Abstract
RNA precursors give rise to mRNA after splicing of intronic sequences traditionally thought to occur in the nucleus. Here, we show that intron sequences are retained in a number of dendritically-targeted mRNAs, using microarray and Illumina sequencing of isolated dendritic mRNA as well as in situ hybridization. Many of the retained introns contain ID elements, a class of SINE retrotransposon. A portion of these SINEs confers dendritic targeting to exogenous and endogenous transcripts showing the necessity of ID-mediated mechanisms for the targeting of different transcripts to dendrites. ID elements are capable of selectively altering the distribution of endogenous proteins, providing a link between intronic SINEs and protein function. As such, the ID element represents the first common dendritic targeting element to be found across multiple RNAs. Retention of intronic sequence is a more general phenomenon then previously thought and plays a functional role in the biology of the neuron, partly mediated by co-opted repetitive sequences.
Introduction
Many mRNAs are targeted to neuronal dendrites and some are translated locally (Aakalu et al., 2001; Crino and Eberwine, 1996; Job and Eberwine, 2001; Miyashiro et al., 1994). While specific localization signals in model organisms such as Xenopus and Drosophila have been described for genes under strict temporal and spatial control in highly polarized cells (Dienstbier et al., 2009), the general mechanisms for targeting specific mRNAs to mammalian neuronal projections remain somewhat unclear (Bramham and Wells, 2007; Miyashiro et al., 2009). The targeting of mRNAs to dendrites requires the recognition of cis dendritic targeting elements (DTEs) by trans-acting RNA-binding proteins (RBPs), usually as part of a larger complex of ribonuclear proteins known as an RNA granule (Elvira et al., 2006; Kanai et al., 2004). Only a small number of DTEs have so far been found, including those associated with CamKIIα (Mayford et al., 1996), MAP2 (Blichenberg et al., 1999), β-actin (Eom et al., 2003), Arc (Kobayashi et al., 2005), and vasopressin (Prakash et al., 1997). DTEs are almost exclusively found in the 3′ untranslated region (UTR) of mRNAs and do not share an obvious consensus sequence (Andreassi and Riccio, 2009). The dearth of known DTEs for the many other localized RNAs (Eberwine et al., 2002) suggests that targeting signals may be transiently associated with the transcript, perhaps as a sequence or structure element that can be removed subsequent to localization.
Introns are known to harbor various regulatory elements, though most of these are presumed to be relevant only for processes occurring inside the nucleus, such as splicing. Intron-containing sequences in the cytoplasm are presumed destined for nonsense-mediated decay. However, recent studies indicate that retention of specific intronic, non-protein-coding sequences within cytoplasmic mRNA (Cytoplasmic Intron sequence-Retaining Transcripts, CIRTs) in mammalian neurons and other cells plays a role in producing functional proteins. The neuronal CIRT KCNMA1i16 contributes to the firing properties of hippocampal neurons and proper channel protein localization to dendrites (Bell et al., 2008). Intron retention within IL1-β mRNA in anucleate platelets has been implicated in governing activity-dependent splicing and translation upon cell activation (Denis et al., 2005). Finally, a retained intron in Tap mRNA contains a transport element that drives nuclear export in human 293T cells, facilitating expression of an alternate Tap protein product (Li et al., 2006).
Here we report that retention of intronic sequence can mediate dendritic localization in neuronal transcripts that contain a specific DTE that arose from neo-functionalization of ID elements, a class of short interspersed repetitive elements (SINE) derived from BC1 RNA, a rodent-specific dendritically localized non-coding RNA (Kim et al., 1994). Using microarray and Illumina sequencing of mRNA isolated from dendrites, in addition to in situ hybridization, we identified a large and diverse group of dendritically-localized CIRTs. by microarray and Illumina sequencing of mRNA isolated from dendrites and in situ hybridization. Computational analysis of the retained intron sequence revealed the enrichment of ID elements. Individual intronic ID elements from different genes were cloned, exogenously expressed in primary neurons, and shown by in situ hybridization to be capable of targeting mRNA to dendrites. Normal dendritic localization of ID-containing transcripts is disrupted when ID-containing transgenes compete for the dendritic targeting machinery, thus showing that ID-mediated localization is an endogenous mechanism. Our findings represent the first example of a general dendritic targeting mechanism for multiple different gene transcripts.
Results
Many CIRTs are detected within dendritic mRNA samples
To determine whether CIRTs are present in dendritic mRNA populations, we focused on a set of 33 candidate genes previously found to localize to dendrites in rat (Eberwine et al., 2002). Three batches of dendritic mRNA, each consisting of 150 to 300 individually dissected dendrites from primary rat hippocampal neurons, were independently aRNA-amplified (Miyashiro et al., 1994) and analyzed using a custom-built microarray consisting of probes generated from the 5′ ends of selected introns from each gene of interest. Three additional batches were subjected to Illumina NextGen sequencing. Sequencing allows us to recover minor, variably expressed CIRTs in the different RNA pools, while augmenting the analysis with microarrays provides additional evidence for a smaller set of hypothesized CIRTs, which may escape detection by sequencing due to low read depth or systematic biases such as nucleotide content (Harismendy et al., 2009).
