Abstract
RecQ-like DNA helicases are conserved from bacteria to humans. They perform functions in the maintenance of genome stability, and their mutation is associated with cancer predisposition and premature aging syndromes in humans. Here, a series of C-terminal deletions and point mutations of Sgs1, the only RecQ-like helicase in yeast, show that the HRDC and Rad51 interaction domain are dispensable for Sgs1’s role in suppressing genome instability, whereas the zinc-binding domain and the helicase domain are required. BLM expression from the native SGS1 promoter had no adverse effects on cell growth, but also was unable to complement any sgs1Δ defects. BLM overexpression, however, significantly increased the rate of accumulating GCRs in a dosage-dependent manner and greatly exacerbated sensitivity to DNA-damaging agents. Co-expressing sgs1 truncations of up to 900 residues, lacking all known functional domains of Sgs1, suppressed HU sensitivity of BLM overexpressing cells, suggesting a functional relationship between Sgs1 and BLM. Indeed, protein disorder prediction analysis of Sgs1 and BLM was used to produce a functional Sgs1-BLM chimera by replacing the N-terminus of BLM with the disordered N-terminus of Sgs1. The functionality of this chimera suggests that it is the disordered N-terminus, a site of protein binding and post-translational modification, that confers species-specificity to these two RecQ-like proteins.
INTRODUCTION
RecQ-like DNA helicases, named after the DNA repair protein RecQ of E. coli 1; 2; 3, are evolutionarily highly conserved. These 3’- to 5’-helicases function at the interface between DNA replication and recombination to maintain genome integrity. Sgs1 is the only known member of this helicase family in Saccharomyces cerevisiae 4. Sgs1-deficient cells show increased sensitivity to the DNA-damaging agents hydroxyurea (HU) and methylmethane sulfonate (MMS), missegregate chromosomes, accumulate gross-chromosomal rearrangements (GCRs) and have a shortened lifespan 5; 6; 7; 8. In contrast, five RecQ-like helicases (RecQL1, BLM, WRN, RecQL4 and RecQL5) are known in humans, and mutations in the BLM, WRN and RECQL4 genes are associated with the rare, cancer-prone Blooms syndrome, Werner syndrome and Rothmund Thompson syndrome, respectively 9; 10; 11; 12; 13. All RecQ-like helicases share a seven-motif helicase domain with Walker A and DEAH motifs. The RQC (RecQ-helicase-conserved) domain, located C-terminal to the helicase domain, is thought to be involved in DNA binding and conferring specificity of binding to DNA structures, such as G4-tetrads 14; 15; 16; 17. The HRDC (Helicase and RNaseD C-terminal) domain is the most C-terminal of the conserved domains and resembles domains in other proteins that are involved in nucleic acid metabolism, such as RNase D and UvrD; but, like the RQC domain, it is not found in all RecQ-like helicases 18; 19. The HRDC domain has been implicated in binding and resolving DNA structures, such as Holliday junctions, and in mediating protein-protein interactions 18; 20; 21; 22; 23. Two acidic regions have also been identified N-terminal of the helicase domain and may be involved in mediating protein-protein interactions 10; 24; 25. Sgs1 is found in a complex with Top3 and Rmi1, and there is also evidence of physical interactions of the N-terminal half of Sgs1 with Top2, Srs2 and Rad16, and interactions of the C-terminus with Mlh1 and Rad51 26; 27; 28; 29; 30; 31.
Defects in BLM, the human RecQ-helicase considered to be most closely related to Sgs1 cause Bloom’s syndrome (BS), an autosomal recessive disorder characterized by chromosome gaps and breaks, elevated sister chromatid exchange, mitotic hyper-recombination, and aberrant DNA replication events 32; 33; 34. Affected individuals suffer from a high incidence and wide variety of cancers, infertility and dwarfism (reviewed in reference 33). BLM catalyses ATP-dependent 3' to 5' DNA unwinding with a preference for DNA structures that may arise spontaneously during DNA replication or as a result of homologous recombination (HR) 35. For example, by unwinding unusual secondary DNA structures, BLM may aid replication fork progression, prevent illegitimate recombination during replication and assist in restarting stalled forks 36; 37; 38; 39. Evidence supporting a role of BLM in maintaining genome integrity has been accumulating. For example, BLM-defective cells exhibit a retarded rate of strand elongation during DNA replication 40, accumulate abnormal replication intermediates 41 and are hypersensitive to agents that impair DNA replication 42. BLM physically interacts with several proteins that play important roles during DNA replication and repair, such as replication protein A (RPA), the flap-endonuclease FEN-1, chromatin assembly factor CAF-1, the mismatch repair protein Mlh1, HR factor Rad51 and topoisomerase III α 43; 44; 45; 46; 47; 48; 49. BLM peaks in S phase and it localizes to replication foci, most likely through its physical interaction with a subunit of DNA polymerase δ 50; 51; 52; 53; 54.
Here we have determined the role of C-terminal domains and protein interaction sites of Sgs1 in suppressing GCR accumulation by expressing point mutants and truncations of Sgs1 lacking as few as 20 and as many as 1428 residues. To investigate BLM’s ability to complement sgs1Δ defects, such as increased genome instability and sensitivity to HU and MMS, human BLM cDNA was expressed under control of the native SGS1 promoter and overexpressed from a galactose-inducible promoter, revealing that BLM could suppress sgs1Δ defects neither in haploid nor in diploid cells. However, using computational protein disorder prediction tools, we have designed a yeast/human chimera that consists of two nonfunctional segments of BLM and Sgs1. The ability of this chimera to suppress all sgs1Δ defects that we tested suggests a functional relationship between BLM and Sgs1, which is also supported by our finding that short N-terminal fragments of Sgs1, which are devoid of all known functional domains for helicase activity and DNA binding, suppress severely detrimental effects of BLM overexpression in yeast.
Materials and Methods
Yeast Strains and Media
All strains are derived from KHSY802, a derivative of S288C. Yeast strains expressing truncations of Sgs1 helicase were constructed by homologous-recombination-mediated integration of PCR products, replacing the desired 3’-segment of SGS1 on chromosome VIII with a myc-epitope coding sequence (from pFA6a-13Myc.His3MX655, gift from Mark Longtine, University of Washington) in frame with the SGS1 coding sequence. Expression of all truncation alleles and the myc-epitope-tagged wildtype allele of SGS1 was confirmed by western blot analysis. All gene replacements, insertions and truncations were performed by the standard LiAc protocol 56, using PCR products with at least 50-nucleotides on each end that matched the chromosomal target locus. To express BLM from the native SGS1 promoter (PSGS1), a PCR fragment containing BLM cDNA (Open Biosystems) and a HIS3 cassette was amplified by PCR from plasmid pKHS293 using primers that include 50-nt homology to the chromosomal SGS1 locus. This PCR product was fused to the native chromosomal SGS1 promoter by homologous-recombination-mediated integration 56. A PCR fragment coding for a 13Myc epitope tag was amplified from pFA6a-13Myc-kanMX6 55 and integrated in-frame at the 3’end of cDNAs or sgs1 alleles for detection of protein expression by western blot analysis. In strain KHSY3350 and KHSY3218, galactose-inducible promoters amplified from plasmids pFA6a-kanMX6-PGAL1 or pFA6a-TRP1-PGAL155, respectively, were used to replace the native SGS1 promoter. To construct KHSY3355, the 3’-terminal 2313 bp of BLM cDNA linked to a HIS3 cassette were amplified by PCR from plasmid pKHS293 and used to replace the 3’-terminal 2400 bp of SGS1 in KHSY802. The accuracy of PCR-derived SGS1 or BLM integrations was confirmed by sequencing. Amino acid changes C1047F and F1056A in Sgs1 were made by site-directed mutagenesis (QuikChange, Stratagene) of pKHS360 and integrated at the sgs1::HIS3 locus in KHSY1338. All yeast strains used in this study are listed in Table S1 (Supplementary Information). Cells were grown in YPD consisting of 10g/l yeast extract (Fisher Scientific), 20 g/l Bacto-peptone (BD Diagnostic Systems), 2% glucose (Fisher Scientific), unless indicated otherwise. For plates, agar (BD Diagnostic Systems) was added at a concentration of 20 g/l.
