Abstract
Eukaryotic and archaeal DRG factors are highly conserved proteins with characteristic GTPase motifs. This suggests their implication in a central biological process, which has so far escaped detection. We show here that the two Saccharomyces cerevisiae DRGs form distinct complexes, RBG1 and RBG2, and that the former co-fractionate with translating ribosomes. A genetic screen for triple synthetic interaction demonstrates that yeast DRGs have redundant function with Slh1, a putative RNA helicase also associating with translating ribosomes. Translation and cell growth are severely impaired in a triple mutant lacking both yeast DRGs and Slh1, but not in double mutants. This new genetic assay allowed us to characterize the roles of conserved motifs present in these proteins for efficient translation and/or association with ribosomes. Altogether, our results demonstrate for the first time a direct role of the highly conserved DRG factors in translation and indicate that this function is redundantly shared by three factors. Furthermore, our data suggest that important cellular processes are highly buffered against external perturbation and, consequently, that redundantly acting factors may escape detection in current high-throughput binary genetic interaction screens.
INTRODUCTION
The developmentally regulated GTP-binding (DRG) proteins subfamily constitutes a deeply rooting branch of the GTPase superfamily whose members are found throughout eukaryotes and in archaea (1). The family DRG is related to OBG proteins whose members are universally found in eubacteria (2). DRG orthologs are highly similar (e.g. on average 66% of identity between human and budding yeast) with two paralogs encoded by most sequenced eukaryotic genomes, while only one is present in archaea (3). The two eukaryotic paralogs, DRG1 and DRG2, are very similar across their entire sequence (i.e. 57% identity between the two human paralogs, this value being 62% for Saccharomyces cerevisiae). Such remarkable evolutionary conservation suggests (an) important role(s) for these proteins in (a) basic biological process(es), yet their molecular functions has resisted previous analyses, including through high throughput approaches in yeast.
Some DRGs have been shown to bind GTP and GDP nucleotides and to hydrolyze GTP, albeit inefficiently (4–6). It was previously suggested that DRGs are involved in the regulation of cell proliferation. Indeed, DRGs are highly expressed in actively growing tissues such as during embryonic development in Xenopus and zebrafish (7,8), or in the developing mouse brain (9); this property forming the basis for the DRG name. Interestingly, DRG1 is also abundantly expressed in the growing and reproductive tissues of Arabidopsis thaliana (10). Concomitantly with their expression in actively growing cells, altered DRG expression interferes with cell proliferation. For example, over-expression of human DRG2 causes arrest in G2/M phase (11,12), whereas human and mouse DRG1 bind the oncogenic SCL/TAL protein and enhance oncogenic transformation (13,14). Expression of DRG2 was also shown to be selectively repressed upon transformation of human fibroblasts (15). Although these observations link the DRG proteins to cell proliferation control, their molecular function(s) remains poorly understood.
Mammalian DRG1 and DRG2 were shown to interact respectively with factors named DFRP1 and DFRP2 (see below). A weak interaction was also observed between DRG1 and DFRP2 upon forced expression of DFRP2 but not between DRG2 and DFRP1 (16). DFRP1 and DFRP2 proteins contain a conserved DRG family regulatory protein (DFRP) domain of ∼60 aa that was originally identified by multiple alignment of sequences from mouse, fly and yeast. The DRFP domain is required for association of DFRP1/2 with DRGs (16). Except for the DFRP domain, DFRP1 and DFRP2 display very different domain architecture. DFRP1 contains a characteristic tandem repeat CCCH zinc finger domain (TZF) with significant similarity to RNA-binding proteins, such as TTP proteins (17). This suggests that DFRP1 function may be linked to RNA metabolism. DFRP2 contains a RWD domain whose name derives from three families of proteins in which this motif was originally identified (RING finger-containing proteins, WD-repeat-containing proteins, and DEAD-like helicases) (18). Also called GI (for Gcn2 and Impact domain) (19), the RWD domain has been shown to mediate protein interaction although additional or alternative function(s) remain possible (20–24). Both DFRP1 and DFRP2 are highly conserved in eukaryotes with respectively 51% and 46% similarity between the human and the yeast orthologs. This conservation level suggests again their implication in important pathways. A screen to isolate new ribosome-associated factors identified the translation machinery associated 46-kDa protein (Tma46), the yeast ortholog of DFRP1 (25). Tma46 is thus associated with polysomes and was found in complex with the ribosome binding GTPase 1 (Rbg1, the yeast ortholog of DRG1). Recently, Gir2 (genetically interacts with ribosomal genes 2, the yeast ortholog of DFRP2) was also identified as a binding partner of Rbg1 using two-hybrid assay and in vitro assays (26). This analysis also indicated that Gir2 interacts with the translational regulator Gcn1 and reported the association of Gir2 to polysomes. Very little is known about Rbg2, the yeast ortholog of DRG2. Rbg2 was shown to interact to Gir2 but does not associate to polysomes (26). Taken together, these recent observations suggest that the yeast orthologs of the DRG1, DFRP1 and DFRP2 proteins are associated to the active translation machinery. Surprisingly, however, neither a strong growth phenotype nor translational defects have been reported for strains carrying deletions of any combination of these nonessential genes. Nevertheless, negative synthetic interactions between some of these factors, as well as between these factors and Gcn20, have been observed in a large-scale analysis (27). The sensitive and quantitative nature of an assay performed in competitive conditions probably explains why a reduced fitness was scored in the latter analysis.
To elucidate the function of these mysterious factors, we analyzed yeast Rbg and Dfrp proteins. Tandem affinity purification and cell fractionation experiments demonstrated unequivocally that Rbg and Dfrp proteins form two distinct RBG complexes that associate either with ribosomes or with factors functionally linked to translation. Most importantly, a genetic screen for triple synthetic interactants identified Slh1, an atypical putative RNA helicase, as functionally redundant with Rbg1 and Rbg2. Polysome formation is strongly affected in mutant backgrounds lacking simultaneously these three factors, demonstrating that the RBG/DFRP proteins are required in the absence of Slh1 for efficient protein synthesis. The availability of this robust yeast genetic assay for Rbgs, Dfrps and Slh1 function allowed us for the first time to conduct a structure/function analysis of these proteins supporting further their role in translation. Beyond these functional results, our observation demonstrates that three factors, including unrelated GTPase and helicase, may compensate for each other for a specific cellular function. We discuss this observation in the context of current high-throughput genetic screens that primarily target binary interactions.
MATERIALS AND METHODS
Strains and plasmids
Yeast strains used in this study were all derived from BMA64 strains (28) and are listed in Supplementary Table S1. Strains containing a single disrupted or epitope-tagged gene were obtained by transformation with KanMX4 or HISMX6 (29), URA3Kl or TRP1Kl, TAP-tag (30), HA-tag or VSV-tag modules (31) carrying short flanking sequences homologous to the targeted gene. Primers sets that were used for that purpose are listed in Supplementary Tables S2 and S3. Integrations were verified to be correct by polymerase chain reaction (PCR) amplification of their two junctions. Mating and sporulation of appropriate strains yielded the strains containing combinations of disrupted or epitope-tagged genes.
Plasmids used in this work are listed in the Supplementary Table S4. They were derived from pFL (32) or YCplac (33) plasmid series and were propagated in the MH1 bacterial strain. Inserts recovered by PCR using genomic DNA as templates, or mutants, were all verified by DNA sequencing (see Supplementary Data for details about plasmid construction).
