Abstract
Peroxiredoxins (Prxs) are a widespread and highly expressed family of cysteine-based peroxidases that react very rapidly with H2O2, organic peroxides, and peroxynitrite. Correct subfamily classification has been problematic since Prx subfamilies are frequently not correlated with phylogenetic distribution and diverge in their preferred reductant, oligomerization state, and tendency towards overoxidation. We have developed a method that uses the Deacon Active Site Profiler (DASP) tool to extract functional site profiles from structurally characterized proteins, to computationally define subfamilies, and to identify new Prx subfamily members from GenBank(nr). For the 58 literature-defined Prx test proteins, 57 were correctly assigned and none were assigned to the incorrect subfamily. The >3500 putative Prx sequences identified were then used to analyze residue conservation in the active site of each Prx subfamily. Our results indicate that the existence and location of the resolving cysteine varies in some subfamilies (e.g. Prx5) to a greater degree than previously appreciated and that interactions at the A interface (common to Prx5, Tpx and higher order AhpC/Prx1 structures) are important for stabilization of the correct active site geometry. Interestingly, this method also allows us to further divide the AhpC/Prx1 into four groups that are correlated with functional characteristics. The DASP method provides more accurate subfamily classification than PSI-BLAST for members of the Prx family and can now readily be applied to other large protein families.
Keywords: functional site profile, mechanistic determinants, function annotation, misannotation, thiol peroxidase, thioredoxin peroxidase, AhpC, Prx, BCP, Tpx
INTRODUCTION
Peroxiredoxins (Prxs or PRDXs) are protective antioxidant enzymes that rapidly detoxify hydrogen peroxide (H2O2), organic hydroperoxides (ROOH), and peroxynitrite (OONO-). This large protein family, which has diverged from thioredoxin-like redox proteins, is widespread across phylogeny and one or more members are typically expressed at high levels in many celltypes 1–4. Prxs regulate H2O2 levels that can cause damage and/or affect signal transduction processes and, thereby, have roles in proliferation, differentiation, and apoptotic pathways through both known and unknown mechanisms 3,5,6.
All Prx proteins contain an absolutely conserved active site cysteine, referred to as the peroxidatic cysteine (CP), which reacts with peroxide to form a cysteine sulfenic acid (R-SOH) and releases water or the corresponding alcohol 7,8. The local sequence motif at the active site, PXXXTXXCP [Figure 1, residue numbers Pro39, Thr43, and Cys46 in the Salmonella typhimurium alkyl hydroperoxide reductase (AhpC), PDB file 1yep], is essentially invariant in known Prx proteins across the otherwise rather diverse family (the T is replaced by S in rare cases) 8–10. In addition, Prx proteins contain a highly conserved Arg (Arg119) that, along with the other three residues and several backbone interactions, activate the CP by stabilizing the deprotonated form of the thiol and position the hydroperoxide oxygens for optimal stabilization of the transition state during -O-O- bond cleavage 7,11.
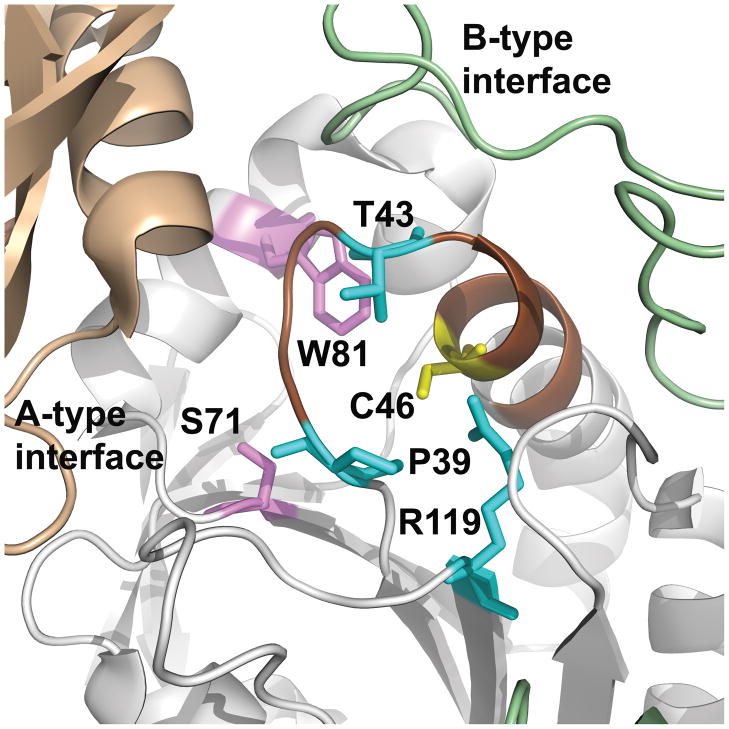
Figure 1. Location of residues conserved across all Prx subfamilies.
The structure of the S. typhimurium AhpC (PDB identifier 1n8j) active site is shown with the location of the CP (Cys46) in yellow (the protein sequence of 1n8j contains a C46S mutation, but for simplicity it is labeled as a Cys). The adjacent monomer across the dimer interface (B interface) is in pale green and the adjacent monomer across the decamer-building interface (A interface) is in tan. In the Tpx and Prx5 subfamilies, the dimer is formed across the A-type interface. Residues conserved across the Prx family are highlighted in cyan (identified previously) and magenta (identified as part of the work described herein). Brown highlights the loop-helix region around the active site that undergoes local unfolding following oxidation to form a disulfide bond between CP and CR in both typical and atypical 2-Cys Prxs. This figure was made using Pymol (http://sourceforge.net/projects/pymol/).
In contrast to the peroxide reduction reaction common to all Prxs, regeneration of the reduced, active form of these enzymes can vary. In many Prxs, a second cysteine, referred to as the resolving cysteine (CR), attacks the R-SOH to form a water molecule and a disulfide bond in the protein. Originally, Prx proteins were classified based on the presence and location of the CR 2,12,13. In the typical 2-Cys Prxs, arguably the earliest Prx group to be recognized, the CR is found in the C-terminus of the partner subunit, yielding an intersubunit disulfide bond upon oxidation. As more examples of Prxs were discovered, the CR participating in the recycling process was recognized to reside in other positions where it generally forms an intrasubunit disulfide bond (atypical 2-Cys Prxs). In 1-Cys Prxs, there is no CR and a thiol from another protein or small molecule presumably takes the place of this residue in the recycling process. While the CR shows up in different locations within the structure (Figure S1), its ability to form a disulfide bond with the CP in a locally unfolded conformation helps promote the next step of reduction by a thiol-containing disulfide reductase system that may vary by organism and Prx subfamily, but is often linked to thioredoxin or a related redox system 7,8. Other features which vary across the Prx family include their oligomeric state, their susceptibility towards inactivation by high peroxide concentrations, and their specificities for both reductants and substrates 14.
The classification of individual Prx proteins into mechanistically informative subfamilies is a key step towards understanding their biological role; however, this has been challenging to accomplish. Though representative Prxs like human Prx1, Escherichia coli thiol peroxidase (Tpx) and human Prx5 have some obvious features distinguishing them as a basis for classification into subgroups, different classification strategies have resulted in as few as 4 or as many as 7 subfamilies described for Prxs 7,10,13,15–20. Moreover, the distinction between 2-Cys and 1-Cys function is not especially useful for global classification because representatives of each type seem to exist within all the subfamilies. Some Prx subfamilies have very narrow distribution across biology (e.g., Tpx) whereas others are broadly distributed (e.g., AhpC/Prx1, Prx6). Indeed, humans express six different Prx proteins from three distinct subfamilies 13, while E. coli express three different Prxs from three different subfamilies 10. While it is early in our understanding of the potential specialization in Prx function to glean why multiple Prx proteins would be needed in a given organism, data from knockout mice provide proof that even very similar Prxs like Prx1 and Prx2 are not redundant in biological or cellular function 21,22.
The challenge of unambiguously defining subfamilies for global classification of Prxs is also exacerbated by the often automated way in which annotations, which may be vague or misleading in the first place, are transferred over to newly discovered proteins (i.e., sequences). Unfortunately for this field, general or confusing terms such as redoxin, thiol-specific antioxidant, thiol peroxidase and thioredoxin peroxidase are often used to denote peroxiredoxins, and none of these terms provide a high level of information regarding mechanism or specificity determinants of use to the biologist. Annotation accuracy for the Prx family is further hindered by underlying weaknesses of the automated computational methods used to annotate many sequences in public databases 23,24. Estimates of annotation errors in molecular functions ascribed to proteins (not just the Prxs) have ranged from 8% to as high as 49% 25–27, due in part to error propagation from annotation transfer 24,27–29 and to “overannotation”, where the level of functional detail transferred is not warranted by the methods 30. It is therefore important to develop improved bioinformatic classification methods that are capable of accurate annotation to the level of family and subfamily for all proteins including Prxs.
To meet the goal of better classifying families of proteins, including the Prxs, our focus in recent years has been to develop a bioinformatic approach called “functional site profiling” (also referred to as active site profiling) that utilizes structural information in the vicinity of the functional site to extract the most relevant sequences from a group of proteins and compare their sequence characteristics 31. This method was originally benchmarked on 193 protein families 31 and has subsequently been used to identify yeast serine hydrolases from yeast genome sequences32. Comparison between the bioinformatic profiling and the experimental activity-based profiling indicated significant complementarity between the bioinformatic and experimental methods. An extension of functional site profiling, called DASP (Deacon Active Site Profiling), has also been implemented as a publicly accessible bioinformatics tool which utilizes the profiles developed from proteins of known structure to search sequence databases; this tool was previously used to identify cyclooxygenases 33. In the work presented herein, DASP was used to generate Prx signatures from structurally-characterized members, signatures were clustered to reveal six distinct subfamilies, and profiles specific to each of the subfamilies were built and used to search the GenBank sequence database. This approach allowed the identification of many new members of each Prx subfamily, and we provide the list of all identified Prxs with their subfamily assignment. With this list in hand, we also analyzed residue conservation, phylogenetic distribution and conservation of the resolving Cys for each subfamily. The AhpC/Prx1 subfamily was also able to be further subdivided into four groups that are correlated with known functional distinctions, emphasizing the function-based focus of this method of protein annotation.