Using these methods, many CIRTs were detected (Table 1). A wide range of expression was observed across the arrays, with intronic loci from CAMK2B and FMR1 among others consistently showing high signal (Figure S1A, Table S1). A similar pattern of intron retention was present in the sequencing data, supported by uniquely aligning end pairs to non-repetitive intronic regions (Figure S1B, Table S2). For some genes such as ADCY4 and GRIK1, sequence reads spanned intron-exon boundaries. Retention of intronic sequence appears to be regulated, as some intronic loci consistently show retention while others do not. Some genes such as CAMK2A and SNCB lacked intron retention despite the confirmed presence of exonic regions in the RNA pool.
Table 1.
Candidate retained intronic loci
| Refseq ID | Gene Symbol | Retained intronic loci |
|---|---|---|
| NM_031007 | ADCY2 | i2, i14, i18 |
| NM_130779 | ADCY3 | i2 |
| NM_019285 | ADCY4 | i11*, i12* |
| NM_022600 | ADCY5 | i17 |
| NM_019288 | APP | i1, i2, i5, i6, i8, i13, i15 |
| NM_147141 | CACNA1B | i18, i31 |
| NM_153814 | CACNA1H | i9 |
| NM_021739 | CAMK2B | i1, i3 |
| NM_012519 | CAMK2D | i2, i3, i11, i18 |
| NM_133605 | CAMK2G | i9, i13, i18 |
| NM_031334 | CDH1 | i15 |
| NM_031017 | CREB1 | i1 |
| NM_052804 | FMR1 | i1, i7, i12 |
| NM_031028 | GABBR1 | i4, i14 |
| NM_017289 | GABRD | i8 |
| NM_080587 | GABRA4 | i8 |
| NM_024370 | GABRG3 | i2, i3, i5, i9 |
| NM_032990 | GRIA3 | i2, i11*, i12, i13*, i15* |
| NM_017263 | GRIA4 | i3, i4, i7, i13* |
| NM_017241 | GRIK1 | i1, i7, i16* |
| NM_017010 | GRIN1 | i8 |
| NM_031040 | GRM7 | i1, i5, i6 |
| NM_031730 | KCND2 | i1 |
| NM_013066 | MAP2 | i4, i7, i9, i10 |
| NM_019169 | SNCA | i2, i4 |
| NM_012700 | STX1B2 | i1 |
Sequencing reads overlapping the splice junction were recovered
The protocol for RNA sample preparation, consisting of mechanical isolation of dendrites from cell bodies, makes it unlikely that the detected intronic sequence arises from genomic DNA or unspliced or non-specifically incompletely spliced nuclear mRNAs. Additionally, the pattern of sequence read coverage is inconsistent with these sources of contamination, as we found no significant enrichment in intergenic reads, nor did we find any systematic pattern of intron presence/absence across all genes. We also analyzed each retained intronic locus using base composition properties and public annotations and found that the majority had no evidence for unannotated alternate exons or overlapping genes (Supplemental Text).
CIRTs are localized to multiple subcellular regions
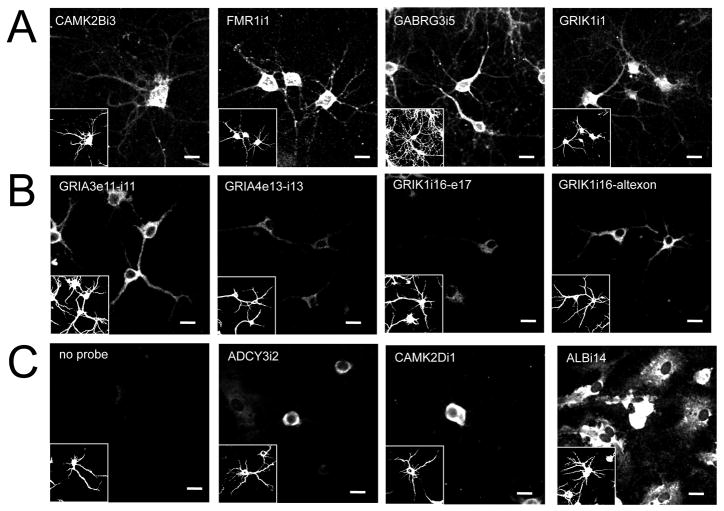
Based on the retained intronic loci detected in the initial screen, we selected several candidates to visualize using in situ hybridization to confirm retention and localization patterns. We assayed intronic probes designed to target microarrayed sequences from RNAs showing varying degrees of intronic sequence retention. Antisense riboprobes were generated and used for in situ hybridization to E18 rat neurons in primary cell culture. Cells were co-stained for MAP2 protein to indicate dendrito-somatic regions of neurons (Figure 1, insets). All sequences tested showed dendritic in situ hybridization signals consistent with microarray results (Figure 1A). In situ hybridization of exonic probes confirmed the dendritic localization patterns of the intron-containing transcripts (Figure S2).
Figure 1.
