Abstract
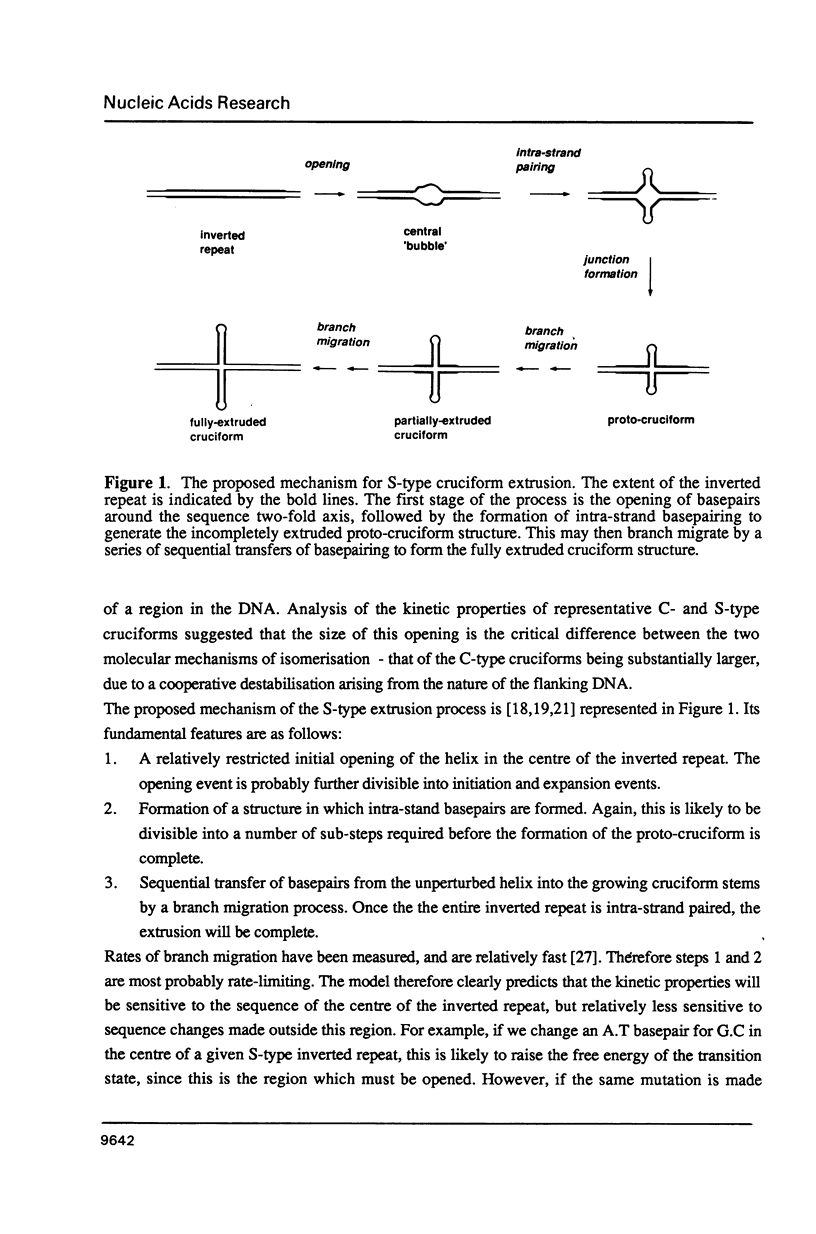
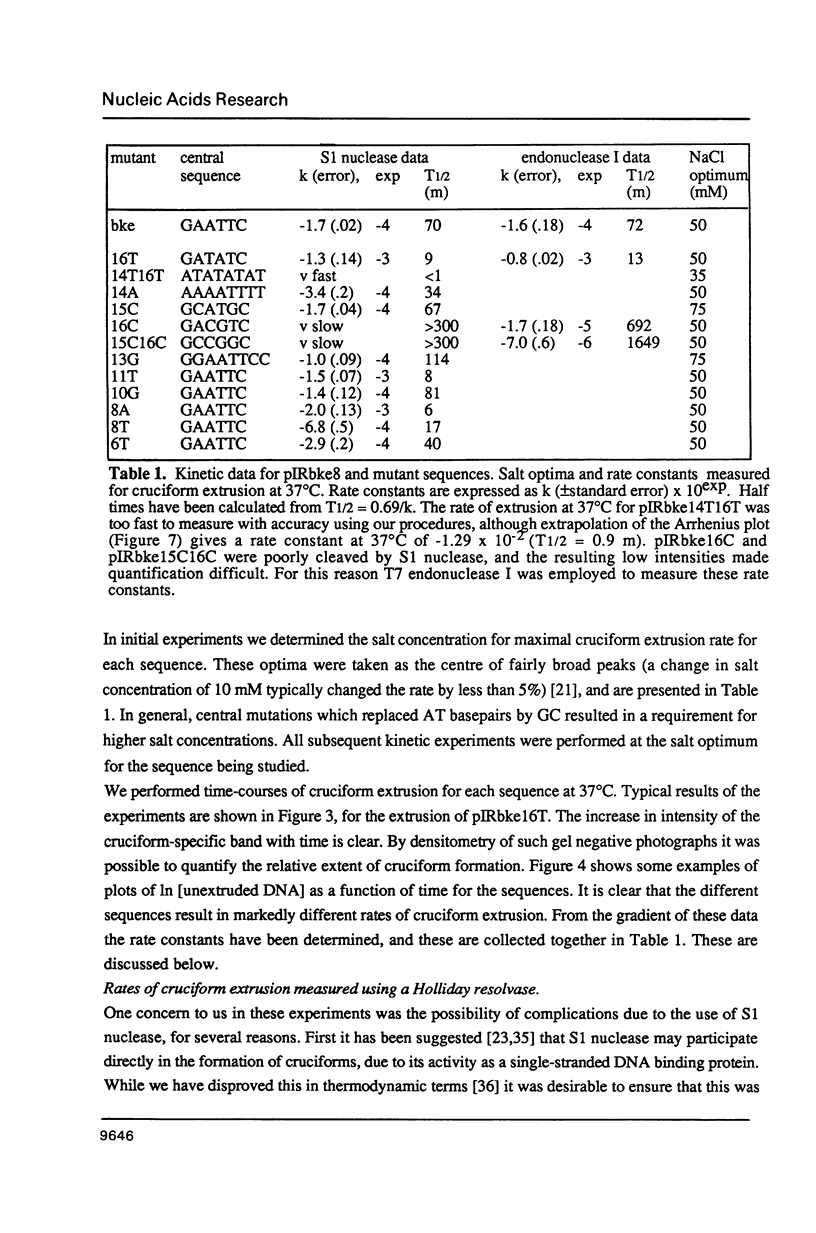
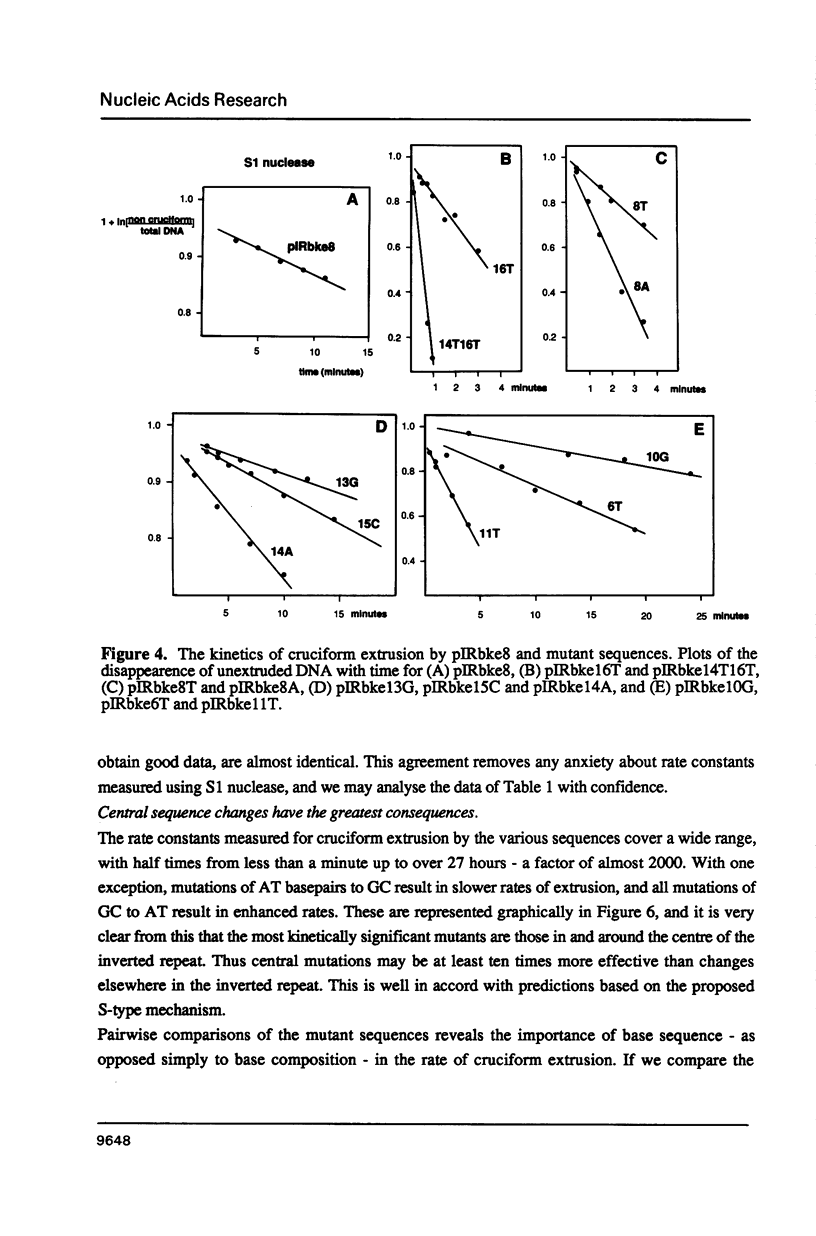
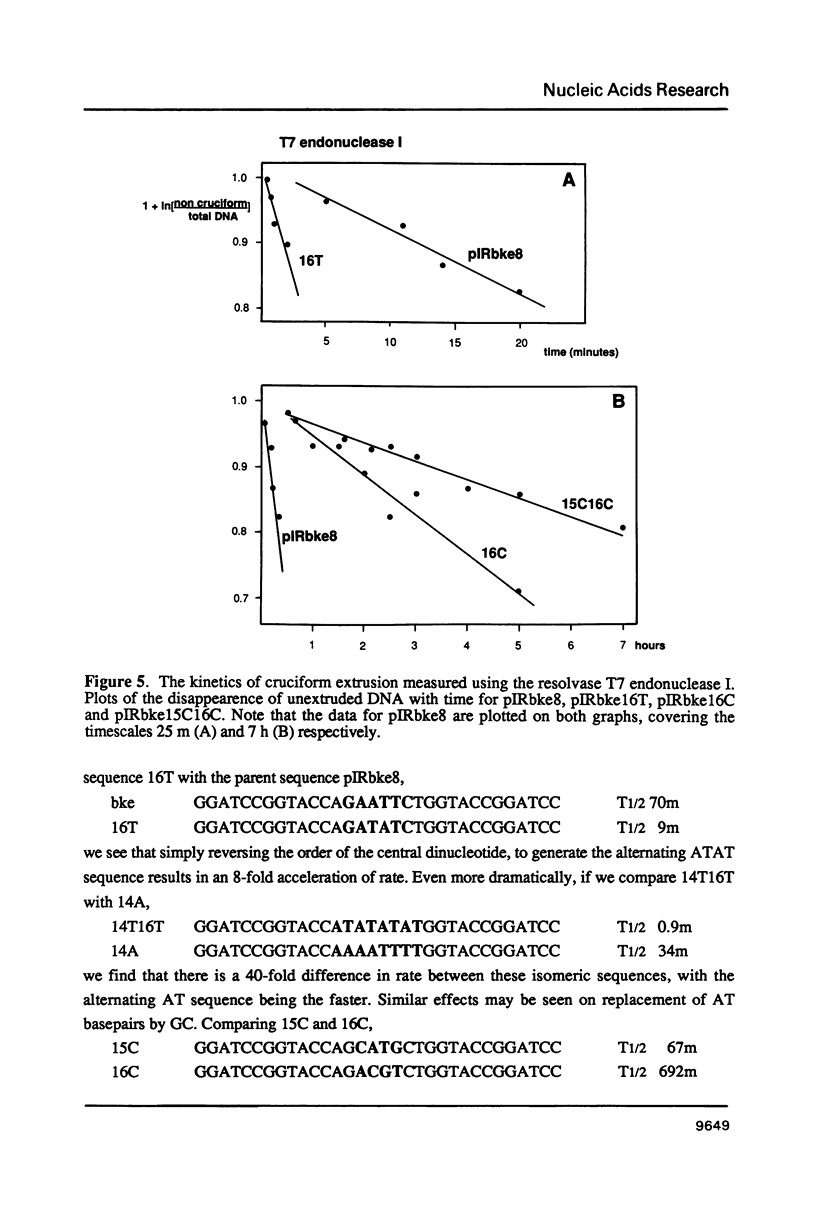
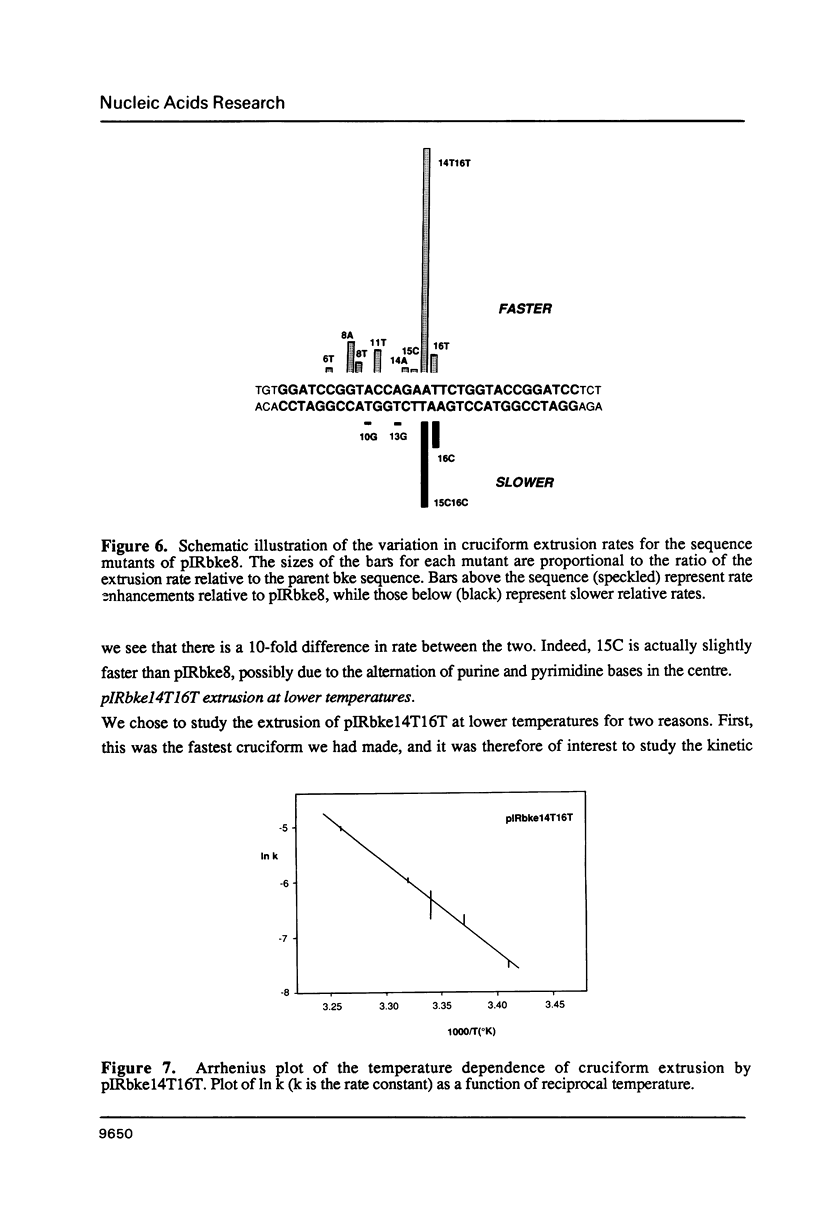
There are two alternative pathways by which inverted repeat sequences in supercoiled DNA molecules may extrude cruciform structures, called C-type and S-type. S-type cruciforms, which form the great majority, are characterised by absolute requirement for cations to promote extrusion, which then proceeds at higher temperatures and with lower activation parameters than for C-type cruciforms. The mechanism proposed for S-type extrusion involves an initial opening of basepairs limited to the centre of the inverted repeat, formation of intra-strand basepairing and a four-way junction, and finally branch migration to the fully extruded cruciform. The model predicts that central sequence changes will be more kinetically significant than those removed from the centre. We have studied the kinetics of cruciform extrusion by a series of inverted repeats related to that of pIRbke8 by either one or two mutations in the symmetric unit. We find that mutations in the central 8 to 10 nucleotides may profoundly affect extrusion