Abstract
We describe a novel 3′-OH unblocked reversible terminator with the potential to improve accuracy and read-lengths in next-generation sequencing (NGS) technologies. This terminator is based on 5-hydroxymethyl-2′-deoxyuridine triphosphate (HOMedUTP), a hypermodified nucleotide found naturally in the genomes of numerous bacteriophages and lower eukaryotes. A series of 5-(2-nitrobenzyloxy)methyl-dUTP analogs (dU.I–dU.V) were synthesized based on our previous work with photochemically cleavable terminators. These 2-nitrobenzyl alkylated HOMedUTP analogs were characterized with respect to incorporation, single-base termination, nucleotide selectivity and photochemical cleavage properties. Substitution at the α-methylene carbon of 2-nitrobenzyl with alkyl groups of increasing size was discovered as a key structural feature that provided for the molecular tuning of enzymatic properties such as single-base termination and improved nucleotide selectivity over that of natural nucleotides. 5-[(S)-α-tert-Butyl-2-nitrobenzyloxy]methyl-dUTP (dU.V) was identified as an efficient reversible terminator, whereby, sequencing feasibility was demonstrated in a cyclic reversible termination (CRT) experiment using a homopolymer repeat of ten complementary template bases without detectable UV damage during photochemical cleavage steps. These results validate our overall strategy of creating 3′-OH unblocked reversible terminator reagents that, upon photochemical cleavage, transform back into a natural state. Modified nucleotides based on 5-hydroxymethyl-pyrimidines and 7-deaza-7-hydroxymethyl-purines lay the foundation for development of a complete set of four reversible terminators for application in NGS technologies.
INTRODUCTION
Next-generation sequencing (NGS) technology has accelerated the field of genomic research by delivering enormous volumes of low-cost sequencing reads (1). Despite these advances, chemistry improvements are still needed, particularly in the production of more accurate read data and longer read-lengths. Our group has focused on developing novel reversible terminators, which ideally halt DNA synthesis after the addition of a single-nucleotide analog by polymerase into the growing primer strand. Recently, we reported a new paradigm in 3′-OH unblocked nucleotide chemistry by attaching a small photocleavable 2-nitrobenzyl group to the N6-position of 2′-deoxyadenosine triphosphate (2). While the results were promising, translation to thymine has proven problematic as an N3-alkylated nucleotide would be expected to interfere with Watson–Crick base pairing, thus reducing nucleotide selectivity. This prompted us to search for alternative thymine analogs that maintain the desired features of base selective, 3′-OH unblocked terminators. A common attachment site for thymine has been at its 5-position coupled with unsaturated amino linkers (3–5), resulting in molecular scars residing on the nucleobase structure upon chemical (6–8) and photochemical (9,10) cleavage reactions. Our goal, therefore, was to identify a thymine analog that could enable the direct coupling of the 2-nitrobenzyl group to the nucleobase, which upon photochemical cleavage would transform back into a natural nucleotide substrate.
5-Hydroxymethyl-2′-deoxyuridine (HOMedU) monophosphate is a natural nucleotide found in the genomes of several Bacillus subtilis bacteriophages (11,12) and dinoflagellates (13). Remarkably, thymidine is completely replaced with HOMedU in B. subtilis phages SP01, SP82G, φ25, SP8, φe, 2C and H1 (11,12). Unlike these phages, however, the degree of HOMedU replacement in the genomes of dinoflagellates (13), B. subtilis phage SP10 (12), Pseudomonas acidovirans phage φW14 (14), and Trypanosoma brucei (15) varies considerably. For B. subtilis phage SP10, P. acidovirans phage φW14 and T. brucei, HOMedU triphosphate (HOMedUTP) is incorporated during DNA synthesis as an intermediate (12,16), and then further modified in their respective genomes as α-glutamylthymidine (12), putrescinylthymidine (14) and β-D-glucosyl-hydroxymethyl-2′-deoxyuridine analogs (15). While roles remain unclear, HOMedU might protect phage DNA against host and phage nucleases and/or provide signals for the regulation of middle- and/or late-phage gene transcription (12,17). Although elusive, modification of HOMedU also serves specific, yet unknown biological roles, with chemical functionalization of HOMedU occurring on the 5-hydroxymethyl group. For our purposes, the 5-hydroxymethyl group could serve as a ‘molecular handle’ to couple directly a 2-nitrobenzyl group, which upon photochemical cleavage with ultraviolet (UV) light, would transform the modified nucleotide back into a natural HOMedU structure. Here, we report novel 3′-OH unblocked reversible terminators based on a core HOMedU nucleotide and determined their incorporation, termination, selectivity and photochemical cleavage properties as candidate reversible terminators for cyclic reversible terminator (CRT) sequencing applications (1,18).
MATERIALS AND METHODS
Reagents and materials
Chemical reagents and solvents were purchased from Alfa Aesar, Sigma-Aldrich or EM Sciences. 2′-Deoxyadenosine triphosphate (dATP), 2′-deoxycytosine triphosphate (dCTP), 2′-deoxyguanosine triphosphate (dGTP), thymidine triphosphate (TTP), 3′-deoxythymidine triphosphate (3′-dTTP) and Q Sepharose Fast Flow anion exchange resin were purchased from GE Healthcare Life Sciences. All DNA polymerases were purchased from New England Biolabs, with the exception of AmpliTaqFS, which was purchased from Applied Biosystems (AB), now Life Technologies. Snake venom phosphodiesterase I was purchased from United States Biochemical (now Affymetrix), and alkaline phosphatase was purchased from Sigma-Aldrich. Oligonucleotides were purchased from Integrated DNA Technologies. M-270 streptavidin-coated magnetic beads, BODIPY-FL SE and BigDye version 3.1 kits were purchased from Life Technologies. Analytical silica gel 60 F254 TLC plates were purchased from Whatman, and silica gel 60 (230–400 mesh) was purchased from EM Sciences. Spheri-5 RP-18 and Aquapore OD-300 columns were purchased from Perkin Elmer.
Nucleosides and nucleotides synthesis
Complete experimental procedures describing the synthesis of the nucleotides used in this work are available in the Supplementary Data.
PEP assays for incorporation
The general procedure for polymerase end-point (PEP) assays has been previously described (2). Interrogation bases for all oligoTemplates described here are underlined and bolded. Dye-labeled primers ending in ‘S’ contain a phosphorothioate linkage between the 3′-end and penultimate nucleotides. For the current study, 5 nM of BODIPY-FL labeled primer-1 (5′-TTGTAAAACGACGGCCAGT) (19) was hybridized with 40 nM of oligoTemplate-4 (5′-TACGGAGCTGAACTGGCCGTCGTTTTACA) to conduct the PEP assays. PEP assays were performed in triplicate for each DNA polymerase/nucleotide analog combination to calculate the average IC50 ± 1 SD.
Weighted-sum assay for termination studies
The weighted-sum assay was designed to examine quantitatively the degree to which a nucleotide could be extended along a homopolymer stretch of complementary template bases. BODIPY-FL-labeled primer-1 (5 nM) was hybridized to 40 nM of oligoTemplate-7 (5′-CCGAAAAAAAAAAACTGGCCGTCGTTTTACAGCCGCCGCCGCCGAACCGAGAC-Biotin), and the primer/template complex was assayed as described above. The IC50 values were determined using PEP assays for dU.I – dU.V using Therminator and Vent(exo−) polymerases with oligoTemplate-7. Primer extensions were then performed at 25× IC50 values for dU.I, dU.II and dU.III with an additional concentration end-point of 3 μM, which is ∼1000× the IC50 value for dU.IV and dU.V. The weighted-sum value was determined from the following formula: 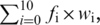 where fi is the fraction of signal for a given band to the overall signal and wi is the weight assigned to discrete positions in the DNA sequence ladder (i.e. for the primer band, w0 = 0, for the n + 1 product, w1 = 1, … for the n+10 product, w10 = 10).
where fi is the fraction of signal for a given band to the overall signal and wi is the weight assigned to discrete positions in the DNA sequence ladder (i.e. for the primer band, w0 = 0, for the n + 1 product, w1 = 1, … for the n+10 product, w10 = 10).