Western blot analysis
To confirm expression of myc-epitope tagged BLM and SGS1 alleles, cells were grown to OD600 = 0.5 in YPD and whole cell extracts were prepared from 5 ml of culture (~ 3.5 × 107 cells) by standard trichloroacetic acid (TCA, Fisher Scientific) extraction 57. Five microliters of TCA extract were separated on 10% polyacrylamide gels, transferred to a PVDF membrane (BioRad), probed with anti-c-myc monoclonal antibody (9E10, Covance Research Products) and visualized by chemiluminescence (ECL Plus, GE Healthcare). To confirm expression of SGS1 and BLM from the GAL1 promoter the same western blot procedure was used, but cells were grown overnight in YP ((10g/l yeast extract (Fisher Scientific), 20 g/l Bacto-peptone (BD Diagnostic Systems)) supplemented with 2% sucrose (Fisher Scientific), then diluted to OD600 = 0.2 either in YP supplemented with 2% sucrose (uninduced sample) or 2% galactose (induced sample) and harvested for TCA extraction when cultures reached OD600 = 0.5. Molecular weight marker (Broad Range) was from BioRad.
Sensitivity to DNA damaging agents HU and MMS
Cell cultures were grown in YPD to OD600 = 0.5 and 10-fold serial dilutions were spotted on YPD supplemented with 0.05% methyl-methanesulfonate (MMS, Sigma Aldrich) or hydroxyurea (HU, US Biologicals) at 50 mM or 100 mM, as indicated. For experiments that included strains expressing BLM or Sgs1 from the GAL1 promoter (Figure 5), cultures were grown in YP-2% sucrose instead of YPD, and spotted on YP-1 % sucrose + 1% galactose (to induce gene expression) supplemented with 100 mM HU, or without HU as the growth control.
Figure 5.
HU sensitivity of diploid cells expressing BLM and mutant alleles of SGS1. (A) Ten-fold dilutions of exponentially growing diploids expressing truncation alleles of SGS1 in the presence of a wildtype allele were spotted on YPD media with and without 100 mM HU. (B) Ten-fold dilutions of exponentially growing diploids expressing truncation alleles of SGS1 in the absence of a wildtype allele were spotted on YPD media with and without 100 mM HU. (C) Ten-fold dilutions of exponentially growing diploids overexpressing BLM from a GAL1 promoter inserted at the native SGS1 locus and expressing truncation alleles of SGS1 under control of the native SGS1 promoter on the other allele were spotted on media containing 1% galactose (to induce gene expression) and 1% sucrose with or without 100 mM HU.
GCR rate measurements
Rates of accumulating gross-chromosomal rearrangements (GCRs) in YPD were determined as previously described 58. For GCR rate measurements of yeast strains expressing BLM or Sgs1 from the GAL1 promoter the same procedure was followed, except that media was supplemented with 2% galactose to induce gene expression. Briefly, 10 ml of YP-2% galactose were inoculated with a single colony, which had been grown on YPD agar for 3 days. After 3 days of growth in liquid media at 30° C with vigorous shaking, cells were plated on GCR plates 58 supplemented with 2% galactose instead of 2% glucose, and 10−6 dilutions were plated on YPD to obtain the viable cell count. Colonies on GCR plates were counted after 5 days of incubation at 30° C. For GCR rate measurements in the presence of varying BLM expression levels (Table 2), 0.1% or 0.5% galactose was added to liquid YP media and to GCR plates instead of 2% galactose, and sucrose was supplemented to reach a total of 2% sugar in the media. 95% confidence intervals were calculated according to Nair 59.
TABLE 2.
Effect of BLM expression on GCR accumulation in the sgs1Δ mutant.
| Relevant Genotype1 | Galactose concentration in media | GCR Rate (Canr 5-FOAr × 10−10) | 95% CI2 (Canr 5-FOAr × 10−10) |
|---|---|---|---|
| wildtype | 0% | 1.1 | < 1 – 6.2 |
| PSGS1BLM | 0% | 70 | 56–151 |
| PGALBLM | 0% | 61 | 30–153 |
| PGALBLM | 0.1% | 335 | 233–576 |
| PGALBLM | 0.5% | 382 | 170–777 |
| PGALBLM | 2% | 1832 | 1090–2910 |
| PGALSGS1 | 2% | < 11 | < 9–12 |
| sgs1Δ | 2% | 54 | 23–104 |
Human BLM cDNA was inserted at the endogenous SGS1 locus, fused to the native SGS1 promoter (PSGS1) or fused to a galactose-inducible promoter (PGAL). In PGALSGS1, the native SGS1 promoter region was disrupted by fusing the SGS1 ORF to a galactose-inducible promoter. If strains expressing BLM or SGS1 genes from the galactose-inducible GAL1 promoter were grown in less than 2% galactose (to lower protein expression levels) media was supplemented with sucrose to reach a total sugar concentration of 2%.
95% confidence intervals (CI) were calculated according to Nair 59.
Random Spore Analysis
Diploids heterozygous for the desired mutant alleles were grown overnight at 30°C in YPD, washed, transferred to 0.1% potassium acetate (Fisher Scientific) and incubated for 5 days at 30°C with vigorous shaking. Asci were incubated in the presence of zymolase (MP Biomedicals, 500 μg/ml) in 1 M sorbitol (Fisher Scientific) for 20 min at 30°C and enriched for haploid spores as previously described 60. Spores were plated on YPD, incubated at 30°C and genotyped by spotting on synthetic drop-out media (US Biologicals) to detect the presence of TRP1 and HIS3 marker cassettes linked to the mutant alleles. Presence of mutant alleles linked to the kanMX6 cassette was detected by the ability of haploids to grow on YPD supplemented with 200 μg/ml G418 (Axxora LLC, San Diego, CA).
RESULTS
Requirement of the RQC domain of Sgs1, but not the HRDC domain, for GCR suppression
Sgs1 contains a conserved DEAH helicase domain, a conserved helicase and RNaseD C-terminal (HRDC) domain, two acid regions (AR1, AR2) and a RecQ-conserved (RQC) domain composed of zinc-binding and winged-helix domains. Several protein interaction sites have also been located in the 1447-amino-acid long protein (Figure 1A). To determine the role of these domains in the maintenance of genome stability, systematic deletions to the 3’ end of the chromosomal SGS1 gene were generated, such that truncations of the C-terminus of Sgs1, ranging from 20 to 1428 amino acids, were expressed as fusions to a myc-epitope. Truncations of up to 80 amino acids were constructed to not affect any known functional domain of Sgs1 while ΔC100 and ΔC200 deletions partially or completely, respectively, removed the HRDC domain and ΔC300 and ΔC400 deletions partially or completely removed the RQC domain. The largest deletions (ΔC700, ΔC800, ΔC900, ΔC1000 and ΔC1100) eliminate the entire helicase domain, including the Walker A motif (803–812 aa), with the ΔC800-ΔC1100 deletions also affecting the part of the N-terminal half of Sgs1 that contains protein interaction sites (e.g., Rad16, residues 421–792; Top2, residues 432–724; Srs2, residues 422–722) and two acid regions (AR1, residues 321–447; AR2, residues 502–648), whereas ΔC500 and ΔC600 deletions partially remove the helicase domain while leaving the Walker A motif intact (Figure 1A). All truncation alleles were stably expressed from the chromosomal SGS1 locus under control of the native SGS1 promoter (Figure 1B). C-terminal fusion to the myc-epitope did not adversely affect Sgs1 function, as indicated by equal sensitivity to HU and MMS of strains expressing tagged and untagged Sgs1 (wildtype) (Figure 2A). The largest deletion, leaving intact only the 19 N-terminal amino acids of Sgs1 (sgs1ΔC1428), was as sensitive to HU and MMS as a complete SGS1 deletion (sgs1Δ), thus behaving like a null allele (Figure 2A). Loss of up to 200 C-terminal amino acids did not increase sensitivity to HU or MMS, whereas loss of 300 or more amino acids led to sensitivity similar to that of the sgs1ΔC1428 and sgs1Δ mutants (Figure 2A). The construction of additional 20-amino-acid truncations extended the C-terminal region that is dispensable for HU/MMS resistance to 240 amino acids (Figure 2B).