MATERIALS AND METHODS
Creation of active site signatures and profiles for Prxs of known structure
The RCSB Protein Data Bank 34 (PDB, release Jan 2008) was searched for all examples of Prx structures. Functional site signatures were created for the active site of each Prx structure, as previously described 31, using the publicly available DASP tool 35 (http://dasp.deac.wfu.edu/). Conserved residues in the PXXX(T/S)XXC motif were chosen as key residues (Pro39, Thr43, and C46 in S. typhimurium AhpC) as well as the conserved Trp or Phe residue (Trp81) (details about selection of the key residues are found in Table SI and Supplemental Results). All residues containing an atom located within 10 Å of the center of geometry of at least one of these key residues were extracted and the sequence fragments containing these residues were concatenated N- to C-terminus, forming the functional site signature for each structure. In cases where the CR forms an intermolecular disulfide bond with the CP (i.e. typical 2-Cys Prxs), the CR is not found in the functional site signatures since DASP only includes residues from a single subunit of the protein in the signature.
The functional site signatures for all Prx structures were then aligned using ClustalW 36,37 to create the functional site profile. The profile is scored using Equation 1:
| Equation 1 |
where SI is the score (=+1.0) for fully conserved positions over n such positions in the profile; SS is the score (=+0.2) for strongly conserved positions over m such positions in the profile; SW is the score (=+0.1) for weakly conserved positions over k such positions in the profile; Sg is the score (=−0.5) for each gap over l gaps along the profile; and N is the number of residues in the profile 31.
Clustering of functional site signatures to determine Prx subfamilies and subgroups
A ClustalW 36,37 alignment of the functional site signatures from Prxs of known structure was created using the Gonnet scoring matrix38. This alignment was read into Matlab and the Jukes-Cantor method (ignoring gaps in signature pairs) was used to calculate pairwise distances. Clustering of the signatures used the unweighted pair group method average (UPGMA) algorithm to generate the dendrogram. The clustering output was used to define the Prx subfamilies, except for the bacterioferritin comigratory protein (BCP)/PrxQ subfamily. The signatures for the BCP/PrxQ subfamily did not group together (Figure 2A), probably due to the sparseness of the data for this subfamily (see Supplemental Results). Based upon structural characterizations indicating these two proteins belong to the BCP/PrxQ subfamily, similar locations of the CR in both proteins, and a significant decrease in DASP scores when either signature was added to another subfamily, 2a4v and 2cx4 were both placed in the BCP/PrxQ subfamily.
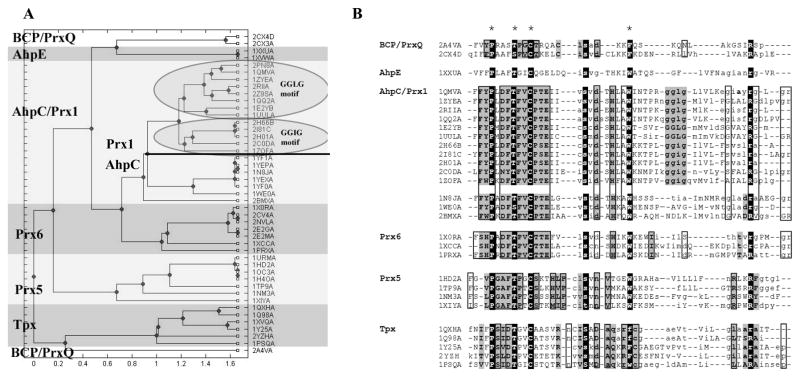
Figure 2. Functional site signatures extracted from all Prx structures identify six Prx subfamilies.
Functional site signatures were created for the active site of each Prx structure in the RCSB protein database (Jan 2008) using the DASP software package 35 at http://dasp.deac.wfu.edu/. (A) The functional site signatures were hierarchically clustered in Matlab using the unweighted pair group method average (UPGMA) algorithm. The dendrogram shown illustrates the resulting organization of functional site signatures and does not imply any evolutionary relationship. The cluster for each Prx subfamily is highlighted and labeled with the subfamily name, taken from one or two prototypical members of that subfamily. (B) Alignment of functional site signatures for Prx proteins of known structure identifies sequence characteristics for each subfamily. Changes between upper and lower case letters across each line denote a change to the next piece of contiguous protein sequence in a signature. Residues that are conserved across all Prxs are highlighted in black and key residues used to create the functional site signatures are starred. Residues conserved across each subfamily based upon analysis of proteins in the PDB database are highlighted in gray; residues found to be conserved within each subfamily following analysis of GenBank(nr) sequences are boxed. Only one signature is shown for any protein with multiple structures in the PDB database. Any engineered mutations, oxidized forms of cysteine, or seleno-methionine residues were changed back to their wild-type residue prior to alignment. This profile was created by first aligning the signatures for each subfamily using ClustalW and then editing by hand to correctly align the key residues across all subfamilies. Because of the variability among the signatures, DASP was unable to score this complete profile.
To further identify the subgroups within the AhpC/Prx1 subfamily, the functional site signatures obtained from the AhpC/Prx1 DASP subfamily search (as described in the next section) were aligned and clustered in Matlab as described above.
It is important to recognize that subfamily analysis that focuses only on active site signatures (and the dendrograms associated with them) should not be used to suggest evolutionary relationships. We do not know if similar functional sites evolved from a common ancestor or were developed by convergent evolution, or a combination of both.
Identification of additional Prxs for each subfamily from sequence databases using the DASP sequence search extension
Functional site profiles were created for each Prx subfamily using the subdivisions determined from clustering (Figure 2A). Information about the structures and key residues used to generate the profiles is found in Table SI.
Additional members of each Prx subfamily were identified using a p-value cutoff of 10−8 and the sequence searching utility of DASP previously developed and published 33,35. This method is described in detail in Supplemental Results and Figure S2. Sequences in each subfamily which lacked the PXXX(T/S)XXCP motif, were identified with a more significant p-value in another subfamily, or that could not be assigned to a single subfamily with any confidence were removed prior to further analyses (Table I).
Table I.
Summary of sequences and annotations of Prx subfamily members identified by DASP from GenBank(nr)
| Subfamily | Total identified by DASP | Total removed no Prx motifa | Total removed due to overlapb | Total after edits | GenBank Correct Annotations (%) | GenBank Incorrect Prx subfamily (%) | GenBank Correct but vaguec (%) | GenBank Incorrect (%) | No annotation in GenBank (%) |
|---|---|---|---|---|---|---|---|---|---|
| BCP/PrxQd | 1130 | 4 | 11 | 1115 | 34e | 1 | 52 | 2 | 11 |
| AhpC/Prx1 | 1073 | 11 | 3 | 1059 | 35f | 0 | 49 | 2 | 14 |
| Prx6 | 507 | 4 | 10 | 493 | 6g | 6 | 66 | 8 | 14 |
| Prx5h | 522 | 5 | 0 | 517 | 10i | 3 | 55 | 3 | 24 |
| Tpx | 308 | 1 | 0 | 307 | 59j | 0 | 34 | 2 | 5 |
| AhpEd | 38 | 0 | 13 | 25 | |||||
| Total or range | 3578 | 25 | 37 | 3516 | 6–59% | 0–6% | 34–66% | 2–8% | 5–24% |
Detailed information about hits with no Prx motif is found in Table SIII
Detailed information about crosshits is found in Table SIV.
Correct but vague annotation includes: peroxiredoxin, AhpC/TSA family protein, antioxidant protein, thiol-specific antioxidant related protein, peroxidase, oxidoreductase, redoxin, 1-Cys Prx, 2-Cys Prx, thioredoxin peroxidase, thiol peroxidase, 9.1 kDa protein, 26,000/30,000 kDa protein, mitochondrial peroxidase, and peroxynitrite reductase.
Numbers reported here for the DASP subfamily searches using engineered profiles for BCP/PrxQ and AhpE subfamilies (see Methods and Supplementary Results). This BCP/PrxQ search was performed on a later version of GenBank(nr) January 2009 than the searches presented for the other subfamilies. The original BCP/PrxQ search using the January 2008 version of GenBank(nr) identified 810 sequences (2 with no Prx motif, 4 that were more significant in another subfamily, and 4 that were equally significant in 2 subfamilies). All other searches were performed on the January 2008 version of GenBank(nr).
Correct annotations for BCP/PrxQ subfamily include BCP, PrxQ, and Dot5.
Correct annotation for the AhpC/Prx1 subfamily includes: AhpC, Prx1, Prx2, Prx3, Prx4, and tryparedoxin peroxidase.
Correct annotations for Prx6 subfamily include Prx6.
Some Prx5 proteins (5%) contain a glutaredoxin domain fused to a Prx5-like domain and are only annotated with glutaredoxin not Prx.
Correct annotations for the Prx5 subfamily include Prx5, type II Prx, PrxD, peroxisomal Prx, and hybrid Prx5.
