Abstract
Long-term survival still eludes most patients with leukemia and non-Hodgkin’s lymphoma. No approved therapies target the hallmark of the B cell, its mIgM, also known as the B-cell receptor (BCR). Aptamers are small oligonucleotides that can specifically bind to a wide range of target molecules and offer some advantages over antibodies as therapeutic agents. Here, we report the rational engineering of aptamer TD05 into multimeric forms reactive with the BCR that may be useful in biomedical applications. Systematic truncation of TD05 coupled with modification with locked nucleic acids (LNA) increased conformational stability and nuclease resistance. Trimeric and tetrameric versions with optimized polyethyleneglycol (PEG) linker lengths exhibited high avidity at physiological temperatures both in vitro and in vivo. Competition and protease studies showed that the multimeric, optimized aptamer bound to membrane-associated human mIgM, but not with soluble IgM in plasma, allowing the possibility of targeting leukemias and lymphomas in vivo. The B-cell specificity of the multivalent aptamer was confirmed on lymphoma cell lines and fresh clinical leukemia samples. The chemically engineered aptamers, with significantly improved kinetic and biochemical features, unique specificity and desirable pharmacological properties, may be useful in biomedical applications.
INTRODUCTION
Non-Hodgkin’s lymphomas (NHLs) are the fifth most common type of cancer in the USA (1). While several effective chemotherapies and immunotherapies are available, cures still elude most patients (2). The addition of more selective strategies with monoclonal antibodies to cell surface targets such as CD20, CD22 or CD23 has improved outcomes and three antibodies to CD20 have been approved by the Food and Drug Administration (FDA) to treat B-cell NHL (3). However, there are no approved agents that target the hallmark of the B cell, the B-cell receptor (membrane Ig: mIgM) or BCR. Development of an antibody to mIgM is challenging because the unique epitopes are close to the cell surface and may be inaccessible to large antibody molecules.
Recent interest in oligonucleotide-based therapeutic agents such as siRNA or microRNA (4) is based on the fundamental advantage of automated synthesis and easy chemical manipulation that allow quality control and modification of function. Technologies that enhance plasma stability by use of unnatural nucleic acids and recent studies characterizing the pharmacokinetics of nucleic acid-based therapeutic agents suggest feasible use of these agents in vivo (5–7).
DNA aptamers can also be developed into therapeutic agents. Aptamers are short oligonucleotide sequences that can specifically bind to a wide range of target molecules, such as drugs, proteins and other inorganic or organic molecules with high affinity and specificity (8,9). Aptamer binding is based on the ability of small oligonucleotides (typically 40–100 mers) to fold into unique three-dimensional structures that can interact with a specific binding region of the target molecule. Aptamers have inherent advantages that merit application as therapeutic agents (10): (i) the ability to withstand high heat and denaturants, (ii) rapid chemical synthesis, (iii) small size (10–20 000 Da versus 150 000 Da for antibodies) and (iv) non-immunogenicity (11). In therapeutic applications, antibodies are limited by large size and the consequent inability to easily diffuse extravascularly or to penetrate large solid tumors (12). Typical monovalent aptamers are potentially limited by reduced retention times on the target cell and lack of crosslinking and subsequent activation of targets. Aptamer-based bivalent ligands, however, have been demonstrated to increase affinity and function compared to the monovalent versions; for example, bivalent aptamers were used to activate thrombin and T cells (13–15).
Recently, selection of a high-affinity DNA aptamer (TD05) reactive with Burkitt’s lymphoma was reported (16). At 4°C, TD05 binds to an epitope on B-cell surface mIgM BCR, exclusively expressed on B cells and most B-cell lymphomas (17). Aptamer TD05 is not useful in vivo, however, because of its lack of affinity and stability at physiological temperatures in human plasma. Moreover, it was not evident that TD05 could reach target B cells in vivo for diagnostic and therapeutic applications if the epitope was also present on circulating IgM, which is found in the plasma at 450–1500 mg/l (18).
To address these issues and to produce aptamers with potential medical applications, we first truncated TD05, and further optimized it by introducing locked nucleic acids (LNA) to increase nuclease resistance and conformational stability. The construct was additionally redesigned into bivalent, trivalent and tetravalent scaffolds in order to improve the affinity, and possibly to create an agent that could crosslink the BCR, which might have the biological effect of modulating the cell surface expression of the BCR, internalizing the complex, or activating or deactivating signaling pathways (19,20). We report the rational engineering of multivalent aptamer scaffolds that show higher thermal and nuclease stability, conformational stability, improved kinetic and biochemical properties at physiological temperatures.
MATERIALS AND METHODS
Cell lines, Ramos (Burkitt’s lymphoma), Daudi (Burkitt’s lymphoma), Raji (Burkitt’s lymphoma), Jeko (mantle cell lymphoma), SKLY-16 (B-cell lymphoma), CRW22R (Prostate cancer), H5V (Endothelial cells), HCT116 (Colorectal carcinoma), HEK293 (human embryonic kidney), HeLa (human adenocarcinoma cervical), K562 (leukemia chronic myelogenous), MOLT (acute lymphoblastic leukemia), SKOV-3 (human adenocarcinoma ovarian), HL60 (acute myelocytic leukemia), Jurkat (T lymphocyte) and SKLY-18 (B-cell lymphoma) were purchased from ATCC except for SKLY16 and 18. All of the cells were cultured in RPMI 1640 medium supplemented with 100 U/ml penicillin–streptomycin and 10% fetal bovine serum (heat-inactivated; Invitrogen). Clinical samples were obtained from patients at Memorial Sloan Kettering Cancer Center or from healthy donors, on IRB approved protocols.
Phosphoramidites including spacer phosphoramidite 18, amino modifier C6-dT, 5′-fluorescein phosphoramidite, Cy3™ phosphoramidite, and all the DNA reagents that are needed for DNA synthesis were purchased from Glen Research. LNA dT and dC were purchased from Exiqon, TetVA.8 S, L-TetVA.8 S were purchased from Trilink Biotechnologies Inc.
All the DNA oligo sequences were chemically synthesized attaching a fluorophore at the 5′ end using standard solid phase phosphoramidite chemistry on an ABI394 DNA synthesizer using either a 0.2 µmol or 1 µmol scale. The completed DNA sequences were de-protected. The crude product was purified using HPLC (Beckman Coulter System Gold Bioessential 125/168 diode-array detection instrument) equipped with a C-18 column (Dyanamax 250 × 10 mm, Varian) using 0.1M TEAA as the mobile phase. The length of each DNA construct was confirmed using 10%-TBE urea polyacrylamide gel electrophoresis. Full-length DNA was quantified by measuring the absorbance at 260 nm and absorbance of the corresponding dye at the 5′ position using a Cary Bio-100 UV-Visible spectrophotometer (Varian). Sequences used in nuclear magnetic resonance (NMR) experiments were further dialyzed overnight with 0.5 mM NaHPO4 buffer using a MWCO 1000-Da dialysis bag.