Mutant and epitope-tagged alleles were obtained by PCR fusion reactions. For some constructs, an intermediate cloning was performed using Zero BluntR TOPOR PCR cloning kits from Invitrogen. Two constructs were obtained by plasmid gap-repair. In this case, suitable yeast cells were co-transformed with digested plasmid DNA and a PCR fragment containing complementary regions to the digested plasmid. After selection, repaired plasmids were recovered from yeast cells and amplified in E. coli. A detailed description of the plasmid constructions is presented in the Supplementary Data.
Tandem affinity purification of yeast protein complexes and protein identification by mass spectrometry
All tandem affinity purification (TAP) purifications were performed as described previously (30,34) from 2 to 4 l of culture grown in YPDA at 30°C. Purified proteins were concentrated by lyophilization, fractionated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on gradient gel (5–20%) and stained with Coomassie blue. Protein identification was performed by Matrix-assisted laser desorption/ionisation-time of flight (MALDI-TOF) analysis at the PAPPSO service of INRA (Jouy en Josas, France).
Genetic screen for mutant displaying a negative synthetic interaction with a Δrbg1Δrbg2 double deletion
A 5-FOA-sensitivity-based assay was used to screen for mutants that require either RBG1 or RBG2 for normal growth (Figure 3). Details of the screen can be found in the Supplementary Data.
Figure 3.
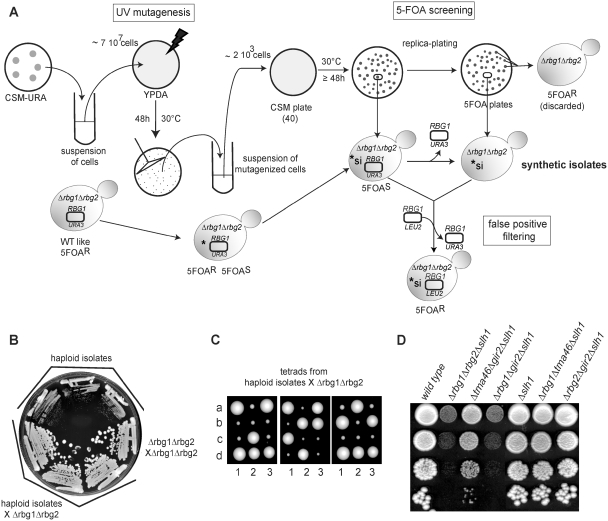
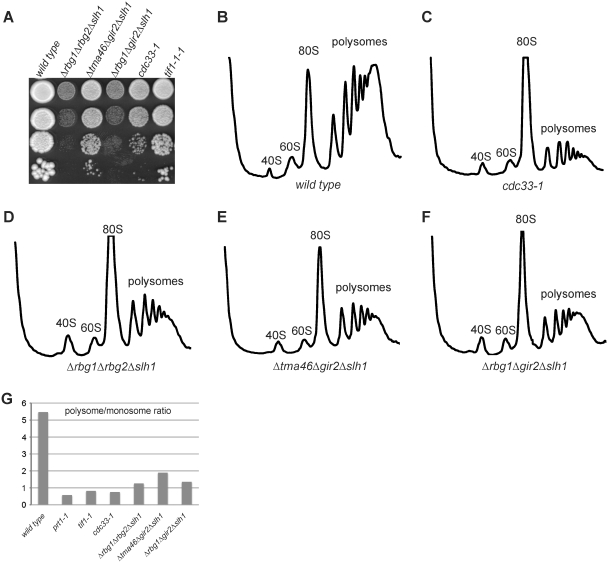
Identification of Slh1 as a genetic partner of the RBG complexes. (A) Description of the screen for gene having a negative synthetic effect in a Δrbg1Δrbg2 strain. A Δrbg1 Δrbg2 ura3-52 yeast strain containing an URA3-plasmid carrying a RBG1 allele was grown on a CSM-URA plate to stationary phase. Colonies from the plate were resuspended in water, plated on YPDA solid medium (7 × 107 cells /plate) and UV-irradiated. After incubation at 30°C for 48 h, colonies were scraped from the irradiated YPDA plates, resuspended in water and plated on CMS plates (2 × 103 cells/plate; 40 plates). After incubation at 30°C for 48 h, CSM plates were replica-plated on 5FOA plates. The original strain grows well upon loss of the URA3-plasmid [WT like] and is thus resistant to 5-FOA [5-FOAR]. Mutants unable to grow (or growing poorly) on 5-FOA plates [5-FOAS] were retained as putative candidates containing a mutation with a negative synthetic interaction (*si) with the double deletion Δrbg1Δrbg2. To eliminate false positive, 5-FOAS candidates were transformed with a LEU2-plasmid carrying a RBG1 allele. Those were plated on 5-FOA plates and only 5-FOAR transformants were kept for further analysis. (B) The three synthetic slow growth strains isolated in the screen carry a recessive mutation. The original isolates and diploids resulting from the cross of these isolates with an isogenic Δrbg1Δrbg2 strain were streaked in parallel with a Δrbg1/Δrbg1; Δrbg2/Δrbg2 isogenic diploid on YPDA plates and incubated 3 days at 30°C to monitor growth. (C) Meiotic segregation analysis indicates that a single gene is responsible for the synthetic slow growth. Diploid obtained by crossing the three synthetic slow growth strains isolated in the screen with a Δrbg1Δrbg2 strain were sporulated and tetrads were dissected on YPDA plates. Pictures were taken after 4 days at 30°C. Tetrads are numbered 1, 2 and 3 and spores are labeled a, b, c and d. (D) Growth phenotype of yeast strains carrying combinations of deletions of RBG1, TMA46, RBG2 and GIR2 and SLH1 genes. Serial dilutions of exponential liquid cultures were spotted on YPDA plates and incubated 3 days at 30°C.
Yeast polysome analysis
Polysomes preparation and analyses were as previously described except that extraction and gradient buffer contained 5 mM MgCl2 (35) Gradient analysis was performed with an ISCO UV-6 gradient collector and continuously monitored at 254 nm. Five-hundred-microliter fractions were collected. For western blot analysis, protein samples from gradient fractions were prepared as follows: 100 µl of TCA 100% 4 mg/ml deoxycholate (in water) were added to 400 µl gradient fraction, incubated 15 min on ice, centrifuged 15 min at 4°C and the supernatant was carefully removed. Protein pellet was washed with pure acetone and resuspended in 25 µl of Laemli sample buffer 1× containing 0.2 M Tris-base.
Western blot analysis
Proteins from gradient fractions or total yeast extract (36) were separated by SDS-PAGE and subsequently transferred to nitrocellulose membrane. TAP- or Protein A-tagged proteins were detected as described earlier (34). HA- or VSV-tagged proteins were detected using mouse anti HA monoclonal antibody (Babco) or mouse anti VSV monoclonal antibody (Sigma) and goat HRP-anti mouse IgG secondary antibody (Pierce) whereas Stm1 and Rpl1A proteins were detected using rabbit polyclonal antibodies and goat HRP-anti rabbit IgG secondary antibody (Pierce). ECL signals were recorded with a LAS3000 device (Fuji).