Correct annotations for the Tpx subfamily include thiol peroxidase, thioredoxin peroxidase, and scavengase p20.
Bootstrap procedure for creating engineered profiles for subfamilies with limited structural coverage
To create more diverse profiles, engineered profiles were created for AhpE and BCP/PrxQ subfamilies. Additional AhpE sequences were found using PSI-BLAST 39 default values and the protein sequence from Mycobacterium tuberculosis AhpE (1xxu, the only known AhpE structure at the time). Pseudo-signatures were created for AhpE proteins from Mycobacterium vanbaaleniim, Nocardia farcinica, and Frankia sp. (GenBank accession numbers 13881989, 90201095, 54015086, and 68234122 respectively). The resulting profile for the AhpE subfamily represented the sequences of four different AhpE proteins.
Pseudo-signatures were created for three examples of characterized BCP/PrxQ proteins: E. coli BCP 40, Populus deltoides PrxQ 41, and Helicobacter pylori BCP 42 (GenBank accession numbers 16130405, 75127599, and 15611194, respectively). When added to the functional site signatures for both redox states of Aeropyrum pernix BCP (2cx3 and 2cx4) and Saccharomyces cerevisiae BCP (2a4v), the resulting engineered profile for the BCP/PrxQ subfamily represented the sequences of four different BCP/PrxQ proteins, one in two different redox states (Figure S3C).
Entropy calculation for evaluation of residue conservation
To evaluate the degree of conservation for each residue within a functional site profile, the full sequence for each protein identified by DASP was extracted from GenBank(nr) and all sequences in each subfamily were aligned using ClustalW36,37. Due to the small number of subfamily members, entropy values were not calculated for the AhpE subfamily. The number of occurrences for each type of amino acid at each position was counted and the entropy value for each residue, Sm, was determined using the formula:
| Equation 2 |
where each possible amino acid identity 1,2,…k is sampled with a frequency f1,f2,…fk and the set fj sums to unity 43. The entropy value for a completely conserved residue is 0, while a completely random distribution of residues was estimated by calculating the entropy value if all residues were present at the same frequency and resulted in an entropy value of 3. The entropy values for each residue position in the profile (shown in Figure 3) were identified based on the alignment of the profile fragments (Figure 2B) in this full multiple sequence alignment. Residues with an entropy value lower than 0.61 (the mean minus one standard deviation, calculated as described in Supplemental Materials and Methods) were considered conserved.
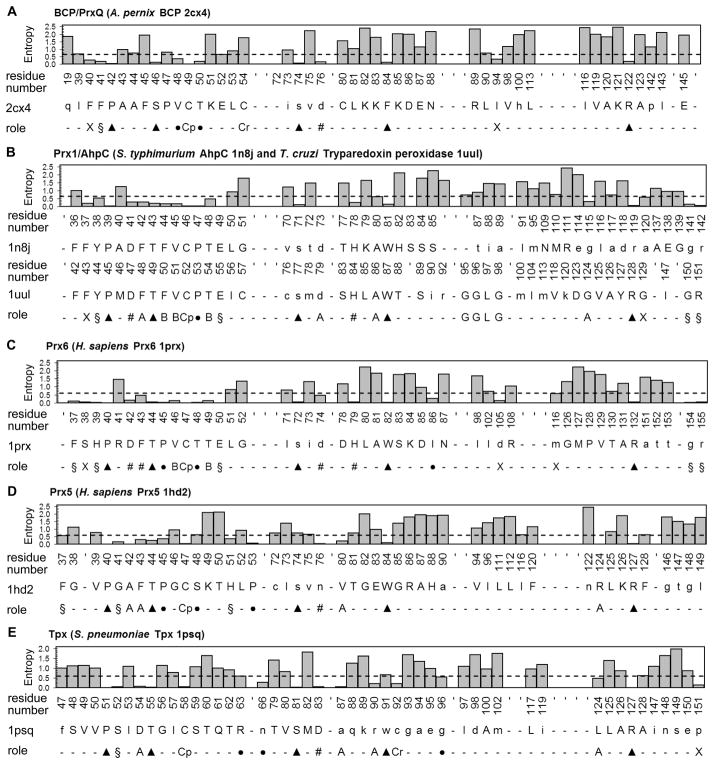
Figure 3. Residue conservation and potential structural/functional role for each residue in Prx subfamily functional site profiles.
The functional site signatures are shown for a representative member of each of the Prx subfamilies. The degree of conservation is shown for each residue after aligning the full sequence for all of the putative members identified from the GenBank(nr) database searches. The actual residues and numbers listed are for (A) Aeropyrum pernix BCP (2cx4), (B) Salmonella typhimurium AhpC (1yep) and Trypanosoma cruzi tryparedoxin peroxidase (1uul), (C) Homo sapiens Prx6 (1prx), (D) H. sapiens Prx5 (1hd2), and (E) Streptococcus pneumoniae Tpx (1psq). Residues with an entropy value below 0.61 (mean minus 1 standard deviation) are considered conserved (dashed line). The potential role for each conserved residue is also represented in the histogram and labeled as follows: the peroxidatic cysteine (Cp), the resolving cysteine (Cr), key residues conserved across all Prx subfamilies (▲), residues involved in forming the active site pocket of the reduced protein (§), residues found in the A-type interface (A), residues found in the B-type interface (B), residues involved in stabilizing the helix containing the CP (●), residues forming a series of H-bonds between the key Thr residue and the region containing the A-type interface (#), and conserved residues that do not fall into any of these groups (X). Hydrogen bonds were analyzed using LIGPLOT 80.
PSI-BLAST searches to identify members of Prx subfamilies
The sequence of a single chain from two PDB structures from each subfamily (except for AhpE, which only has one) were used as a query to search the Genbank(nr) sequence database with PSI- BLAST 39 (1xxu, AhpE; 1xiy and 1hd2, Prx5; 2cx3 and 2av4, BCP; 1psq and 1xvq, Tpx; 1xcc and 2cv4, Prx6; and 1qmv and 1yep, AhpC/Prx1). One set of results were obtained using the default scoring parameters on the National Center for Biotechnology Information (NCBI)/Blast server. Another set of PSI-BLAST searches were done using a more stringent cutoff. The structurally characterized proteins were retrieved at scores more significant than e-40 in the default PSI-BLAST searches; thus, a cutoff of e-40 was selected to determine which sequences were added to the PSSM after each iteration in the stringent PSI-BLAST searches.
Following three complete iterations, the top 5000 sequences identified using either “default” or “stringent” parameters for each subfamily were exported to Excel and then imported into Microsoft Outlook Access database tables. These data were queried using the sequence GenBank identification numbers in order to identify hits found in multiple PSI-BLAST searches. For each search, the e-values were determined for all test proteins belonging to the same subfamily and the least significant score was set as a cutoff for that search (cutoff scores for each search are listed in Table II). For analysis of PSI-BLAST hits containing no Prx motif and subfamily specificity, only sequences with more significant scores than the cutoff were analyzed.
Table II.
DASP and PSI-BLAST subfamily assignments of literature defined test proteins
| Search | Number of test proteins identified in each search with a more significant p/e-value than the cutoff | |||||
|---|---|---|---|---|---|---|
| BCP/PrxQ | AhpC/Prx1 | Prx6 | Prx5 | Tpx | ||
| DASP | p-value cutoff | (11 test proteins) | (14 test proteins) | (17 test proteins) | (10 test proteins) | (6 test proteins) |
| BCP/PrxQ | 1 × 10−8 | 11 | 0 | 0 | 0 | 0 |
| AhpC/Prx1 | 1 × 10−8 | 0 | 14 | 0 | 0 | 0 |
| Prx6 | 1 × 10−8 | 0 | 0 | 17 | 0 | 0 |
| Prx5 | 1 × 10−8 | 0 | 0 | 0 | 9a | 0 |
| Tpx | 1 × 10−8 | 0 | 0 | 0 | 0 | 6 |
| PSI-BLAST (default) | e-value cutoffb | |||||
| BCP – 2cx3 | 9 × 10−29 | 11 | 14 | 17 | 0 | 4 |
| BCP – 2a4v | 7 × 10−39 | 11 | 5 | 0 | 0 | 0 |
| AhpC/Prx1 - 1qmv | 4 × 10−33 | 0 | 13c | 7 | 0 | 0 |
| AhpC/Prx1 - 1yep | 4 × 10−36 | 0 | 13c | 2 | 0 | 0 |
| Prx6 – 2cv4 | 8 × 10−40 | 0 | 10 | 17 | 0 | 0 |
| Prx6 – 1xcc | 2 × 10−30 | 0 | 11 | 17 | 0 | 0 |
| Prx5 – 1hd2 | 1 × 10−13 | 0 | 0 | 0 | 9d | 0 |
| Prx5 – 1xiy | 2 × 10−15 | 0 | 0 | 1 | 10 | 0 |
| Tpx – 1psq | 1 × 10−29 | 8 | 12 | 9 | 0 | 6 |
| Tpx – 1xvq | 1 × 10−29 | 7 | 12 | 6 | 0 | 6 |
| PSI-BLAST (stringent) | e-value cutoffb | |||||
| BCP – 2cx3 | 2 × 10−12 | 11 | 12 | 6 | 0 | 2 |
| BCP – 2a4v | 1 × 10−7 | 11 | 0 | 1 | 0 | 0 |
| AhpC/Prx1 - 1qmv | 2 × 10−65 | 0 | 13c | 0 | 0 | 0 |
| AhpC/Prx1 - 1yep | 4 × 10−69 | 0 | 13c | 0 | 0 | 0 |
| Prx6 – 2cv4 | 2 × 10−73 | 0 | 0 | 17 | 0 | 0 |
| Prx6 – 1xcc | 7 × 10−61 | 0 | 0 | 17 | 0 | 0 |
| Prx5 – 1hd2 | 1 × 10−8 | 0 | 1 | 0 | 10 | 0 |
| Prx5 – 1xiy | 4 × 10−18 | 0 | 1 | 0 | 10 | 0 |
| Tpx – 1psq | 1 × 10−52 | 0 | 0 | 0 | 0 | 6 |
| Tpx – 1xvq | 1 × 10−45 | 0 | 0 | 0 | 0 | 6 |
The DASP search of the Prx5 subfamily did not identify Saccharomyces cerevisiae Ahp1p.