Nucleotide selectivity assays for TTP, HOMedUTP and dU.I–dU.V
OligoTemplate-4 was substituted with either 40 nM of oligoTemplate-2 (5′-TACGGAGCTGTACTGGCCGTCGTTTTACA) or 40 nM of oligoTemplate-5 (5′-TACGGAGCAGCACTGGCCGTCGTTTTACA). For the T—G mismatch, 5 nM of BODIPY-FL-labeled primer-3 (5′-TATGACCATGATTACGCC) was hybridized with 40 nM of oligoTemplate-8 (5′-TACGGAGCACGGGCGTAATCATGGTCATA, and the primer/template complexes were analyzed using PEP assays.
Photochemical cleavage studies
dU.I–dU.V were incorporated, as described for the PEP assays, at a final concentration of 100 nM, using the BODIPY-FL labeled primer-1/oligoTemplate-4 complex (10 nM/80 nM ratio). The incorporated reactions were then quenched with either 10 μl of FBD solution [20% aqueous deionized formamide; 10 mM Na2EDTA, pH 8.0; 16.6 mg/ml Blue Dextran, MW 2 000 000, (2)] or 50 mM sodium azide solution, exposed to 365 nm UV light for various time-points using our custom-designed UV deprotector [see Supplementary Figure S1 in Ref. (2)], and then placed on ice. Twenty μl of stop solution (98% deionized formamide; 10 mM Na2EDTA, pH 8.0; 25 mg/ml Blue Dextran, MW 2 000 000) was added, and samples were analyzed using an AB model 377 DNA sequencer. Cleavage assays were performed in triplicate to calculate the average DT50 value ± 1SD (i.e. the time at which 50% of 2-nitrobenzyl groups were photochemically cleaved from the extended primer/template complex).
PCR and Sanger sequencing
For PCR experiments, exons 2 and 10 from the TCF1 gene were amplified [see Supplementary Table S1 in Ref. (20) for primer sequences]. Approximately 50 ng of genomic DNA was amplified with 0.4 µM of HNF1a_2.1 (F/R) or HNF1a_10.1 (F/R) primer pairs and one unit of Vent(exo−) polymerase in 1× ThermoPol buffer [20 mM Tris–HCl, pH 8.8; 10 mM (NH4)2SO4; 10 mM KCl; 2 mM MgSO4; 0.1% Triton X-100; New England BioLabs], 1 M betaine (21,22) and 200 μM each of dATP, dCTP, dGTP and either TTP or HOMedUTP. Cycling conditions were 4 min at 95°C, then 35 cycles of 95°C for 30 s, 59°C for 30 s and 72°C for 90 s with a final step at 72°C for seven min. PCR products were purified using a Qiagen QIAquick gel extraction kit and sequenced using a ‘1/16’ dilution of BigDye version 3.1 chemistry. Sequencing reactions were cycled using the following conditions: 96°C for 1 min, then 25 cycles of 96°C for 10 s, 50°C for 5 s and 60°C for 4 min. Reaction products were purified by ethanol precipitation, resuspended in 30 µl of HPLC water, and then loaded onto an AB model 3100 DNA sequencer.
Primer extension assays
As described above for PEP assays, primer extensions for Therminator and Vent(exo−) polymerases were performed using 5 nM of BODIPY-FL-labeled primer-1S hybridized to 40 nM of oligoTemplate-7. TTP and HOMedUTP were extended at 25× their IC50 value and 3 μM (∼1000× their IC50 values) and analyzed using an AB model 377 DNA sequencer.
Ten cycle CRT sequencing
The binding of a biotinylated template to M-270 beads and the hybridization of BODIPY-FL-labeled primer-2S (5′-GGCGGCGGCGGCTGTAAAACGACGGCCAG-s-T) have been previously described (2), except oligoTemplate-7 was substituted for oligoTemplate-3. The bead bound primer/template complex was incubated with 0.5 units of Therminator polymerase in 1× ThermoPol buffer on ice for 5 min.
Incorporation step
dU.V (3 µM) in 1× ThermoPol buffer (reaction volume: 20 µl) was added to the polymerase bound nucleic acid complex and incubated at 65°C for 5 min, and then placed on ice. dU.V-incorporated beads were washed three times with 50 μl W10 washing solution (10 mM Tris–HCl, pH 8.0; 10 mM Na2EDTA; 0.1% Triton X-100).
Photochemical cleavage step
The beads were resuspended in 20 µl cleavage solution (50 mM sodium azide), exposed to 365 nm UV light with an intensity of ∼0.7 W/cm2 for 4 min (i.e. 4× 1-min exposures interrupted with a 15 s mixing step to ensure good resuspension of the beads) using the customized UV deprotector, then washed three times with 50 μl 1× ThermoPol buffer.
The entire cycle was then repeated from the incorporation step. Final reactions were washed three times with 50 μl W10 washing solution, quenched with 10 μl of stop solution, heated to 50°C for 30 s and placed on ice. The extension products were analyzed on a 10% Long Ranger polyacrylamide gel using an AB model 377 DNA sequencer.
Nucleoside composition analysis after prolonged UV exposure
The primer/template duplex was quantitatively analyzed for potentially damaging effects from UV exposure, similar to those described in reference (23). Unlabeled primer-2 (10 nmole) was hybridized to 10 nmole of oligoTemplate-7 in 1× ThermoPol buffer (reaction volume: 35 μl) at 80°C for 30 s, 57°C for 30 s and then cooled to 4°C. Thirty-five µl of 100 mM sodium azide solution was then added to the duplex solution. Samples were exposed to 365 nm UV light with an intensity of ∼1 W/cm2 at 0, 30, 60, 90, 120 and 150 min time increments. The duplex was denatured by heating to 80°C for 30 s and then placed on ice. Following the addition of 1.5 μl of 1.2 M sodium acetate, pH 5.3, 4U of snake venom phosphodiesterase I were added directly to the duplex solution and incubated at 37°C for 16 h. Four units of alkaline phosphatase were then added and incubated at 37°C for 1 h. Digested samples were analyzed by reverse-phase high-performance liquid chromatography (RP-HPLC) using a 4.6 mm × 250 mm Spheri-5 RP-18 column. A linear gradient of 0% Buffer B to 13.34% Buffer B over 30 min was used to separate the nucleosides at a flow rate of 1.5 ml per min. Buffer A contained 20 mM ammonium acetate, and Buffer B contained 20 mM ammonium acetate, 40% acetonitrile (v/v).
RESULTS
Nucleotides synthesis
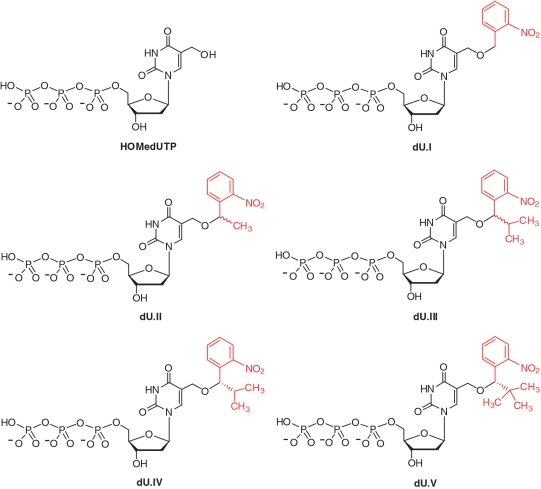
In this report, HOMedUTP, a parent 5-(2-nitrobenzyloxy)methyl-dUTP (dU.I), and four 5-(α-substituted-2-nitrobenzyloxy)methyl-dUTP analogs (dU.II, dU.III, dU.IV and dU.V) were synthesized and characterized (Figure 1). Our initial motivation in creating α-methylene carbon substituted dU.II and dU.III was based on the achievement of better photocleavage product yields (24,25). Upon photochemical cleavage, these α-substituted analogs produced less reactive ketone intermediates compared with that of a benzaldehyde by-product formed from a parent 2-nitrobenzyl group. Subsequent reports have also shown that α-methyl-2-nitrobenzyl ester (26) and α-methyl-2-nitrobenzyloxycarbonyl (27) linkages enhanced photochemical cleavage rates by 5-to-10 fold. Unexpectedly, we discovered that increasing the size from α-methyl to α-isopropyl improved the termination properties of these 2-nitrobenzyl alkylated HOMedUTP analogs (see below). We then synthesized the α-tert-butyl analog to further tune its termination properties. The S configuration of the α-substitutions for dU.IV and dU.V was chosen arbitrarily, based on previous reports (28,29). The design and synthesis of dU.I–dU.V, therefore, gave us the opportunity to explore photocleavage product yields and rates, and termination properties of the parent and α-substituted 2-nitrobenzyl ether linkages. To avoid confusion, the term ‘5-(2-nitrobenzyloxy)methyl-dUTP analogs’ is used to describe both the parent and α-substituted nucleotides shown in Figure 1.