In situ hybridization results for intronic probes on cultured rat hippocampal neurons. Panels are labeled according to intronic sequence detected using biotinylated (A) riboprobes or (B) oligo probes. Negative controls (C) for cells labeled in absence of probe (No probe) and with probes to non-retained introns are also included for reference (exon targeted and sense probe results found in Figure S2). Glia cells (MAP2 negative) are clearly positive for ALBi14 as shown. Riboprobe sequences are identical to those printed on intron microarrays (see Table S2). Oligo probes are 24-mers corresponding to genomic exon/intron splice junctions found in Illumina sequencing results for dendrites. Cells were also immunostained for MAP2 to identify neuronal soma and dendrites (insets). Scale bars = 20μm.
Further, oligo probes to intron-exon junctions with sequencing support successfully confirmed that each region was within the dendritic compartment by in situ hybridization (Figure 1B). Interestingly, GRIK1 shows a higher dendritic signal for intron 16 joined with an alternate exon than to the canonical exon 17, suggesting an interaction between intronic sequence and the isoforms of the transcript in localization (Bell et al., 2010).
Retained intronic sequences contain common sequences, including ID elements
Given the widespread occurrence of CIRTs, we hypothesized that sequence elements important for mRNA regulation may be embedded in the retained intronic sequences and searched for putative regulatory sequences. We found several sets of sequences shared among different introns, including a large number of BC1 RNA-like ID elements.
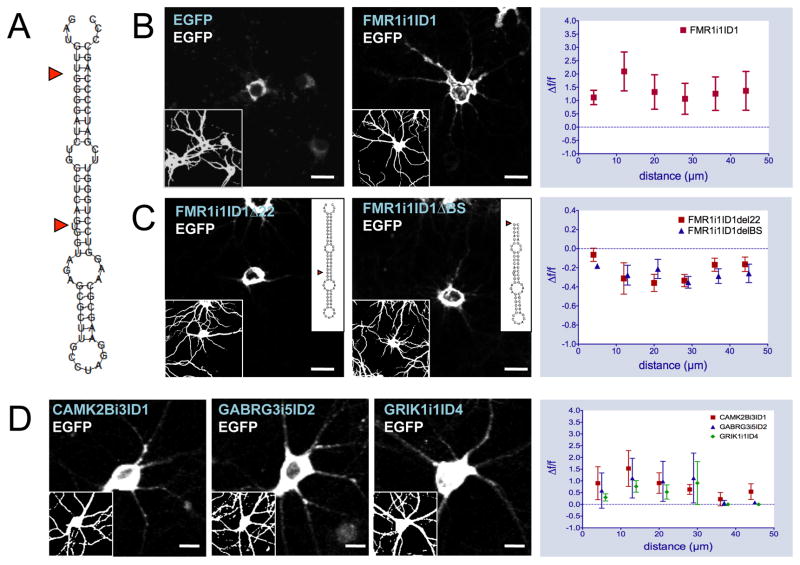
While ID elements are not unique to retained introns, many are found in the dendritic introns detected by microarray and sequencing. Among these intronic ID elements, we found that many retain motifs previously identified as BC1 localization signals that confer targeting to microinjected mRNA (Muslimov et al., 2006) as evidenced by their predicted secondary structures (Figure 2A). A total of 308 blocks of ID-derived sequence were found. Of these, 70 elements appearing in 46 introns across 23 genes were determined to possess mRNA targeting potential - occurring in the sense orientation and forming a hairpin structure with a basal-medial unbranched helix, a uracil at position 22, and at least 90 percent sequence identity to the BC1 5′ domain (Table S3).
Figure 2.
Intronic ID element sequences confer dendritic localization to reporter gene mRNA. Secondary structure of FMR1i1ID1 wild-type (A, inset for mutant sequences). In situ hybridizations with antisense biotinylated EGFP riboprobe on primary hippocampal neurons transfected with pEGFP-N1, FMR1i1ID1-EGFP wild-type (B), FMR1i1ID1 22-EGFP, and FMR1i1ID1 BS-EGFP mutant constructs (C). Blue text indicates transfected DNA construct, white text indicates in situ probe sequence. Insets represent MAP2 immunostaining (left) or secondary structure of FMR1i1ID1 mutants (right). Graphs at right represent in situ signal F/F (relative fluorescence signal difference) against distance from soma for FMR1i1ID1-EGFP wild-type versus pEGFP-N1 (top, expanded in Figure S3A–B), FMR1i1ID1 22-EGFP, and FMR1i1ID1 BS-EGFP mutants versus FMR1i1ID1-EGFP wild-type (bottom). (D) In situ hybridizations with antisense biotinylated EGFP riboprobe on hippocampal neurons transfected with CAMK2Bi3ID1ID1-EGFP, GABRG3i5ID2-EGFP, and GRIK1i1ID4-EGFP constructs. Graphs at right represent in situ signal F/F against distance from soma for each ID-EGFP construct versus pEGFP-N1. Scale bars = 20μm.