All the in vitro experiments were done using a binding buffer composed of RPMI 1640 and 4.5 g/l glucose (Sigma-Aldrich) and 5 mM MgCl2 for monomers, 20 mM MgCl2 for multimers (Sigma-Aldrich), 100 mg/l tRNA (Sigma-Aldrich), 100 mg/l single-stranded DNA (Sigma-Aldrich), 100 mg/l BSA (Sigma-Aldrich) and binding was analyzed using flow cytometry (Acurri C6) using wash buffer. We used 20 mM Mg+2 for optimal folding of the aptamer in vitro and 5 mM Mg+2 for experiments in vivo. The binding buffer consists of tRNA and single-stranded DNA that are capable of scavenging Mg+2, thereby reducing the effective concentration of Mg ions. The concentrations of MgCl2 used did not show any toxicity towards the cells. Wash buffer was composed of RPMI1640 with 20 mM MgCl2,0.5% BSA.
Cell binding assays
Affinity of each construct was evaluated by incubating Ramos cells (2.5 × 105) with a series of fluorescein isothiocyanate (FITC)- or Cy3-labeled constructs in a 50 µl of binding buffer on ice for 45 min. Cells were then washed with 1 ml of wash buffer at 4°C and resuspended in 100 µl of wash buffer. The binding of the constructs was analyzed using flow cytometry by counting 10 000 events for each concentration. As a positive control, a similar assay was performed using an FITC-labeled anti-IgM antibody (1 µg, Goat anti human, Invitrogen) along with an isotype control (1 µg, Goat anti mouse IgG2a, Invitrogen). Binding at respective concentrations and absolute fluorescence intensity difference was used for the binding curves. When calculating the relative binding constant, binding curves were fitted with median fluorescence intensity observed for each histogram and observed Bmax/2 was used as the binding constant. Control random sequences with each fluorophore were synthesized separately and used to compare nonspecific background binding. Assays were done using flow cytometry in which each data point reported corresponds to the median of 10 000 counted events. Investigation of the multimeric aptamer binding with clinical chronic lymphocytic leukemia (CLL) and peripheral blood mononuclear cell (PBMC) samples were done by incubating FITC-labeled multimeric aptamer (0.5 µM) or FITC-labeled random control along with APC labeled anti-CD19 (0.5 µg), Cy5.5-labeled anti-CD45 (50 ng) with 1 × 106 cells (cultured or CLL) on ice. After 45 min, the cells were washed with wash buffer and analyzed for binding using flow cytometry. Investigation of aptamer interaction of mIgM was done in 20% or 50% or 100% human serum supplemented with 20 mM MgCl2 for multimeric aptamers, or 5 mM MgCl2 for monomeric aptamers using a similar protocol as above.
Nuclease stability
Aliquots of 50 pmol of multivalent constructs labeled with a fluorophore were incubated at 37°C in a final volume of 20 µl in human serum and in 20 ul of phosphate-buffered saline (PBS) buffer for 0, 0.5 1, 2, 4, 6, 8, 10, 24 h. At the end of each time point, the reactions were terminated by adding 20 µl 2X nucleic acid loading buffer (Bio-Rad) and stored in −80°C. Full-length and digested DNA were analyzed by 10%-TBE urea polyacrylamide gel electrophoresis and fluorescence was quantified using FUJI FILM multi gauge V2.2 software. The average half-lives were calculated with Prism V fitting to exponential decay.
Trypsin digestion experiments
Cold PBS washed 5 × 105 Ramos cells were incubated with 1 mg/ml trypsin (TPCK treated, Sigma T1426) in 500 µl of 0.05% Trypsin EDTA 1X in HBSS (Cellgro 25-052-Cl) for 40 min at 37°C. The incubation time was optimized by initial pilot experiments with varying incubation times from 10 to 40 min. The trypsin cleavage site is masked (17); therefore, longer incubation times were needed to observe optimal cleavage. In earlier time points, we did not observe significant reduction in binding with cells by either the aptamer or anti-IgM antibody. After incubation, cells were pelleted, washed with cold PBS, and aliquots were incubated with 1 µM of TD05.1, 1 µM of Random DNA, anti-CD20 and FITC-labeled goat anti-human IgM antibody (1 µg) along with an isotype control (1 µg, Goat anti mouse IgG2a, Invitrogen) for 45 min at 4°C. Then the cells were washed and resuspended in cold PBS containing 20 mM MgCl2 and 0.5% BSA, and the binding was detected using flow cytometry (Becton Dickinson FacsCalibur 2002 model).
In vivo binding assay
Female athymic nude mice, 4–8 weeks of age (Taconic, Germantown, NY) were inoculated i.p. with 7 × 106 Ramos cells in 0.5 ml of saline. After 10 min, 0.5 ml of a 1 µM solution of TetVA.8 S or random aptamer in saline was introduced i.p. Animals were sacrificed after 1 h and the intraperitoneal cavity was flushed by 10 ml PBS and collected the cells. The cells were divided into three aliquots and incubated with binding buffer supplemented with (i) 0.5% BSA, (ii) 10 µg/ml of anti-CD19 and (iii) 200 µg/ml of anti-IgM antibody for 30 min in ice. The cells were then washed using 1 ml of washing buffer and binding of the aptamer and the antibody was investigated using flow cytometry counting 10 000 events (Becton Dickinson FacsCalibur 2002 model). Prior to the experiment, animals were housed in filter-top cages and provided with sterile food, water and bedding. Animal protocols were approved by the Animal Care and Use Committee at Memorial Sloan-Kettering Cancer Center.
RESULTS
Modification of TD05
Truncation
TD05 was unstable at physiological conditions and unable to bind its target on Ramos B-cell lymphoma (Figure S1 and S2). The predicted secondary structure of the TD05 using m-fold was a ‘hairpin’ structure (Figure 1 and Supplementary Figure S1; m-fold is a software application used to predict secondary structure of DNA) (21). Structural studies of a stem identical to the palindromic region of TD05 and full-length TD05 (Supplementary Figure S3) using NMR showed predominant formation of the stem at 37°C. Broadening of the peaks with decreasing temperature suggested the loss of homogeneity of the secondary structure. At lower temperatures therefore, TD05 might be forming multiple folds that lead to a more heterogeneous mixture, with only a few structures that fit the target epitope.