rates--the fastest being 2000-fold faster than the slowest, whereas mutations further from the centre affect rates to a much smaller extent, typically up to ten-fold. These data support the proposed mechanism for extrusion via central opening.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Selsing E. Structures for the polynucleotide complexes poly(dA) with poly (dT) and poly(dT) with poly(dA) with poly (dT). J Mol Biol. 1974 Sep 15;88(2):509–521. doi: 10.1016/0022-2836(74)90498-7. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Burd J. F., Wartell R. M., Dodgson J. B., Wells R. D. Transmission of stability (telestability) in deoxyribonucleic acid. Physical and enzymatic studies on the duplex block polymer d(C15A15) - d(T15G15). J Biol Chem. 1975 Jul 10;250(13):5109–5113. [PubMed] [Google Scholar]
- Cantor C. R., Efstratiadis A. Possible structures of homopurine-homopyrimidine S1-hypersensitive sites. Nucleic Acids Res. 1984 Nov 12;12(21):8059–8072. doi: 10.1093/nar/12.21.8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Wang J. C. Cruciform formation in a negatively supercoiled DNA may be kinetically forbidden under physiological conditions. Cell. 1983 Jul;33(3):817–829. doi: 10.1016/0092-8674(83)90024-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA structural variations in the E. coli tyrT promoter. Cell. 1984 Jun;37(2):491–502. doi: 10.1016/0092-8674(84)90379-9. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Ohmori H., Tomizawa J. DNA gyrase and DNA supercoiling. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):35–40. doi: 10.1101/sqb.1979.043.01.007. [DOI] [PubMed] [Google Scholar]
- Gellert M., O'Dea M. H., Mizuuchi K. Slow cruciform transitions in palindromic DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5545–5549. doi: 10.1073/pnas.80.18.5545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh O. Prediction of melting profiles and local helix stability for sequenced DNA. Adv Biophys. 1983;16:1–52. doi: 10.1016/0065-227x(83)90007-2. [DOI] [PubMed] [Google Scholar]
- Greaves D. R., Patient R. K., Lilley D. M. Facile cruciform formation by an (A-T)34 sequence from a Xenopus globin gene. J Mol Biol. 1985 Oct 5;185(3):461–478. doi: 10.1016/0022-2836(85)90064-6. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Pulleyblank D. E. Transition of a cloned d(AT)n-d(AT)n tract to a cruciform in vivo. Nucleic Acids Res. 1985 Jun 25;13(12):4343–4363. doi: 10.1093/nar/13.12.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Hallam L. R. Thermodynamics of the ColE1 cruciform. Comparisons between probing and topological experiments using single topoisomers. J Mol Biol. 1984 Nov 25;180(1):179–200. doi: 10.1016/0022-2836(84)90436-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Markham A. F. Dynamics of cruciform extrusion in supercoiled DNA: use of a synthetic inverted repeat to study conformational populations. EMBO J. 1983;2(4):527–533. doi: 10.1002/j.1460-2075.1983.tb01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The kinetic properties of cruciform extrusion are determined by DNA base-sequence. Nucleic Acids Res. 1985 Mar 11;13(5):1443–1465. doi: 10.1093/nar/13.5.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. A pH-dependent structural transition in the homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1985 Oct;3(2):327–338. doi: 10.1080/07391102.1985.10508420. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McClellan J. A., Lilley D. M. A two-state conformational equilibrium for alternating (A-T)n sequences in negatively supercoiled DNA. J Mol Biol. 1987 Oct 20;197(4):707–721. doi: 10.1016/0022-2836(87)90477-3. [DOI] [PubMed] [Google Scholar]
