 31.Uchida W, Matsunaga S, Sugiyama R, Kawano S. Interstitial telomere-like repeats in the Arabidopsis thaliana genome. Genes Genet. Syst. 2002;77:63–67. doi: 10.1266/ggs.77.63. [DOI] [PubMed] [Google Scholar]
- 32.Shakirov E, Shippen D. Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell. 2004;16:1959–1967. doi: 10.1105/tpc.104.023093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzgerald M, Riha K, Gao F, Ren S, McKnight T, Shippen D. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallego M, White C. RAD50 function is essential for telomere maintenance in Arabidopsis. Proc. Natl Acad. Sci. USA. 2001;98:1711–1716. doi: 10.1073/pnas.98.4.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gámez-Arjona F, López-López C, Vaquero-Sedas M, Vega-Palas M. On the organization of the nucleosomes associated with telomeric sequences. Biochim. Biophys. Acta. 2010;1803:1058–1061. doi: 10.1016/j.bbamcr.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 36.Vannier J, Depeiges A, White C, Gallego M. ERCC1/XPF protects short telomeres from homologous recombination in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000380. doi: 10.1371/journal.pgen.1000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riha K, Watson J, Parkey J, Shippen D. Telomere length deregulation and enhanced sensitivity to genotoxic stress in Arabidopsis mutants deficient in Ku70. EMBO J. 2002;21:2819–2826. doi: 10.1093/emboj/21.11.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernatavichute Y, Zhang X, Cokus S, Pellegrini M, Jacobsen S. Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS One. 2008;3:e3135. doi: 10.1371/journal.pone.0003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan S, Chen H, Henderson I, Shinn P, Pellegrini M, Jacobsen S, et al. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 40.Zhang X, Bernatavichute Y, Cokus S, Pellegrini M, Jacobsen S. Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 2009;10:R62. doi: 10.1186/gb-2009-10-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krauss V. Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica. 2008;133:93–106. doi: 10.1007/s10709-007-9184-z. [DOI] [PubMed] [Google Scholar]
- 42.Millar C, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell. Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 43.Vrbsky J, Akimcheva S, Watson J, Turner T, Daxinger L, Vyskot B, Aufsatz W, Riha K. siRNA-mediated methylation of Arabidopsis telomeres. PLoS Genet. 2010;6:e1000986. doi: 10.1371/journal.pgen.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gendrel A, Colot V. Arabidopsis epigenetics: when RNA meets chromatin. Curr. Opin. Plant Biol. 2005;8:142–147. doi: 10.1016/j.pbi.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Henderson I, Jacobsen S. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 46.Vaughn M, Martienssen R. It’s a small RNA world, after all. Science. 2005;309:1525–1526. doi: 10.1126/science.1117805. [DOI] [PubMed] [Google Scholar]
- 47.Gustafson A, Allen E, Givan S, Smith D, Carrington J, Kasschau K. ASRP: the Arabidopsis small RNA project database. Nucleic Acids Res. 2005;33:D637–D640. doi: 10.1093/nar/gki127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watson J, Riha K. Comparative biology of telomeres: where plants stand. FEBS Lett. 2010;584:3752–3759. doi: 10.1016/j.febslet.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin K, Yan J. Endings in the middle: current knowledge of interstitial telomeric sequences. Mutation Res. 2008;658:95–110. doi: 10.1016/j.mrrev.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 50.Peng J, Karpen G. Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.