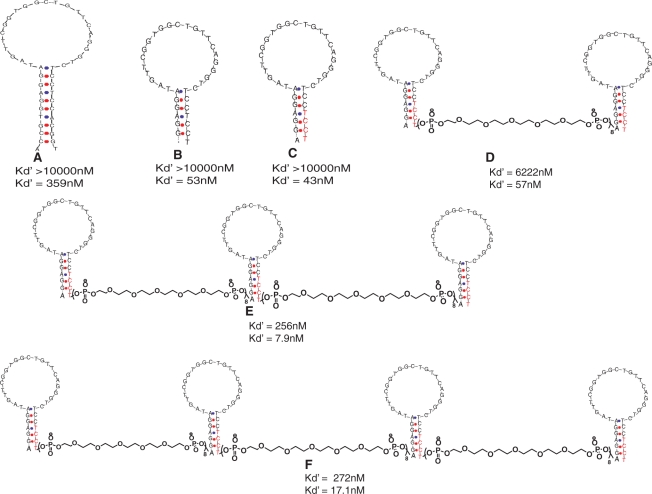
Figure 1.
Optimization of monomeric and multimeric scaffolds and their Bmax/2 (Kd′) at 37°C (top number) and at 4°C (bottom number). (A) original TD05 sequence. (B) Truncated TD05.1. (C) LNA modified TD05.1 (TD05.17). (D) Bivalent TD05.17 (L-BVA.8S). (E) Trivalent TD05.17 (L-TVA.8S). (F) Tetravalent TD05.17 (L-TetVA.8S). The constructs were synthesized with PEG at the 5′- and 3′-ends; Cy3 or FITC was added at the 5′-end. Red = LNA bases.
The original TD05 aptamer consisted of 48 nt and a later truncated version of 44 nt was reported by removing bases from the ends (16,17,22). Because affinity was retained in the truncated version, in which the stem was shortened, it is likely that bases in the central loop region of the aptamer play the most important role in binding. We investigated the possibility of further truncating TD05 with the aim of increasing the affinity by stabilizing the secondary structure of TD05. We truncated the bases starting from the 3′ and 5′ region (Figure 1). Shorter versions, e.g. TD05.1, showed higher affinity than the original version even after 10 nt were removed (Table 1). We speculated that the reduction of the length of the TD05 stem could be leading to a population with more stable secondary structure containing a favorable fold that better fit the protein-binding site. In addition, shorter sequences could also be synthesized with higher yields and lower costs, which was particularly important for a strategy in which multimeric aptamers were planned.
Table 1.
Modifications of TD05 alter binding at 4°C
| Name | Figure 1 structure | Deletions/changes | Bmax/2 at 4°C (nM) |
|---|---|---|---|
| TD05 | A | ACCGTGGAGGATAGTTCGGTGGCTGTTCAG GGTCTCCTCCACGGT | 359 |
| TD05.7 | XXCGTGGAGGATAGTTCGGTGGCTTCAGGGTCTCCTCCCGXX | 148 | |
| TD05.1 | B | XXXXXAGGAG GATAGTTCGGTG GCTGTTCAG GGTCTCCTCCTXXXXX | 53 |
| TD05.10 | GGAGGANAGTTCGGTGGCTGTTCAGGGTCTCCTCC | >1000 | |
| TD05.11 | GGAGGATAGTTCGGTNGCTGTTCAGGGTCTCCTCC | >1000 | |
| TD05.12 | GGAGGATAGTTCGGTGGCTGTNCAGGGTCTCCTCC | >1000 | |
| TD05.16 | +A+G+G +A GGATAGTTCGGTGGCTGTTCAGGGTCTCC+T+C+C+T | 368 | |
| TD05.17 | C | AGGAGGATAGTTCGGTGGCTGTTCAGGGTCTCC+T+C+C+T | 43 |
| TD05.18 | +A+G+G+A GGATAGTTCGGTGGCTGTTCAGGGTCTCCTCCT | 819 |
+N = LNA; italic, bold N = 2′OMe substituted nucleotides; X = deletions.
LNA incorporation
The incorporation of LNA bases to a stem region of a stem–loop structure has been shown to increase the melting temperature, nuclease stability and overall stability of the secondary structure of aptamers (23–25). This is mainly because LNA modified oligonucleotides, derived from the constrained sugar moiety with 3′-endo conformation resulting from the methylene link between the 2′ oxygen and 4′ carbon of the ribose ring exhibit increased affinity toward their cDNA (26). We postulated that the further stabilization of the stem by inclusion of LNA would also increase the in vivo stability of the secondary and tertiary structures. We incorporated LNA into the stem region of TD05.1 in three different combinations by substituting purines and pyrimidines (TD05.16), pyrimidines only (TD05.17), and purines only (TD05.18) (Table 1). The low binding of TD05.18 suggests LNA substitution of purines A1G2G3A4 had a profound effect on the secondary and tertiary structure (Table 1). Substitution of the pyrimidines T32C33C34T35 with LNA further increased the dissociation constant, indicating that these bases may not be involved in binding, but may aid in stabilizing the stem region. Attempts to introduce nuclease-resistant 2′-OMe bases into the loop region impaired binding, yielding a Bmax/2 values higher than the TD05.1, suggesting that the loop region cannot be modified with nuclease-resistant 2′OMe (Table 1). Difficulties in introducing the modified bases within an aptamer sequence have been reported before, and the loss of binding is probably due to changes of the favorable fold of the aptamer (24).
Multivalent TD05.1 constructs and optimization of the linker lengths
One strategy to decrease the dissociation rates is to increase local concentration by multimerization of the ligand. The resulting decreased dissociation rates might also be useful in vivo. In therapeutic applications, lower retention times would likely decrease the therapeutic index. Therefore, based on the improved affinity and stability of TD05.1, we designed multivalent analogs with PEG linkers in an attempt to increase affinity at physiological temperatures (Figure 1).
Although several approaches to multivalent designs are possible, we focused on linear molecular assembly of TD05.1 using poly-ethylene-glycol (PEG) linkers, because this would also more likely allow cross-linking of BCR on the cell surface, which might in turn lead to internalization of the complex for delivery of cytotoxic cargo or to modulation of BCR signal transduction pathways. PEG phosphoramidite is commercially available for solid-state synthesis of multivalent analogs, so this linker was chosen to construct various linear multivalent aptamer forms. Linker length between aptamer binding sites was optimized to avoid steric hindrance and to promote binding. Most naturally occurring antibodies are bivalent; therefore, first, we designed a bivalent (BV) TD05.1. In order to mimic dimensions of an antibody and provide appropriate spatial flexibility to promote binding of a bivalent aptamer, linker lengths of 6, 8 and 12 (lengths correspond to 12.6 , 16.8 and 25.2 nm) were designed and their binding to Ramos cells was evaluated using flow cytometery. The PEG spacer was ∼2.1 nm in length, which has been reported elsewhere (13), and this approximation was assumed for this study. Observed binding for bivalent analogs was similar to monomeric TD05.1 at 37°C; however, at 4°C BV.8 S showed slightly higher binding than BV.6 S and BV.12S, indicating that bivalent design with eight linkers, which corresponds to 16.8 nm, is more favorable for the bivalent design. Therefore, we evaluated the linker length of 16.8 nm on the bivalent version of TD05.17 (i.e. L-BVA.8S), assuming that increased structural stability of the stem along with the dimerization might play additive roles in increasing avidity. L-BVA.8S did show a significant increase in the Bmax/2 at 37°C (Table 2). However, this increment was likely still inadequate for in vivo applications. This suggested that increasing valency by two, alone, was not enough to substantially increase avidity. Subsequently, eight linkers were used to design trivalent (TVA.8S) and tetravalent (TetVA.8S) TD05.1-based aptamers to further investigate the binding avidity (Figure 1).