Figure 1.
Structures of HOMedUTP and 5-(2-nitrobenzyloxy)methyl-dUTP analogs.
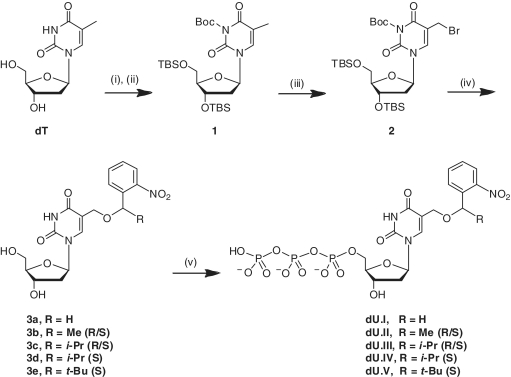
We envisioned that nucleophilic coupling of the allylic bromide 2 with 2-nitrobenzyl or α-substituted-2-nitrobenzyl alcohols would provide the desired nucleoside analogs 3a–3e (Scheme 1). Intermediate 2 was obtained via bromination of the fully protected thymidine 1, according to Anderson et al. (30), although we found that N3 protection increased the stability of the bromomethyl product (data not shown). 2-Nitrobenzyl alkylated HOMedU products 3a–3e were obtained when 2 was heated neat with their corresponding 2-nitrobenzyl alcohols. 2-Nitrobenzyl alcohol is commercially available, and the synthesis of the racemic α-methyl-2-nitrobenzyl alcohol has been reported (31). Racemic α-isopropyl- and α-tert-butyl-2-nitrobenzyl alcohols were synthesized using a Grignard reaction with 2-nitrophenyl-magnesium chloride (generated in situ from 1-iodo-2-nitrobenzene and phenylmagnesium chloride) and isobutyraldehyde or trimethylacetaldehyde, respectively (Scheme 2).
Scheme 1.
Synthesis of 5-(2-nitrobenzyloxy)methyl-dUTP analogs. Reagents and conditions: (i) TBSCl, imidazole, anhydrous DMF, room temperature, overnight, 95%; (ii) Boc2O, DMAP, anhydrous DMF, room temperature, 16 h, 78%; (iii) N-bromosuccinimide, benzoyl peroxide, CCl4, reflux, 1 h, 43%; (iv) 2-nitrobenzyl or α-substituted-2-nitrobenzyl alcohols, 100–115°C, 15–60 min, (5–22% for 3a-3e); (v) POCl3, proton sponge, (MeO)3PO, 0°C, 2 h; (n-Bu3NH)2H2P2O7, n-Bu3N, DMF, 30 min; 1 M HNEt3HCO3, 1 h.
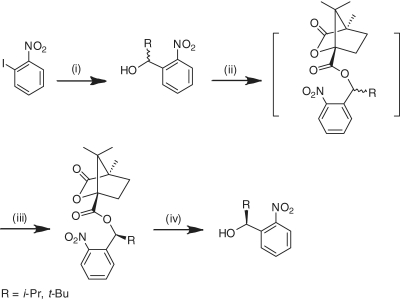
Scheme 2.
Synthesis of (S) α-substituted 2-nitrobenzyl alcohol. Reagents and conditions: (i) PhMgCl, anhydrous THF minus 40°C; RCHO, minus 40°C to room temperature, 99% (R = i–Pr), 72% (R = t–Bu); (ii) (1S)-(-)-camphanic acid chloride, anhydrous pyridine, room temperature; (iii) fractional crystallization from methanol, 36% (R = i–Pr), 36% (R = t–Bu); (iv) K2CO3, MeOH, reflux, 97% (R = i–Pr), 92% (R = t–Bu).
The 1H NMR spectra of compounds 3b and 3c showed a 50:50 mixture of both diastereomers resulting from the chirality of α-substitution. Because the mixture could not be separated by standard chromatographic means, we set out to synthesize single diastereomeric 3d and 3e analogs via neat coupling of 2 with enantio-pure (S)-α-isopropyl- and (S)-α-tert-butyl-2-nitrobenzyl alcohols, respectively. (S)-α-Isopropyl- and (S)-α-tert-butyl-2-nitrobenzyl alcohols were resolved by fractional crystallization of their respective diastereomeric (1S)-camphanates, according to Corrie et al. (32). The absolute stereochemistry of the camphanates was determined by X-ray crystallography (Supplementary Figures S1 and S2). The enantio-pure (S)-α-isopropyl- and (S)-α-tert-butyl-2-nitrobenzyl alcohols were recovered by saponification of the diastereomeric (1S)-camphanates with potassium carbonate in methanol in near quantitative yields (Scheme 2).
Triphosphate syntheses were performed using the ‘one-pot’ procedure, as described by Ludwig (33), to yield dU.I–dU.V. HOMedUTP was obtained by photochemical cleavage of dU.II with 365 nm UV light. All triphosphates were purified by Q sepharose FF anion-exchange chromatography followed by RP-HPLC.
5-(2-nitrobenzyloxy)methyl-dUTP analogs are active with Family B DNA polymerases
Initially, dU.I was synthesized and tested for base-specific incorporation by eight different DNA polymerases using the PEP assay described in reference (2). TTP, HOMedUTP, and 3′-dTTP were also examined (Supplementary Table S1), providing a benchmark for determining the incorporation bias (i.e. the modified nucleotide IC50 value divided by the natural nucleotide IC50 value) for the 5-(2-nitrobenzyloxy)methyl-dUTP analogs. As shown, dU.I was incorporated by Bst and Klenow(exo−) polymerases showing incorporation biases of 45 and 30, respectively, compared with HOMedUTP (Table 1). Taq and TaqFS polymerases poorly incorporated dU.I, extending the primer <50% before revealing inhibitory effects. In contrast to these Family A polymerases, all Family B polymerases examined, that is Therminator, Therminator II, Vent(exo−) and DeepVent(exo−), incorporated dU.I efficiently revealing only slight incorporation biases that ranged from 0.8 to 1.8. These data provide good evidence that Family B polymerases incorporate dU.I similarly to that of its natural nucleotide counterpart.
Table 1.
PEP assay for compounds dU.I–dU.III
| Polymerase | IC50 values |
Incorporation bias |
||||
|---|---|---|---|---|---|---|
| dU.I | dU.II | dU. III | dU.I/HOMedUTP | dU.II/HOMedUTP | dU.III/HOMedUTP | |
| Bst | 98 ± 13 nM | 3.5 ± 0.2 μM | Not Activea | 45 | 1600 | ND |
| Klenow(exo−) | 48 ± 4 nM | 2.4 ± 0.1 μM | Not Activea | 30 | 1500 | ND |
| Taq | 50 μMb | Not Activea | Not Activea | ND | ND | ND |
| TaqFS | 25 μMc | Not Activea | Not Activea | ND | ND | ND |
| Therminator | 1.8 ± 0.1 nM | 1.7 ± 0.2 nM | 2.3 ± 0.1 nM | 1.1 | 1.0 | 1.4 |
| Therminator II | 5.7 ± 0.4 nM | 14 ± 1 nM | 0.14 ± 0.01 μM | 0.8 | 1.9 | 20 |
| Vent(exo−) | 2.0 ± 0.1 nM | 2.2 ± 0.2 nM | 5.9 ± 0.7 nM | 1.4 | 1.6 | 4.2 |
| DeepVent(exo−) | 4.9 ± 1.0 nM | 17 ± 1 nM | 0.22 ± 0.02 μM | 1.8 | 6.3 | 80 |
Values in italics represent µM concentrations to contrast those IC50 values in the nM range.
aNo visible +1 band observed in 377 gel image at 100 μM.
bConcentration that yielded highest percent incorporation: 45 ± 1%.
cConcentration that yielded highest percent incorporation: 35 ± 4%;
ND: Not determined.
dU.II and dU.III were then examined against the battery of DNA polymerases. The IC50 values for dU.II markedly increased for Bst and Klenow(exo−) polymerases, and Taq and TaqFS polymerases showed no activity at all (Table 1). For dU.III, none of the Family A polymerases showed any activity up to a final concentration of 100 μM. In contrast, the Family B polymerases incorporated dU.II efficiently with IC50 values being slightly higher over those for dU.I (i.e., incorporation biases ranged from 1.0 to 6.3). dU.III was also incorporated by all Family B polymerases with Therminator and Vent(exo−) polymerases showing the least bias of incorporation of 1.4 and 4.2, respectively. Based on these findings, 5-(2-nitrobenzyloxy)methyl-dUTP analogs were further characterized with Therminator and Vent(exo−) polymerases.