PSI-BLAST e-value cutoff was selected after data collection by selecting the least significant e-value for all of the literature defined test proteins belonging to the same subfamily as the search sequence. Yellow highlights indicate all test proteins were identified in a search from the correct subfamily and orange highlights indicate that one test protein was missed. Gray highlights indicate test proteins that are identified in an incorrect subfamily search.
PSI-BLAST searches using default parameters did not identify Schistosoma mansoni Prx2 in either the AhpC/Prx1-1yep or the AhpC/Prx1-1qmv searches. S. mansoni Prx2 was identified above the e-value cutoff by the BCP-2cx3 PSI-BLAST search with default parameters. S. mansoni Prx2 was not identified in any PSI-BLAST search using stringent parameters and was identified correctly by DASP in the AhpC/Prx1 search.
The Prx5-1hd2 PSI-BLAST search using default parameters did not identify Pisum sativum Prx IIF.
Phylogenetic analysis
The phylogenetic distribution of each subfamily was calculated by first extracting the organism name for each Prx sequence identified by DASP. A house-written Java script was then used to query the NCBI Taxonomy databases to identify the complete lineage for each organism. This information was imported to an Excel file and genus and species numbers were calculated. To prevent results being biased by oversampling of sequences from multiple bacterial strains, multiple strains of the same species were only counted once for each subfamily. Each species was also only counted once in each subfamily even if multiple protein sequences were identified.
RESULTS
Clustering of functional site signatures identifies six Prx subfamilies and clearly distinguishes the AhpC/Prx1 subfamily from the Prx6 subfamily
Functional site profiling 31, as implemented in the DASP application 35, involves selecting a few key residues from proteins of known structure based upon their functional importance. All residues within 10 Å of the key residues are then extracted and placed in order from N- to C-terminus, forming what is referred to as the signature for each functional site. The signature thus contains the information regarding the features in the structural vicinity of the functional site, but puts that information into a format that can be used for sequence-based alignment and searching. The functional site profile for a given protein family is the alignment of all signatures generated from each of the protein structures within the family.
Functional site signatures were created for each Prx structure listed in Table SI using the residues equivalent to CP (Cys46), Pro39, Thr43, and Trp81 as key residues (numbering for S. typhimurium AhpC, Figure 1). (Key residue selection criteria are described in Supplemental Results.) To identify the subfamily divisions suggested by this active site information, the signatures in the profile were hierarchically clustered and analyzed (Figure 2A). Six subfamilies (AhpC/Prx1, Prx6, Prx5, Tpx, BCP/PrxQ, and AhpE) were identified and named after one or two canonical subfamily members. The signatures from each cluster (Figure 2B) were aligned to create the functional site profile for that subfamily. Scores for each subfamily ranged from 0.06 (for AhpC/Prx1) to 0.33 (Prx6) (see Supplemental Results for more details). These subfamilies are consistent with those identified by previously reported sequence- and structure-based methods 7,13,14,16. Previous studies have shown that functional site profile scores for 193 families ranged from 0.04–1.0. Higher profile scores are correlated with more similarity at the functional site 31, suggesting that the active sites in the Prx subfamilies are relatively diverse compared to other protein families. DASP was unable to generate a profile score for the entire Prx family, further suggesting that the residues found in different Prx active sites vary greatly.
Because both the AhpE and BCP/PrxQ subfamilies contained very few structurally characterized representatives at the time of this study, the original profile used for searching did not represent the diversity of these subfamilies; therefore, a procedure was developed to create engineered profile in each case. Description of this method can be found in Methods and verification of this method with the BCP/PrxQ subfamily is provided in Supplementary Results. These engineered profiles were used in all subsequent searches and analyses.
Identification and subfamily classification of known and new Prx proteins from the sequence database using DASP provides highly specific subfamily assignments
The functional site profiles created from the known structures (or engineered profiles for the BCP/PrxQ and AhpE subfamilies) for each Prx subfamily were used to search the GenBank(nr) database (January, 2008 release, and January, 2009 release for BCP), following previously described methods 33. To do this, a position-specific scoring matrix (PSSM) 44 is generated for each continuous fragment (motif) and the PSSM is used to search the non-redundant (nr) protein sequences of GenBank 45 for other subfamily members. A final p-value is calculated that represents the statistical significance of matching all fragments in a profile to a given sequence. A detailed description of how DASP accomplishes this is found in Supplemental Methods and in Figure S2. Using a p-value cutoff of 10−8 (selection of this cutoff is described in Supplemental Results), a list of 3578 putative Prx sequences was generated with each Prx assigned to one of the six subfamilies. The quality of these subfamily assignments was assessed in three ways, asking (1) if biochemically characterized Prxs were placed into the experimentally-determined subfamily, (2) if the conserved PXXX(T/S)XXCP motif was present in the returned sequences, and (3) whether the returned sequences were specific to a single subfamily, or if some sequences were identified in more than one subfamily search.
To determine how well DASP identified experimentally-determined proteins in each subfamily, a “test set” of Prx proteins was identified that (1) had not been used to create the functional site profile, and (2) were assigned to a particular Prx subfamily based upon literature data. The identity, literature reference, and scores for all DASP searches for the 58 test proteins (11 BCP/PrxQ, 14 AhpC/Prx1, 17 Prx6, 10 Prx5, and 6 Tpx) are shown in Table SII. At a p-value cutoff of 10−8, DASP correctly identified the Prx subfamily for all but one of the test proteins (57 out of 58, a 98% true positive rate) and did not assign any of the test proteins to an incorrect Prx subfamily (a 0% false positive rate). DASP did not assign the Saccharomyces cerevisiae Ahp1p protein (GenBank accession no. 6323138) to any subfamily (a 1.7% false negative rate), although literature reports assign this protein to the Prx5 subfamily 46. Previous analyses have grouped Prx6 with AhpC/Prx1 subfamily members16; however, DASP searches correctly identified the subfamily for the 14 AhpC/Prx1 and 17 Prx6 proteins (Table II), suggesting that these subfamilies can be cleanly separated based on their active site features.
As a second assessment of the quality of the DASP-assigned subfamilies, we asked how many sequences were lacking the PXXX(T/S)XXCP motif, which is invariant in the Prx family 8–10. Although DASP weights this motif heavily in the PSSM due to its conservation, the residues are not required to be invariant. Of the 3578 Prx proteins, only 25 (0.7 %) identified by DASP with a p-value cutoff of 10−8 did not contain the PXXX(T/S)XXCP motif (Table I, proteins listed in Table SIII). Of these, nine were incomplete sequences lacking the Prx motif region, one is a known sequencing error 47, and one sequence looks like a Prx only in the C-terminal portion of the protein (a potential frameshift error). Either the P or the T was not conserved within the motif in fourteen other sequences for unknown reasons.
Finally, the specificity of subfamily searches was explored. Specificity was assessed by determining whether any sequences were identified in more than one subfamily search. DASP results, summarized in Table II, exhibited a high degree of specificity overall. Only 37 sequences (1.0 %) were assigned to two subfamilies and no sequences were assigned to three subfamilies (proteins listed in Table SIV). Of the 37 sequences, 33 were identified by one subfamily search with a much more significant score; we consider these 33 sequences to be members of the subfamily with the most significant DASP score. The four remaining sequences (0.1% of the original 3578 sequences identified) were identified by two subfamilies searches, both with less significant DASP p-values of 10−9; because these four sequences could not be confidently assigned to a single subfamily, they were removed prior to residue conservation and phylogenetic analysis.
The Tpx and Prx5 subfamilies were highly specific; no proteins identified in these searches were identified in other searches (Table I), suggesting that the active sites for Prx5 and Tpx are distinct from one another and the other Prx subfamilies. The BCP/PrxQ subfamily exhibited some overlap with AhpE, Prx6, and AhpC/Prx1 subfamilies (3, 4 and 4 cross-hits, respectively, Table SIV), suggesting that these subfamilies share some similarity in the region surrounding the active site. These data support the hypothesis by Copley et al that the BCP/PrxQ subfamily (referred to in their paper as class 1 Prxs) was the first evolutionarily and that other subfamilies arose through adaptations to this subfamily.
Analysis of cross-hits also allows for clarification of the relationship between the Prx6, Prx1, and AhpC subfamilies, which has varied in the literature: sequence-based comparison combined Prx6, Prx1, and AhpC into a single subfamily 16, while structure-based expert comparisons identified Prx6 as a separate subfamily 7. Clustering of DASP functional site signatures for Prxs of known structure also suggests a significant distinction between the Prx6 and AhpC/Prx1 subfamilies (Figure 2A). Only 12 sequences were identified in both the Prx6 search and the AhpC/Prx1 search using DASP (0.77% of the total identified in both searches) and, in every case, one DASP score was much more significant (Table SIV). These results indicate that Prx6 proteins are readily distinguished from AhpC/Prx1 subfamily members based on their active site features.