Figure 1.
C-terminal truncations of Sgs1 used in this study. (A) Full-length Sgs1 contains a DEAH-helicase domain, an RQC domain and an HRDC domain in its C-terminal half and acidic regions AR1 and AR2 in its N-terminal half; interaction sites with Top3, Top2, Srs2, Rad51 and Rad16 are indicated. C-terminal truncations ranging in size from 200 residues to 1428 residues were constructed by fusion to a myc-epitope tag. All truncations were introduced at the endogenous SGS1 locus on chromosome VIII. (B) Expression of wildtype Sgs1 and truncation alleles from the endogenous SGS1 promoter (PSGS1) was confirmed by western blotting, using a myc-antibody. Molecular weights (MW) are indicated on the left.
Figure 2.
Sensitivity of cells expressing Sgs1 truncation alleles to the DNA damaging agents HU and MMS. Ten-fold dilutions of exponentially growing cultures (OD600 = 0.5) were spotted on YPD for viable cell count and on YPD containing 100mM HU or 0.05% MMS, followed by incubation at 30° C. (A) Haploid cells expressing sgs1 alleles lacking 300 or more residues from the C-terminus are as sensitive to HU and MMS as the null allele. (B). Additional incremental 20-amino-acid deletions reveal that cells expressing sgs1 alleles lacking up to 240 residues are as resistant to HU and MMS as wildtype cells whereas those lacking 260 or more residues are as sensitive as the sgs1Δ mutant. (C) Spores from diploids heterozygous for an srs2Δ deletion and heterozygous either for the sgs1-ΔC200, sgs1Δ-C260 or sgs1-ΔC300 were spread on YPD to allow for growth of spores of all possible genotypes. Similar sized colonies obtained from the spores of the diploid heterozygous for sgs1ΔC200 and srs2Δ mutations (left) indicate that the sgs1ΔC200 srs2Δ mutant grows as well as the single mutants, suggesting that deletion of the C-terminal 200 amino acid residues does not negatively affect growth of the srs2Δ mutant. In contrast, spores from diploids heterozygous for the srs2Δ mutation and the sgs1ΔC260 allele (middle) or the sgs1ΔC300 allele (right), grew into a mixture of normal-sized colonies (corresponding to single mutants and wildtype) and small-sized colonies (corresponding to srs2Δ sgs1ΔC260 or srs2Δ sgs1ΔC300 mutants as determined by genotyping), demonstrating that an intact RQC domain in Sgs1 is required for the viability of the srs2Δ mutant.
It was previously shown that cells lacking the DNA helicase Srs2 (srs2Δ) depend on functional Sgs1 for their viability 61. To assess the ability of sgs1 truncation alleles to support growth of the srs2Δ mutant, we constructed diploid strains heterozygous for the srs2Δ deletion and heterozygous for the sgs1ΔC200, sgs1ΔC260 or sgs1ΔC300 alleles. The meiotic products of the sporulated diploids were spread on nonselective, rich media (YPD), allowing all spores to grow (Figure 2C). Diploids heterozygous for the srs2Δ deletion and the sgs1ΔC200 truncation yielded spores that grew into colonies of the same size, suggesting that the C-terminal 200 amino acid residues of Sgs1, which harbor the HRDC domain and an interaction site with the homologous recombination factor Rad51, are not required for the viability of the srs2Δ mutant. In contrast, sporulation of diploids heterozygous for the srs2Δ deletion and sgs1ΔC260 or sgs1ΔC300 alleles yielded mixtures of normal-sized and small colonies. Genotyping revealed that the small colonies were srs2Δ sgs1ΔC260 or srs2Δ sgs1ΔC300 mutants whereas the normal-sized colonies corresponded to wildtype spores or single mutants. Thus, Sgs1 that lacks 260 or more C-terminal residues and therefore does not contain a complete RQC domain cannot support normal growth of cells lacking Srs2.
When we tested the effect of the C-terminal deletions on the accumulation of GCRs, we found that the C-terminal 240 amino acids were dispensable for maintaining genome integrity, whereas deleting as little as an additional 20 amino acids (sgs1ΔC260) caused the GCR rate to increase to that exhibited by the null mutant without a discernable intermediate phenotype (Table 1). Combining the sgs1ΔC300 truncation allele with a deletion of the DNA-damage checkpoint sensor MEC3 led to a synergistic GCR rate increase, while, as expected, combining the sgs1ΔC200 allele with a mec3Δ mutation did not. Thus, these findings show that the HRDC domain and the previously reported C-terminal interaction with Rad51 are not required for Sgs1’s role in preventing the accumulation of GCRs and supporting normal growth of the srs2Δ mutant, whereas the integrity of the RQC domain, which has been suggested to span amino acids 1075 to 1207 based on the alignment of three-dimensional structures 62, is essential.
TABLE 1.
Accumulation of gross-chromosomal rearrangements in cells expressing mutant alleles of SGS1.
| Relevant Genotypei | GCR Rate (Canr 5-FOAr × 10−10) | 95% CIii (Canr 5-FOAr × 10−10) |
|---|---|---|
| wildtype | 1.1 | < 1 – 6.2 |
| sgs1Δ | 251 | 80–310 |
| sgs1ΔC200 | 7 | <6–23 |
| sgs1ΔC220 | 31 | 5–41 |
| sgs1ΔC240 | 10 | <6–27 |
| sgs1ΔC260 | 159 | 85–362 |
| sgs1ΔC280 | 244 | 166–387 |
| sgs1ΔC300 | 145 | 76–204 |
| sgs1ΔC400 | 106 | 60–180 |
| sgs1ΔC500 | 102 | 53–252 |
| sgs1ΔC600 | 152 | 26–283 |
| sgs1ΔC700 | 189 | 49–271 |
| sgs1ΔC800 | 133 | 71–225 |
| sgs1ΔC1428 | 206 | 97–273 |
| sgs1-C1047F | 64 | 35–131 |
| sgs1-F1056A | <16 | <10–26 |
| mec3Δ sgs1ΔC200 | 11 | < 7 – 22 |
| mec3Δ sgs1ΔC300 | 1003 | 691–1500 |
| mec3Δ sgs1ΔC800 | 758 | 645–895 |
| mec3Δ sgs1ΔC800-blmΔN647 c | 361 | 330–419 |
All sgs1 truncations (sgs1ΔC) are C-terminally fused to a myc-epitope tag.
95% confidence intervals (CI) were calculated according to Nair 59.
The sgs1ΔC800-blmΔN647 allele expresses a chimeric protein that consists of the N-terminal 647 residues of Sgs1 and the C-terminal 770 residues of human BLM.