RESULTS
Rbg1 and Rbg2 form two distinct protein complexes
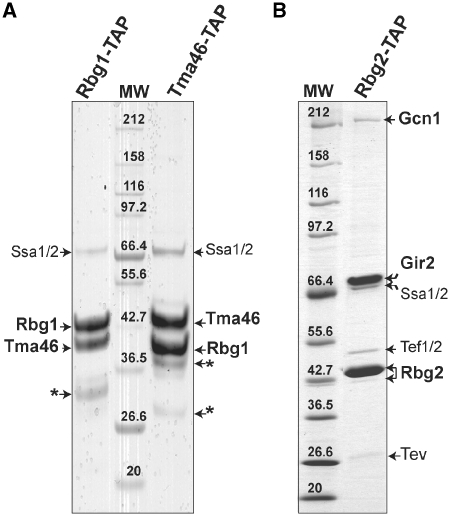
While the existence of two separate human DRG complexes has been reported (16,37), the nature of the homologous yeast complexes is still contradictory (25,26,38). To better characterize these complexes, we performed TAPs of Rbg1 and Rbg2 proteins fused at their C-terminus to the TAP tag (30,34). Purification using Rbg1-TAP resulted in a simple pattern of two proteins in apparent stoichiometric amounts. Those were identified as Rbg1 and Tma46 by mass spectrometry. Reciprocal experiment with Tma46-TAP as bait confirmed this result (Figure 1A). It is noteworthy that purification performed with reduced stringency washes (lower salt concentration) resulted in the recovery of a significant amount of ribosomal proteins from the small and large subunits (data not shown). Purification of Rbg2-TAP recovered two proteins in apparent 1:1 stoichiometric amount that were identified as Rbg2 and Gir2. In addition, lower quantities of two others proteins identified as the translation elongation factor Tef1/2 (translational elongation factor EF-1 alpha with identical proteins encoded by the Tef1 and Tef2 genes) and the large translational regulator Gcn1 was found associated to Rbg2 (Figure 1B). No trace of Gir2 was detected in the Rbg1 purification, neither of Rbg2 with Tma46 nor of Tma46 with Rbg2. Thus, Rbg1 and Rbg2 do not interact with the same partners and form two distinct complexes, Rbg1/Tma46 and Rbg2/Gir2. These factor complexes are later named respectively RBG1 and RBG2 to differentiate them from their human homologs. Moreover, both of these complexes appear to copurify with factors whose functions are linked to translation.
Figure 1.
Rbg1 and Rbg2 are not associated with the same protein partners in vivo. Protein profiles observed after TAP purification of the Rbg1-TAP or Tma46-TAP fusions (A) or Rbg2-TAP fusion (B). TAP purified proteins were fractionated on a 5–20% gradient SDS-PAGE gel and stained with Coomassie blue. Molecular weight markers are indicated (MW). Proteins identified by mass-spectrometry are indicated. Gir2 is a highly acidic 31-kDa protein and has an anomalous electrophoretic behavior (38). The protein chaperones, Ssa1 and Ssa2 also observed in other TAP purified purifications, are likely to be nonspecific interactants. Some remaining TEV protease was detected in the gel shown in (B). An asterisk indicates bands identified as degradation products of Rbg1 or Tma46 proteins.
The RBG1 complex is associated with translating ribosomes
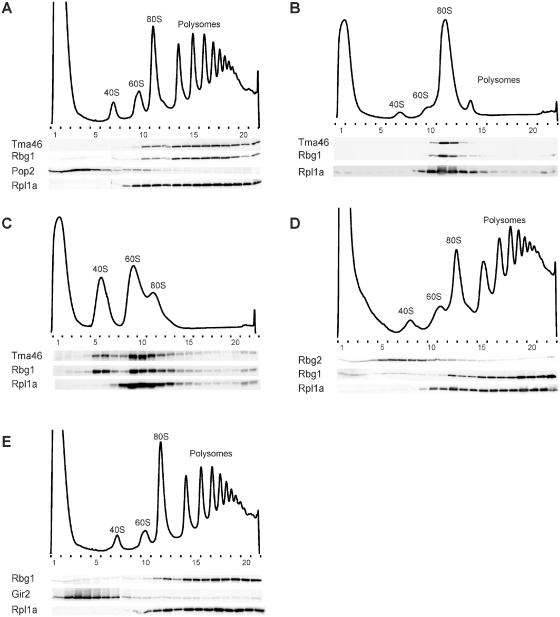
Tma46, Rbg1 and Gir2, but not Rbg2, were previously reported to associate with the translation machinery (25,26). These conclusions were partly contradictory with the existence of two distinct RBG complexes in vivo. To assay for association of each subunit of the two RBG complexes with ribosomes, we performed polysomes analyses using epitope tagged version of these factors. (Note that all epitope tagged proteins used in this study were shown by complementation to be functional, see below). Whole extracts from cycloheximide-treated cells were fractionated on discontinuous sucrose density gradients. Western blot detections of epitope-tagged Rbg1 and Tma46 show co-fractionations and exclusive association with monosome- and polysome-containing fractions (Figure 2A). Micrococcal nuclease treatment of cell lysates prior to sucrose gradient sedimentation disrupted most of the polysomes into 80S monosomes (Figure 2B) and resulted in Tma46 and Rbg1 relocating to the 80S monosome fractions, indicating that the RBG1 complex is specifically associated to translating ribosomes. Chelation of Mg2+ from micrococcal nuclease-treated cell lysates by addition of EDTA dissociated most 80S monosomes into free 40S and 60S ribosomal subunits (Figure 2C). Interestingly, in these conditions, Tma46 and Rbg1 appeared mostly associated to 40S and 60S ribosomal subunits and remnant 80S monosomes.
Figure 2.
Factors Rbg1 and Tma46, but not Rbg2 and Gir2, co-fractionate with translating ribosomes. Polysomes extracts were prepared from cells expressing several epitope-tagged proteins. Tagged proteins were expressed from their genomic loci to avoid overproduction, except when otherwise stated. Polysomes were resolved by density sedimentation in 10–50% sucrose gradient. The UV absorbance trace (254 nm) obtained by continuous monitoring during fractionation is shown with the position of the 40S, 60S, 80S and polysomes peaks indicated. Fractions (numbered) were analyzed by western blotting to detect the TAP, HA and/or VSV tags. The 60S ribosomal protein Rpl1a was used as a positive control for polysome/ribosome/60S subunit association and detected with specific rabbit polyclonal antibodies. The Pop2 deadenylase fused to the VSV tag was used as a marker for nonpolysomal association. To demonstrate the specificity of the association of factors to polysomes or ribosomes, those were dissociated by treating extracts with micrococcal nuclease or micrococcal nuclease and EDTA prior to fractionation on the sucrose gradient. (A) Distribution of Tma46-TAP, Rbg1-HA, Pop2-VSV and RPL1A. (B) Distribution of Tma46-TAP, Rbg1-HA and Rpl1a after micrococcal nuclease treatment of the extract. (C) Distribution of Tma46-TAP, Rbg1-HA and Rpl1a after micrococcal nuclease treatment of the extract followed by EDTA addition. (D) Distribution of Rbg1-HA, Rbg2-TAP and Rpl1a. (E) Distribution of Rbg1-TAP, Gir2-HA and Rpl1a. For this experiment the Gir2-HA protein was encoded by a centromeric (LEU2) plasmid (pMCD-G2) and expressed in a Δgir2 strain that also carried a genomic RBG1-TAP fusion.
In contrast to the RBG1 complex, analysis of the distribution of Rbg2 and Gir2 revealed that both proteins fractionated throughout the gradient with the vast majority migrating in the fractions lighter than the one corresponding to polysomes (Figure 2D and E). As ATP has been reported to stimulate association of Gcn1 to polysomes (39), we tested whether inclusion of ATP stimulated an indirect association of Rbg2/Gir2 with polysomes through Gcn1. No effect detectable was observed (Supplementary Figure S1), possibly owing to the small fraction of Rbg2/Gir2 apparently associated with Gcn1 (Figure 1), or because specific conditions are required to trigger association of Rbg2/Gir2/Gcn1 with polysomes.
In conclusion, the RBG1 complex is clearly associated to translating ribosomes, whereas the RBG2 complex seems mainly not. This differential behavior confirms the existence of two distinct complexes and suggests that those may have only partially overlapping roles.