Clustering of the functional site signatures of proteins of known structure indicated that Prx1 and AhpC might also be distinguishable (indicated by division in Figure 2A dendrogram), Despite this fact, DASP searches using separate profiles for Prx1 and AhpC were unable to distinguish the two subfamilies (e.g., 95% of the AhpC representatives were also returned in the Prx1 search). All further analyses were therefore undertaken using a combined AhpC/Prx1 subfamily profile.
Overall, the three assessments—comparison to experimentally-determined subfamily assignments, presence of the PXXX(T/S)XXCP motif, and analysis of search specificity by counting cross-hits—indicate that the DASP-assigned subfamily assignments are specific and are of high quality. Sequences were removed which lacked the PXXX(T/S)XXCP motif, were identified with a more significant p-value in another subfamily, or could not be assigned to a single subfamily with any confidence, resulting in 3516 sequences that were used for all further analyses (Table SV). The final total membership within each of the six Prx subfamilies was: BCP/PrxQ (1115), AhpC/Prx1 (1059), Prx6 (493), Prx5 (517), Tpx (307), and AhpE (25). All results, including detailed information about the identified sequences, are reported in Table SV.
The functional site focused approach used by DASP is more specific for Prx subfamily assignments than the full sequence approach of PSI-BLAST
PSI-BLAST 39 is a commonly used tool for family and subfamily-specific annotation and, during its iterative process, develops a PSSM that is used to search the sequence database in a manner similar to DASP. Unlike DASP, which focuses on sequences found at the functional site, PSI-BLAST utilizes the entire sequence of a single query (or seed) to initiate its search. To determine how well DASP classified Prxs compared to PSI-BLAST, two sequences for each Prx subfamily were used to performed three iterations of PSI-BLAST (AhpE was not considered).
Using default parameters, PSI-BLAST identified all of the 58 experimentally identified proteins as Prxs, but the proteins in PSI-BLAST were not cleanly assigned to a specific subfamily (Table II). In contrast, DASP identified 57 test proteins and assigned each to just one subfamily. PSI-BLAST was able to assign Prx5 and Tpx subfamily members to the correct subfamily (and no other) using a search-specific cutoff score based on the least significant score for test proteins in the same subfamily; however, many of the other literature defined test proteins were still assigned to multiple subfamilies (Table II). One protein, Schistosoma mansoni Prx2 (GI# 5163492) was incorrectly identified by PSI-BLAST in one of the BCP/PrxQ searches, but was not identified in either AhpC/Prx1 search; in contrast, this protein was correctly assigned to the AhpC/Prx1 subfamily by DASP.
To determine if more stringent parameters would allow PSI-BLAST to assign Prxs to the correct subfamily, only sequences with p-values more significant than 10−40 from each iteration were selected for inclusion in the next iteration, for a total of three iterations. These stringent PSI-BLAST parameters proved more specific than the default parameters (Table II); however, the subfamily assignments were still not as accurate as DASP. The PSI-BLAST search results were highly dependent on the protein chosen to seed the search. For example, the Prx5 subfamily member Pisum sativum PrxII F (GI# 118721272) was identified with default parameters using 1xiy as the seed sequence, but not 1hd2 (Table II). The cutoff scores also differed from search to search (Table II), making it difficult to use the scores to evaluate the best subfamily assignment. DASP identified fewer proteins with no Prx motif than did PSI-BLAST, though the difference was not statistically significant; DASP always identified <1% of hits with no Prx motif while PSI-BLAST identified between <1%– 4.5%, depending on the seed sequence.
These results indicate that, with appropriate parameterization and score cutoff, PSI-BLAST searches can be used to identify Prx proteins, but cannot be used to confidently determine subfamily assignment for some of the 58 literature-defined test proteins. In contrast, the functional site-focused approach utilized by DASP appears to identify differences at the active site that might be obscured by PSI-BLAST’s use of full-sequence information and is expected to be more accurate than PSI-BLAST for molecular function-based subfamily assignments.
DASP-based subfamily annotations provide more detailed subfamily information than the annotations currently found in GenBank
We then explored how our subfamily classification compared with the annotations currently found in the GenBank(nr) database. Depending on the subfamily, only 11–58% of the Genbank annotations were correctly annotated to the subfamily level assigned by DASP (Table I, Correct). For a large number (5%–24%) of the sequences we identified, no previous function (“hypothetical protein” or “unknown protein”) had been assigned. In addition, numerous examples of “correct, but vague” annotation were identified, ranging from 34% (Tpx subfamily) to 66% (Prx6 subfamily). These GenBank annotations typically identified a redox-based function (though not necessarily as a Prx) but not the correct subfamily and included terms like redoxin, peroxidase, thiol specific antioxidant, AhpC/thiol specific antioxidant family protein, and peroxiredoxin that are technically correct, but provide limited functional information. Protein annotations such as “thiol peroxidase” and “thioredoxin peroxidase” are used as both a general term for all Prxs and a specific subfamily name; these were classified as correct but vague for all subfamilies except for Tpx, where they were considered correct. Previous work by Babbitt and colleagues has suggested that misannotations can be due to overuse or incorrect use of annotation transfer 30. Among the Prxs, a small percent of sequences were either assigned to a subfamily different from the one assigned by DASP (Table I, Incorrect Prx subfamily) or not annotated as a Prx at all (Table I, Incorrect). Thus, while over-annotation has been a small problem with annotation in the Prx family (perhaps due to a misunderstanding of subfamily identification terms), the lack of accurate subfamily information or sometimes even the lack of annotation as a Prx or peroxidase are the most common problems. DASP classification, therefore, provides a significant improvement over the current GenBank annotations with regard to subfamily information.
Phylogenetic analysis shows that all Prx subfamilies are found in bacteria and the AhpC/Prx1 and BCP/PrxQ subfamilies are widely represented
We next assessed the phylogenetic distribution of each Prx subfamily (Table III). As has been noted previously 10,12, the AhpC/Prx1 and Prx6 subfamilies are widely distributed among archaea, bacteria, and eukaryotes. Prx5, which has been predominantly characterized in eukaryotes, is also widely distributed; 66% of the species containing a Prx5 subfamily member are bacterial. While the majority of the BCP/PrxQ subfamily proteins were found in bacteria (84.7%), putative BCP/PrxQ proteins were also observed in archaea (6.2%) and eukaryotes (9.1% including plant PrxQs). The subfamily that was the smallest and most restricted in phylogenetic distribution was AhpE. Only 25 proteins in 22 species were identified as potential AhpE-type proteins, all from aerobic gram-positive bacteria in the order Actinomycetales.
Table III.
Phylogenetic distribution for Prx subfamilies
| Number of Speciesa | |||||||
|---|---|---|---|---|---|---|---|
| Subfamily | Archaeal | Bacterial | Eukaryotic | ||||
| total | Fungi | Plant | Animal | Other | |||
| BCP/PrxQ | 34 | 537 | 55 | 30 | 15 | 10 | |
| AhpC/Prx1 (total) | 5 | 437 | 153 | 20 | 22 | 72 | 39 |
| Group 1 (Prx1) | 2 | 187 | 152 | 20 | 22 | 71 | 39 |
| Group 2 (AhpC) | 3 | 171 | 1 | 1b | |||
| Group 3 | 59 | ||||||
| Group 4 | 20 | ||||||
| Prx6 | 39 | 181 | 111 | 38 | 15 | 49 | 9 |
| Prx5 | 197 | 95 | 34 | 22 | 31 | 8 | |
| Tpx | 224 | 1 | 1 | ||||
| AhpE | 22 | ||||||
Each species was only counted once even if multiple protein sequences were identified within each subfamily. For instance, H. sapiens would only be counted once in the AhpC/Prx1 subfamily even though H. sapiens contains four distinct Prx proteins that are members of this subfamily. Multiple strains of the same species were only counted once.
The only group2 AhpC identified from eukaryotes (gi number 110768962) is from Apis mellifera, a species of honeybee. This protein is 80% identical to a group2 AhpC/Prx1 from Marinobacter aquaeolei, a type of marine bacteria.
Tpx subfamily members are found almost exclusively in bacteria (Table III). Interestingly, two Tpx sequences (GI# 123501795 and 123457052; 74% identical to each other) were observed in a eukaryote, Trichomonas vaginalis, an anaerobic flagellated parasite that is the most common cause of sexually transmitted infections in industrialized countries 48. Detailed sequence analyses of these proteins suggest that they are indeed Tpxs (e.g., the CR is in the correct location). Based upon alignment of the full sequence, the T. vaginalis Tpxs are most similar (~70% identity) to Tpxs from multiple species of Bacteroides, anaerobic bacteria found in the gut and colon of humans that are also frequently responsible for infections. We speculate that these species might have been present in the same organism under conditions expected to lead to increased reactive oxygen species levels due to the immune response, and that a lateral transfer of the gene encoding Tpx may have occurred and provided an advantage to T. vaginalis.
Analysis of residue conservation in each Prx subfamily identifies conserved residues and indicates that many of the conserved residues in each subfamily are located in the functional site profile
The Prx active site contains features that both stabilize the deprotonated form of the CP and support a very high reaction rate with peroxides (> 107 M−1s−1 at 20 °C) 11,49,50. Because very few residues are conserved across all Prxs, our goal was to identify residues conserved in, and unique to, each Prx subfamily. To assess residue conservation within each subfamily, entropy values were calculated at each residue position across the full sequence alignment. Anywhere from 35% (Prx5) to 67% (AhpC/Prx1) of the residues in the profile are conserved. In contrast, 9–20% of the residues are conserved across the entire protein sequence (Table SVI). This indicates that approximately 50% of the conserved residues within each subfamily are located within the functional site profile, reflecting the functional importance of this region. Note that this conservation is not observed using a contiguous sequence fragment, but rather with several discontinuous fragments that are proximal in three-dimensional space to the Prx active site. Entropy values across the functional site profiles for each subfamily are shown in Figure 3.