Bloom’s Syndrome Associated RQC Domain Mutations Cause Loss of Sgs1 Function in vivo
Of the 32 exonic base substitutions that are causative of Bloom’s syndrome, thirteen are missense mutations 9; 13; 63; 64, with six of these mutations affecting conserved residues that have been shown in vitro to participate in zinc binding and G-tetrad DNA binding activity (Figure 3A). Studies, however, have been limited to biochemical and biophysical analyses of mutant proteins and were hampered by the inability to purify some mutant BLM proteins 16; 17; 65. Since the cysteine residues are highly conserved between RecQ-like helicases, including Sgs1, we replaced the corresponding cysteine residue in Sgs1 with the BS-associated mutation (sgs1-C1047F). Unlike BLM with mutations in any of the three conserved cysteine residues C1036, C1063 or C1066, which degraded upon purification and could therefore not be characterized 65, the sgs1-C1047F mutant allele was stably expressed in vivo from the native SGS1 locus (Figure 3B). The sgs1-C1047F mutant showed increased HU and MMS sensitivity, which, however, did not reach the level of the sgs1Δ allele, and exhibited levels of GCR accumulation comparable to the sgs1Δ mutant, demonstrating that the C1047F mutation severely impairs Sgs1 function (Figure 3C, Table 1). In addition to conserved cysteine residues and immediately adjoining arginine (R1037) and aspartic acid (D1064) residues, ClustalW2 alignments showed F1056 to be the only other fully conserved amino acid residue in the zinc-binding domain of Sgs1 (Figure 3A). Although the corresponding residue in BLM (F1045) is not associated with a BS mutation, the BLM-F1045A mutation has been shown to cause a severe helicase defect and ssDNA binding deficiency in vitro 65. When we introduced the corresponding mutation into Sgs1 (F1056A), however, the mutant was no more sensitive to HU and MMS than wildtype cells (Figure 3C), but instead appeared fully functional with a wildtype GCR rate (Table 1).
Figure 3.
Effect of zinc-binding domain mutations on Sgs1 function in vivo. (A) Zinc-binding domain is conserved from bacterial to human RecQ-like DNA helicases. Protein sequences were aligned with ClustalW2 84. The alignment of RecQL1 was manually adjusted. Amino acid residues identical in all sequences are highlighted in gray and indicated by '*' below the alignment, conserved substitutions are indicated by ':' below the alignment, and cysteine residues thought to be involved in zinc-binding are shown in red. At least six different missense mutations in the zinc-binding domain are associated with Bloom’s syndrome. (B) C1047F and F1056A mutations were introduced into Sgs1 and expression was confirmed by western blot using antibody against the C-terminal myc-epitope. Molecular weights (MW) are indicated in kDa to the left. (C) Mutation of the highly conserved F1056 does not impair Sgs1 function whereas the C1047F mutation leads to an increase in sensitivity to HU and MMS, but not to the level seen in the sgs1Δ mutant.
Expression of human BLM cDNA from the endogenous SGS1 promoter does not complement Δsgs1 defects
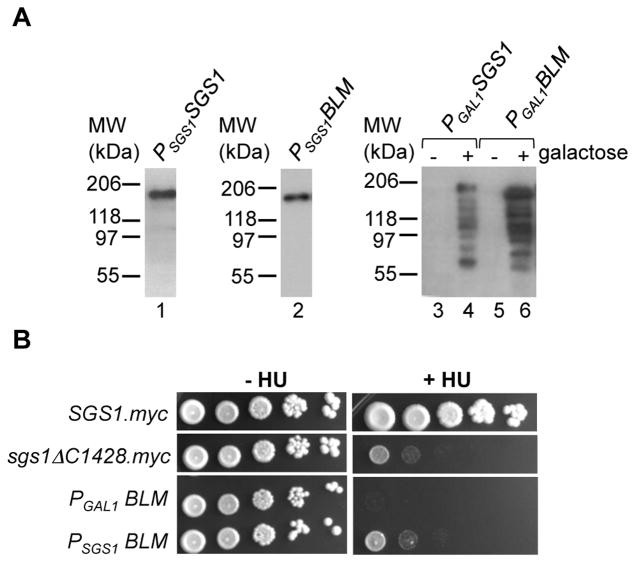
RecQ-like DNA helicases are evolutionarily conserved from bacteria to humans. Since cells from BS patients share defects seen in sgs1Δ cells, including increased sensitivity to DNA-damaging agents, increased levels of aberrant genetic exchange and reduced life-span, it has been suggested that RecQ-like DNA helicases from different phyla or even kingdoms might complement each other, thus allowing the development of simple model organisms for the functional and mutational characterization of disease-associated human RecQ-like helicases, such as BLM and WRN 66. Thus, to assess the ability of BLM to suppress genome instability in the sgs1Δ mutant, BLM cDNA was inserted in-frame with the start codon of SGS1 at its chromosomal locus (PSGS1BLM). We reasoned that insertion at the wildtype SGS1 locus would promote cell-cycle-dependent regulation of BLM expression and expression levels similar to those previously shown for Sgs167. Stable expression of BLM was confirmed by western blot analysis, using a yeast strain expressing myc-tagged BLM (Figure 4A); however, all subsequent experiments were carried out with untagged BLM. Expression of a single copy of BLM (PSGS1 BLM ) did not lead to a statistically significant difference in the GCR rate compared to the sgs1Δ mutant (Table 1, Table 2), or alleviate HU sensitivity (Figure 4B), demonstrating that BLM can be successfully expressed in yeast under control of the native SGS1 promoter without detrimental effects on cell growth, but is unable to complement the tested sgs1Δ defects to any extent.
Figure 4.
BLM expression does not suppress sgs1Δ defects and BLM overexpression is detrimental to yeast cells. (A) Expression of myc-epitope tagged Sgs1 (lane 1) and BLM (lane 2) from the native chromosomal SGS1 locus or galactose-inducible overexpression of myc-epitope tagged Sgs1 (lane 4) and BLM (lane 6) in yeast cells grown in YP supplemented with 1% sucrose and 1% galactose (to induce expression, lanes 4 and 6) or without galactose (lanes 3 and 5). Both BLM and Sgs1 show signs of degradation upon overexpression (lanes 4 and 6) whereas expression from the native SGS1 promoter is stable (lanes 1 and 2). Molecular weights (MW) are indicated in kDa on the left. (B) Cells expressing BLM from the SGS1 promoter on chromosome VIII are as sensitive to HU as cells lacking Sgs1 (Δsgs1). Replacement of the natural SGS1 promoter with a galactose-inducible GAL1 promoter induces BLM overexpression and leads to increased HU sensitivity. Ten-fold dilutions of cells were spotted on media containing 1% sucrose and 1% galactose (to induce BLM overexpression) with and without 100 mM HU.
Overexpression of BLM leads to increased sensitivity to DNA damaging agents and rapid accumulation of GCRs
Since a single copy of BLM (PSGS1BLM) did not complement Δsgs1 defects, we examined the effect of increasing BLM expression levels on sgs1Δ mutant phenotypes. For this purpose, the native SGS1 promoter was replaced with a GAL1 promoter and galactose-dependent expression of BLM was verified by fusing BLM to a myc-epitope tag (Figure 4A). Overexpression of BLM did not compensate for the lack of Sgs1 when cells were exposed to HU, but instead led to a further increase in sensitivity to HU compared to the sgs1ΔC1428 cells or cells expressing BLM under the SGS1 promoter (Figure 4B). We found that maximum induction of BLM expression led to a 1665-fold increase in the GCR rate compared to wildtype and a 34-fold increase compared to the sgs1Δ mutant assayed under the same conditions (Table 2). In contrast, overexpression of Sgs1 from the GAL1 promoter did not lead to GCR accumulation (Table 2). The GCR rate increase upon BLM overexpression was dependent on induction levels, with the GCR rate gradually decreasing to that of the sgs1Δ mutant as the galactose concentration in the media decreased (Table 2). Thus, sgs1Δ defects cannot be complemented by any level of BLM expression; in fact, increasing BLM expression levels induce higher sensitivity to DNA damaging agents and significantly higher genome instability compared to the sgs1Δ mutant.
N-terminus of Sgs1 suppresses detrimental effects of BLM overexpression
Since Sgs1 is important for the suppression of illegitimate recombination between identical sequences, such as those found in related genes, on homologous chromosomes and sister-chromatids, we tested HU sensitivity of diploid strains expressing truncated sgs1 alleles in the presence or absence of the SGS1 wildtype allele (Figure 5). HU sensitivity was fully suppressed for all alleles if a single copy of wildtype SGS1 was expressed from the other allele (Figure 5A), demonstrating that the sgs1 truncation alleles did not have a dominant effect. As in haploid cells, only the sgs1ΔC200 allele complemented HU sensitivity of the sgs1Δ diploid completely (Figure 5B); however, cells expressing the sgs1ΔC300 to sgs1ΔC900 alleles were less sensitive than diploids that expressed larger truncations or the sgs1ΔC1428 null allele (Figure 5B). This ability of sgs1Δ300 to sgs1ΔC900 truncation alleles to at least partially suppress HU sensitivity indicates that there may be N-terminal segments in Sgs1 that contribute to HU resistance.