Size of the α-carbon group influences termination of 5-(2-nitrobenzyloxy)methyl-dUTP analogs
We developed the weighted-sum assay to evaluate quantitatively the termination property of nucleotide analogs being extended along a homopolymer stretch of complementary template bases (oligoTemplate-7). A weighted-sum value of 1.0 indicates the addition of a single-nucleotide analog for a given concentration. IC50 values for dU.I–dU.V were determined using oligoTemplate-7, which in some cases varied from those calculated for oligoTemplate-4 (Supplementary Table S2). dU.I gave weighted-sum values of 3.7 and 3.0 for Therminator and Vent(exo−) polymerases, respectively, at a concentration of 25× their IC50 values (Table 2). Unexpectedly, the (R/S) α-methyl (dU.II) and (R/S) α-isopropyl (dU.III) nucleotide analogs’ weighted-sum values at the same concentration decreased to 1.7 and 1.0 for Therminator polymerase and to 1.2 and 1.0 for Vent(exo−) polymerase, respectively. The (S) α-isopropyl (dU.IV) and (S) α-tert-butyl (dU.V) nucleotide analogs displayed weighted-sum values of 1.0 at 25× their IC50 values. Nucleotide analogs, however, are typically utilized at higher concentrations in CRT experiments (see below). Increasing the dU.IV concentration to 3 μM, which is ∼1000× its IC50 value, increased the weighted-sum values to 1.9 and 1.7 for Therminator and Vent(exo−) polymerases, respectively. The bulkier α-tert-butyl group of dU.V, however, maintained weighted-sum values of 1.0 for both polymerases (Table 2). These data highlight the role of the size of the α-substituent in ‘tuning’ the termination properties of 3′-OH unblocked 5-(2-nitrobenzyloxy)methyl-dUTP analogs.
Table 2.
Weighted-sum values for dU.I–dU.V
| Nucleotide analog | Therminator |
Vent(exo−) |
||
|---|---|---|---|---|
| 25 × IC50 | 3 μM | 25 × IC50 | 3 μM | |
| dU.I | 3.7 ± 0.0 | ND | 3.0 ± 0.1 | ND |
| dU.II | 1.7 ± 0.1 | ND | 1.2 ± 0.0 | ND |
| dU.III | 1.0 ± 0.0 | ND | 1.0 ± 0.0 | ND |
| dU.IV | 1.0 ± 0.0 | 1.9 ± 0.2 | 1.0 ± 0.0 | 1.7 ± 0.1 |
| dU.V | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.0 | 1.0 ± 0.0 |
ND: Not determined.
Size of the α-carbon group improves selectivity of 5-(2-nitrobenzyloxy)methyl-dUTP analogs
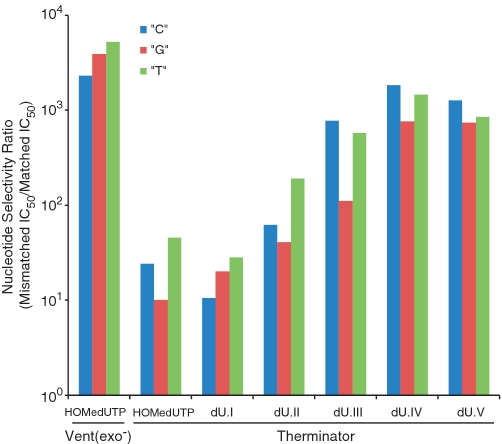
We next examined the nucleotide selectivity of incorporation using the PEP discrimination assay (2). Vent(exo−) polymerase revealed a selectivity ratio (i.e. the mismatched IC50 value divided by the matched IC50 value) for TTP (Supplementary Table S3) and HOMedUTP (Figure 2 and Supplementary Table S3) greater than three orders of magnitude for each mismatch combination. dU.I, dU.II or dU.III incorporated poorly against pyrimidine template bases, extending the primer to ∼50% or less before revealing inhibitory effects (Supplementary Table S4). The distinction here is that unlike natural nucleotides that completely extend the primer by misincorporation at micromolar concentrations, Vent(exo−) polymerase is more selective towards 5-(2-nitrobenzyloxy)methyl-dUTP analogs as it does not efficiently extend the primer up to a concentration of 100 μM. Selectivity ratios of 900 and 4900 were determined for dU.I and dU.II, respectively, against a ‘G’ template base, with dU.III poorly extending the primer before revealing inhibitory effects. These results provide good evidence that 5-(2-nitrobenzyloxy)methyl-dUTP analogs are base-specific nucleotides using Vent(exo−) polymerase and, with exception of the dU.I—G mismatch, showed higher selectivity against mismatch incorporation than natural nucleotides.
Figure 2.
Histogram plot of nucleotide selectivity ratios for HOMedUTP [Vent(exo−) and Therminator polymerases] and dU.I–dU.V (Therminator polymerase). Nucleotide selectivity ratios (i.e. the mismatched IC50 value divided by the matched IC50 value) were calculated based on PEP discrimination assay results, see Supplementary Tables 3–6.
In contrast, selectivity ratios for Therminator polymerase were approximately two orders of magnitude lower for TTP (Supplementary Table S5) and HOMedUTP (Figure 2 and Supplementary Table S5) compared with Vent(exo−) polymerase. Lower selectivity ratios are expected to increase error rates. These data are consistent with a study by Ichida et al. (34) who reported an error rate ∼100-fold higher for Therminator polymerase compared with Taq polymerase; both Vent(exo−) and Taq polymerases have similar error rates (35). Comparable selectivity results were obtained for dU.I using Therminator polymerase. The selectivity ratios for dU.II and dU.III, however, increased with Therminator polymerase in a size-dependent manner based on the α-substituent group (Figure 2 and Supplementary Table S6). The improvement results from the marked increase in mismatch IC50 values, while those match values remain relatively constant. The single diastereomer dU.IV gave even higher selectivity ratios when compared with its mixed diastereomeric counterpart dU.III. Both dU.IV and dU.V showed similar selectivity results. Overall, selectivity ratios for dU.IV and dU.V were at least 30-fold higher than the parent dU.I analog and approached selectivity ratios of almost three orders of magnitude for Therminator polymerase.
5-(2-nitrobenzyloxy)methyl-dUTP analogs are cleaved quickly and efficiently
The UV light spectrum can be divided into UVC (100–280 nm), UVB (280–315 nm) and UVA (315–400 nm) regions. Both UVB and UVC light have been reported to cause pyrimidine dimers (36), whereas UVA does not appear to cause these dimers in any significant amount (37). UVA light can generate hydroxide radicals (•OH) among other reactive oxygen species, which can damage DNA by oxidative base modifications [i.e. 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxo-dG)] (38) and single-strand breaks (39). Powerful oxygen radical scavengers, such as acetate, azide, EDTA, formate and mannitol, have been used effectively in neutralizing these damaging effects (40–42). Here, we compared a 50 mM sodium azide solution with our previously described cleavage FBD solution (see ‘Materials and Methods’ section). DT50 values for dU.I, dU.II, and dU.III were approximately three-fold lower (i.e., faster) in sodium azide compared with the FBD solution (Table 3). dU.II and dU.III were 33-60% faster in the azide solution compared with the parent dU.I analog, although no difference was observed in the FBD solution. Single diastereomeric dU.IV and dU.V showed the fastest DT50 values in the azide solution of 1.8 s and 1.3 s, respectively. All 5-(2-nitrobenzyloxy)methyl-dUTP analogs were photochemically cleaved to 100% efficiency within 60 s at 365 nm UV light exposure with an intensity of ∼0.7 W/cm2 in azide solution (Supplementary Figure S3). These results prompted us to test the UV protective effects of azide solution in CRT sequencing.