Other than the PXXX(T/S)XXCP motif 8 and Arg1192,8,10 (numbering for S. typhimurium AhpC), our analysis identified only three other sites of conservation across all Prx functional site signatures (Figure 2B, highlighted in black). The locations of these residues in representative Prxs are shown in Figure 4 (residues in pink). The first is Trp81; this residue is replaced with a Phe in some Prxs, particularly in the BCP/PrxQ and Tpx subfamilies. The second residue, Ser71, is conserved across all Prx structures and most of the signatures (Figure 3). This residue is located between the active site and the A-type interface and is part of a hydrogen bonding network with other conserved residues. Although Ser71 (Figure 4, pink) is conserved across all subfamilies except Prx5, the remaining residues involved in this network differ from subfamily to subfamily. The third residue, Glu49 in S. typhimurium AhpC, is not conserved across all subfamilies, but all subfamilies contain a conserved residue at this position that is capable of forming a hydrogen bond with the stringently conserved Arg. These observations suggest that the ability of this residue to hydrogen bond to Arg119 is important and that it may indirectly influence the pKa of the CP. More details and discussion about the role of each of these residues is found in Supplemental Results.
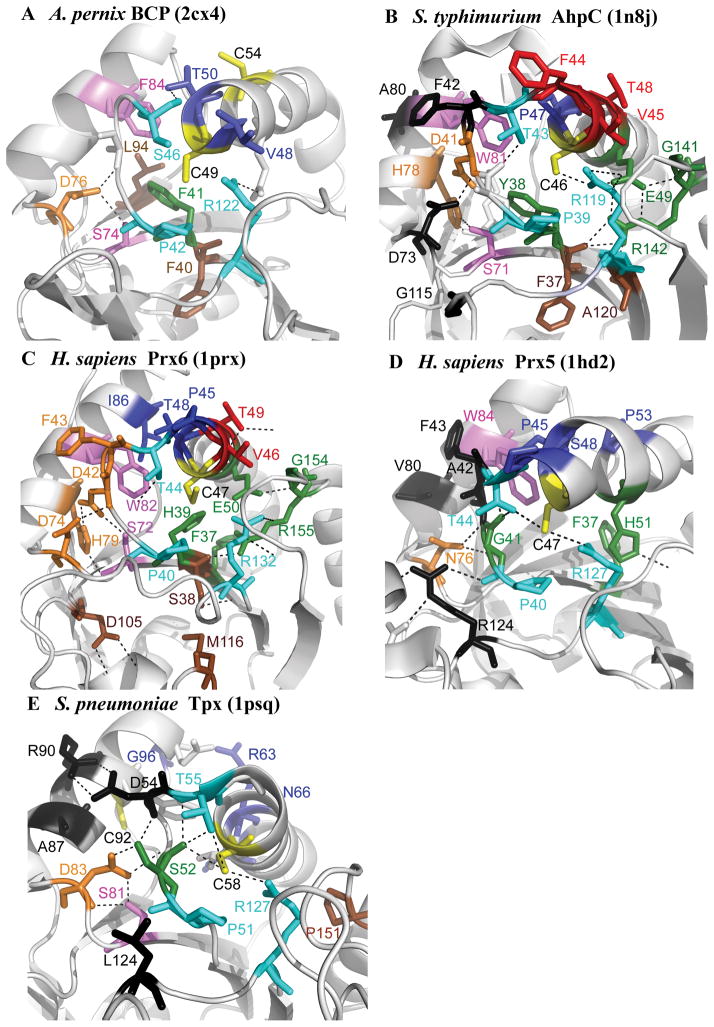
Figure 4. The location of conserved residues mapped to the structure for each subfamily.
Structures are shown for (A) Aeropyrum pernix BCP (2cx4), (B) Salmonella typhimurium AhpC (1n8j), (C) Homo sapiens Prx6 (1prx), (D) H. sapiens Prx5 (1hd2), and (E) Streptococcus pneumoniae Tpx (1psq). The CP and CR are in yellow, residues conserved across all Prx subfamilies in magenta and cyan, residues involved in forming the active site pocket of the reduced protein in green, residues found in the A-type interface in black, residues found in the B-type interface in red, residues involved in stabilizing the helix containing the CP in deep blue, residues forming a series of H-bonds between the key Thr residue and the region containing the A-type interface in orange, and conserved residues that do not fall into any of these groups in brown. Figure was made using Pymol (http://sourceforge.net/projects/pymol/).
Mapping of conserved residues to Prx structures indicates the importance of A-interface interactions for three Prx subfamilies
Residues found to be conserved in each subfamily (Figure 3) were identified and their locations were mapped to the structure of a representative member of each subfamily (Figure 4). One interesting group of conserved residues is located on helix α2, which contains the CP (Figures 1, brown helix, and S1). These residues form hydrogen bonds or hydrophobic interactions with residues in the neighboring helix α3, potentially stabilizing the fully folded conformation and promoting enzymatic activity when the CP is reduced [Figure 4, residues in deep blue; Figure 3, residues marked with (•)].
With the exception of BCP/PrxQ proteins, which are reportedly monomeric 7,40,41,51, Prx proteins form two distinct types of oligomeric interfaces that are both close to the active site (Figure 1). Members of the AhpC/Prx1, and Prx6 subfamilies dimerize utilizing the “B interface” (denoting the beta strand interactions; Figure 1, pale green) to form an extended 10- to 14-strand beta sheet 7. Members of the Tpx and Prx5 subfamilies, which are typically dimeric, associate across the “A interface” (for either alternate or ancestral); this interface involves helix α3 packing against its counterpart in the other chain (Figure 1, tan). In many Prxs that form dimers across the B interface, the dimers further associate in a redox-sensitive manner to form decamers (or in a few cases, octamers or dodecamers) across the A interface 17.
Interestingly, many of the conserved residues appear to be located either directly in an interface (Figure 3, residues marked with A; Figure 4, residues in black) or in the portion of the protein structure that bridges the CP and the A-type interface (Figure 3, residues marked with #; Figure 4, residues in orange). Of the subfamilies that have been shown to utilize the A interface for oligomerization (AhpC/Prx1, Prx6, Tpx, and Prx5), 30–41% of the conserved residues are located either in the interface or within the H-bonding network bridging the interface and the active site loop (Figure 4B, A interface in black, residues in bridge region in orange), suggesting that interactions at the A interface are important for stabilization of the correct active site geometry in these subfamilies. This linkage between features of the active site and the A interface interactions is in agreement with experimental work linking oligomeric state to redox state and activity in S. typhimurium AhpC 52. Destabilization of the A interface through a single amino acid substitution was shown to increase the Km for H2O2, suggesting that dimeric AhpC is a less efficient catalyst of peroxide reduction than the decameric protein 49. In addition, computational analysis of another member of the AhpC/Prx1 subfamily, Trypanosoma cruzi tryparedoxin peroxidase, indicated that interactions at both the A and B interfaces are critical for lowering the pKa of the CP 53.
The position of CR is highly conserved in the AhpC/Prx1 and Tpx subfamilies, but not in BCP/PrxQ or Prx5 subfamilies
Prxs are most commonly characterized based upon the presence and location of the CR (Figure S1) 2; thus, we analyzed the location of CR in each subfamily identified by DASP (Table IV). These data indicate that the CR location is highly conserved within, and characteristic of, the AhpC/Prx1 and Tpx subfamilies, although there are some exceptions even in these subfamilies 8. In contrast, the presence and location of the CR in the BCP, Prx5, and possibly AhpE subfamilies is more variable.
Table IV.
Location of CR, if present, in each Prx subfamily
| Canonical location of CR | Subfamily members with CR in canonical location (%) | Subfamily members with only 1 Cys in sequence (%)a | Subfamily members with alternative locations for CR (location, %) | Uncertainb (%) | |
|---|---|---|---|---|---|
| BCP/PrxQ | Helix α2c (CPXXXXCR) | 54 | 13 | Helix α3 7 | 26 |
| AhpC/Prx1 | C-terminusd | 99.1 | 0.4 | - | 0.5 |
| Prx6 | No CR | - | 40 | C-terminus 1.6 Helix α3 0.8 |
57 |
| Prx5 | Helix α5 | 17 | 14 | - | 69e |
| Tpx | Helix α3f | 96.4 | 1.0 | - | 2.6 |
| AhpEg | No CR | - | 28 | Helix α2 56 | 16 |
If no CR is present, the resolving thiol must come from another protein or a small molecule thiol like glutathione.
These sequences contain more than one Cys, however the other Cys are not in a location shown to serve as a CR. Many of these proteins are expected to function as 1-Cys Prxs, but this cannot be proven without experimental verification.
Structural designations come from Hall et al 201016.
The CR for the AhpC/Prx1 subfamily is found near the C-terminus where it forms a disulfide bond across the B-interface with the CP on its partner subunit. Mycobacterium tuberculosis AhpC contains a third Cys that is present two residues after the canonical CR that may in some cases act as a replacement for the CR 78. While 6.7% of the AhpC/Prx1 proteins contain the CRXC motif, 2.2% contained only the second Cys, providing support for its ability to serve a functional role.
This category includes the 16% of Prx5 subfamily members that are fused to a Grx domain.