A genetic screen identifies the putative RNA helicase Slh1 as a factor functionally related to the RBG complexes
Despite evidence indicating that RBG complexes associate with the translation apparatus [(25,26) and reference therein], single or double deletion mutants of RBG1, RBG2, TMA46 and GIR2 genes (Supplementary Figure S2) were not observed to impair polypeptide synthesis nor to modulate cells sensitivity to translation inhibitors. The reduced fitness of Δrbg1Δgir2, Δrbg1Δrbg2 and Δtma46Δgir2 strains observed in a large-scale analysis relying on a sensitive assay suggested that RBG complexes might have partially overlapping roles (27). We reasoned that additional functional partners of these complexes might have escaped detection because of the binary nature of the assays used. Therefore, we performed a screen for synthetic enhancers in a Δrbg1Δrbg2 double deletion background (Figure 3A). Out of 60 000 colonies tested after ultraviolet (UV) mutagenesis, we isolated three independent candidates exhibiting synthetic slow growth (Figure 3B) but no synthetic lethal mutants. Diploid heterozygous strains generated by crossing these isolates to a Δrbg1Δrbg2 strain had a wild-type growth rate (Figure 3B) and the slow growth phenotype segregated 2:2 after sporulation (Figure 3C), demonstrating that a single recessive mutation was responsible for the observed phenotype. Complementation analyses indicated that the three candidates were alleles of the same gene (data not shown). Transformation of one of this strain with a yeast genomic library prepared from a Δrbg1Δrbg2 strain yielded one clone complementing the growth defect. Sequencing inserts ends and further mapping demonstrated that mutations in SLH1 (for Ski2-like helicase 1) were responsible for the synthetic slow growth. Slh1 is a nonessential protein with homologs conserved throughout the eukaryotic kingdom. It belongs to the Ski2-like subfamily of the superfamily 2 (SF2) helicase but atypically contains a tandem repeat of the helicase domain in a single polypeptide, a feature only shared with the splicing factor Brr2 (40,41). Slh1 was first identified by its role in the repression of dsRNA viruses in yeast, a function performed in collaboration with Ski2. Moreover, the simultaneous deletion of Slh1 and Ski2 was reported to dramatically increase translation of poly(A)- mRNAs (42,43). However, the molecular function of Slh1 remains elusive.
We constructed a deletion of SLH1 in the W303 genetic background. This does not affect growth in standard conditions. By crossing this mutant with RBGs and DFRPs deletion strains, we observed that Δslh1Δrbg1Δtma46 and Δslh1Δrbg2Δgir2 strains grow like wild-type, indicating that loss of one RBG complex in the absence of Slh1 is not deleterious for cells. By contrast, we observed that a Δslh1Δrbg1Δrbg2 strain exhibited severe growth defects (Figure 3D). This result validates the screen identifying Slh1 as a functional partner of Rbg1 and Rbg2. We also observed that Δslh1Δrbg1Δgir2, and to a lesser extent, Δslh1Δtma46Δgir2 strains also grew poorly (Figure 3D), indicating that the two RBG complexes are required, together with Slh1, to perform an important cellular function
Slh1 and RBG complexes are essential for efficient translation
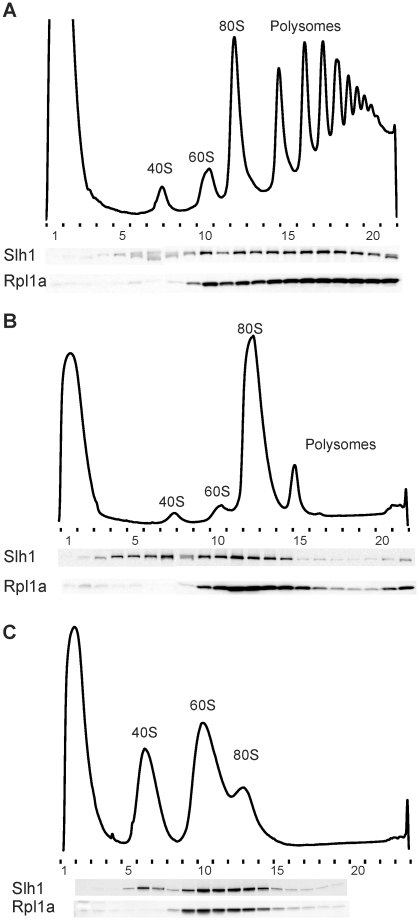
Slh1 has been shown to limit translation of poly(A)- mRNAs (42,43). Polysome profiles from Δslh1 cells are similar to that of wild-type cells (data no shown). To determine whether Slh1 may interact directly with the translation machinery, we analyzed its distribution in polysome gradients. Slh1 was mainly detected in the fractions corresponding to 80S monosomes and polysomes (Figure 4A). To exclude that Slh1 associated with complex with high sedimentation properties different from translating ribosomes, the sedimentation of Slh1 in the gradient was examined after disruption of polysomes into monosomes by micrococcal nuclease treatment. Under these conditions, Slh1 shifted to the fractions corresponding to the peak of 80S monosomes and to lighter fractions (Figure 4B) and, like Rbg1 and Tma46, remained associated with 40S and 60S ribosomal subunits after chelation of Mg2+ from microccal nuclease-treated cell lysate (Figure 4C). Thus, like the RBG1 complex, Slh1 is associated with actively translating ribosomes.
Figure 4.
Slh1 protein is associated with translating ribosomes. (A) Whole cell extracts were fractionated on 10–50% sucrose gradient and analyzed as described in Figure 3. Extracts were prepared from cells expressing the Slh1–TAP fusion protein expressed from its natural genomic locus to avoid over expression. In (B), the extract was treated with micrococcal nuclease before loading onto the gradient. In (C), micrococcal nuclease treatment of the extract was followed by EDTA addition before loading onto the gradient.
This observation reinforces the idea that RBG complexes also affect translation. To test whether the severe growth defects of Δslh1Δrbg1Δrbg2, Δslh1Δrbg1Δgir2 and Δslh1Δtma46Δgir2 strains could result from impaired translation, we analyzed polysome profiles in these genetic contexts. An overall decrease of polysomes amount coupled to a shift toward polysomes with a lower number of ribosomes and a large accumulation of the 80S peak was detected for these strains (Figure 5D–F) compared to wild type (Figure 5B). Control experiments demonstrated that these effects did not result from altered ribosome levels or impaired ribosomal RNA processing (data not shown). Interestingly, the polysome pattern presents similarities with the ones observed with translation initiation mutants, such as cdc33-1 strain (Figure 5C). Indeed, under conditions of limiting initiation of translation, ribosomes terminate translation, disengage from mRNAs and accumulate as free 80S awaiting recruitment into new rounds of translation (44). This is characterized by a nearly complete loss of polysomes and a consequently large increase of the 80S peak corresponding to translating monosomes and mostly inactive 80S particles not bound to mRNA. This defect can be quantified by the polysome-to-monosome ratio (P/M). This value is low for Δslh1Δrbg1Δrbg2, Δslh1Δrbg1Δgir2 and Δslh1Δtma46Δgir2 compared to wild type, and similar to the ones observed for the three strains carrying mutation in translation initiation factors eIF4A (tif1-1), eIF4E (cdc33-1) and eIF3 (prt1-1) that we analyzed in parallel (Figure 5G). Overall, our data support the functional interplay of the two RBG complexes and Slh1. The presence of at least one of these three factors is required for efficient translation. Our polysome analysis suggests that these factors may act at the level of initiation but additional experiments will be required to characterize the precise block occurring in the cognate triple mutants.