Table 3.
Cleavage results for dU.I–dU.V
| Nucleotide analog | DT50 values (s) |
|
|---|---|---|
| FBD solution | 50 mM NaN3 | |
| dU.I | 8.7 ± 0.9 | 3.2 ± 0.1 |
| dU.II | 7.4 ± 1.2 | 2.0 ± 0.1 |
| dU.III | 8.8 ± 0.8 | 2.4 ± 0.2 |
| dU.IV | ND | 1.8 ± 0.1 |
| dU.V | ND | 1.3 ± 0.1 |
Multiple incorporations of HOMedUTP in PCR amplified and synthetic templates
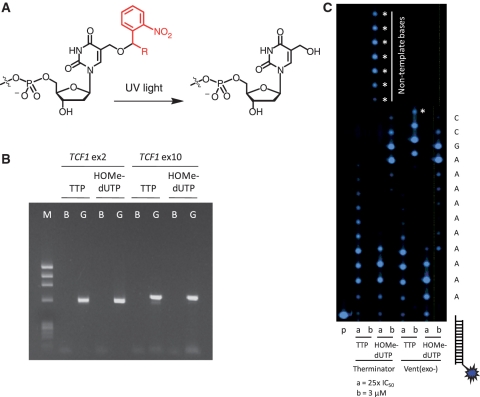
Following incorporation, the photochemical cleavage of 5-(2-nitrobenzyloxy)methyl-dUTP analogs produces a HOMedU monophosphate in the primer strand (Figure 3A), and with subsequent CRT cycles, the accumulation of HOMedU monophosphate residues. We investigated the effects on the polymerase incorporating HOMedUTP using PCR and primer extension experiments. We only conducted PCR experiments with Vent(exo−) polymerase (Figure 3B) because of the low selectivity of Therminator polymerase with TTP or HOMedUTP. Comparable yields and product size were obtained from exons 2 and 10 of the TCF1 gene (20), amplified using standard PCR conditions in the presence of dATP, dCTP, dGTP and either TTP or HOMedUTP. Automated Sanger sequencing data were indistinguishable for PCR products containing either HOMedUTP or TTP, verifying the integrity of the HOMedUTP amplicons (Supplementary Figure S4). These data show that not only can multiple HOMedUTP be incorporated into the primer strand, but HOMedU can act as a complementary templating base in subsequent PCR cycles and in automated Sanger sequencing.
Figure 3.
Polymerase extension assays with HOMedUTP. (A) Chemical scheme showing UV cleavage of a 5-(α-substituted-2-nitrobenzyloxy)methyl-2′-deoxyuridine monophosphate into its naturally occurring HOMedU counterpart. (B) PCR results comparing TTP with HOMedUTP in reactions containing equimolar concentrations of dATP, dCTP and dGTP using Vent(exo−) polymerase. Reactions were initiated with either HPLC water (blank) or 50 ng human genomic DNA (gDNA) as described in the ‘Materials and Methods’ section. The expected PCR size for TCF1 exon 2 is 602 bp with the region of interest flanking the primers being 564 bp, of which 103 thymine bases (18.3% T) were present, and the expected PCR size for TCF1 exon 10 is 670 bp with the region of interest flanking the primers being 633 bp, of which 129 thymine bases (20.4% T) were present. (C) Linear extension on a synthetic poly(dA)10 template hybridized with BODIPY-FL-labeled (blue star) primer-1S. OligoTemplate-7 served as the template for either TTP or HOMedUTP incorporation at 25× their IC50 values (lane ‘a’) and 3 μM (lane ‘b’) using Vent(exo−) or Therminator polymerase. The addition of non-template directed nucleotides are marked by asterisk.
At 25× IC50 values, Therminator polymerase extended both TTP and HOMedUTP farther (weighted-sum values of 4.0 and 2.6, respectively) compared with Vent(exo−) polymerase (weighted-sum values of 3.3 and 1.9, respectively; Figure 3C). These data also suggest that TTP is extended more efficiently than HOMedUTP using either polymerase. At a concentration of 3 μM, Therminator and Vent(exo−) polymerases efficiently extended both nucleotides along oligoTemplate-7, upon which TTP, but not HOMedUTP, extended farther in a non-template directed manner, see ‘*’ in Figure 3C. Collectively, these data provide evidence that HOMedUTP can be incorporated into a growing primer strand of at least 600 bp in length and can be extended consecutively through a homopolymer stretch of at least ten complementary template bases.
Ten-base CRT sequencing with dU.V
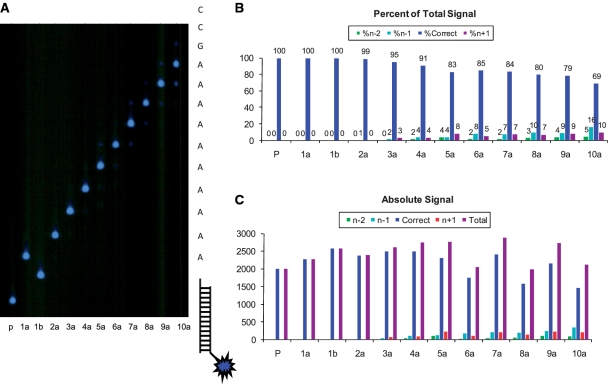
Based on the incorporation, termination, nucleotide selectivity, and photochemical cleavage experiments described above, we tested dU.V as a reversible terminator in CRT sequencing using Therminator polymerase. A ten-base experiment was performed using a biotinylated template containing a poly(dA) stretch (oligoTemplate-7) attached to streptavidin-coated magnetic beads. Therminator polymerase was bound to the primer/template complex and underwent multiple cycles of incorporation (5 min) with 3 μM dU.V in 1× ThermoPol buffer and UV cleavage (4 min) in 50 mM sodium azide (Figure 4A). The gel image in Figure 4A was analyzed further by quantifying the fluorescent bands at different CRT cycles (Figure 4B). During the first cycle, the product of incorporation efficiency (1a: 100%) and cleavage efficiency (1b: 100%) resulted in an initial cycle efficiency (Ceff) of 100% on a solid support (18). The signal-to-noise ratio decreased slightly in subsequent cycles because of lagging and leading dephasing signals (1). The overall signal of the reaction products remained relatively constant over the 10 CRT cycles, indicating that the primer/template or Therminator polymerase did not undergo any significant damage due to UV exposure (Figure 4C). The bound Therminator undergoing multiple cycles of incorporation, washing, UV exposure and washing again showed good activity in all cycles. To further examine the primer/template complex for UV damage, the duplex in azide solution was exposed to 365 nm UV light, with an intensity of ∼1 W/cm2, up to 150 min and then characterized by RP-HPLC analysis. Quantitative analysis of the nucleoside composition revealed no significant alteration of the primer/template duplex after prolonged UV exposure (Supplementary Figure S5). This represents a significant improvement over our previous work with the N6-(2-nitrobenzyl)-dATP analog (2), which showed a 50% signal drop by the fifth cycle. We attribute this finding to the α-substituted 2-nitrobenzyl group forming the less reactive ketone intermediate and the addition of the azide radical scavenger, thereby, minimizing adverse conditions in the sequencing reaction (24,25).
Figure 4.
Ten-base CRT sequencing with dU.V. (A) 377 DNA sequencer gel image of a 10-cycle CRT experiment. Therminator polymerase was bound to primer/template complex and subjected to multiple cycles of incorporation (5 min) with 3 μM dU.V in 1× ThermoPol buffer, plus UV cleavage (4 min) in 50 mM sodium azide. P, dye-labeled primer (blue star represents BODIPY-FL label); 1a, first incorporation; 1b, first cleavage; 2a–10a, subsequent cleavage and incorporation cycles on dye-labeled primer. (B) Histogram plot of quantified gel bands in (A). Cycle efficiency was determined as a product of incorporation efficiency (1a: 100%) and cleavage efficiency (1b: 100%). Dephasing signals were also quantified as ‘%n–2’ (incomplete incorporation for the current cycle), ‘%n–1’ (incomplete UV cleavage from the previous cycle, extension of the ‘%n–2’ from the previous cycle and/or 3′-exonuclease activity followed by incorporation in the current cycle) and ‘%n+1’ [natural (photochemically cleaved) nucleotide carryover from the previous cycle]. (C) Histogram plot including the total signal for each cycle, representing the sum of the correct signal plus all dephasing signals.