Sequencing data provided evidence that many of these ID-containing loci were present in the dendritic RNA pools. We estimate a 20-fold enrichment in ID element sequence compared to other SINEs in dendritic RNA (Table S4), though due to the high copy number of ID-related sequence in the rat genome, it is not possible to establish unambiguous genomic positions for most of the ID-derived reads. However, more than half of the 70 potentially functional ID loci fall within introns with uniquely mapping sequence reads, including six cases in which the ID elements themselves are spanned by end pairs uniquely aligning to neighboring non-repetitive sequence (Table S3).
Exogenous expression of ID element reporter sequences leads to dendritic targeting of mRNA
To test targeting efficacy of intron-derived ID elements, PCR products consisting of ID elements plus flanking sequence from retained intron regions were cloned into pEGFP-N1 expression vectors with the ID region placed upstream of the EGFP coding sequence. ID-EGFP transcripts are generated upon transfection into primary rat hippocampal neurons and detected by in situ hybridization targeted to the EGFP portion of the sequence. pEGFP-N1 transfected cells were used as a control for ID-independent RNA localization (Figure 2B). The in situ results show that ID elements from the retained introns do indeed confer dendritic targeting to the transgene mRNA (Figure 2B, 2D). Versions of the construct with selective mutations to the ID element sequence significantly disrupted dendritic targeting (Figure 2C, Supplemental Text). Similarly, targeting was not observed for a construct containing an FMR1i1-isolated B2 SINE instead of an ID element, confirming that general structured intronic sequence is insufficient to confer localization (Figure S3C).
To quantify the extent of targeting of the fusion constructs, we developed a custom program using Igor (WaveMetrics, Inc.) to measure probe intensity along curves drawn in the in situ images through the dendritic processes, originating at the somal end based on MAP2 immunostaining. For each of the assays described below, three dendrites were quantified per cell and 8 to 10 cells were quantified for each probe. A greater signal can be seen in ID-EGFP versus EGFP transfected cells at further distances away from the cell body for all four ID elements. Transcripts were present at distances of ~50–80 μm from the cell soma (>2x the diameter of the soma) (Figure 2B).
Actively transported RNAs are expected to have greater ISH intensity and a shallower gradient along the length of the dendrite, while non-actively transported RNA is expected to have less intensity and steeper gradients. We tested the intensity level differentials in 8 μm intervals along the dendrites out to a distance of approximately 50 μm from the soma and found that all test probes showed significantly greater signal intensity compared to the EGFP control (p < 1E-10, Fisher’s combined p-value for Bonferroni-corrected t-tests from each interval, see Supplemental Text). As a control for over-expression artifacts, we also used diffusion modeling to show fundamental differences in the targeting of exogenous EGFP transcripts relative to ID-EGFP transcripts and confirmed that the test probe in situs showed evidence of active transport of the mRNA (Supplemental Text).
Despite similar expression levels, different distribution patterns along the length of the dendrites are observed for different ID constructs, which can be described as diffuse (GABRG3i5ID2), punctate (CAMK2Bi3ID1) and intense (FMR1i1ID1). These findings suggest the existence of different targeting mechanisms for ID-containing sequences that may be governed by flanking sequence or subtle sequence/structural variations.
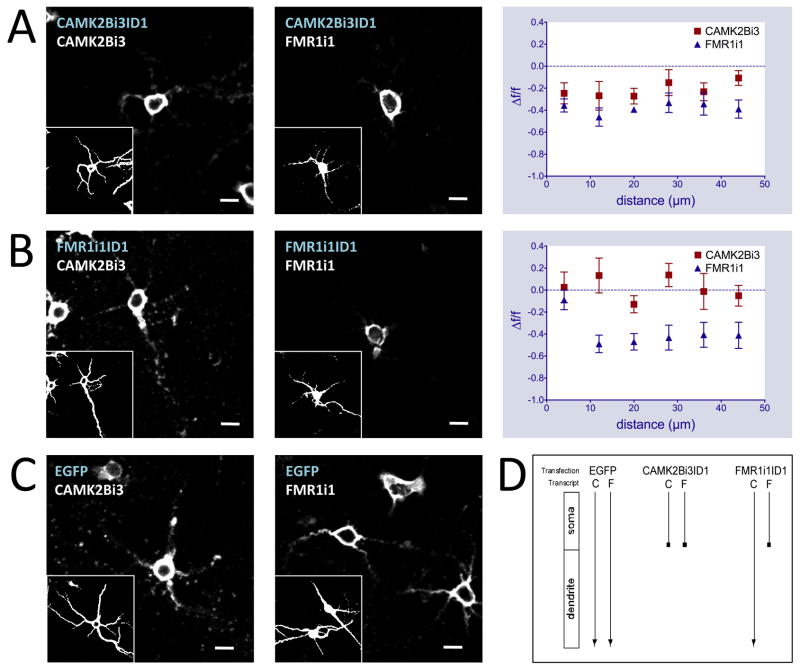
Transgene intronic ID elements compete with their endogenous intron-containing counterparts for targeting machinery
Using a stringent in vivo competition assay, we found that exogenous expression of ID elements can block the targeting of endogenous transcripts to dendrites. This results from transcript competition for localization machinery analogous to the competition between Drosophila I factor and gurken mRNA (Van De Bor et al., 2005). Forty-eight hours post-transfection with ID constructs, in situ hybridization was performed using probes directed at intronic regions absent from the transfected ID-EGFP transcripts. Only endogenous transcripts containing the intronic sequence would be detected, allowing the study of the ID-EGFP transcript’s effect on endogenous intron-containing mRNA (Figure 3). In all cases tested, ID-EGFP transfection significantly disrupted the localization of analogous endogenous intron-retaining transcripts, as measured by signal intensity differential along the length of the dendrite as above (p < 1E-6, Fisher and Bonferroni analysis, Supplemental Text). These data show that the endogenous localization mechanism is ID element dependent.