- McClellan J. A., Palecek E., Lilley D. M. (A-T)n tracts embedded in random sequence DNA--formation of a structure which is chemically reactive and torsionally deformable. Nucleic Acids Res. 1986 Dec 9;14(23):9291–9309. doi: 10.1093/nar/14.23.9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Panyutin I., Klishko V., Lyamichev V. Kinetics of cruciform formation and stability of cruciform structure in superhelical DNA. J Biomol Struct Dyn. 1984 Jun;1(6):1311–1324. doi: 10.1080/07391102.1984.10507522. [DOI] [PubMed] [Google Scholar]
- Platt J. R. POSSIBLE SEPARATION OF INTERTWINED NUCLEIC ACID CHAINS BY TRANSFER-TWIST. Proc Natl Acad Sci U S A. 1955 Mar 15;41(3):181–183. doi: 10.1073/pnas.41.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulleyblank D. E., Haniford D. B., Morgan A. R. A structural basis for S1 nuclease sensitivity of double-stranded DNA. Cell. 1985 Aug;42(1):271–280. doi: 10.1016/s0092-8674(85)80122-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Shakked Z., Rabinovich D., Kennard O., Cruse W. B., Salisbury S. A., Viswamitra M. A. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C). J Mol Biol. 1983 May 15;166(2):183–201. doi: 10.1016/s0022-2836(83)80005-9. [DOI] [PubMed] [Google Scholar]
- Sinden R. R., Pettijohn D. E. Cruciform transitions in DNA. J Biol Chem. 1984 May 25;259(10):6593–6600. [PubMed] [Google Scholar]
- Suggs J. W., Wagner R. W. Nuclease recognition of an alternating structure in a d(AT)14 plasmid insert. Nucleic Acids Res. 1986 May 12;14(9):3703–3716. doi: 10.1093/nar/14.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan K. M., Lilley D. M. A dominant influence of flanking sequences on a local structural transition in DNA. Cell. 1986 Dec 5;47(5):817–827. doi: 10.1016/0092-8674(86)90524-6. [DOI] [PubMed] [Google Scholar]
- Sullivan K. M., Lilley D. M. Influence of cation size and charge on the extrusion of a salt-dependent cruciform. J Mol Biol. 1987 Jan 20;193(2):397–404. doi: 10.1016/0022-2836(87)90227-0. [DOI] [PubMed] [Google Scholar]
- Symington L. S., Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Peck L. J., Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- West S. C., Körner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy B., Studier F. W., Dorgai L., Appelbaum E., Weisberg R. A. Enzymes and sites of genetic recombination: studies with gene-3 endonuclease of phage T7 and with site-affinity mutants of phage lambda. Cold Spring Harb Symp Quant Biol. 1984;49:715–726. doi: 10.1101/sqb.1984.049.01.081. [DOI] [PubMed] [Google Scholar]