DISCUSSION
Natural bases are not limited to adenine, cytosine, guanine and thymine, as there are numerous examples of biological species that utilize hypermodified bases as part of the normal life cycle. HOMedU is one such example found in the genomes of numerous organisms, acting both as an incorporating nucleotide and a complementary templating base. Here, we describe a novel 3′-OH unblocked reversible terminator, based on a core HOMedUTP, which exhibits excellent enzymatic properties of incorporation, single-base termination, nucleotide selectivity, and efficient photochemical cleavage. The hydroxymethyl group at the 5-position of HOMedU acts as a molecular handle to which a photocleavable, terminating group can be attached. This contrasts our previous work describing N-alkylation of dATP with a parent 2-nitrobenzyl group, as the extrapolation of this example to thymidine would lead to N3-(2-nitrobenzyl)-TTP (2). We predicted that an N3-alkylated thymidine analog would interfere with Watson–Crick base pairing, adversely affecting nucleotide selectivity. We have confirmed this hypothesis by synthesizing and characterizing N3-(2-nitrobenzyl)-TTP (T.I). Of the eight DNA polymerases tested, only Therminator polymerase incorporated T.I, albeit indiscriminately against all mismatched template bases (Supplementary Table S7).
The use of HOMedU as a core nucleoside fits within our overall strategy of creating reversible terminator reagents that transform back into a natural state (2), representing an important distinction between our nucleotide reagents and those described by others (7,10,43). In addition to our work, Ju and colleagues have reported the synthesis of 3′-OH unblocked nucleotides using a photocleavable 2-nitrobenzyl group (9). There are notable differences in the overall structure and biological utility of these nucleotide analogs compared with those described in this report. For example, to the 5-position of 2′-deoxyuridine is attached an 3-amino-1-propargyl (AP3) linker (4) to which an α-methyl-2-nitrobenzyloxycarbonyl linker with a fluorescent dye is joined. The authors showed that single nucleotides could be incorporated using ThermoSequenase™ (44), a Taq polymerase containing the same F667Y amino acid variant (45) as TaqFS polymerase, using the non-repetitive template sequence 5′-TCTGATATCAGT. A single nucleotide addition strategy (18) was used to demonstrate DNA sequencing feasibility, whereby, the polymerase is paused only until addition of the next complementary nucleotide. In their companion paper, the authors stated that the 3′-OH requires blocking with a chemical moiety, such as an 3′-O-allyl group, to effect the termination property of these nucleotides (10). We note that 3′-O-allyl-dATP is not efficiently incorporated by Vent(exo−) polymerase (46). Ju and colleagues have reported, however, that their dye-labeled 3′-O-allyl-dNTPs were incorporated with Therminator II polymerase, which contains the Y409V amino acid variant in addition to the A485L variant found in the Therminator polymerase. The 409 amino acid residue of Therminator acts as a steric gate for incorporation of ribonucleotides (NTPs) (47–49). In contrast, the 3′-OH unblocked 5-[(S)-α-tert-butyl-2-nitrobenzyloxy]methyl-dUTP dU.V is an efficient terminator with both Vent(exo−) and Therminator polymerases, but is not incorporated with TaqFS polymerase. Proximity of the 2-nitrobenzyl group to the nucleobase and size of the alkyl group attached to its α-methylene carbon are important structural features that confer the unique properties of termination and selectivity upon 3′-OH unblocked 5-(2-nitrobenzyloxy)methyl-dUTP analogs.
Vent(exo−) and Therminator are highly homologous polymerases, showing ∼77% identity and ∼89% similarity by protein sequence alignment. It is, therefore, not surprising that both perform with similar efficiencies in incorporation and termination assays. Our nucleotide selectivity assays, however, revealed significantly lower ratios for TTP and HOMedUTP using Therminator polymerase compared with Vent(exo−) polymerase. Therminator polymerase contains the A485L variant, which is analogous to the modified Vent(exo−) A488L polymerase. Gardner and Jack (49) suggested that the modified Vent(exo−) A488L polymerase is more tolerant of base-modified nucleotides, but with the trade-off of lower nucleotide selectivity. Here, Therminator and wild-type Vent(exo−) polymerases incorporated 5-(2-nitrobenzyloxy)methyl-dUTP analogs with similar efficiencies, providing evidence that the respective L485 and A488 residues do not play a major role in their incorporation. In the selection process for the correct nucleotide, however, these residues appear to play a major role as increasing the size of the α-carbon substituent of the 2-nitrobenzyl group improved nucleotide selectivity. In a broader sense, the use of variant polymerases in NGS technologies (7,43) potentially raises concern that their use contributes to higher error rates observed in sequencing read data. To our knowledge, this is the first example of base-modified nucleotides improving the nucleotide selectivity of DNA polymerases over that of natural nucleotides. This point is exemplified by comparing the performance of Therminator polymerase, which uncontrollably extended TTP along the entire sequence of oligoTemplate-7, misincorporating the last three template bases and adding nucleotides in a non-template directed manner (Figure 3C) compared with the efficient step-wise addition of dU.V along the same template sequence (Figure 4A). Our expectation is that use of 2-nitrobenzyl alkylated hydroxymethyl-dNTP analogs in NGS technologies should yield more accurate read data.
The molecular basis for the improved nucleotide selectivity of the 5-(α-substituted-2-nitrobenzyloxy)methyl-dUTP analogs (dU.II, dU.III, dU.IV and dU.V) with Vent(exo−) and Therminator polymerases is unknown. As a whole, DNA polymerases play different cellular roles, range in replication fidelities, and exhibit different activities to modified substrates. Despite this diversity, they have the extraordinary property of nucleotide selectivity that cannot be explained simply by free energy differences between matched and mismatched Watson–Crick base pairs (50–52). Crystal structure studies of several unrelated DNA polymerases have revealed a common conformational change from the open to closed state, resulting in the assembly of the catalytic complex (53). For high-fidelity polymerases, the resulting ‘tight fit’ around the correctly formed base pair within the active site may preclude mismatched base pairs due to steric effects (53–56). Low fidelity polymerases, such as those involved in rapid production of highly related genomes (57) or in repair and lesion bypass processes (58) may have a looser fit to accommodate the larger geometric shapes of mismatched or damaged base-pairs. Three-dimensional modeling of site-directed mutants of human immunodeficiency virus type 1 reverse transcriptase have led authors to propose that nucleotide selectivity may be a function of active site flexibility, with a more rigid catalytic pocket providing higher DNA synthesis fidelity (59). Neither model is mutually exclusive, as both the size and rigidity of the active site may play a role in nucleotide selectivity (60). Based on the crystal structure of the RB69 polymerase, the A488L variant in Vent(exo−), analogous to the A485L variant in Therminator, is located in the α-helix of the fingers domain, known to play a role in binding of the incoming nucleotide. The 488/485 amino acid sites, however, are predicted to face away from the active site, suggesting no direct interaction within the nucleotide binding pocket (49). Even if the amino acid residue did directly contact the correctly formed base pair, it is counterintuitive that a larger leucine residue would increase the size of the active site. These observations suggest that nucleotide behavior with the L485 variant of Therminator polymerase favors the flexible active site hypothesis, resulting in lower nucleotide selectivity of natural nucleotides. In this model, increasing the size of the alpha substituted group of 2-nitrobenzyl-dUTP analogs would stretch the limits of the catalytic pocket, resulting in a tighter geometric fit for correctly paired bases and, therefore, higher nucleotide selectivity against mismatch incorporation. Our data, however, do not completely rule-out the role of active site size, as improved nucleotide selectivity of the 5-(α-substituted-2-nitrobenzyloxy)methyl-dUTP analogs was also observed with Vent(exo−) polymerase. Ongoing kinetic studies are underway to shed more light on the relative roles of active site size and flexibility.