Figure 3.
Intronic ID element sequences disrupt dendritic localization patterns of endogenous mRNA. In situ hybridizations with antisense biotinylated intron riboprobes on primary hippocampal neurons transfected with CAMK2Bi3ID1-EGFP (A), FMR1i1ID1-EGFP (B), pEGFP-N1 (C) constructs (GABRG3i5ID2-EGFP and GRIK1i1ID4 results found in Figure S4). Blue text indicates transfected DNA construct, white text indicates in situ probe sequence. Graphs at right represent in situ signal F/F against distance from soma for ID-EGFP constructs versus pEGFP-N1 using CAMK2Bi3 (left) and FMR1i1 (right) riboprobes. Insets represent MAP2 immunostaining. Scale bars = 20μm. (D) Schematic of ID cross competition results. Transfection labels indicate transfected DNA constructs, transcript labels indicate endogenous intron-retaining transcripts: C is CAMK2Bi3, F is FMR1i1. Arrows indicate endogenous intron-retaining transcript targeting from soma to dendrites.
Transgene intronic ID elements selectively cross-compete with other endogenous intron-containing mRNAs for targeting
To determine whether targeting mechanisms are specific to particular ID element variants or common to all targeted transcripts containing ID elements, we performed cross-competition experiments to assess the capacity of an ID element from one gene’s transcript to disrupt the localization of a CIRT from a different gene. This was tested using probes to introns from genes that do not contain the particular ID element being exogenously expressed.
CAMK2Bi3ID1, when transfected into neurons, disrupts dendritic localization of endogenous CAMK2Bi3 transcripts and is also capable of disrupting FMR1i1 localization (Figure 3A), at a magnitude equal to or greater to that caused by FMR1i1ID1 (p < 1E-11, Fisher and Bonferroni analysis, Supplemental Text). This shows that the CAMK2Bi3-derived ID element can cross compete for targeting machinery with the endogenous FMR1 intron-retaining transcript, suggesting a dendritic localization mechanism that accepts both CAMK2Bi3 and FMR1i1 as substrates.
Conversely, transfection of FMR1i1ID1 disrupted the intronic in situ pattern of FMR1i1 transcripts while dendritic targeting of CAMK2Bi3 transcripts was only minimally affected (Figure 3B, p < 1E-5, Fisher and Bonferroni analysis). This indicates that a more selective mechanism exists for FMR1i1ID1-mediated targeting, for which CAMK2Bi3 is not a substrate, and that the FMR1i1ID1 sequence can target to dendrites in a distinct manner from CAMK2Bi3. Additionally, transfection of either GABRGi5ID or GRIK1i1ID had no effect on either FMR1i1 or CAMK2Bi3 localization (Figure S4) confirming the specificity of the FMR1i1ID1 and CAMK2Bi3ID1 driven mechanisms.
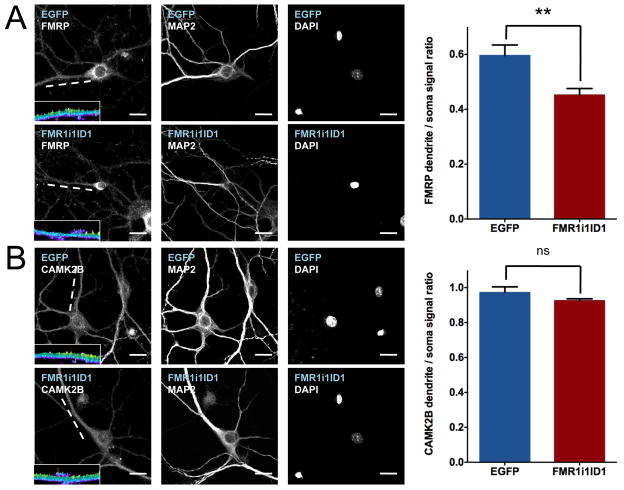
Transgene intronic ID element containing RNAs modify the subcellular distribution of endogenous FMRP protein
To assess whether transcript localization affects the location of the encoded protein product, we visualized the subcellular distribution of FMRP, which is encoded by FMR1, using immunofluorescence. FMRP is normally distributed throughout both the cell soma and dendrites of neurons, as was the case for cells transfected with EGFP (Figure 4A). Upon transfection with FMR1i1ID1-EGFP, the relative amount of FMRP in the dendrites decreases, with FMRP concentrating at the outer boundaries of the soma (Figure 4A). In contrast, subcellular distribution of CAMK2B protein is unaffected by FMR1i1ID1-EGFP transfection (Figure 4B). Thus, the function of this ID-element containing CIRTs is consistent with a role in regulating dendritically localized FMRP protein levels and subsequent function in the dendrite.