Figure 5.
Mutant strains lacking the Rbg1, Rbg2 and Slh1 proteins exhibit polysome profiles indicative of translation initiation defects. (A) Growth behavior of yeast strains carrying combinations of deletions of RBG1, TMA46, RBG2 and GIR2 and SLH1 genes is compared to the growth behavior of cells carrying conditional mutations of translation initiation factors. These phenotypes can be correlated with the polysome profiles of the same strains (B–F). Serial dilutions of exponential liquid cultures were spotted on YPDA plates and incubated 3 days at 30°C. Polysomes extracts from wild type (B), cdc33-1 (eIF4E) (C), Δrbg1Δrbg2Δslh1 (D), Δtma46Δgir2Δslh1 (E) and Δrbg1Δgir2Δslh1 (F) strains were resolved by sedimentation in 10–50% sucrose gradient. The UV absorbance trace (254 nm) is drawn and the position of the 40S, 60S, 80S and polysomes peaks is indicated. (G) Histograms of polysomes/monosomes ratios derived from the polysomes profiles of the indicated strains.
Conserved motifs of Rbg1, Tma46, Slh1 and Gir2 are critical for function
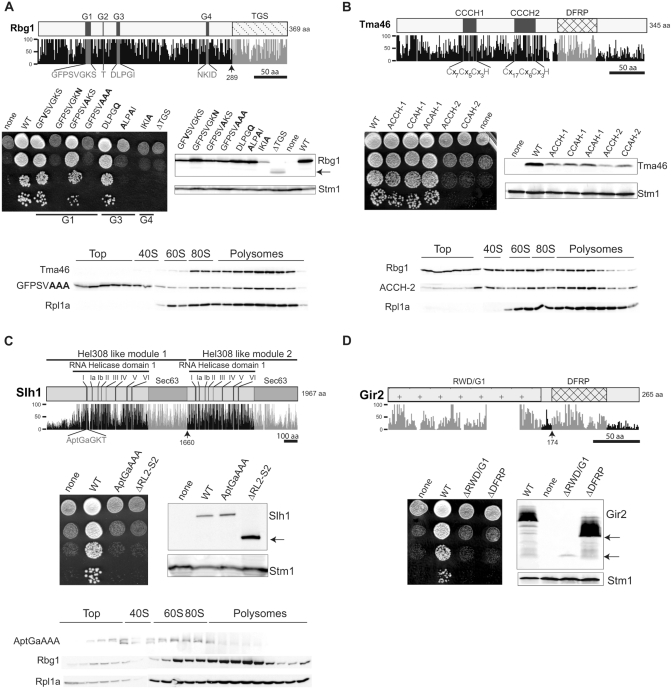
Subunits of the RBG complexes and Slh1 contain conserved sequence motifs suggesting specific functions. The role(s) of these conserved elements has(have) not been analyzed so far in the absence of adequate assays. Our observation that at least one RBG complex or Slh1 is absolutely required for wild-type growth offered us the unique possibility to test the contribution of some of these motifs to the function of these factors. First, we verified whether plasmid-borne epitope-tagged construction encoding the wild-type protein complemented the growth defect of strains with triple deletions affecting the two RBG complex and Slh1. Next, mutations targeting defined sequence motifs were introduced in these constructs by site directed mutagenesis and the resulting mutants tested for their ability to complement appropriate triple mutant strains. Finally, using the epitope tag, we verified that nonfunctional mutant proteins were expressed correctly to exclude that noncomplementation was due to instability of the mutant factor.
We first tested the impact of mutations expected to impair GTP binding and hydrolysis by Rbg1 (1). We replaced several amino acids in three of the conserved motifs of the core GTPase domain (Figure 6A). The P-loop/Walker A/G1 motif (consensus [A/G]X4GK[S/T]) was shown to participate to ATP/GTP binding. The Walker B loop corresponds to the G3 motif (consensus DxxG) and is involved in binding of the gamma phosphate of GTP, while motif G4 is the guanine-ring binding sequence (NKXD).
Figure 6.
Structurally conserved motifs of Rbg1, Tma46, Slh1 and Gir2 are critical for function. Schematic representations of the Rbg1 (A), Tma46 (B), Slh1 (C) and Gir2 (D) protein sequences with a plot of the percentage of conservation to the human, mouse, chicken, zebrafish, nematode, drosophila and arabidopsis orthologs. [Percentage conservation was derived from ClustalX alignments (38).] Boxes depict some characteristic domains of each protein and the corresponding plots are in light gray. The characteristic residues displaying 100% of conservation among eukaryote species are plotted in dark gray. Black lines indicate amino acid scales. Arrows indicate the position of truncations used in the structure/function analysis. To monitor cellular growth, serial dilutions of mutant strains (Δrbg1Δrbg2Δslh1 for (A) and Δtma46Δgir2Δslh1 for (B–D) expressing none, wild type (WT) or mutant proteins, were spotted on YPDA plates and incubated at 30°C for 2–3 days. Amounts of the WT or mutant protein were determined by western blotting. The ribosome associated Stm1 protein was used as a loading control. Distributions of mutant proteins in polysomes sucrose gradient were analyzed by western blotting. Polysomes extracts were prepared from: (A) Δrbg1 cells expressing a Tma46-TAP fusion encoded in the genome and the HA-Rbg1 (GFPSVAAA) mutant encoded by a plasmid. (B) Δtma46 cells expressing a Rbg1-TAP fusion encoded in the genome and HA-Tma46 (ACCH-2) mutant encoded by a plasmid. (C) Δslh1 cells expressing a Rbg1-TAP fusion encoded in the genome and Slh1-ProtA (AptGaAAA) mutant encoded by a plasmid. Positions of the 40S, 60S, 80S monosomes and the polysomes are indicated.
Several substitutions in motifs G1 (GFPSVGKA, GFPSVAAA), G3 (ALPAI) or G4 (IKIA) previously shown to affect nucleotide binding and GTPase activities of several GTPases (45) were unable to restore growth of the Δrbg1Δrbg2Δslh1 strain. By contrast, the GFVSVGKS and GFPSVAKS substitutions in motif G1 restored wild-type growth. The latter mutations are equivalent to those previously shown to inhibit members of the Ras-protein family of GTPases but this inhibition appears to be specific for the Ras subfamily of GTPase without affecting the DRG protein family.
The GFPSVAAA Rbg1 mutant protein was also analyzed on polysome gradients prepared from lysates of Δrbg1 strains. Interestingly, significant amounts of the mutant protein shifted to the top of the gradient indicating that polysomal association was affected (Figure 6A). Notably, the distribution of Tma46 remained unchanged, suggesting either that formation of the RBG1 complex or its stability depends upon a fully functional enzymatic activity. Thus, the GTPase activity of Rbg1 is required for RBG complex function.
We also analyzed a deletion of Rbg1 TGS domain whose function has not been experimentally determined but that is also present in tRNA synthetase and bacterial SpoT (46). Deletion of this domain (ΔTGS) did not complement growth of the Δrbg1Δrbg2Δslh1 strain, possibly because the truncated protein (similar to the IKIA mutant, see above) is produced at insufficient levels to allow viability (Figure 6A). However, the same mutants expressed from multicopy plasmids under the control of the strong Gal promoter still failed to complement the growth phenotype, suggesting that they are not functional (data not shown).