The CR in the Tpx subfamily is located immediately following the highly conserved Trp used as a key residue to create the functional site signatures – Cys92 in Fig 3E.
The canonical AhpE from M. tuberculosis has no CR, but a majority of the subfamily members identified here have a putative CR five residues after the CP, similar to the BCP/PrxQ subfamily.
Although Prx5 proteins have generally been considered to be atypical 2-Cys proteins, our analysis indicates that only 17% of Prx5 subfamily members contain a Cys residue in the same location as the CR in H. sapiens Prx5. While Prx5 subfamily members are distributed across bacteria and eukaryotes (Table III), all but one metazoan Prx5 sequence (from Pyrocoelia rufa) contained a CR in the same location as that in human Prx5. Other members of the Prx5 group have been reported to use a 1-Cys mechanism 20; of the Prx subfamily members identified by DASP, 14% contain only one cysteine and, thus, must utilize a 1-Cys recycling mechanism. Haemophilus influenzae Prx5 (1nm3) contains a 1-Cys Prx5 domain fused to a glutaredoxin (Grx) domain; it has been proposed that the CP is reduced by the CXXC motif in the Grx domain 54. We found that 16% of the Prx5 subfamily members contained a CXXC-containing, Grx-like domain, all from bacteria.
These data highlight the significant diversity in the CR location in most Prx subfamilies and indicate that the presence and structural location of CR within most subfamilies is not critical as it has arisen multiple times during the course of evolution. These data also highlight the fact that the nomenclature of 1-Cys, typical 2-Cys, and atypical 2-Cys cannot be used to designate subfamily assignments; the only exception is the designation of typical 2-Cys, which is specific for the AhpC/Prx1 subfamily.
The AhpC/Prx1 subfamily can be further subdivided into four groups that are correlated with functional attributes
The identification of a large number of representatives of each Prx subfamily using GenBank searches offers the opportunity to further cluster each subfamily individually to identify functionally relevant groups within each subfamily. We explored this possibility with the AhpC/Prx1 subfamily because it is arguably the best characterized of the Prx subfamilies and because subfamily members have distinctive characteristics that have been described in the literature. For instance, some subfamily members have been shown to be much more resistant to inactivation through hyperoxidation of CP than others 55. Also, while thioredoxin (Trx) is the preferred reductant for many Prx proteins, some members of this subfamily are reduced by specialized reductants including AhpF and tryparedoxin 2,56. Further subdivision of this subfamily was supported by the DASP profile scores, which were more significant when separate profiles were created for AhpC (0.33) and Prx1 (0.13) than when they were combined (0.06). Despite this, we were not able to distinguish the separate AhpC and Prx1 groups during GenBank searches with these profiles, as noted above.
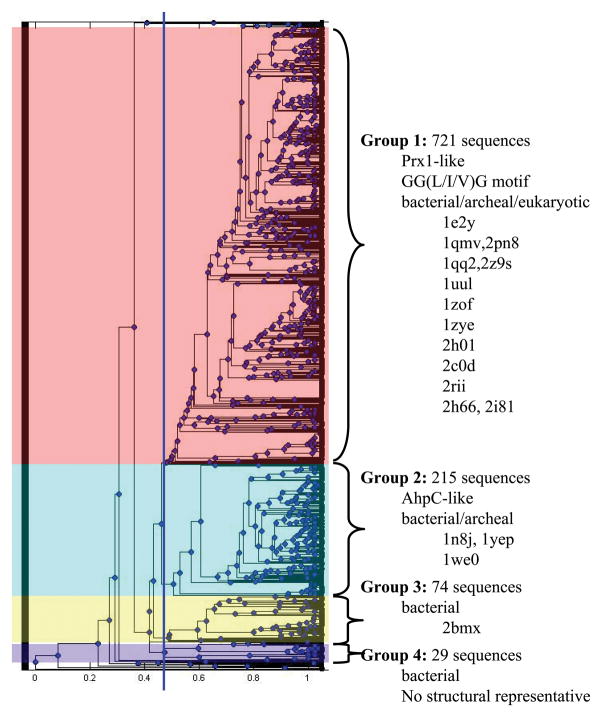
Hierarchical clustering of the AhpC/Prx1 functional site signatures identified during the GenBank(nr) search suggested 4 major groups (Figure 5) which are, indeed, consistent with known biological function. The largest group, Group 1, contained 721 sequences, including human Prxs 1 through 4, and was comprised of both eukaryotic and bacterial proteins. Of the species in group 1, 52% were bacterial (Table III). Characterized members of this subgroup are reduced by Trx or by organism-specific reductants like tryparedoxin 57 or the glutaredoxin-like Cp9 in Clostridium pasteurianum 58. The GGLG motif or a variation of it is observed in 91% of the sequences in group 1; leucine (52%), valine (8 %) or isoleucine (31%) are all observed in the third position of this motif, indicating the importance of an aliphatic side chain at this position. The GGLG motif was originally noted as a feature of eukaryotic and potentially “sensitive” Prxs that were more prone to CP overoxidation by peroxide than their more robust bacterial counterparts 55; later analysis pointed out that some pathogenic bacteria contain a Prx with a GGIG motif 5. The GGLG motifs in Group 1 proteins may be associated with hypersensitivity toward overoxidation, but features at the C-terminal end are also important and this characteristic has been measured for only a few proteins within this subfamily.
Figure 5. The AhpC/Prx1 subfamily can be subdivided into four distinct groups.
The functional site signatures obtained from the GenBank(nr) search for AhpC/Prx1 subfamily members were hierarchically clustered in Matlab. The dendrogram shown illustrates the resulting organization of functional site signatures and does not imply any evolutionary relationship. A cluster cutoff was identified (blue line in the dendrogram) and the subfamily was subdivided into four groups. Characteristics and structural representatives for each group are listed to the right.
Group 2 contains 215 proteins from 176 species (Figure 5), including S. typhimurium and Amphibacillus xylanus AhpC. This group represents the canonical AhpC proteins and is predominantly bacterial (95%) with some archaeal representatives (Table III). Because the AhpF coding sequence is typically found immediately downstream of the canonical ahpC gene, this genetic linkage was assessed to test whether the presence of a gene for this putative reductant is limited to and/or characteristic of the Group 2 proteins. PSI-BLAST was used with the default parameters and the full-length sequence of S. typhimurium AhpF to identify organisms that could express AhpF. Hits were considered significant if they had a p-value of 10−5 or more significant. Entrez Gene was then used to identify the AhpC sequence genetically associated with each AhpF. All of the AhpC-like proteins associated with an AhpF were found in Group 2, accounting for at least 30% of the organisms encoding a group 2 AhpC.
Group 3 (74 sequences; 60 species; Table III) is exclusively bacterial and contains Prxs from Mycobacterium, Bordetella, and Streptomyces species. AhpC activity has also been linked, both genetically and functionally, to a downstream coding sequence for AhpD 59,60, and all of the AhpC-like proteins associated with an AhpD (determined as described above for AhpF) are found in Group 3.
Group 4 (29 sequences; 22 species) is also exclusively bacterial and contains Prxs from the genera Flavobacteria and Chlamydia. There are currently no structural representatives for Group 4 (Figure 5); therefore, these present a good potential target for exploring the structural and mechanistic diversity of this Prx subfamily.
Overall, the AhpC/Prx1 groups identified by hierarchical clustering of the GenBank-derived AhpC/Prx1 subfamily members appear to be associated with at least a few well-established functional differences.
DISCUSSION
This study is the first large-scale example of the application of a methodology to identify and characterize protein subfamilies by focusing on information at the protein active site. Because the sequence database was searched with profiles generated from structural information, we identify and classify over 3500 putative Prx sequences into one of six subfamilies with a high degree of specificity. The majority of proteins identified from GenBank(nr) were previously annotated either as a peroxiredoxin or with a more general function (redoxin, peroxidase, etc), and the current results suggest that more specific subfamily membership could be added to the annotation. All software is publicly accessible (http://dasp.deac.wfu.edu/), the user interface is amenable to use by those without a strong computational background and detailed usage guidelines have been published 35.
The assignment of a significant number of members to each Prx subfamily allowed the calculation of residue conservation at each location within the functional site signature (Figure 3) and the mapping of these residues to the structure of a representative subfamily member (Figure 4). This analysis provides significant new insights into potential roles for conserved residues, and an opportunity for designing hypothesis-driven experiments aimed at identifying mechanistic and specificity determinants for these proteins. The utility of these data is supported by recent analysis of Prx active sites with bound substrate or substrate analogues which revealed that different Prx subfamilies exhibit two different conformations for the stringently conserved Arg 11. In AhpC/Prx1, Prx6, and AhpE subfamily members, this Arg is oriented through a Arg-Glu-Arg hydrogen bonding network; all of these residues were identified by our analysis to be conserved in these three subfamilies (Arg119, Glu49, Arg142 in S. typhimurium AhpC, Figure 3B and 4B). When the second Arg is not present (i.e. in most BCP, Prx5, and Tpx subfamily members, Figure 3A, D, and E), the conserved Arg adopts another conformation. Although the Arg is shifted by ~1 Å, it is still apparently able to form hydrogen bonds to the peroxide oxygen proximal to the cysteine thiolate as well as the thiolate itself 11. Arg128, Glu55, and Arg151 (numbering for T. cruzi tryparedoxin peroxidase, 1uul; Figure 3B) were also identified as contributing to the lowered pKa of the CP in a separate study using molecular dynamics (MD) 53. This MD study also identified a hydrogen-bonding network which extends from the active site to the dimer-dimer (A-type) interface and includes residues (Phe43, Tyr44, and Phe50) identified as conserved in this paper (Figure 3B).