Figure 4.
Intronic ID element sequences modify subcellular distributions of endogenous proteins. Immunofluorescence with antibodies to FMRP (A) and CAMK2B (B) on primary hippocampal neurons transfected with pEGFP-N1 and FMR1i1ID1-EGFP constructs. Blue text indicates transfected DNA construct, white text indicates antibody target. Graphs at right represent immuno signal as ratios of dendrite to soma with SEM indicated. Mean dendritic to somal signal ratio +/− SEM for FMRP immunostaining: EGFP 0.56 +/− 0.04 (n=10), FMR1i1ID1 0.45 +/− 0.02 (n=13), t-test p = 0.0031. Mean dendritic to somal signal ratio +/− SEM for CAMK2B immunostaining: EGFP 0.97 +/− 0.03 (n=7), FMR1i1ID1 0.93 +/− 0.01 (n=9), t-test p = 0.16. Insets are three-dimensional topographical plots of signal intensity for the indicated dendrites (white lines) on a 0 (dark blue) to 2000 (yellow) pseudocolor range, showing a decreased signal for FMRP along the length of the FMR1i1ID1-transfected dendrite compared to the EGFP-transfected dendrite; no such difference is observed for CAMK2B. Scale bars = 20μm.
Discussion
Using three independent methods of detection in multiple cell cultures, we have described a large number of previously unreported intronic sequences in the dendritically localized mRNA of primary rat hippocampal neurons. These Cytoplasmic Intron sequence-Retaining Transcripts (CIRTs) represent a novel class of transcript that has important cellular function in neurons including involvement in dendritic targeting of mRNAs via co-opted retrotransposons. This is the first dendritic targeting element described outside the transcript UTR or coding region and the first common element to be found in more than two different gene transcripts. Our in situ hybridization results show a variety of dendritic distribution patterns, suggesting that localization is a complex process that likely involves multiple ID-element-dependent and independent mechanisms. The fact that exogenous expression of any particular intronic ID element does not necessarily disrupt targeting of all intron-retaining transcripts suggests the existence of multiple variant targeting mechanisms -- if only a single mechanism existed, transfection of any intronic ID element would block the targeting all endogenous intron-retaining transcripts containing an ID element. Our data reflect at least three targeting mechanisms for intron-retaining transcripts in dendrites: one that is distinct for FMR1i1ID1, one that is common to CAMK2Bi3iD1 and FMR1i1ID1, and at least one that is ID element independent. Further, we observed that FMRP localization is directly affected by the disruption of endogenous ID element-mediated FMR1i1 targeting, suggesting that CIRTs may be critically important in the presumptive role of FMRP in modulation of local dendritic protein synthesis (Huber et al., 2002; Weiler et al., 1997).
BC1-like ID elements have been implicated in brain-specific gene regulation since their discovery (Milner et al., 1984), and while it is believed that most of these sequences are removed from pre-mRNA via intron splicing, we show that some are retained in dendritically-localized mRNA and facilitate dendritic targeting of the host transcript. Previous work with, transgenic mice having ID elements fused to the 3′UTR of EGFP showed that these sequences were not sufficient for dendritic targeting (Khanam et al., 2007). Additionally, ID elements occurring endogenously in the 3′UTRs of neuronally expressed genes also showed no evidence of dendritic localization. In contrast, earlier work showed that microinjected BC1-containing chimeric RNAs were successfully targeted to dendrites (Muslimov et al., 1997). Our results suggest that the discrepancy may be sequence-position related and due to a requirement for partial nuclear processing of the nascent transcripts. If localization is coupled to splicing or nuclear export, it could be position dependent, such that 3′UTR-placement of ID elements (as was the case for the Khanam, et al., transgene constructs) is not favorable for driving localization, while ID elements in upstream regions (as in our constructs or endogenous CIRTs) are targeting competent. This is an intriguing idea given that the majority of known DTEs are in fact 3′UTR elements, suggesting unique regulation of ID element DTEs. There is evidence that specific targeting mechanisms can depend on intronic sequence; for example in Drosophila, correct localization of oskar mRNA to the posterior pole of a developing oocyte requires the presence of an intron (Hachet and Ephrussi, 2004).
Additionally, rats and mice are distinct with regard to the distribution of genomic regulatory elements. ID elements have undergone great expansion in rats, with approximately 150,000 well-formed instances of the 5′ targeting domain according to our analysis, while the mouse genome contains two orders of magnitude less (approximately 1000 instances). These numbers are consistent with a previous survey of ID elements in rodents, which suggested a wide variety of genomic distributions (Kass et al., 1996). This suggests that species-specific novel retroelement expansion may play a functional role in neuronal physiology in rodents and other lineages including primates, where BC200, a functional analog of BC1 RNA, is thought to have arisen from Alu retrotransposon functionalization (Tiedge et al., 1993). It is reasonable to speculate that the acquisition of some of these functional roles has been mediated by regulated novel processing of retained intronic sequences. Transposable elements have long been hypothesized to play a role in eukaryotic gene regulation (McClintock, 1950) and functionalization of retroelements has been suggested to provide a dynamic reservoir of rapid genome evolution (Kazazian, 2004). Here, we provide evidence for evolutionarily rapid functionalization of a mobile element.