We next analyzed the effect of mutations in the tandem repeat of CCCH-type zinc finger (TZF) domain of Tma46 that is likely to contact RNA (47) (Figure 6B). Although the first zinc finger, CCCH-1, closely resembles the consensus (C-x8-C-x5-C-x3-H), the second, CCCH-2 (amino acids 219–248), is characterized by a larger number of residues (X17) between the first and the second cysteine (Figure 6B). We substituted the first or the third cysteine of each CCCH motifs with alanine as these mutations have already been shown to impair the function of other ZF containing proteins [e.g. ref. (48)]. Remarkably, any substitution in the second zinc finger (ACCH-2 and CCAH-2) failed to restore growth of the Δtma46Δgir2Δslh1 mutant strain, whereas all substitutions in the first zinc finger (ACCH-1, CCAH-1 and ACAH-1) were fully functional despite a reduced protein accumulation compared to wild type. Thus, the TZF domain appears to be critical for the function of Tma46 but the contribution of the two zinc fingers is not identical. Notably, the distribution of Tma46-ACCH-2 in polysome gradients from Δtma46 cells complemented by this mutant was altered leading to a reduced polysomal association of Tma46 and Rbg1 (Figure 6B).
Slh1 structural organization consists in a tandem repeat of two putative Hel308 helicase-like modules both potentially endowed with ATPase and RNA unwinding activity (40,41). A Slh1 mutant carrying substitution of highly conserved amino acids within the walker A (P loop) motif (AptGaAAA) of the first Hel308-like module was unable to rescue growth of a Δtma46Δgir2Δslh1 strain (Figure 6C). Similarly, the complete deletion of the second Hel308-like module (ΔRLII) resulted in a noncomplementing allele, despite a clear accumulation of the truncated protein. Additionally, when the slh1-AptGaAAA allele was expressed as the sole copy in a Δslh1 strain, we observed a markedly reduced association of the mutant protein to translating ribosomes (Figure 6C). We conclude that the ATPase activity of Slh1 is most likely required for Slh1 function in translation, and that both helicase domains are involved in this process.
Finally, we analyzed deletion of either the DFRP (ΔDFRP) or the RWD/GI (ΔRWD/GI) domain of Gir2 (16,18,19). Full-length Gir2p and the RWD/GI domain were both expressed to similarly high levels and displayed an anomalous electrophoretic behavior likely due to a high content of acidic amino acid residues (38), whereas accumulation of the C-terminal part containing the DFRP domain was very poor (Figure 6D). None of the truncated alleles were able to sustain normal growth of a Δgir2Δtm46Δslh1 strain, indicating that both domains are essential to Gir2 function or stability (Figure 6D).
DISCUSSION
Our biochemical analysis of the yeast DRG factors Rbg1 and Rbg2 indicates that they form two distinct complexes. Rbg1 interacts with Tma46 and this complex associates with translating ribosomes, while Rbg2 binds to Gir2. The latter complex does not appear to associate directly with ribosomes. However, it interacts with Gcn1, which itself has been shown to bind to ribosome in specific starvation conditions. Interaction of Rbg1 with Tma46 and Rbg2 with Gir2 in yeast was previously reported but our observation differs significantly from the conclusion reached earlier (26). It is noteworthy that, in the latter study, an interaction between Rbg1 and Gir2 was detected in a two hybrid assays in which proteins fused to heterologous domains are overexpressed and targeted to the nuclear compartment. These authors confirmed this observation by co-precipitation of tagged proteins, which were again overexpressed in yeast. These data, obtained upon artificial expression conditions, together with the conservation of the DFRP domain, suggest that overexpressed Rbg1 is endowed with the capacity to interact with Gir2 even though this interaction is not detectable in cells probably because Rbg1 has higher affinity for Tma46 while Rbg2 associates preferentially with Gir2, especially when these factors are in competition. Consistent with this interpretation, we observed that joint deletions of rbg2 and tma46 did not produce a slow growth phenotype when combined with a slh1 deletion (data not shown) supporting the possible formation of a functional Rbg1-Gir2 complex in the absence of competing factors (e.g. their natural partners). As our TAP purification results are consistent with the cellular co-fractionation of Rbg1 with Tma46 and Rbg2 with Gir2, our data leave little doubt that two distinct complexes are present in yeast cells. More importantly, all of our observations are in total agreement with the results obtained with human factors (16,37). This is also in agreement with the extremely high conservation of these factors in eukaryotic cells.
Consistent with previous results, we observed association of Rbg1 and Tma46 with ribosomes (25,26). Contradictory results had been reported for the association of Gir2 with Gcn1 and with ribosomes (26,38). We observe that Gcn1 copurifies with Rbg2 probably by interacting with Gir2, but we do not detect a co fractionation of Gir2 with translating ribosomes. Again, the latter result parallel the one obtained in mammals (37) supporting the high functional conservation of these factors. Although Gir2 and its binding partner Rbg2 do not appear to be associated with translating ribosomes in actively growing cells, it is likely that they can interact with ribosomes through Gcn1, which, itself, is known to bind directly to ribosomes together with its partner Gcn20 under specific conditions (49). Overall, these observations were consistent with a role of the yeast DRGs Rbg1 and Rbg2 in translation. Yet, deletion of these two proteins, independently or simultaneously, did not generate strong growth phenotypes (Supplementary Figure S2) nor alteration of polysome profiles (data not shown). Reduced cell fitness upon the simultaneous inactivation of rbg1 and rbg2, rbg1 and gir2, or tma46 and gir2 was only detected using a sensitive competitive assay (27). Interestingly, this study also reported the same phenotype resulting from inactivation of rbg1 or tma46 together with gcn1 or gcn20. The latter observation supports the idea that Rbg2 acts through the Gcn1/Gcn20 complex, consistent with the observation that Rbg2/Gir2 interact with Gcn1. This association most likely occurs through the RWD domain of Gir2 that is highly similar to the homologous domain of Gcn2, which is known to mediate interaction with Gcn1 (19,21,23,24,50). In standard laboratory conditions, the synthetic interactions detected between Rbg1/Tma46, on the one hand, and Rbg2/Gir2/Gcn1/Gcn20, on the other hand, have no significant impact on cell growth (Supplementary Figure S2). The lack of clear function for DRG proteins, opposed to their strong sequence conservation, was highly surprising. Thus, we performed a genetic screen to identify factors that would act redundantly with Rbg1 and Rbg2, and identified Slh1. Synthetic interactions suggest that Rbg1, Rbg2 and Slh1 most likely act in parallel pathways. Indeed, synthetic interactions occur between cells carrying complete deletions of these genes, which is not consistent with the additive impairment resulting from combination of partially active alleles acting in the same pathway usually detected with conditional genes. Thus, it is likely that one pathway requires Rbg1 and Tma46, the second Rbg2, Gir2 and possibly Gcn1 and Gcn20, while the third would require Slh1. It is unlikely that additional factors specifically join Slh1 for this task as we recovered different alleles of SLH1 in our screen (judged from their growth phenotype) but failed to identify any other genes redundant with RBG1 and RBG2. Moreover, genetic screens for gene dosage-dependent or genomic suppressors of Δrbg1Δrbg2Δslh1 growth defect failed so far to uncover any additional factor (data not shown), suggesting again that the absence of additional players able to mediate the same function. Like for the Rbg-dependent pathways, the Slh1-dependent pathway is likely to be conserved in eukaryotes given the good sequence conservation of this factor across the eukaryotic kingdom. In this situation, it will be of interest to test whether the simultaneous inhibition of Drg1, Drg2 and Slh1 also result in translation inhibition in other eukaryotic species.