Diploids expressing BLM from native SGS1 promoters on both alleles were as sensitive to HU as diploids not expressing Sgs1, whereas diploids overexpressing BLM from one allele or from both alleles were severely HU-sensitive, with the highest expression level lacking any growth on 100 mM HU (Figure 5C), reflecting the severe HU sensitivity of haploid cells expressing the PGALBLM allele (Figure 4B). Diploids overexpressing BLM also appeared to grow more slowly than any other diploid tested here (Figure 5C). Remarkably, expression of a single copy of SGS1 from its endogenous promoter (SGS1/PGALBLM) completely eliminated the severe HU sensitivity conferred by overexpression of BLM. To determine if full-length Sgs1 was required for this suppression, we crossed the haploid strain overexpressing BLM with haploids expressing various Sgs1 truncations. We found that a single copy of the sgs1ΔC200 allele was as sufficient as wildtype Sgs1 in suppressing HU sensitivity and slow growth of the BLM overexpressing strain, and as few as the N-terminal 547 amino acids remaining in the sgs1ΔC900 allele were sufficient for significant suppression of HU sensitivity and slow growth caused by BLM overexpression (Figure 5C). These findings suggest that none of the known enzymatic activities or functional and conserved domains are required for suppressing the HU sensitivity of the BLM overexpressing diploids, but that the N-terminal 547 amino acids are sufficient for suppressing the detrimental effects of BLM overexpression in a diploid. That the sgs1ΔC1000 and sgs1ΔC1100 alleles were clearly less effective at suppressing HU sensitivity shows that the N-terminal 447 amino acids, which contain the Top3 interaction site, are necessary but not sufficient for complementation.
Design of a functional Sgs1-BLM chimera
Sgs1 and BLM share about 21% of their amino acid residues in a pair-wise alignment of the full-length proteins (ClustalW2), with most of the identical residues in the helicase domain. In fact, the N-terminal segment of Sgs1 expressed by the sgs1ΔC800 allele, which is able to suppress the HU sensitivity of BLM-overexpressing diploids, shares only 11% with the corresponding N-terminal segment of BLM. Devoid of conserved domains and known enzymatic activities, the N-terminus of Sgs1 has been shown to be required for physical interactions with Top3, Top2, Srs2 and Rad16 6; 26; 27; 28; 29; 30. Using IUPred, an algorithm for the prediction of intrinsically disordered proteins, we found that the N-terminal 650 residues contain a similar distribution of ordered and intrinsically disordered segments (Figure 6A, B). In disorder prediction algorithms, such as IUPred 68; 69, a score of > 0.5 predicts a disordered amino acid residue and a score of < 0.5 predicts an ordered residue, with 30 consecutive disordered amino acids commonly being used as a lower limit for detecting disorder in whole proteome searches 68; 69; 70; 71. The helicase domains of Sgs1 and BLM coincide with the predicted ordered regions in both proteins, starting at around residue 648, and are surrounded by a long N-terminal and a short C-terminal segment, which contain mostly disordered residues. In fact, using the IUPred output scores, 83% of the 648 N-terminal residues of Sgs1 (538/648) are disordered, with 70% of all 648 residues being located in segments of more than 30 consecutive disordered residues, whereas only 16% of the C-terminal 800 residues of Sgs1 are predicted to be disordered, with only a single disordered segment that is longer than 30 residues (residues 1396–1447). Based on the IUPred prediction, BLM can also be divided into a disordered N-terminus and an ordered C-terminus (Figure 6A, B). For BLM, 52% of the N-terminal 648 residues are predicted to be disordered but only 15% of these residues are found in stretches of more than 30 disordered residues. The difference in the pattern of disorder predicted for the N-terminal segments of Sgs1 and BLM led us to hypothesize that this region may be involved in conferring species-specificity to BLM and Sgs1 function and, thus, prevent BLM from functioning in yeast. This hypothesis is supported by the fact that the N-terminus of Sgs1 is sufficient for complementation of the HU sensitivity induced by overexpression of BLM. To test this hypothesis, we constructed a yeast-human chimera in which the N-terminal 647 residues of BLM were replaced by the N-terminal 647 residues of Sgs1 (sgs1ΔC800-blmΔN647) (Figure 6C). To express this chimera from the native SGS1 promoter we replaced nucleotides 1941 to 4344 of the endogenous SGS1 gene with nucleotides 1941 to 4254 of BLM cDNA (Figure 6E). Remarkably, the chimera was nearly as effective as wildtype SGS1 in conferring resistance to HU, whereas the N-terminal segment of Sgs1 by itself was ineffective (Figure 6D). Moreover, when we combined the chimeric allele with a mec3Δ mutation, GCRs accumulated at a significantly lower rate than in the mec3Δ mutant carrying the GCR-deficient sgs1ΔC300 or sgs1ΔC800 alleles, albeit not at the low rate of the mec3Δ mutant carrying the GCR-proficient sgs1ΔC200 allele, signifying partial functionality of the chimerical protein in the suppression of chromosomal rearrangements (Table 1). Finally, besides Srs2, the sgs1Δ mutant also requires the DNA helicase Rrm3 for viability. Synthetic lethality between sgs1Δ and rrm3Δ mutations is suppressed by disrupting HR factors such as Rad51 and Rad55, suggesting that the lethality is due accumulation of aberrant HR intermediates 72; 73; 74. To assess if the Sgs1-BLM chimera was capable of preventing the accumulation of lethal levels of aberrant recombination intermediates we constructed a diploid heterozygous for the rrm3Δ mutation and heterozygous for the sgs1ΔC800-blmΔN647 allele, expressing the Sgs1-BLM chimera. Spreading of spores from this diploid on YPD, which allows all spores to grow, showed that the rrm3Δ mutant expressing the chimera grows normally with the diameter of double mutant colonies measuring approximately 90% of that of the single mutants (Figure 6F). These findings indicate that the Sgs1-BLM chimera is functional and, while not capable of fully suppressing chromosomal rearrangements, prevents the accumulation of lethal levels of aberrant recombination intermediates when Rrm3 helicase is absent.
Figure 6.
Construction of a functional chimerical protein composed of the N-terminus of Sgs1 and the C-terminus of BLM. (A – B) Protein disorder prediction of Sgs1 (red) and BLM (black) using the IUPred algorithm. Values above 0.5 indicate a disordered residue whereas values below 0.5 indicate ordered residues; amino acid residue numbers (1–1447) are indicated on the abscissa. Black lines above the graph show a simplified order and disorder distribution along the length of the protein with values above 0.5 being assigned a “1” and values below 0.5 being assigned a “0”. The vertical red line indicates the site in Sgs1, BLM and the chimera where the disordered N-terminal segment transitions into the ordered helicase domain at residue 647/648. This site was chosen as the fusion site for the chimera. The approximate location of Sgs1 domains is indicated above panel A. (C) Disorder prediction for the Sgs1-BLM chimera in which the N-terminal 647 residues of BLM (black) were replaced with the N-terminal 647 residues of Sgs1 (red). (D) Ten-fold dilutions of exponentially growing haploids were spotted on YPD with or without 100 mM HU. (E) The C-terminus of the Sgs1-BLM chimera was fused to a myc-epitope tag and expression was confirmed by western blotting. Molecular weight marker bands (kD) are indicated on the left (F) A diploid heterozygous for the rrm3Δ mutation and the sgs1ΔC800-blmΔN647 allele expressing the chimera was sporulated and random spores were plated on YPD to allow all spores to grow. An open circle indicates the haploid double mutant, and the open square and pentagon indicate haploid sgs1ΔC800-blmΔN647 and rrm3Δ single mutants, respectively.