Table 2.
Optimization of linker length and linear assembly of TD05.1
| Name | Structure in Figure 1 | Sequence | Bmax/2@37°C (nM) |
|---|---|---|---|
| TD05.1 | B | AGGAG GATAGTTCGGTG GCTGTTCAG GGTCTCCTCCT | >10 000 |
| BV.6S | TD05.1 -(sp18)6 - TD05.1 | >10 000 | |
| BV.8S | TD05.1 -(sp18)8 - TD05.1 | >10 000 | |
| BV.12S | TD05.1 -(sp18)12 - TD05.1 | >10 000 | |
| BV.16S | TD05.1 -(sp18)16- TD05.1 | 2030 | |
| TVA.8S | TD05.1 -(sp18)8 - TD05.1 -(sp18)8 - TD05.1 | 490 | |
| TetVA.8S | TD05.1 -(sp18)8 - TD05.1 -(sp18)8 - TD05.1-(sp18)8 - TD05.1 | 425 | |
| TD05.17 | C | AGGAGGATAGTTCGGTGGCTGTTCAGGGTCTCC+T+C+C+T | >10 000 |
| L-BVA.8S | D | FITC-sp18-TD05.17-(sp18)8 - TD05.17-sp18 | 6222 |
| L-TVA.8S | E | Cy3-Sp18-TD05.17 -(sp18)8 - TD05.17 -(sp18)8 - TD05.17-Sp18 | 256 |
| L-TetVA.8S | F | Cy3-sp18-TD05.17-(sp18)8-TD05.17-(sp18)8-TD05.17-(sp18)8-TD05.17-sp18 | 272 |
+N = LNA; Sp18 = spacer 18.
At 37°C, a trivalent aptamer (TVA.8S) using eight linkers between each monomeric aptamer, showed increased binding (Table 2). In order to determine whether the improved TVA.8S binding is a result of interaction of each of the three aptamers with epitopes versus an improvement based on either length or geometry, we designed a hetero-trimeric analog of the homo-trimeric TD05.1 in which the loop of the central monomer was randomized and linked to distal monomers using eight PEG units (TVSR). At 4°C, the binding of the heteromeric TVSR was three to four times lower than that of the TVA.8S at a fixed concentration, suggesting that the internal monomer in the trivalent aptamer plays a role in increasing the avidity of the trimeric molecular assembly, either by altering structure or by adding an additional binding site. The decrease in the binding of TVSR also might be due to intramolecular interactions of the randomized region with the distal aptamers. We also designed dimers with 16 and 20 linkers to generate a dimeric version with a length similar to the TVA.8S. Binding at 4°C of the long dimer BVA.20S was less than the TVA.8S, suggesting that the longer linkers may be leading to unfavorable conformations. Alternatively, the longer length between the two monomers could make the dimer spatially unsuitable for binding. BVA.16S showed a Bmax/2 of 2030 nM at 37°C; however, affinity of the dimer was not as high as the corresponding trivalent design. In addition, the tetravalent analog of TD05.1 showed no significant increment in binding. This might be due to the density of receptors on the cell surface limiting the increase in the dissociation constant. Alternatively, the binding interaction might be linker independent beyond 16.8 nm.
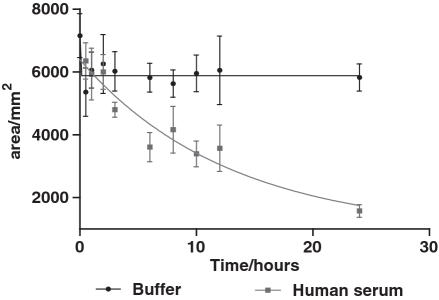
To further investigate the improved affinity, the tri- and tetra- valent scaffolds were re-synthesized using TD05.17 (containing LNA) as the monomer. The affinity of each construct was increased ∼2-fold suggesting that the locked version of the aptamer aided in stabilizing the conformation of the stem (Table 2). The LNA-modified multimeric versions were further modified with PEG spacers at the 3′ and 5′ ends to avoid 3′ and 5′exonuclease activity. The original unmodified TD05 showed a half-life of <1 h, whereas L-TVA.8S showed an estimated half-life of 8.75 h, demonstrating that the modifications introduced for TVA.8S significantly enhanced the nuclease stability in serum (Figure 2). L-BVA.8S showed a half-life of 7.87 h suggesting that multimerization and PEGylation contributes to increase stability (Supplementary Figure S4).
Figure 2.
Analysis of nuclease stability of L-TVA.8S in human serum at physiological temperature. Aptamers were separated using poly-acrylamide gel electrophoresis and fluorescence intensity of full-length DNA/area (mm2) was plotted as a function of time (hours).
Cell specificity
The specificity of the divalent scaffold for B cells and B-cell lymphoma was investigated using fresh mononuclear cells from healthy donors, patients with chronic lymphocytic leukemia, cultured B-cell lymphoma cells expressing or not expressing mIgM and cultured non-B-cell cancers, using FITC-labeled L-BVA.8S and quantified using flow cytometry. An analogous randomized aptamer sequence linked with eight PEG units was used as the isotype control. There was generally little nonspecific binding to cells not expressing surface mIgM, whereas B-cell lymphoma cell lines expressing mIgM and B cells gated from clinical samples were usually positive (Table 3). Interestingly, B cells obtained from healthy individuals only sometimes stained positively for L-BVA.8S aptamer. CD3-positive T cells rarely bound to this aptamer, further confirming the specificity (Figure 3). However, several of the mIgM expressing B-cell lymphoma cell lines and clinical samples did not bind the aptamers (Table 3), perhaps because the epitope was masked or the expression levels were low, suggesting that while specificity was high, sensitivity was lower (Table 3). In addition, the TVA.8S displayed poor binding to fresh samples as compared to the L-BVA.8S, although competition analyses (data not shown) demonstrated that both bivalent and trivalent aptamers competed for binding of the monomeric aptamer to Ramos. These data suggest that subtle differences in the local environment of the epitope may be affecting the ability of the different-sized aptamers to bind to fresh samples.