We have presented in vitro evidence that HOMedUTP can substitute for TTP in PCR without compromise of product yield, size, or integrity using Vent(exo−) polymerase (Figure 3 and Supplementary Figure S4). We note that a PCR assay is a more stringent test in demonstrating that HOMedU nucleotides do not alter normal polymerase action in extending long stretches of nucleic acids. This is due to the HOMedU nucleotide serving the additional role of complementary templating base during subsequent PCR cycles. Demands for the CRT method require only the accumulation of hydroxymethylated nucleotides into the growing primer strand as the template strand is not regenerated. Work is ongoing to demonstrate by in vitro assays that read-lengths are not adversely affected by the accumulation of all four hydroxymethyl nucleoside monophosphates into the growing primer strand. We argue that this demonstration will be important in modeling longer read-lengths for use in NGS technologies. To this end, we have now developed a complete set of reversible terminators based on the core nucleoside triphosphates of 7-deaza-7-hydroxymethyl-2′-deoxyadenosine, 5-hydroxymethyl-2′-deoxycytidine, 7-deaza-7-hydroxymethyl-2′-deoxyguanosine, and 5-hydroxymethyl-2′-deoxyuridine (61,62), the results of which are being prepared for publication. The dye-labeled version of these 2-nitrobenzyl alkylated nucleotides are called Lightning Terminators™, structures of which include the recently reported 5-(α-isopropyl-2-nitrobenzyloxy)methyl-dUTP derivative (1).
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
National Institutes of Health (R01 HG003573). Funding for open access charge: Direct charges to Baylor College of Medicine interim funding grant.
Conflict of interest statement. We declare that LaserGen plans on commercializing these compounds, along with their derivatives. No other conflicts have been declared.
ACKNOWLEDGEMENTS
We authors thank Sherry Metzker and David Hertzog from LaserGen; Joseph Reibenspies from Texas A&M University for critical reading of the article; Andy Gardner for helpful discussions; and Diane Scaduto for performing automated Sanger sequencing experiments.
REFERENCES
- 1.Metzker ML. Sequencing technologies — the next generation. Nature Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Wu W, Stupi BP, Litosh VA, Mansouri D, Farley D, Morris S, Metzker S, Metzker ML. Termination of DNA synthesis by N6-alkylated, not 3'-O-alkylated, photocleavable 2'-deoxyadenosine triphosphates. Nucleic Acid Res. 2007;35:6339–6349. doi: 10.1093/nar/gkm689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langer PR, Waldrop AA, Ward DC. Enzymatic synthesis of biotin-labeled polynucleotides: novel nucleic acid affinity probes. Proc. Natl Acad. Sci. USA. 1981;78:6633–6637. doi: 10.1073/pnas.78.11.6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prober JM, Trainor GL, Dam RJ, Hobbs FW, Robertson CW, Zagursky RJ, Cocuzza AJ, Jensen MA, Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987;238:336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- 5.Lee LG, Connell CR, Woo SL, Cheng RD, McArdle BF, Fuller CW, Halloran ND, Wilson RK. DNA sequencing with dye-labeled terminators and T7 DNA polymerase: effect of dyes and dNTPs on incorporation of dye-terminators and probability analysis of termination fragments. Nucleic Acids Res. 1992;20:2471–2483. doi: 10.1093/nar/20.10.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitra RD, Shendure J, Olejnik J, Edyta-Krzymanska-Olejnik, Church GM. Fluorescent in situ sequencing on polymerase colonies. Anal. Biochem. 2003;320:55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 7.Ju J, Kim DH, Bi L, Meng Q, Bai X, Li Z, Li X, Marma MS, Shi S, Wu J, et al. Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators. Proc. Natl Acad. Sci. USA. 2006;103:19635–19640. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turcatti G, Romieu A, Fedurco M, Tairi A-P. A new class of cleavable fluorescent nucleotides: synthesis and optimization as reversible terminators for DNA sequencing by synthesis. Nucleic Acids Res. 2008;36:e25. doi: 10.1093/nar/gkn021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seo TS, Bai X, Kim DH, Meng Q, Shi S, Ruparel H, Li Z, Turro NJ, Ju J. Four-color DNA sequencing by synthesis on a chip using photocleavable fluorescent nucleotides. Proc. Natl Acad. Sci. USA. 2005;102:5926–5931. doi: 10.1073/pnas.0501965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruparel H, Bi L, Li Z, Bai X, Kim DH, Turro NJ, Ju J. Design and synthesis of a 3'-O-allyl photocleavable fluorescent nucleotide as a reversible terminator for DNA sequencing by synthesis. Proc. Natl Acad. Sci. USA. 2005;102:5932–5937. doi: 10.1073/pnas.0501962102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemphill HE, Whiteley HR. Bacteriophages of Bacillus subtilis. Microbiol. Mol. Biol. Rev. 1975;39:257–315. doi: 10.1128/br.39.3.257-315.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Warren RAJ. Modified bases in bacteriophage DNAs. Ann. Rev. Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- 13.Rae PMM. Hydroxymethyluracil in eukaryote DNA: a natural feature of the Pyrrophyta (Dinoflagellates) Science. 1976;194:1062–1064. doi: 10.1126/science.988637. [DOI] [PubMed] [Google Scholar]
- 14.Kropinski AMB, Bose RJ, Warren RAJ. 5-(4-Aminobutylaminomethyl)uracil, an unusual pyrimidine from the deoxyribonucleic acid of bacteriophage ϕW-14. Biochemistry. 1973;12:151–157. doi: 10.1021/bi00725a025. [DOI] [PubMed] [Google Scholar]
- 15.Gommers-Ampt JH, Leeuwen FV, deBeer ALJ, Vliegenthart JFG, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. β-D-glucosyl-hydroxymethyluracil: a novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- 16.Gommers-Ampt JH, Teixeira AJR, van de Werken G, van Dijk WJ, Borst P. The identification of hydroxymethyluracil in DNA of Trypanosoma brucei. Nucleic Acids Res. 1993;21:2039–2043. doi: 10.1093/nar/21.9.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gommers-Ampt JH, Borst P. Hypermodified bases in DNA. FASEB J. 1995;9:1034–1042. doi: 10.1096/fasebj.9.11.7649402. [DOI] [PubMed] [Google Scholar]
- 18.Metzker ML. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. doi: 10.1101/gr.3770505. [DOI] [PubMed] [Google Scholar]
- 19.Metzker ML, Lu J, Gibbs RA. Electrophoretically uniform fluorescent dyes for automated DNA sequencing. Science. 1996;271:1420–1422. doi: 10.1126/science.271.5254.1420. [DOI] [PubMed] [Google Scholar]
- 20.Haaland WC, Scaduto DI, Maldonado MR, Mansouri DL, Nalini R, Iyer D, Patel S, Guthikonda A, Hampe CS, Balasubramanyam A, et al. The "A-β-" subtype of Ketosis-Prone Diabetes (KPD) is not predominantly a monogenic diabetic syndrome. Diabetes Care. 2009;32:873–877. doi: 10.2337/dc08-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baskaran N, Kandpal RP, Bhargava AK, Glynn MW, Bale A, Weissman SM. Uniform amplification of a mixture of deoxyribonucleic acids with varying GC content. Genome Res. 1996;6:633–638. doi: 10.1101/gr.6.7.633. [DOI] [PubMed] [Google Scholar]
- 22.Henke W, Herdel K, Jung K, Schnorr D, Loening SA. Betaine improves the PCR amplification of GC-rich DNA sequences. Nucleic Acids Res. 1997;25:3957–3958. doi: 10.1093/nar/25.19.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crean C, Uvaydov Y, Geacintov NE, Shafirovich V. Oxidation of single-stranded oligonucleotides by carbonate radical anions: generating intrastrand cross-links between guanine and thymine bases separated by cytosines. Nucleic Acids Res. 2008;36:742–755. doi: 10.1093/nar/gkm1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patchornik A, Amit B, Woodward RB. Photosensitive protecting groups. J. Am. Chem. Soc. 1970;92:6333–6335. [Google Scholar]