In addition to harboring targeting elements, retained introns may also encode other regulatory structures with functions specific to the dendritic compartment. In searching for structured RNA segments within our focus genes, we found that retained introns are more likely to contain structures with low minimum-free energy z scores (MFEZ) (Clote et al., 2005) compared to introns with no retention evidence (p < 8.4 e-6, Wilcoxon rank-sum test on retained vs. non-retained repeat-masked introns), suggesting that retained introns may be enriched in functionally significant elements. An intriguing possibility is that microRNAs (miRNAs), a class of posttranscriptional expression regulators widely found in introns that can be co-transcribed with their host genes (Baskerville and Bartel, 2005; Kim and Kim, 2007), may act through cytoplasmic splicing of CIRTs (Glanzer et al., 2005). We have identified several candidate miRNAs within the retained introns that score favorably when evaluated by different miRNA-gene finding protocols and merit further investigation (Table S5), though whether these candidates are processed (nuclearly or cytoplasmically) is unclear at this stage.
From these observations, an appealing model emerges for transcript localization, in which a fraction of a gene’s transcripts are non-canonically spliced and participate in regulatory modulation. Processing of these transcripts to remove non-coding sequence post-transport – e.g. by activation upon cell stimulation by external signals – produces a translatable transcript in addition to potentially other intron-encoded RNAs that may further regulate either their own host transcript or a different gene’s products. Thus, incorrect cytoplasmic localization or processing may produce any of a number of downstream effects that may ultimately lead to brain disfunction. Recently Gage and colleagues have shown that L1 retrotransposon activities are increased in the absence of MeCP2 in rodents, and that human Rett syndrome patients carrying MeCP2 mutations have increased susceptibility for L1 retrotransposition (Marchetto et al., 2010; Muotri et al., 2010); previous work from the same group showed that L1 activity is an important component of brain development. Misregulation of L1 activity may also induce SINE activity, which may lead to mislocalization of critical RNA products in subcellular compartments of neurons. While we do not know whether SINE elements are involved in RNA localization in systems other than rat, our data provide an intriguing hypothesis that mechanistically connects retroviral element activity to cellular neurophysiology, with implications for viral etiology of neuropsychiatric diseases. As the diversity of evolutionarily novel targeting mechanisms come to be understood, insight into their regulation promises to provide important information about maintaining and enhancing brain tissue viability and function.
Materials and Methods
Culturing conditions
Hippocampi were harvested from embryonic day 18 rat pups, dispersed and plated at 100,000 cells per ml of Neurobasal medium and B27 (Invitrogen). Neurons are grown in culture for 14 days on 12mm round German Spiegelglas coverslips (Bellco) coated with poly-L lysine (Peptide Institute).
mRNA amplification
Independent samples of approximately 150–300 dendrites were harvested by mechanical isolation from primary rat hippocampal neurons and used as template material for aRNA amplification (as described in Eberwine et al., 2002).
Microarray preparation
PCR products corresponding to intron loci of interest were amplified and spotted onto glass slides for hybridization to labeled dendritic aRNA material. Signal intensities were background corrected, normalized, and compared to arrays hybridized with random hexamer sequence as a compositional control.
Illumina sequencing
Libraries were constructed from fragmented dendritic aRNA material and sequenced using Illumina II short read technology. Reads were aligned to RefSeq genes using Bowtie (Langmead et al., 2009).
In situ hybridization and imaging
Labeled antisense riboprobes were produced as runoff transcripts of subcloned PCR products or synthesized oligomers and hybridized to paraformaldehyde fixed and permeabolized primary rat hippocampal neurons at 42°C with 10ng/ul (for EGFP probes) or 20ng/ul (for intron probes) probe concentrations. Samples were visualized by confocal microscopy using either slit width (Olympus fluoview 1000, 60x N.A.1.2 or 20x N.A.0.7) or Meta detector (Zeiss 510 meta, 40x N.A 1.0). Collected images were processed using Metamorph software (Molecular Devices) and extracted information from regions of interest was used to quantify signals.
Immunostaining and imaging
Cells were immunostained for MAP2, FMRP (1c3 antibody, Greenough lab), and CAMK2B (ab89197 antibody, Abcam) proteins as indicated. Images were measured by drawing line paths from the center of the DAPI labeled nucleus into the dendrites based on MAP2 protein signal. Measurements were made in the FMRP or CAMK2B signal channel and used for calculations. Dendrite signal was defined by the soma proximal 20 terminal pixels measured (approximately 120 pixels from the center of the nucleus) and cell body signal was defined as the 20 pixels proximal to but not overlapping the DAPI signal.
Computational sequence analysis
Intronic ID elements were identified using BLAST (Altschul et al., 1990) and Repeat Masker (Smit, 1996–2004) and folded using Vienna RNA fold (Hofacker, 2003).