DISCUSSION
Yeast cells that lack Sgs1 exhibit upregulated and aberrant recombination in mitosis, increased sensitivity to DNA damaging agents, accumulation of GCRs, synthetic lethality with mutations in other DNA metabolic genes, such as the SRS2 and RRM3 helicase genes, and meiotic defects that lead to poor spore viability 8; 30; 61; 67; 73; 75; 76; 77; 78; 79. Sgs1 contains several conserved domains (DEAD-helicase, RQC, HRDC, AR1 and AR2) and protein interaction sites (Top2, Top3, Srs2, Rad16, Rad51, Mlh1) have been identified by two-hybrid screens 27; 29; 30; 46; 80. How the integrity of these conserved motifs and protein-protein interaction sites affects the role of Sgs1 in suppression of aberrant genome rearrangements has not been determined. The requirement of some domains and/or protein interaction sites, but not others, may shed light on the poorly understood mechanism(s) by which Sgs1 contributes to the maintenance of genome integrity in yeast. Here, we find that the C-terminal 240 amino-acid segment, which contains Rad51 and Mlh1 interaction sites as well as the conserved HRDC domain thought to be involved in DNA binding and in recognition and processing of double Holliday junctions 20; 23, is dispensable for Sgs1’s role in suppressing GCRs. The integrity of the RQC domain, however, is essential for GCR suppression. That zinc-binding is crucial for Sgs1 activity, and loss of function of the C-terminal truncation allele was not due to disruption of protein structure/function because of such a large deletion, was further confirmed by the finding that the point mutation of a conserved zinc-coordinating cysteine, which has also been observed in BS patients 64, led to loss of Sgs1’s ability to suppress HU sensitivity and GCR accumulation. This loss of function was not due to degradation of the mutant protein as had been previously observed for some cysteine mutants of BLM during attempts at overexpression and purification from E. coli. However, we cannot exclude the possibility that the loss of function resulted from intracellular mislocalization of the mutant protein. Previously, modeling of the zinc-binding domain of BLM and instability of purified mutant proteins had indicated that hydrogen bonds between three conserved residues, Y1029 (Y1040 in Sgs1), R1037 (R1048 in Sgs1) and D1064 (D1070 in Sgs1), are required for folding of the zinc-binding domain and overall protein stability 17. Although F1056 of Sgs1 does not appear to be involved in this zinc-domain stabilization and the Sgs1-F1056A mutant protein appears stable in this study, F1056 is the only other fully conserved residue in the zinc-binding domain of RecQ-like helicases, suggesting functional significance. However, introduction of the F1056A mutation had no effect on Sgs1 function in vivo when we assessed HU sensitivity, consistent with a previous study 81, or GCR accumulation. That in a previous in vitro study 65 the corresponding BLM mutation (F1045A) had severely impaired helicase and ssDNA binding activities could either be due to differences in the importance of this residue for enzymatic activity of BLM and Sgs1 or, more likely, be due to the fact that only the helicase-core segment of BLM, lacking 769 residues of N- and C-termini, was purified. The in vitro function of this isolated domain could be more strongly affected by a mutation than the in vivo function of the full-length Sgs1 mutant protein assessed here. Although nearly half of all BLM alleles that are associated with single-amino-acid changes (7 of 17 alleles) in BS patients are located in the RQC domain 9; 13; 63; 64 none affect F1045, consistent with our finding that mutation of this conserved residue may not be associated with significant loss of function in vivo.
We find that Sgs1 retains partial functionality even when it lacks the HRDC, RQC and DEAH helicase domains, as demonstrated by the greater HU resistance of diploids that only express the N-terminal 547 amino acid residues compared to those alleles expressing fewer than 447 residues of Sgs1. One explanation for this finding could be that protein-protein interactions conferred by the N-terminus could contribute to the structural stability of multi-protein complexes, such as the Sgs1/Top3/Rmi1 31 complex or, even more relevant to HU resistance, DNA-damage-specific complexes with Srs2 and Mre11 29. In these multi-protein complexes, enzymatic activity of Sgs1 may be dispensable. Indeed, sgs1 alleles with point mutations in the helicase domain have been shown to be capable of performing some functions of the wildtype allele, including those carried out during meiosis and checkpoint activation 79; 82.
In contrast to two previous reports 66; 83, which both used the same yeast strain that constitutively expressed BLM from a GAPDH promoter and showed partial suppression of some sgs1Δ defects, including HU sensitivity, we found that neither BLM expression under control of the natural SGS1 promoter nor varying levels of BLM expression under control of a galactose-inducible promoter had any positive effect on the sgs1Δ mutant. That a single copy of BLM, when expressed under control of the native SGS1 promoter, cannot alleviate sgs1Δ defects initially suggested to us that BLM had no functionality in yeast. In fact, the strong increase in genome instability, accompanied by severe HU sensitivity and some growth retardation upon overexpression of BLM, indicated that BLM expression is detrimental to yeast cells. The absence of any GCR accumulation upon Sgs1 overexpression suggests that increased accumulation of GCRs in BLM overexpressing cells is not simply due to increased unwinding. Rather, we propose that BLM may possess helicase activity in yeast, leading to increased unwinding upon overexpression, but fails to elicit proper downstream responses, for example due to lack of proper N-terminal protein-protein interactions, which ultimately leads to an overabundance of aberrantly repaired lesions. That endogenous levels of N-terminal segments of Sgs1 as short as 547 residues suppressed the slow growth phenotype and the severe HU sensitivity of BLM-overexpressing cells argues in favor of a functional relationship between Sgs1 and BLM. For example, co-expression of Sgs1 and BLM could alleviate HU sensitivity in BLM overexpressing cells by acting as a bridge between BLM and Top3 (and/or other protein complexes interacting with the Sgs1 N-terminus), thereby linking enzymatic activity to appropriate upstream and downstream events. Remarkably, even relatively short N-terminal fragments of Sgs1 are sufficient for the suppression of the increased HU sensitivity of BLM-overexpressing cells, further supporting the importance of the Sgs1 N-terminus with its role in mediating interaction with other DNA metabolic factors. HU resistance comparable to wildtype cells and significantly reduced GCR accumulation of cells expressing a chimeric fusion of the Sgs1 N-terminus, which is devoid of enzymatic function and dispensable for helicase activity and ssDNA binding in vitro, and the BLM C-terminus, which contains helicase/RQC and HRDC domains, is consistent with helicase activity of BLM in yeast and a biologically significant, functional interaction between BLM and Sgs1. That not only fusion of the Sgs1 and BLM segments provides HU resistance, but also co-expression of BLM and Sgs1 polypeptides from separate alleles in the same cell may indicate that the N-terminus of Sgs1 can physically interact with BLM. Our findings also suggest that it is the inability of the N-terminus of BLM to interact with or be modified by yeast proteins that leads to the inability of BLM to function in yeast. A previous report that BLM expression in yeast alleviates several sgs1Δ phenotypes, including partial suppression of HU sensitivity 66; 83, could be explained by the fact that in the earlier study BLM was expressed from a GAPDH promoter, whereas here it was expressed either from the native SGS1 promoter or from a galactose-inducible promoter. However, in light of the findings presented here, there could also be an alternative explanation. Since the GAPDH-promoter-BLM construct appears to have been inserted into the middle of the wildtype SGS1 gene, an N-terminal segment of Sgs1 could have been expressed from the native SGS1 promoter in addition to BLM being expressed from the GAPDH promoter. As shown here for haploids expressing the chimera and for diploids co-expressing the N-terminus of Sgs1 and full length BLM, such co-expression of an Sgs1-N-terminal segment from the native SGS1 promoter and BLM from the GAPDH promoter could be the an explanation for the reported increase in HU resistance of BLM-expressing cells compared to sgs1Δ cells.