Analyses of polysomes assembled in vivo demonstrate that Rbg1, Rbg2 and Slh1 are required for efficient translation. Taken together with their association with ribosomes, this new result indicates that these proteins are new translation factors with redundant functions. However, given that the triple mutant still grows, albeit slowly, this function is not essential for protein production. A possibility is that Rbg1, Rbg2 and Slh1 are involved in quality control pathways. Indeed, such a process would not be essential but its inactivation could result in impaired translation. Polysome profiles of the triple mutant strain are highly altered, indicating that in this context the translation of most or all mRNA is affected. This suggests that Rbg1, Rbg2 and Slh1 do not control the expression of a subset of specific mRNAs but rather that they have a more general function. The profiles that we observed are consistent with altered translation initiation defects. Interestingly, Slh1 was first identified by its role in the inhibition of translation of mRNAs lacking poly(A) tails (43) and the effect of the poly(A) tail on translation was proposed to be mediated during initiation (51), at least in part by favoring 60S subunit joining (42). Further analyses will be required, however, to characterize the precise molecular function of Rbg1, Rbg2 and Slh1 in translation.
Although DRGs have long been known to contain GTPase signature sequences, the role of this domain remained unclear in the absence of a functional test. The observation that triple mutant altering Rbg1, Rbg2 and Slh1 (or partners thereof) grow slowly offered the possibility to assay this in vivo. Hence, it appears that the GTP-binding site of Rbg1 is required for its function. Similarly, mutant analysis supports the idea that RNA binding by Tma46 and association of Gir2 with Gcn1 and Rbg2 is necessary for their activity. Finally, the function of Slh1 appears to require both of its helicase domains and most likely its ability to hydrolyze ATP. GTPases and helicases have been implicated as switches, or through their capacities to remodel RNPs, at many steps of the translation process. If some GTPase and helicase have been shown to cooperate in translation, as for Vasa and eIF5B (52,53), they had up-to-now distinct dedicated functions, including partners and binding sites. How can we imagine Rbg1, Rbg2 and Slh1 being redundant? While it is easy to envisage that the two Rbg factors have similar molecular mechanisms of action, it is more difficult to conceive that Slh1 would act through the same binding site and partners. It is more likely that these proteins act on related substrates through different molecular mechanisms. Hence, if Rbg1, Rbg2 and Slh1 are involved in quality control reactions, it is easy to imagine that an aberrant substrate may be recognized through different means, and redirected to a normal status or discarded through different pathways. In this vein, it is noteworthy that both some GTPases and helicases have been shown to participate to quality control processes. This includes, for example, the precise selection of cognate tRNA during translation (54) or the targeting of mRNA containing premature stop codons to decay (55).
While further experiments will be required to delineate the functions of yeast DRGs and Slh1, an important orthogonal conclusion of our work is that cellular processes are very robust owing to the presence of redundant factors that protect cells from aggression through their buffering effects. Indeed, the simultaneous inactivation of Rbg1, Rbg2 and Slh1 is required to produce a clear growth inhibition phenotype; in standard laboratory conditions strains containing any combination of double mutant behaved as wild type. In this context, it is interesting to note that even for an extensively analyzed organism such as S. cerevisiae, many genes remain classified as orphan for function. It is not unlikely that some of those have redundant roles with several other proteins. Such a situation would however not be detected by current large-scale analyses that only address effects of binary interactions. These analyses indeed suffer from technical limitations as the number of combinations to be tested to assay for triple interactions is currently far beyond our capacity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
La Ligue contre le Cancer ‘Equipe Labellisée 2008', IGBMC, the CNRS, the European Union 6th Framework programs ‘3D-Repertoire’ (LSHG-CT-2005-512028). Funding for open access charge: Ligue Nationale contre le Cancer (Equipe Labellisée 2008).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Céline Henry from the PAPPSO service at INRA for protein identification and T. Villa for critical reading of the manuscript.
REFERENCES
- 1.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 2.Caldon CE, March PE. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 2003;6:135–139. doi: 10.1016/s1369-5274(03)00037-7. [DOI] [PubMed] [Google Scholar]
- 3.Li B, Trueb B. DRG represents a family of two closely related GTP-binding proteins. Biochim. Biophys. Acta. 2000;1491:196–204. doi: 10.1016/s0167-4781(00)00025-7. [DOI] [PubMed] [Google Scholar]
- 4.O'Connell A, Robin G, Kobe B, Botella JR. Biochemical characterization of Arabidopsis developmentally regulated G-proteins (DRGs) Protein Expr. Purif. 2009;67:88–95. doi: 10.1016/j.pep.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Sazuka T, Tomooka Y, Ikawa Y, Noda M, Kumar S. DRG: a novel developmentally regulated GTP-binding protein. Biochem. Biophys. Res. Commun. 1992;189:363–370. doi: 10.1016/0006-291x(92)91567-a. [DOI] [PubMed] [Google Scholar]
- 6.Sommer KA, Petersen G, Bautz EK. The gene upstream of DmRP128 codes for a novel GTP-binding protein of Drosophila melanogaster. Mol. Gen. Genet. 1994;242:391–398. doi: 10.1007/BF00281788. [DOI] [PubMed] [Google Scholar]
- 7.Kumar S, Iwao M, Yamagishi T, Noda M, Asashima M. Expression of GTP-binding protein gene drg during Xenopus laevis development. Int. J. Dev. Biol. 1993;37:539–546. [PubMed] [Google Scholar]
- 8.Wei D, Yao J, Yang X, Cheng L, Lu D, Xue J. Molecular cloning and expression of two closely related GTP-binding proteins from zebrafish. DNA Seq. 2004;15:246–250. doi: 10.1080/10425170400002439. [DOI] [PubMed] [Google Scholar]
- 9.Sazuka T, Kinoshita M, Tomooka Y, Ikawa Y, Noda M, Kumar S. Expression of DRG during murine embryonic development. Biochem. Biophys. Res. Commun. 1992;189:371–377. doi: 10.1016/0006-291x(92)91568-b. [DOI] [PubMed] [Google Scholar]
- 10.Devitt ML, Maas KJ, Stafstrom JP. Characterization of DRGs, developmentally regulated GTP-binding proteins, from pea and Arabidopsis. Plant Mol. Biol. 1999;39:75–82. doi: 10.1023/a:1006178710443. [DOI] [PubMed] [Google Scholar]
- 11.Ko MS, Lee UH, Kim SI, Kim HJ, Park JJ, Cha SJ, Kim SB, Song H, Chung DK, Han IS, et al. Overexpression of DRG2 suppresses the growth of Jurkat T cells but does not induce apoptosis. Arch. Biochem. Biophys. 2004;422:137–144. doi: 10.1016/j.abb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 12.Song H, Kim SI, Ko MS, Kim HJ, Heo JC, Lee HJ, Lee HS, Han IS, Kwack K, Park JW. Overexpression of DRG2 increases G2/M phase cells and decreases sensitivity to nocodazole-induced apoptosis. J. Biochem. 2004;135:331–335. doi: 10.1093/jb/mvh040. [DOI] [PubMed] [Google Scholar]
- 13.Mahajan MA, Park ST, Sun XH. Association of a novel GTP binding protein, DRG, with TAL oncogenic proteins. Oncogene. 1996;12:2343–2350. [PubMed] [Google Scholar]
- 14.Zhao XF, Aplan PD. SCL binds the human homologue of DRG in vivo. Biochim. Biophys. Acta. 1998;1448:109–114. doi: 10.1016/s0167-4889(98)00129-3. [DOI] [PubMed] [Google Scholar]