Supplementary Material
Acknowledgments
We thank M. Maronski for help with cell cultures, and C. Garner for the MAP2 antibody. This work was funded in part by HRF funds from the Commonwealth of Pennsylvania (JYK), DOE Computational Science Graduate Fellowship, DE-FG02-97ER25308 (MTL) and NIH AG9900 (JE).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TJ, Miyashiro KY, Sul JY, Buckley PT, Lee MT, McCullough R, Jochems J, Kim J, Cantor CR, Parsons TD, Eberwine JH. Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21152–21157. doi: 10.1073/pnas.1015264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell TJ, Miyashiro KY, Sul JY, McCullough R, Buckley PT, Jochems J, Meaney DF, Haydon P, Cantor C, Parsons TD, Eberwine J. Cytoplasmic BK(Ca) channel intron-containing mRNAs contribute to the intrinsic excitability of hippocampal neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1901–1906. doi: 10.1073/pnas.0711796105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blichenberg A, Schwanke B, Rehbein M, Garner CC, Richter D, Kindler S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J Neurosci. 1999;19:8818–8829. doi: 10.1523/JNEUROSCI.19-20-08818.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8:776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- Clote P, Ferre F, Kranakis E, Krizanc D. Structural RNA has lower folding energy than random RNA of the same dinucleotide frequency. RNA. 2005;11:578–591. doi: 10.1261/rna.7220505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino PB, Eberwine J. Molecular characterization of the dendritic growth cone: regulated mRNA transport and local protein synthesis. Neuron. 1996;17:1173–1187. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23:1546–1558. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochemical research. 2002;27:1065–1077. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- Eom T, Antar LN, Singer RH, Bassell GJ. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J Neurosci. 2003;23:10433–10444. doi: 10.1523/JNEUROSCI.23-32-10433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanzer J, Miyashiro KY, Sul JY, Barrett L, Belt B, Haydon P, Eberwine J. RNA splicing capability of live neuronal dendrites. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:16859–16864. doi: 10.1073/pnas.0503783102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, Frazer KA. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Job C, Eberwine J. Identification of sites for exponential translation in living dendrites. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13037–13042. doi: 10.1073/pnas.231485698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kass DH, Kim J, Deininger PL. Sporadic amplification of ID elements in rodents. J Mol Evol. 1996;42:7–14. doi: 10.1007/BF00163205. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Khanam T, Raabe CA, Kiefmann M, Handel S, Skryabin BV, Brosius J. Can ID repetitive elements serve as cis-acting dendritic targeting elements? An in vivo study. PLoS ONE. 2007;2:e961. doi: 10.1371/journal.pone.0000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Martignetti JA, Shen MR, Brosius J, Deininger P. Rodent BC1 RNA gene as a master gene for ID element amplification. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:3607–3611. doi: 10.1073/pnas.91.9.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Yamamoto S, Maruo T, Murakami F. Identification of a cis-acting element required for dendritic targeting of activity-regulated cytoskeleton-associated protein mRNA. Eur J Neurosci. 2005;22:2977–2984. doi: 10.1111/j.1460-9568.2005.04508.x. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bor YC, Misawa Y, Xue Y, Rekosh D, Hammarskjold ML. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:13250–13255. doi: 10.1073/pnas.93.23.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The origin and behavior of mutable loci in maize. Proceedings of the National Academy of Sciences of the United States of America. 1950;36:344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner RJ, Bloom FE, Lai C, Lerner RA, Sutcliffe JG. Brain-specific genes have identifier sequences in their introns. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:713–717. doi: 10.1073/pnas.81.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro K, Dichter M, Eberwine J. On the nature and differential distribution of mRNAs in hippocampal neurites: implications for neuronal functioning. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:10800–10804. doi: 10.1073/pnas.91.23.10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashiro KY, Bell TJ, Sul JY, Eberwine J. Subcellular neuropharmacology: the importance of intracellular targeting. Trends Pharmacol Sci. 2009;30:203–211. doi: 10.1016/j.tips.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Iacoangeli A, Brosius J, Tiedge H. Spatial codes in dendritic BC1 RNA. The Journal of cell biology. 2006;175:427–439. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J Neurosci. 1997;17:4722–4733. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash N, Fehr S, Mohr E, Richter D. Dendritic localization of rat vasopressin mRNA: ultrastructural analysis and mapping of targeting elements. Eur J Neurosci. 1997;9:523–532. doi: 10.1111/j.1460-9568.1997.tb01629.x. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2004. [Google Scholar]
- Tiedge H, Chen W, Brosius J. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J Neurosci. 1993;13:2382–2390. doi: 10.1523/JNEUROSCI.13-06-02382.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev Cell. 2005;9:51–62. doi: 10.1016/j.devcel.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Weiler IJ, Irwin SA, Klintsova AY, Spencer CM, Brazelton AD, Miyashiro K, Comery TA, Patel B, Eberwine J, Greenough WT. Fragile X mental retardation protein is translated near synapses in response to neurotransmitter activation. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:5395–5400. doi: 10.1073/pnas.94.10.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.