Table 3.
Analysisa of specificity of dimeric aptamers with cultured cells and clinical samples
| Cell line | Cell type | Bivalent aptamer (structure D) staining |
|---|---|---|
| Ramos | B-Lymphoma, Burkitt’s IgM+ | + |
| SKLY-16 | B-lymphoma IgM+ | + |
| Daudib | B-lymphoma IgM+ | + |
| Raji | B-lymphoma, Burkitt’s IgM+ | − |
| Jeko | Mantle cell lymphoma (B) | − |
| Bjab | B-Lymphoid leukemia | − |
| SKLY 18 | B-lymphoma IgM- | − |
| AL67 | Mouse fibroblast | − |
| CRW22R | Prostate cancer | − |
| H5V | Endothelial cell lines (heart) | − |
| HCT116 | Carcinoma colon | − |
| HEK293 | Human embryonic kidney | − |
| HeLa | Human adenocarcinoma (cervical) | − |
| K-562 | Leukemia, chronic myelogenous | − |
| MOLT | T-Leukemia, acute lymphoblastic | − |
| SKOV-3 | Ovarian | − |
| HL60 | Leukemia, acute promyelocytic | − |
| Jurkat | T-Leukemia, acute | − |
| CMLc | Clinical sample | − |
| CLLd | Clinical samples | + |
| HCLe | Clinical sample | + |
| Normal B cellsf | Donors | +/− |
| Normal T cellsg | Donors | − |
aMedian of the fluorescence intensity of FITC-labeled aptamer/ median of the fluorescence intensity of FITC-labeled random sequence ≥2.0 equals positive (+).
Experiments were done at 4°C.
bVariable binding to Daudi was observed.
cOne sample. CML is chronic myeloid leukemia, a non-B-cell neoplasm.
dFourteen samples; CLL is B-chronic lymphocytic leukemia; in two of the samples, the gated CD19-negative population showed positive signal with the aptamer
eOne sample. HCL is hairy cell leukemia, a B-cell neoplasm.
fNine samples; five of the normal B-cell samples were negative and three were only weakly positive.
gTwenty-three samples; a small subpopulation of one of the normal T-cell samples was weakly positive.
Figure 3.
Binding of LNA-modified bivalent aptamer binding with B cells and T cells. (Upper left) Bivalent aptamer binding to CD19-positive B cells. (Upper right) Bivalent aptamer does not bind to CD3-positive, CD19-negative cells. (Lower left) Bivalent randomized control aptamer does not bind to CD19-positive cells. (Lower right) Bivalent randomized aptamer does not bind to CD3-positive cells.
As previously reported, the monomeric aptamer TD05 competed with the anti-IGHM (anti-Immunoglobin Heavy Mu Chain) antibody for binding to Ramos cells, indicating that the aptamer binding site was located on or near the IgM heavy chain (17). We determined whether the multimeric aptamer could retain binding in the presence of the anti-IGHM antibody. In the presence of anti-IgM antibody, fluorescence shifts to the background level for the multimeric aptamer scaffolds, which demonstrated that anti-IgM competes with the aptamer (Supplementary Figure S5). This suggests that multimerization does not change the specificity. These data in sum lead to the conclusion that the multimeric aptamer interaction is selective for an epitope on the mIgM itself.
Aptamers recognize membrane bound mIgM, but not soluble IgM
Soluble Ig is found in high concentrations in plasma. Hence, one of the major drawbacks in using anti-Ig antibodies as therapeutic vehicles for the treatment of lymphoma is that these antibodies interact with the soluble Ig in serum, leading to immune complexes that are cleared. Thus, binding is significantly limited at tumor targets. mIgM contains an additional 41 amino acids that are not present in the soluble IgM, which are present in or near the membrane. Approximately 15 of these amino acids are found extracellularly (27–29).
If the aptamer binds only to the mIgM, we postulated that these additional extracellular amino acids might play a significant role in defining the epitope specificity. Interestingly, a trypsin cleavage site is present at amino acid 430, at the border of the sequence difference between soluble and membrane bound mIgM (28). Thus, the membrane-associated sequences proximal to amino acid 430 are present only in the membrane-bound form. Trypsinization to cleave off the domains of the membrane bound mIgM distal to this site left only the most proximal region present on the cell membrane. At high concentrations of trypsin, there is a dramatic reduction in binding of goat anti-IgM antibody with Ramos cells but TD05.1 does not change its binding significantly (Figure 4). These data indicate that TD05.1 predominately interacts with the unique extra amino acid sequences of mIgM and not with the distal portions of IgM that are present in soluble, circulating IgM. CD20 is a non-glycolysated phosphoprotein expressed on early and mature normal B cells at developmental stages that are the source of a variety of B-cell neoplasms, including B-cell NHL and CLL (30). Therefore, CD20 was used as a positive control for the enzymatic cleaving experiment.
Figure 4.
Binding of TD05.1 to mIgM after trypsin treatment. Cells were treated with trypsin for 40 min and binding of FITC-labeled (A) TD05.1, (B) anti-CD20 and (C) anti-IgM antibody was evaluated and compared with untreated control.
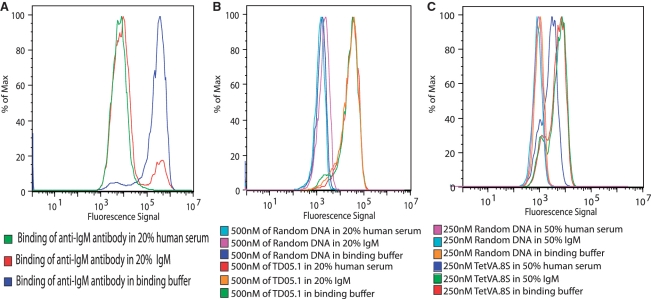
We further investigated whether the epitope of the aptamer was restricted to the surface mIgM by measuring the binding of TD05.1 in the presence of soluble IgM. The aptamers were incubated with Ramos cells in the presence of a large excess of purified soluble IgM in binding buffer or in the presence of 20% human serum (containing ∼90–300 mg/l IgM). An antibody specific for IgM heavy chain showed a dramatic reduction in binding to the cell membrane when free soluble pentameric IgM was present in the reaction (Figure 5A). The aptamer binding was unaffected (Figure 5B) showing that the epitope of the aptamer was restricted to cell membrane bound IgM. This specificity is critical to the feasibility of using this aptamer as a targeting vehicle in vivo. In addition to the monovalent aptamer, TetVA.8S was also assayed for its ability to specifically recognize the ‘membrane–bound’ heavy chain of the IgM when excess amount of soluble IgM (50%) was present in the binding buffer. The TetVA.8S binding to Ramos cells was unaffected when excess soluble IgM was present; however, a decrease in binding was observed in 50% human serum (Figure 5C). This decrease might be due to nonspecific interactions with serum proteins other than soluble IgM which would decrease the effective concentration of the TetVA.8S.