- 15.Schenker T, Lach C, Kessler B, Calderara S, Trueb B. A novel GTP-binding protein which is selectively repressed in SV40 transformed fibroblasts. J. Biol. Chem. 1994;269:25447–25453. [PubMed] [Google Scholar]
- 16.Ishikawa K, Azuma S, Ikawa S, Semba K, Inoue J. Identification of DRG family regulatory proteins (DFRPs): specific regulation of DRG1 and DRG2. Genes Cells. 2005;10:139–150. doi: 10.1111/j.1365-2443.2005.00825.x. [DOI] [PubMed] [Google Scholar]
- 17.Carrick DM, Lai WS, Blackshear PJ. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doerks T, Copley RR, Schultz J, Ponting CP, Bork P. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 2002;12:47–56. doi: 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubota H, Sakaki Y, Ito T. GI domain-mediated association of the eukaryotic initiation factor 2alpha kinase GCN2 with its activator GCN1 is required for general amino acid control in budding yeast. J. Biol. Chem. 2000;275:20243–20246. doi: 10.1074/jbc.C000262200. [DOI] [PubMed] [Google Scholar]
- 20.Carbia-Nagashima A, Gerez J, Perez-Castro C, Paez-Pereda M, Silberstein S, Stalla GK, Holsboer F, Arzt E. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1alpha during hypoxia. Cell. 2007;131:309–323. doi: 10.1016/j.cell.2007.07.044. [DOI] [PubMed] [Google Scholar]
- 21.Garcia-Barrio M, Dong J, Ufano S, Hinnebusch AG. Association of GCN1-GCN20 regulatory complex with the N-terminus of eIF2alpha kinase GCN2 is required for GCN2 activation. EMBO J. 2000;19:1887–1899. doi: 10.1093/emboj/19.8.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubota H, Ota K, Sakaki Y, Ito T. Budding yeast GCN1 binds the GI domain to activate the eIF2alpha kinase GCN2. J. Biol. Chem. 2001;276:17591–17596. doi: 10.1074/jbc.M011793200. [DOI] [PubMed] [Google Scholar]
- 23.Pereira CM, Sattlegger E, Jiang HY, Longo BM, Jaqueta CB, Hinnebusch AG, Wek RC, Mello LE, Castilho BA. IMPACT, a protein preferentially expressed in the mouse brain, binds GCN1 and inhibits GCN2 activation. J. Biol. Chem. 2005;280:28316–28323. doi: 10.1074/jbc.M408571200. [DOI] [PubMed] [Google Scholar]
- 24.Sattlegger E, Swanson MJ, Ashcraft EA, Jennings JL, Fekete RA, Link AJ, Hinnebusch AG. YIH1 is an actin-binding protein that inhibits protein kinase GCN2 and impairs general amino acid control when overexpressed. J. Biol. Chem. 2004;279:29952–29962. doi: 10.1074/jbc.M404009200. [DOI] [PubMed] [Google Scholar]
- 25.Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wout PK, Sattlegger E, Sullivan SM, Maddock JR. Saccharomyces cerevisiae Rbg1 protein and its binding partner Gir2 interact on Polyribosomes with Gcn1. Eukaryot. Cell. 2009;8:1061–1071. doi: 10.1128/EC.00356-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle JC, Fromont-Racine M, Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl Acad. Sci. USA. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baudin-Baillieu A, Guillemet E, Cullin C, Lacroute F. Construction of a yeast strain deleted for the TRP1 promoter and coding region that enhances the efficiency of the polymerase chain reaction-disruption method. Yeast. 1997;13:353–356. doi: 10.1002/(SICI)1097-0061(19970330)13:4<353::AID-YEA86>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 29.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 30.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 31.De Antoni A, Gallwitz D. A novel multi-purpose cassette for repeated integrative epitope tagging of genes in Saccharomyces cerevisiae. Gene. 2000;246:179–185. doi: 10.1016/s0378-1119(00)00083-4. [DOI] [PubMed] [Google Scholar]
- 32.Bonneaud N, Ozier-Kalogeropoulos O, Li GY, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 33.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 34.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 35.Daugeron MC, Linder P. Characterization and mutational analysis of yeast Dbp8p, a putative RNA helicase involved in ribosome biogenesis. Nucleic Acids Res. 2001;29:1144–1155. doi: 10.1093/nar/29.5.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kushnirov VV. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 37.Ishikawa K, Akiyama T, Ito K, Semba K, Inoue J. Independent stabilizations of polysomal Drg1/Dfrp1 complex and non-polysomal Drg2/Dfrp2 complex in mammalian cells. Biochem. Biophys. Res. Commun. 2009;390:552–556. doi: 10.1016/j.bbrc.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Alves VS, Castilho BA. Gir2 is an intrinsically unstructured protein that is present in Saccharomyces cerevisiae as a group of heterogeneously electrophoretic migrating forms. Biochem. Biophys. Res. Commun. 2005;332:450–455. doi: 10.1016/j.bbrc.2005.04.151. [DOI] [PubMed] [Google Scholar]
- 39.Marton MJ, Vazquez de Aldana CR, Qiu H, Chakraburtty K, Hinnebusch AG. Evidence that GCN1 and GCN20, translational regulators of GCN4, function on elongating ribosomes in activation of eIF2alpha kinase GCN2. Mol. Cell. Biol. 1997;17:4474–4489. doi: 10.1128/mcb.17.8.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pena V, Jovin SM, Fabrizio P, Orlowski J, Bujnicki JM, Luhrmann R, Wahl MC. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol. Cell. 2009;35:454–466. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat. Struct. Mol. Biol. 2009;16:731–739. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Searfoss A, Dever TE, Wickner R. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Searfoss AM, Wickner RB. 3′ poly(A) is dispensable for translation. Proc. Natl Acad. Sci. USA. 2000;97:9133–9137. doi: 10.1073/pnas.97.16.9133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fringer JM, Acker MG, Fekete CA, Lorsch JR, Dever TE. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol. Cell. Biol. 2007;27:2384–2397. doi: 10.1128/MCB.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paduch M, Jelen F, Otlewski J. Structure of small G proteins and their regulators. Acta Biochim. Pol. 2001;48:829–850. [PubMed] [Google Scholar]
- 46.Wolf YI, Aravind L, Grishin NV, Koonin EV. Evolution of aminoacyl-tRNA synthetases–analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 47.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat. Struct. Mol. Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 48.Prouteau M, Daugeron MC, Séraphin B. Regulation of ARE transcript 3′ end processing by the yeast Cth2 mRNA decay factor. EMBO J. 2008;27:2966–2976. doi: 10.1038/emboj.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sattlegger E, Hinnebusch AG. Polyribosome binding by GCN1 is required for full activation of eukaryotic translation initiation factor 2{alpha} kinase GCN2 during amino acid starvation. J. Biol. Chem. 2005;280:16514–16521. doi: 10.1074/jbc.M414566200. [DOI] [PubMed] [Google Scholar]
- 50.Nameki N, Yoneyama M, Koshiba S, Tochio N, Inoue M, Seki E, Matsuda T, Tomo Y, Harada T, Saito K, et al. Solution structure of the RWD domain of the mouse GCN2 protein. Protein Sci. 2004;13:2089–2100. doi: 10.1110/ps.04751804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Preiss T, Hentze MW. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature. 1998;392:516–520. doi: 10.1038/33192. [DOI] [PubMed] [Google Scholar]
- 52.Carrera P, Johnstone O, Nakamura A, Casanova J, Jackle H, Lasko P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- 53.Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- 54.Daviter T, Gromadski KB, Rodnina MV. The ribosome's response to codon-anticodon mismatches. Biochimie. 2006;88:1001–1011. doi: 10.1016/j.biochi.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 55.Neu-Yilik G, Kulozik E. NMD: multitasking between mRNA surveillance and modulation of gene expression. Adv. Genet. 2008;62:185–243. doi: 10.1016/S0065-2660(08)00604-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.