Of the five human RecQ-like DNA helicases, BLM is considered to be most closely related to Sgs1. Even though we show here that BLM cannot suppress any defects of the sgs1Δ mutant, the functional chimera does provide evidence for a functional relationship between the two RecQ-like helicases and provides a model system for the further characterization of BLM functional domains in yeast. In fact, all BS-associated missense mutations and numerous polymorphisms are located within the 770-residue C-terminal fragment of BLM that is part of the chimera, so that they are now accessible to further functional and mutational characterization in yeast. The in vivo functionality of the Sgs1-BLM chimera also demonstrates the remarkable utility of protein disorder prediction as a tool for the construction of functional mutants. It will be interesting to see whether domains of any of the other human RecQ-like helicases will, like BLM, be able to form functional chimeras with the Sgs1 N-terminus.
Supplementary Material
Acknowledgments
Funding
This work was supported by National Institutes of Health grant 5R01GM081425 to K.H.S.
We are thankful to Gary Daughdrill (Florida Center of Excellence for Biomolecular Identification and Targeted Therapeutics (FCoE-BITT), USF) for helpful discussions about protein disorder prediction and to Lillian Doerfler, Cynthia Toughlian and Kent Seeley for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hegde SP, Qin MH, Li XH, Atkinson MA, Clark AJ, Rajagopalan M, Madiraju MV. Interactions of RecF protein with RecO, RecR, and single-stranded DNA binding proteins reveal roles for the RecF-RecO-RecR complex in DNA repair and recombination. Proc Natl Acad Sci U S A. 1996;93:14468–73. doi: 10.1073/pnas.93.25.14468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol Cell. 2003;11:1337–47. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 3.Ivancic-Bace I, Salaj-Smic E, Brcic-Kostic K. Effects of recJ, recQ, and recFOR mutations on recombination in nuclease-deficient recB recD double mutants of Escherichia coli. J Bacteriol. 2005;187:1350–6. doi: 10.1128/JB.187.4.1350-1356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fricke WM, Brill SJ. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 2003;17:1768–78. doi: 10.1101/gad.1105203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mullen JR, Kaliraman V, Brill SJ. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–14. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–6. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt KH, Wu J, Kolodner RD. Control of Translocations between Highly Diverged Genes by Sgs1, the Saccharomyces cerevisiae Homolog of the Bloom's Syndrome Protein. Mol Cell Biol. 2006;26:5406–20. doi: 10.1128/MCB.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–66. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 10.Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–52. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- 11.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–4. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 12.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–62. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 13.German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom's syndrome registry. Hum Mutat. 2007 doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- 14.Lee JW, Kusumoto R, Doherty KM, Lin GX, Zeng W, Cheng WH, von Kobbe C, Brosh RM, Jr, Hu JS, Bohr VA. Modulation of Werner syndrome protein function by a single mutation in the conserved RecQ domain. J Biol Chem. 2005;280:39627–36. doi: 10.1074/jbc.M506112200. [DOI] [PubMed] [Google Scholar]
- 15.von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA. Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem. 2003;278:52997–3006. doi: 10.1074/jbc.M308338200. [DOI] [PubMed] [Google Scholar]
- 16.Huber MD, Duquette ML, Shiels JC, Maizels N. A conserved G4 DNA binding domain in RecQ family helicases. J Mol Biol. 2006;358:1071–80. doi: 10.1016/j.jmb.2006.01.077. [DOI] [PubMed] [Google Scholar]
- 17.Guo RB, Rigolet P, Zargarian L, Fermandjian S, Xi XG. Structural and functional characterizations reveal the importance of a zinc binding domain in Bloom's syndrome helicase. Nucleic Acids Res. 2005;33:3109–24. doi: 10.1093/nar/gki619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitano K, Yoshihara N, Hakoshima T. Crystal structure of the HRDC domain of human Werner syndrome protein, WRN. J Biol Chem. 2007;282:2717–28. doi: 10.1074/jbc.M610142200. [DOI] [PubMed] [Google Scholar]
- 19.Morozov V, Mushegian AR, Koonin EV, Bork P. A putative nucleic acid-binding domain in Bloom's and Werner's syndrome helicases. Trends Biochem Sci. 1997;22:417–8. doi: 10.1016/s0968-0004(97)01128-6. [DOI] [PubMed] [Google Scholar]
- 20.Wu L, Chan KL, Ralf C, Bernstein DA, Garcia PL, Bohr VA, Vindigni A, Janscak P, Keck JL, Hickson ID. The HRDC domain of BLM is required for the dissolution of double Holliday junctions. Embo J. 2005;24:2679–87. doi: 10.1038/sj.emboj.7600740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Killoran MP, Keck JL. Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ. Nucleic Acids Res. 2008;36:3139–49. doi: 10.1093/nar/gkn143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernstein DA, Keck JL. Conferring substrate specificity to DNA helicases: role of the RecQ HRDC domain. Structure. 2005;13:1173–82. doi: 10.1016/j.str.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Macias MJ, Bottomley MJ, Stier G, Linge JP, Nilges M, Bork P, Sattler M. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure. 1999;7:1557–66. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- 24.Miyajima A, Seki M, Onoda F, Ui A, Satoh Y, Ohno Y, Enomoto T. Different domains of Sgs1 are required for mitotic and meiotic functions. Genes Genet Syst. 2000;75:319–26. doi: 10.1266/ggs.75.319. [DOI] [PubMed] [Google Scholar]
- 25.Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. Embo J. 2009;28:915–25. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett RJ, Noirot-Gros MF, Wang JC. Interaction between yeast sgs1 helicase and DNA topoisomerase III. J Biol Chem. 2000;275:26898–905. doi: 10.1074/jbc.M003137200. [DOI] [PubMed] [Google Scholar]
- 27.Duno M, Thomsen B, Westergaard O, Krejci L, Bendixen C. Genetic analysis of the Saccharomyces cerevisiae Sgs1 helicase defines an essential function for the Sgs1-Top3 complex in the absence of SRS2 or TOP1. Mol Gen Genet. 2000;264:89–97. doi: 10.1007/s004380000286. [DOI] [PubMed] [Google Scholar]
- 28.Fricke WM, Kaliraman V, Brill SJ. Mapping the DNA topoisomerase III binding domain of the Sgs1 DNA helicase. J Biol Chem. 2001;276:8848–55. doi: 10.1074/jbc.M009719200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiolo I, Carotenuto W, Maffioletti G, Petrini JH, Foiani M, Liberi G. Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol Cell Biol. 2005;25:5738–51. doi: 10.1128/MCB.25.13.5738-5751.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–60. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 31.Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. Embo J. 2005;24:2024–33. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaganti RS, Schonberg S, German J. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci U S A. 1974;71:4508–12. doi: 10.1073/pnas.71.11.4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hojo ET, van Diemen PC, Darroudi F, Natarajan AT. Spontaneous chromosomal aberrations in Fanconi anaemia, ataxia telangiectasia fibroblast and Bloom's syndrome lymphoblastoid cell lines as detected by conventional cytogenetic analysis and fluorescence in situ hybridisation (FISH) technique. Mutat Res. 1995;334:59–69. doi: 10.1016/0165-1161(95)90031-4. [DOI] [PubMed] [Google Scholar]
- 35.Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–9. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu Rev Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 37.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem. 2006;281:22839–46. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 38.Hanada K, Hickson ID. Molecular genetics of RecQ helicase disorders. Cell Mol Life Sci. 2007;64:2306–22. doi: 10.1007/s00018-007-7121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–33. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