Figure 5.
Binding of aptamer in the presence of soluble IgM or human serum. The FITC-labeled monomeric and tetrameric aptamer was incubated with Ramos cells in the presence of soluble IgM/human serum for 30 min, and subsequently washed and binding was analyzed using flow cytometry. (A) Positive control showing blocking of anti-IgM by serum or soluble IgM. (B) Monomeric aptamer binding is not affected when serum or excess soluble IgM is present. (C) Tetrameric aptamer binding is not significantly affected when serum and excess soluble IgM is present.
The trypsin cleavage experiments and cross-blocking with anti-IgM are most consistent with the aptamer binding epitope localizing to a segment of the mIgM unique to the proximal membrane portion of the molecule. However, these data could also be explained by conformational changes in the soluble IgM that hide the epitope at its proximal terminus. It is also possible that accessory molecules in the vicinity of the epitope on the cell surface allow for binding of the aptamer.
In vivo binding of TetVA.8S with Ramos cells
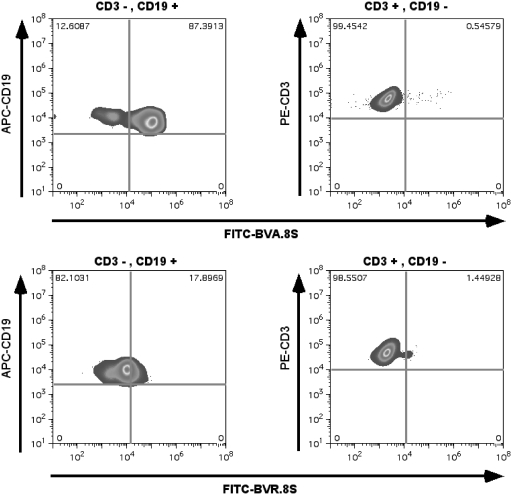
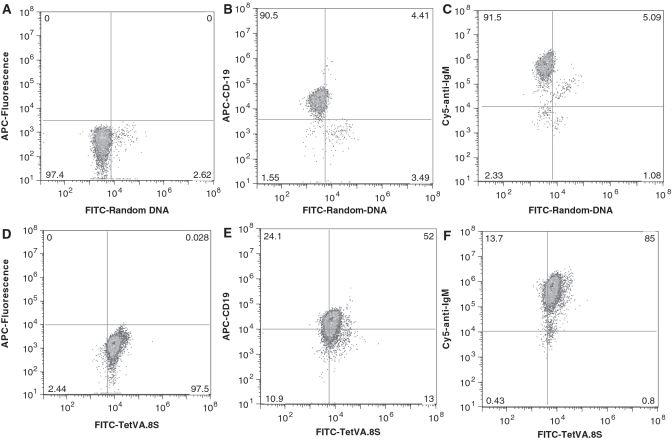
The TetVA.8S was administered i.p. into mice bearing Ramos cells in their intraperitoneal cavities to investigate binding in vivo. TetVA.8S selectively recognized Ramos cells in the intraperitoneal cavity of live mice, suggesting the feasibility of using this aptamer as a therapeutic carrier or agent (compare Figure 6A and D). Flow cytometry confirmed the specificity of the TetVA.8S binding to mIgM expressing B cells in vivo using excess anti-IgM antibody and anti-CD19 antibody as controls (Figure 6B–E and F). Although the intraperitoneal model does not exactly resemble intravascular model, this experiment demonstrates at physiological conditions in a live mouse that the multivalent construct of the aptamer TD05 specifically recognizes its target epitope on Ramos cells.
Figure 6.
Binding of TetVA.8S with Ramos cells in the intraperitoneal cavity. 1 µM of either TetVA.8S or random DNA in saline was injected into intraperitoneal cavity. Ramos cells were withdrawn from the intraperitoneal cavity and co-stained with control (BSA alone), cy5-labeled anti-IgM antibody or APC-labeled anti-CD19. (A) FITC-random sequence injected i.p. (ex vivo BSA control). (B) FITC-random sequence injected i.p.; co-stained with anti-CD19. (C) FITC-random injected i.p.; co-stained with anti-IgM. (D) FITC-TetVA.8S injected i.p. (ex vivo BSA control). (E) FITC-TetVA.8S injected i.p.; co-stained with anti-CD19, F: FITC-TetVA.8S injected i.p.; co-stained with cy-5-anti-IgM.
DISCUSSION
The BCR is a unique feature of cells of B-cell lineage, including most B-cell NHL. Therefore, the BCR is an attractive therapeutic target for lymphoma or disorders of the immune system. Approaches to this target, however, have been limited by the lack of an appropriate reagent that could selectively bind to an epitope on the BCR that was not also present on the circulating, soluble IgM, which is found at high concentrations in human plasma. One difficulty in identifying an antibody-based reagent to the mIgM may be that the unique epitopes near the cell surface are inaccessible to large antibody molecules. As the activation of the BCR in B cells is directly related to B-cell growth and function, the multivalent forms of agents that specifically bind to the BCR, such as the aptamers described here, might be useful not only as vehicles for therapeutic cargo, but also as modulators of B-cell function. Therefore, aptamers might have therapeutic applications in autoimmune diseases, immune deficiency diseases, in immunosuppression or in vaccination strategies.
A previously described aptamer, TD05, appeared to bind to the mIgM, but was not able to bind its target under physiologic conditions, nor was its selectivity for the mIgM versus soluble IgM known. So far, attempts to develop antibodies for mIgM have been limited due to interactions with the soluble versions (31). The optimized aptamer described here is unique due to its interaction with the mIgM but not with soluble IgM, significantly increasing the potential therapeutic application as a BCR cross-linker or as a drug delivery agent.
An important goal our study intended to use was rational engineering to prepare a new aptamer, derived from TD05, which can selectively recognize mIgM under physiological conditions. Here, we aimed at improving aptamer TD05 to make it suitable for in vivo applications by addressing issues pertaining to specificity, affinity and in vivo stability. First, truncation of the aptamer TD05 resulted in improved binding, presumably by creating a compact fold which better fit with the epitope. Second, in order to increase the structural stability, we introduced LNA bases in a favorable combination by substituting pyrimidines on the stem. The substitution of the purines in the stem has impaired binding, suggesting that the binding site of the aptamer is affected by these residues. However, the pyrimidine substitution stabilized the structure leading to a higher-affinity constant than the original structure. As LNAs are resistant to nuclease digestion, LNA modification of the stem not only stabilizes the secondary structure by forming a strong stem, but will also increase the stability against nuclease attack. The use of LNA to increase the aptamer stability has been demonstrated before. For example, LNA modification in the stem region of the anti-tenascin and sgc8 aptamer showed increased thermal and nuclease stability (22,25). Third, PEGylation of the aptamer ends was employed to improve stability and pharmacologic performance. Fourth, the multimerization of monomer significantly increase the avidity at physiological temperature.