- 25.Kaplan JH, Forbush B, Hoffman JF. Rapid photolytic release of adenosine 5'-triphosphate from a protected analog: utilization by the sodium:potassium pump of human red blood cell ghosts. Biochemistry. 1978;17:1929–1935. doi: 10.1021/bi00603a020. [DOI] [PubMed] [Google Scholar]
- 26.Reichmanis E, Smith BC, Gooden R. o-Nitrobenzyl photochemistry: solution vs. solid-state behavior. J. Polymer Sci. 1985;23:1–8. [Google Scholar]
- 27.Hasan A, Stengele K-P, Giegrich H, Cornwell P, Isham KR, Sachleben RA, Pfleiderer W, Foote RS. Photolabile protecting groups for nucleosides: synthesis and photodeprotection rates. Tetrahedron. 1997;53:4247–4264. [Google Scholar]
- 28.Wenter P, Fürtig B, Hainard A, Schwalbe H, Pitsch S. Kinetics of photoinduced RNA refolding by real-time NMR spectroscopy. Angew. Chem. Int. Ed. 2005;44:2600–2603. doi: 10.1002/anie.200462724. [DOI] [PubMed] [Google Scholar]
- 29.Höbartner C, Silverman SK. Modulation of RNA tertiary folding by incorporation of caged nucleotides. Angew. Chem. Int. Ed. 2005;44:7305–7309. doi: 10.1002/anie.200502928. [DOI] [PubMed] [Google Scholar]
- 30.Anderson AS, Hwang J-T, Greenberg MM. Independent generation and reactivity of 2'-deoxy-5-methyleneuridin-5-yl, a significant reactive intermediate produced from thymidine as a result of oxidative stress. J. Org. Chem. 2000;65:4648–4654. doi: 10.1021/jo000271s. [DOI] [PubMed] [Google Scholar]
- 31.Dong Z-R, Li Y-Y, Chen J-S, Li B-Z, Xing Y, Gao J-X. Highly efficient iridium catalyst for asymmetric transfer hydrogenation of aromatic ketones under base-free conditions. Org. Lett. 2005;7:1043–1045. doi: 10.1021/ol047412n. [DOI] [PubMed] [Google Scholar]
- 32.Corrie JET, Reid GP, Trentham DR, Hursthouse MB, Mazid MA. Synthesis and absolute stereochemistry of the two diastereoisomers of P3-1-(2-nitrophenyl)ethyl adenosine triphosphate (‘caged’ ATP) J. Chem. Soc., Perkin Trans. 1992;1:1015–1019. [Google Scholar]
- 33.Ludwig J. A new route to nucleoside 5′-triphosphates. Acta Biochem. Biophys. Acad. Sci. Hung. 1981;16:131–133. [PubMed] [Google Scholar]
- 34.Ichida JK, Horhota A, Zou K, McLaughlin LW, Szostak JW. High fidelity TNA synthesis by Therminator polymerase. Nucleic Acids Res. 2005;33:5219–5225. doi: 10.1093/nar/gki840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling LL, Keohavong P, Dias C, Thilly WG. Optimization of the polymerase chain reaction with regard to fidelity: modified T7, Taq, and Vent DNA polymerases. Genome Res. (formerly PCR Methods Appl.) 1991;1:63–69. doi: 10.1101/gr.1.1.63. [DOI] [PubMed] [Google Scholar]
- 36.Kawanishi S, Hiraku Y, Oikawa S. Mechanism of guanine-specific DNA damage by oxidative stress and its role in carcinogenesis and aging. Mutat. Res. 2001;488:65–76. doi: 10.1016/s1383-5742(00)00059-4. [DOI] [PubMed] [Google Scholar]
- 37.Cadet J, Vigny P. The photochemistry of nucleic acids. In: Morrison H, editor. Bioorganic Photochemistry: Photochemistry and the Nucleic Acids. New York: John Wiley & Sons; 1990. [Google Scholar]
- 38.Cadet J, Douki T, Ravanat J-L. Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells. Acc. Chem. Res. 2008;41:1075–1083. doi: 10.1021/ar700245e. [DOI] [PubMed] [Google Scholar]
- 39.Peak JG, Pilas B, Dudek EJ, Peak MJ. DNA breaks caused by monochromatic 365 nm ultraviolet-A radiation or hydrogen peroxide and their repair in human epitheloid and xeroderma pigmentosum cells. Photochem. Photobiol. 1991;54:197–203. doi: 10.1111/j.1751-1097.1991.tb02007.x. [DOI] [PubMed] [Google Scholar]
- 40.Blazek ER, Peak MJ. The role of hydroxyl radical quenching in the protection by acetate and ethylenediaminetetraacetate of supercoiled plasmid DNA from ionizing radiation-induced strand breakage. Int. J. Radiat. Biol. 1988;53:237–247. doi: 10.1080/09553008814550591. [DOI] [PubMed] [Google Scholar]
- 41.Peak MJ, Peak JG. Hydroxyl radical quenching agents protect against DNA breakage caused by both 365-nm UVA and by gamma radiation. Photochem. Photobiol. 1990;51:649–652. [PubMed] [Google Scholar]
- 42.Bordin F, Conconi MT, Capozzi A. Certain singlet oxygen quenchers affect the photoreaction between 8-MOP and DNA. Photochem. Photobiol. 1987;46:301–304. doi: 10.1111/j.1751-1097.1987.tb04770.x. [DOI] [PubMed] [Google Scholar]
- 43.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vander Horn PB, Davis MC, Cunniff JJ, Ruan C, McArdle BF, Samols SB, Szasz J, Hu G, Hujer KM, Domke ST, et al. Thermo Sequenase™ DNA polymerase and T. acidophilum pyrophosphatase: new thermostable enzymes for DNA sequencing. Biotechniques. 1997;22:758–765. doi: 10.2144/97224pf02. [DOI] [PubMed] [Google Scholar]
- 45.Tabor S, Richardson CC. A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc. Natl Acad. Sci. USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Metzker ML, Raghavachari R, Richards S, Jacutin SE, Civitello A, Burgess K, Gibbs RA. Termination of DNA synthesis by novel 3'-modified deoxyribonucleoside triphosphates. Nucleic Acids Res. 1994;22:4259–4267. doi: 10.1093/nar/22.20.4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao G, Orlova M, Georgiadis MM, Hendrickson WA, Goff SP. Conferring RNA polymerase activity to a DNA polymerase: a single residue in reverse transcriptase controls substrate selection. Proc. Natl Acad. Sci. USA. 1997;94:407–411. doi: 10.1073/pnas.94.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Astatke M, Ng K, Grindley NDF, Joyce CM. A single side chain prevents Escherichia coli DNA polymerase I (Klenow fragment) from incorporating ribonucleotides. Proc. Natl Acad. Sci. USA. 1998;85:3402–3407. doi: 10.1073/pnas.95.7.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gardner AF, Jack WE. Determinants of nucleotide sugar recognition in an archaeon DNA polymerase. Nucleic Acids Res. 1999;27:2545–2553. doi: 10.1093/nar/27.12.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loeb LA, Kunkel TA. Fidelity of DNA synthesis. Ann. Rev. Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 51.Goodman MF. Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl Acad. Sci. USA. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunkel TA, Bebenek K. DNA Replication Fidelity. Ann. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 53.Doublié S, Sawaya MR, Ellenberger T. An open and closed case for all polymerases. Structure. 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 54.Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Ann. Rev. Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 55.Kool ET. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 56.Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- 57.Scaduto DI, Brown JM, Haaland WC, Zwickl DJ, Hillis DM, Metzker ML. Source identification in two criminal cases using phylogenetic analysis of HIV-1 DNA sequences. Proc. Natl Acad. Sci. USA. 2010;107:21242–21247. doi: 10.1073/pnas.1015673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang W. Portraits of a Y-family DNA polymerase. FEBS Lett. 2005;579:868–872. doi: 10.1016/j.febslet.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 59.Harris D, Kaushik N, Pandey PK, Yadav PNS, Pandey VN. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J. Biol. Chem. 1998;273:33624–33634. doi: 10.1074/jbc.273.50.33624. [DOI] [PubMed] [Google Scholar]
- 60.Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase. J. Biol. Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 61.Wu W, Litosh VA, Stupi BP, Metzker ML. 3′-OH unblocked, nucleotides and nucleosides, base-modified with photocleavable, terminating groups, and methods for their use in DNA sequencing. 2008 US Patent Application 11/567,189. Filed Dec 5, 2006; Notice of Allowance received Oct 29, 2010. [Google Scholar]
- 62.Litosh VA, Hersh MN, Stupi BP, Wu W, Metzker ML. Nucleotides and nucleosides and methods for their use in DNA sequencing. 2010 US Patent Application 12/483,080. Filed Jun 11, 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.