- 40.Hand R, German J. A retarded rate of DNA chain growth in Bloom's syndrome. Proc Natl Acad Sci U S A. 1975;72:758–62. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lonn U, Lonn S, Nylen U, Winblad G, German J. An abnormal profile of DNA replication intermediates in Bloom's syndrome. Cancer Res. 1990;50:3141–5. [PubMed] [Google Scholar]
- 42.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Molecular and Cellular Biology. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiao RJ, Bachrati CZ, Pedrazzi G, Kuster P, Petkovic M, Li JL, Egli D, Hickson ID, Stagljar I. Physical and functional interaction between the Bloom's syndrome gene product and the largest subunit of chromatin assembly factor 1. Molecular and Cellular Biology. 2004;24:4710–4719. doi: 10.1128/MCB.24.11.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson FB, Lombard DB, Neff NF, Mastrangelo MA, Dewolf W, Ellis NA, Marciniak RA, Yin Y, Jaenisch R, Guarente L. Association of the Bloom syndrome protein with topoisomerase IIIalpha in somatic and meiotic cells. Cancer Res. 2000;60:1162–7. [PubMed] [Google Scholar]
- 45.Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–73. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L, Davies SL, Levitt NC, Hickson ID. Potential role for the BLM helicase in recombinational repair via a conserved interaction with RAD51. J Biol Chem. 2001;276:19375–81. doi: 10.1074/jbc.M009471200. [DOI] [PubMed] [Google Scholar]
- 47.Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J Biol Chem. 2000;275:23500–8. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 48.Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM., Jr Stimulation of flap endonuclease-1 by the Bloom's syndrome protein. J Biol Chem. 2004;279:9847–56. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 49.Langland G, Kordich J, Creaney J, Goss KH, Lillard-Wetherell K, Bebenek K, Kunkel TA, Groden J. The Bloom's syndrome protein (BLM) interacts with MLH1 but is not required for DNA mismatch repair. J Biol Chem. 2001;276:30031–5. doi: 10.1074/jbc.M009664200. [DOI] [PubMed] [Google Scholar]
- 50.Selak N, Bachrati CZ, Shevelev I, Dietschy T, van Loon B, Jacob A, Hubscher U, Hoheisel JD, Hickson ID, Stagljar I. The Bloom's syndrome helicase (BLM) interacts physically and functionally with p12, the smallest subunit of human DNA polymerase delta. Nucleic Acids Res. 2008;36:5166–79. doi: 10.1093/nar/gkn498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J Cell Biol. 2001;153:367–80. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom's syndrome DNA helicase. Oncogene. 2000;19:2731–8. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- 53.Sanz MM, Proytcheva M, Ellis NA, Holloman WK, German J. BLM, the Bloom's syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet Cell Genet. 2000;91:217–23. doi: 10.1159/000056848. [DOI] [PubMed] [Google Scholar]
- 54.Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc Natl Acad Sci U S A. 2000;97:5214–9. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 56.Gietz RD, Woods RA. Yeast transformation by the LiAc/SS Carrier DNA/PEG method. Methods Mol Biol. 2006;313:107–20. doi: 10.1385/1-59259-958-3:107. [DOI] [PubMed] [Google Scholar]
- 57.Foiani M, Liberi G, Piatti S, Plevani P. Saccharomyces cerevisiae as a model system to study DNA replication. In: Cotterill S, editor. Eukaryotic DNA Replication. A Practical Approach. Oxford University Press; Oxford, UK: 1999. pp. 185–200. [Google Scholar]
- 58.Schmidt KH, Pennaneach V, Putnam CD, Kolodner RD. Analysis of gross-chromosomal rearrangements in Saccharomyces cerevisiae. Methods Enzymol. 2006;409:462–76. doi: 10.1016/S0076-6879(05)09027-0. [DOI] [PubMed] [Google Scholar]
- 59.Nair KR. Table of confidence intervals for the median in samples from any continuous population. Sankhya. 1940;4:551–558. [Google Scholar]
- 60.Rockmill B, Lambie EJ, Roeder GS. Spore enrichment. Methods Enzymol. 1991;194:146–9. doi: 10.1016/0076-6879(91)94012-2. [DOI] [PubMed] [Google Scholar]
- 61.Lee SK, Johnson RE, Yu SL, Prakash L, Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–42. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- 62.Kitano K, Kim SY, Hakoshima T. Structural basis for DNA strand separation by the unconventional winged-helix domain of RecQ helicase WRN. Structure. 18:177–87. doi: 10.1016/j.str.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 63.Barakat A, Ababou M, Onclercq R, Dutertre S, Chadli E, Hda N, Benslimane A, Amor-Gueret M. Identification of a novel BLM missense mutation (2706T>C) in a Moroccan patient with Bloom's syndrome. Hum Mutat. 2000;15:584–5. doi: 10.1002/1098-1004(200006)15:6<584::AID-HUMU28>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 64.Foucault F, Vaury C, Barakat A, Thibout D, Planchon P, Jaulin C, Praz F, Amor-Gueret M. Characterization of a new BLM mutation associated with a topoisomerase II alpha defect in a patient with Bloom's syndrome. Hum Mol Genet. 1997;6:1427–34. doi: 10.1093/hmg/6.9.1427. [DOI] [PubMed] [Google Scholar]
- 65.Janscak P, Garcia PL, Hamburger F, Makuta Y, Shiraishi K, Imai Y, Ikeda H, Bickle TA. Characterization and mutational analysis of the RecQ core of the bloom syndrome protein. J Mol Biol. 2003;330:29–42. doi: 10.1016/s0022-2836(03)00534-5. [DOI] [PubMed] [Google Scholar]
- 66.Yamagata K, Kato J, Shimamoto A, Goto M, Furuichi Y, Ikeda H. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc Natl Acad Sci U S A. 1998;95:8733–8. doi: 10.1073/pnas.95.15.8733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- 68.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–39. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 69.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–4. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 70.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 71.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–45. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 72.Ooi SL, Shoemaker DD, Boeke JD. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat Genet. 2003;35:277–86. doi: 10.1038/ng1258. [DOI] [PubMed] [Google Scholar]
- 73.Schmidt KH, Kolodner RD. Requirement of Rrm3 helicase for repair of spontaneous DNA lesions in cells lacking Srs2 or Sgs1 helicase. Mol Cell Biol. 2004;24:3213–26. doi: 10.1128/MCB.24.8.3213-3226.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Torres JZ, Schnakenberg SL, Zakian VA. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: viability of rrm3 cells requires the intra-S–phase checkpoint and fork restart activities. Mol Cell Biol. 2004;24:3198–212. doi: 10.1128/MCB.24.8.3198-3212.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cobb JA, Bjergbaek L, Gasser SM. RecQ helicases: at the heart of genetic stability. FEBS Lett. 2002;529:43–8. doi: 10.1016/s0014-5793(02)03269-6. [DOI] [PubMed] [Google Scholar]
- 76.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1–Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–11. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27:113–6. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 78.Versini G, Comet I, Wu M, Hoopes L, Schwob E, Pasero P. The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. Embo J. 2003;22:1939–49. doi: 10.1093/emboj/cdg180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miyajima A, Seki M, Onoda F, Shiratori M, Odagiri N, Ohta K, Kikuchi Y, Ohno Y, Enomoto T. Sgs1 helicase activity is required for mitotic but apparently not for meiotic functions. Mol Cell Biol. 2000;20:6399–409. doi: 10.1128/mcb.20.17.6399-6409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saffi J, Feldmann H, Winnacker EL, Henriques JA. Interaction of the yeast Pso5/Rad16 and Sgs1 proteins: influences on DNA repair and aging. Mutat Res. 2001;486:195–206. doi: 10.1016/s0921-8777(01)00093-3. [DOI] [PubMed] [Google Scholar]
- 81.Ui A, Satoh Y, Onoda F, Miyajima A, Seki M, Enomoto T. The N-terminal region of Sgs1, which interacts with Top3, is required for complementation of MMS sensitivity and suppression of hyper-recombination in sgs1 disruptants. Mol Genet Genomics. 2001;265:837–50. doi: 10.1007/s004380100479. [DOI] [PubMed] [Google Scholar]
- 82.Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. Embo J. 2005;24:405–17. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heo SJ, Tatebayashi K, Ohsugi I, Shimamoto A, Furuichi Y, Ikeda H. Bloom's syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells. 1999;4:619–25. doi: 10.1046/j.1365-2443.1999.00288.x. [DOI] [PubMed] [Google Scholar]
- 84.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.