Generation of multimeric ligands for targeted delivery and imaging is well established (32). For example, Fab fragments have been chemically engineered to increase their avidity (33), and RGD multimerization has been introduced to increase the avidity of the monomeric RGD ligand (34). In addition, multimeric agents such as mAb and various nano-materials such as nanoparticles, nanotubes, dendrimers and biodegradable polymers are being used as diagnostic and therapeutic platforms (35). Interaction of the multivalent ligands with its target can be explained by increased effective concentrations of the ligand at the target site without losing the entropic penalty after initial ligand–receptor contact. The subsequent interaction of the unbound ligand becomes exclusively intramolecular, contributing to the increase in the affinity by decreasing the dissociation rates.
Although larger-sized nano-materials achieve multivalency, these constructs can be limited due to poor pharmacological properties (36). On the other hand, the linear assembly of small aptamers to synthesize multivalent scaffolds has been shown to improve therapeutic properties. For example, the linear assembly of thrombin aptamers has been shown to enhance clotting function compared to monomeric forms (12). Here, we demonstrated, using a linear assembly, a novel multimeric scaffold with increased avidity. These constructs were readily synthesized using PEG phosphoramidite and automated DNA synthesis to synthesize the multivalent scaffold. The direct synthesis minimized tedious and low yield bio-conjugation reactions and poly-dispersed products. The use of long flexible linkers aided in promoting the proper conformation and avoided loss of binding due to steric hindrance.
The design of a multivalent scaffold for multiple targets is often challenging, due to complexity of the binding patterns. Here, the binding of low-valency scaffolds mainly depends on the protein density on the cell surface. The dependence on the cell surface density on binding is problematic for the optimization of the linker lengths. We found that linker lengths from 12.6 to 25.2 nM did not show a significant difference in the binding constant at 4°C, which may be due to different patterns of binding in which the binding of the ligands on either one site or in two different sites in two different proteins can accommodate the aptamer. The changes of the avidity at 37°C are more significant with increasing valency. While the avidity increased with increased valency, the nature of the interactions with variable linkers will likely vary with mIgM density on the membrane; thus, a direct comparison of aptamers of different valency for affinities is difficult. The combination of truncation, LNA modification and multimerization has shown additive effects in making the avidity of the tri- and tetrameric scaffolds more than 40-fold higher than the corresponding original monomeric aptamer.
The observed lack of binding of the original aptamer TD05 at 37°C may be due to changes of the aptamer structure with increasing temperature. We predict that the bi-loop structure (Supplementary Figure S1) instead of a stem–loop region in TD05 melts with increasing temperature, yielding a low-affinity structure B at physiological temperatures that cannot compete with the thermal instability, resulting in dissociation of the complex. Moreover, it has been reported that, the lateral angle of the mIgM in the membrane changes with temperature due to changes of the hydrocarbon structure of the membrane of B lymphocytes (37). As the nature of the aptamer–epitope interaction is nonlinear, the aforementioned changes in both aptamer and the membrane of the lymphocytic cells with temperature might negatively affect the complex, leading to dissociation.
The use of DNA aptamers as therapeutic carriers may be effective for several reasons. The small size of the aptamers should yield desirable pharmacokinetic properties, allowing higher signal to background ratio from more rapid penetration to extra-vasculature tumor sites, as well as more rapid clearance. The molecular weights of the aptamers described are 52 kDa for the tetravalent aptamer and 39 kDa for the trivalent aptamer, which is below the estimated molecular weight cutoff for kidney clearance. Rapid clearance may be critical to keeping toxicity low when using these aptamers as radiologic or chemotherapeutic drug delivery agents.
A key step in increasing the selectivity of current therapeutic approaches is the development of novel molecular constructs that target specific epitopes expressed in diseased cells. The B-cell receptor, which is the cell membrane Ig and the hallmark of the B cell, is an attractive target for therapeutic regulation of normal and neoplastic B-cell function. In addition, the BCR may also serve as a target for directed cytotoxicity, for example, via a ligand or an antibody for example. However, there are no approved antibody agents that target the BCR, mainly due to interference by the large quantities of circulating forms of secreted Ig in the plasma. In conclusion, here we report the development of an LNA-stabilized multimeric DNA aptamer that specifically binds to the membrane IgM (BCR) on neoplastic B cells, possibly on the unique 15 aa region expressed on mIgM near the cell surface. The lack of sensitivity of binding to all B cells that express the mIgM, and the differences in binding by the different sized aptamers, suggest that there may be structures in the local environment near the epitope that affect binding. Rational chemical engineering was used to design the multimeric versions, including (i) the linear assembly of the monomeric version using PEG linkers—linear assembly allowed the use of automated synthesis, eliminating the bio-conjugation procedures typically used in developing multivalent constructs; (ii) systematic truncation of the aptamer coupled with (iii) modification with LNA to increase conformational stability and nuclease resistance: (iv) dimeric, trimeric and tetrameric versions with (v) optimized polyethyleneglycol (PEG) linker lengths to yield high avidity at physiological temperatures both in vitro and in vivo. So far, to our knowledge there are no reports of a molecule that targets the mIgM in BCR, at physiological temperatures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health R01 CA55349, National Institutes of Health P01 33049, National Institutes of Health R21 CA128406; R25T CA096945; Laurie Straus Leukemia Foundation; a Lymphoma Research Foundation Research Fellowship grant; and the Lymphoma Foundation and the Tudor and Glades Funds. Funding for open access charge: NIH R01 and PO1.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors acknowledge Dr. O. Ouerfelli, Chemistry core facility, MSKCC for providing us the DNA synthesizer, Mr. Michael Curcio for assistance with cell culture, Mr. C. Villa, Mr. F. E. Escorcia, for helpful discussions. P.R.M. designed research, performed experiments analyzed results, made the figures and wrote the article; J.R.G. performed clinical experiments, analyzed data, made figures. A.R., V.K., and W.F.M. performed experiments and analyzed results, M.R.McD., M.L.H., D.J.P. and D.A.S. designed research and wrote the article.
REFERENCES
- 1.Clarke CA, Glaser SL, Dorfman RF, Bracci PM, Eberle E, Holly EA. Expert review of non-Hodgkin's lymphomas in a population-based cancer registry: reliability of diagnosis and subtype classifications. Cancer Epidemiol. Biomarkers Prev. 2004;13:138–143. doi: 10.1158/1055-9965.epi-03-0250. [DOI] [PubMed] [Google Scholar]
- 2.Winter MC, Hancock BW. Ten years of rituximab in NHL. Expert Opin. Drug Saf. 2009;8:223–235. doi: 10.1517/14740330902750114. [DOI] [PubMed] [Google Scholar]
- 3.Morris JC, Waldmann TA. Antibody-based therapy of leukaemia. Expert Rev. Mol. Med. 2009;11:e29. doi: 10.1017/S1462399409001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petri A, Lindow M, Kauppinen S. MicroRNA silencing in primates: towards development of novel therapeutics. Cancer Res. 2009;69:393–395. doi: 10.1158/0008-5472.CAN-08-2749. [DOI] [PubMed] [Google Scholar]
- 5.Braasch DA, Constantinescu ZP, Ren G, Oz OK, Mason RP, Corey DR. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg. Med. Chem. Lett. 2004;14:1139–1143. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 6.Stenvang J, Kauppinen S. MicroRNAs as targets for antisense-based therapeutics. Expert Opin. Biol. Ther. 2008;8:59–81. doi: 10.1517/14712598.8.1.59. [DOI] [PubMed] [Google Scholar]
- 7.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 9.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 10.Hnatowich DJ, Nakamura K. The influence of chemical structure of DNA and other oligomer radiopharmaceuticals on tumor delivery. Curr. Opin. Mol. Ther. 2006;8:136–143. [PubMed] [Google Scholar]
- 11.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 12.Samaranayake H, Wirth T, Schenkwein D, Räty JK, Ylä-Herttuala S. Challenges in monoclonal antibody-based therapies. Ann. Med. 2009;41:322–331. doi: 10.1080/07853890802698842. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Cao Z, Tan W. Molecular assembly for high-performance bivalent nucleic acid inhibitor. Proc. Natl Acad. Sci. USA. 2008;105:5664–5669. doi: 10.1073/pnas.0711803105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E. Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. Clin. Investig. 2008;118:376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dollins CM, Nair S, Boczkowski D, Lee J, Layzer JM, Gilboa E, Sullenger BA. Assembling OX40 aptamers on a molecular scaffold to create a receptor-activating aptamer. Chem. Biol. 2008;15:675–682. doi: 10.1016/j.chembiol.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 17.Mallikaratchy P, Tang Z, Meng L, Shangguan D, Kwame S, Tan W. Aptamer directly evolved from live cells recognizes membrane bound immunoglobin heavy mu chain in Burkitt's lymphoma cells. Mol. Cell. Proteomics. 2007;6:2230–2238. doi: 10.1074/mcp.M700026-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Furst DE. Serum immunoglobulins and risk of infection: how low can you go? Semin. Arthr. Rheum. 2009;39:18–29. doi: 10.1016/j.semarthrit.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Irish J, Czerwinski DK, Garry P, Nolan XR, Levy R. Altered B-cell receptor signaling kinetics distinguish human follicular lymphoma B cells from tumor infiltrating nonmalignant B cells. Blood. 2006;108:3135–3142. doi: 10.1182/blood-2006-02-003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signaling. Nat. Rev. Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 21.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;13:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mallikaratchy P, Tang Z, Tan W. Cell specific aptamer photosensitizer conjugates as a molecular tool in photodynamic therapy. Chem. Med. Chem. 2008;3:425–427. doi: 10.1002/cmdc.200700260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hicke BJ, Marion C, Chang YF, Gould T, Lynott CK, Parma D, Schmidt PG, Warren S. Tenascin-C aptamers are generated using tumor cells and purified protein. J. Biol. Chem. 2001;276:48644–48654. doi: 10.1074/jbc.M104651200. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, Friebe M, Dinkelborg L, Erdmann VA. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shangguan D, Tang Z, Mallikaratchy P, Xiao Z, Tan W. Optimization and modifications of aptamers selected from live cancer cell lines. Chembiochem. 2007;8:603–606. doi: 10.1002/cbic.200600532. [DOI] [PubMed] [Google Scholar]
- 26.Kaur H, Babu BR, Maiti S. Perspectives on chemistry and therapeutic applications of locked nucleic acid (LNA) Chem. Rev. 2007;107:4672–4697. doi: 10.1021/cr050266u. [DOI] [PubMed] [Google Scholar]
- 27.Cambier JC, Campbell KS. Membrane immunoglobin and its accomplices: new lessons from an old receptor. FASEB J. 1992;6:3207–3217. doi: 10.1096/fasebj.6.13.1397843. [DOI] [PubMed] [Google Scholar]
- 28.Rogers J, Early P, Carter C, Calame K, Bond M, Hood L, Wall R. Two mRNAs with different 3 ends encode membrane-bound and secreted forms of immunoglobulin chain. Cell. 1980;20:303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- 29.Friedlander RM, Nussenzweig MC, Leder P. Complete nucleotide sequence of the membrane form of the human IgM heavy chain. Nucleic Acids Res. 1990;18:4278. doi: 10.1093/nar/18.14.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meerten TV, Hagenbeek A. CD20-targeted therapy: the next generation of antibodies. Semin. Hematol. 2010;47:199–210. doi: 10.1053/j.seminhematol.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Thielemans K, Maloney DG, Meeker T, Fujimoto J, Doss C, Warnke RA, Bindl J, Gralow J, Miller RA, Levy R. Strategies for production of monoclonal anti-idiotype antibodies against human B cell lymphomas. J. Immunol. 1984;133:495–501. [PubMed] [Google Scholar]
- 32.Joshi A, Vance D, Rai P, Thiyagarajan A, Kane RS. The design of polyvalent therapeutics. Chem. Eur. J. 2008;14:7738–7747. doi: 10.1002/chem.200800278. [DOI] [PubMed] [Google Scholar]
- 33.Albrecht H, DeNardo SJ. Recombinant Antibodies: from the laboratory to the clinic. Cancer Biother. Radiopharmaceut. 2006;21:285–304. doi: 10.1089/cbr.2006.21.285. [DOI] [PubMed] [Google Scholar]
- 34.Garanger E, Boturyn D, Dumy P. Tumor targeting with RGD peptide ligands-design of new molecular conjugates for imaging and therapy of cancers. Anticancer Agents Med. Chem. 2007;7:552–558. doi: 10.2174/187152007781668706. [DOI] [PubMed] [Google Scholar]
- 35.Scheinberg DA, Villa CH, Escorcia FE, McDevitt MR. Conscripts of the infinite armada: systemic cancer therapy using nanomaterials. Nat. Rev. Clin. Oncol. 2010;7:266–276. doi: 10.1038/nrclinonc.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine. 2008;3:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krolick KA, Wisnieski BJ. Sercarz, EE. Differential leteral mobility of IgM and IgG receptors in mouse B lymphocyte membranes. Proc. Natl Acad. Sci. USA. 1977;74:4595–4599. doi: 10.1073/pnas.74.10.4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.