More detailed analysis of the AhpC/Prx1 subfamily has identified four groups based on clustering of the signatures in the profile (Figure 5) and these can be correlated with functional differences between the members of each subgroup. Analysis of Group 2 and Group 3 indicated that AhpF- and AhpD-associated Prxs were all located in Groups 2 and 3, respectively. While it cannot replace experimental evidence, knowing the group for members of the AhpC/Prx1 subfamily can guide the selection of appropriate conditions (e.g., reductants) for initial biochemical analysis.
Babbitt and coworkers propose that over-annotation is a significant cause of mis-annotation 30. Our data suggest that over-annotation is more of a hazard using full sequence comparisons and that more detailed subfamily assignment can be accomplished by focusing on the specific features around the appropriate functional site(s). The correlation of the clustering of the Prx profiles (Figure 2) with subfamilies previously identified from sequence and structural analysis, the correct assignment of the Prx test proteins (Table II and Table SII), and the low number of sequences assigned by DASP to multiple subfamilies (Table I) argue that our method does, indeed, identify subfamily members specifically, provided the original profile and resulting PSSM is specific for that subfamily and includes a representative selection of proteins. The majority of proteins identified from GenBank(nr) were previously annotated either as a peroxiredoxin or with a more general function (redoxin, peroxidase, etc), and the current results suggest that more specific subfamily membership could be added to the annotation.
A p-value cutoff of 10−8 for DASP was used to identify Prx sequences presented in Table SV based on preliminary data looking at scores for structurally characterized Prxs (described in supplemental results). We note that a cutoff of 10−10 would have resulted in no proteins being identified in more than one subfamily search; however, it would have also increased the number of test proteins not identified in any DASP subfamily search from 1 to 4 (false negative rates of 1.7% and 6.9% for p-value cutoffs of 10−8 and 10−10, respectively). The DASP cutoff value should be explored for other protein families and subfamilies to which this approach is applied.
Use of profiles and PSSMs for sequence searching is not new. Profiles have long been used for sequence and structure alignment and searching 39,61,62, protein fold prediction 63,64, membrane protein topology prediction 65,66, fitness of folds for designed proteins 67, and protein function prediction 68–71. Indeed, our specific approach is based on the pioneering work on profiling by Gribskov and coworkers 44,72. PSSMs have been applied to full sequences, as in PSI-BLAST, and some have composed PSSMs that are protein family specific, such as PRIAM 73. DASP differs because DASP focuses on short protein fragments, thus avoiding the need to align across complete sequences. This is more similar to other approaches that use profiles to identify conserved sequence motifs (PROSITE 74, PRINTS 75, BLOCKS 76) or structural motifs (RIGOR, 77). But, unlike PROSITE and RIGOR, DASP does not identify and create profiles for all conserved sequence motifs within a family, because this would include some motifs far from a given molecular functional site. DASP also does not focus on individual short structural motifs; instead, DASP identifies fragments that are proximal in structure to a specific molecular functional site and builds profiles based on all of those fragments that are longer than two residues. This is a unique focus, aimed not at global classification, but at identifying mechanistic and specificity determinants associated with a particular functional site. The results presented here comparing whole sequence (PSI-BLAST) and active site motif (DASP) subfamily assignments indicate the advantages of the latter approach.
Although DASP subfamily searches are able to classify Prx proteins with greater specificity than PSI-BLAST, there are limitations to the method. Searching the sequence databases requires more than one or two representative structures, so that the profile (and the resulting PSSM) is not overly specific. In this contribution, this limitation was overcome for the AhpE and BCP/PrxQ subfamilies by creating engineered profiles (see Methods and Supplemental Results), which add additional signatures to the profile based on a multiple sequence alignment. The current method also requires expert knowledge of the protein function to select multiple key residues that are critical for defining the appropriate protein function. In this work, we observed that when only the CP was used as a key residue, signatures were too small to be specific for each subfamily. In addition, the current method using the PSSM to search sequences weights all residues in the signature equally. The results presented here suggest that it might be beneficial to develop a more specific PSSM-based approach for sequence searching that would utilize differential weights among the signature residues based on expert knowledge. Finally, for this paper, residue conservation was defined simply by residue identity; it is possible that some of the key features within the peroxiredoxin subfamily active sites are more dependent on conservation of characteristics (e.g. charge or hydrogen bonding features) rather than identity. This suggests that including biophysical characteristics, such as electrostatic contributions, to the PSSM scoring algorithm would provide greater information.
As an important output of the current work, we have provided a carefully constructed and curated list of members of each subfamily and current annotations (Table SV). This information provides considerable clarity as to the identity and subfamily membership of putative Prxs in the GenBank database. We have developed a web-accessible database based on this information (http://www.csb.wfu.edu/prex/) that allows users to search the data found in Table SV 78. We are also collaborating with the developers of the Structure-Function Linkage Database (SFLD; http://sfld.rbvi.ucsf.edu 79) and plan to include the Prx family and the subfamily assignments identified here within that more comprehensive resource, which would also be made available to the larger community.
Supplementary Material
Acknowledgments
Grants: This investigation was supported by a NSF grant to JSF (MCB0517343). These calculations were performed on Wake Forest University’s DEAC cluster, http://www.deac.wfu.edu, supported by a SUR grant from IBM for storage hardware and by the Wake Forest IS department. NIH grants to K.J.N. (F32 GM074537) and L.B.P. (RO1 GM050389) are also acknowledged.
We would like to thank William Turkett for his help in creating a Java script to query taxonomic information and Derek Parsonage for editorial contributions.
Abbreviations
- Prx
peroxiredoxin
- H2O2
hydrogen peroxide
- ROOH
hydroperoxide
- Cp
peroxidatic cysteine
- R-SOH
sulfenic acid
- AhpC
alkyl hydroperoxide reductase “C” protein
- CR
resolving cysteine
- Tpx
thiol peroxidase
- DASP
Deacon Active Site Profiler
- PDB
Protein Data Bank
- UPGMA
unweighted pair group method average
- BCP
bacterioferritin comigratory protein
- PSSM
position-specific scoring matrix
- NCBI
National Center for Biotechnology Information
- Grx
glutaredoxin
- Trx
thioredoxin
- SFLD
Structure-Function Linkage Database
Footnotes
AUTHOR SUMMARY
Genome sequencing projects have resulted in tremendous quantities of sequence information, but experimental characterization of protein function has been performed on only a small fraction of sequences. Although numerous computational methods exist that provide functional classification for many uncharacterized proteins, misannotation is a significant problem, since most sequence-focused methods are unable to distinguish the features of individual subfamilies. Our recently developed method called Deacon Active Site Profiling (DASP) is able to extract the features located near the functional site of structurally characterized proteins and utilize this information to identify other proteins in the sequence database that share similar functional site characteristics. In this paper, we used this method to analyze the widely distributed and moderately well-characterized peroxiredoxin protein family; family members detoxify hydrogen peroxide and other oxidized molecules in the cell. We identified over 3500 putative peroxiredoxin sequences from the sequence database and classified them into one of six subfamilies. Subfamily searches using DASP were highly specific and allowed identification of key features at the active site of each subfamily, providing a number of experimentally testable hypotheses. This work paves the way for assignment of sequences from other large protein families to subfamilies with a reasonable degree of accuracy.
References
- 1.Link AJ, Robison K, Church GM. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis. 1997;18:1259–313. doi: 10.1002/elps.1150180807. [DOI] [PubMed] [Google Scholar]
- 2.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 3.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–86. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 5.Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins--structures, mechanisms and functions. Febs J. 2009;276:2469–77. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phalen TJ, Weirather K, Deming PB, Anathy V, Howe AK, van der Vliet A, Jönsson TJ, Poole LB, Heintz NH. Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J Cell Biol. 2006;175:779–89. doi: 10.1083/jcb.200606005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karplus PA, Hall A. Structural Survey of the Peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. New York: Springer; 2007. pp. 41–60. [DOI] [PubMed] [Google Scholar]
- 8.Poole LB. The Catalytic Mechanism of Peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. New York: Springer; 2007. pp. 61–81. [DOI] [PubMed] [Google Scholar]
- 9.Fomenko DE, Gladyshev VN. Identity and functions of CxxC-derived motifs. Biochemistry. 2003;42:11214–25. doi: 10.1021/bi034459s. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol Chem. 2002;383:347–64. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 11.Hall A, Parsonage D, Poole LB, Karplus PA. Structural Evidence that Peroxiredoxin Catalytic Power Is Based on Transition-State Stabilization. J Mol Biol. 2010;402:194–209. doi: 10.1016/j.jmb.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain: alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci U S A. 1994;91:7017–21. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoops B, Loumaye E, Van Der Eecken V. Evolution of the peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. New York: Springer; 2007. pp. 27–40. [DOI] [PubMed] [Google Scholar]
- 14.Hall A, Nelson KJ, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3624. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cha MK, Hong SK, Kim IH. Four thiol peroxidases contain a conserved GCT catalytic motif and act as a versatile array of lipid peroxidases in Anabaena sp. PCC7120. Free Radic Biol Med. 2007;42:1736–48. doi: 10.1016/j.freeradbiomed.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Copley SD, Novak WR, Babbitt PC. Divergence of function in the thioredoxin fold suprafamily: evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry. 2004;43:13981–95. doi: 10.1021/bi048947r. [DOI] [PubMed] [Google Scholar]
- 17.Hall A, Nelson K, Poole LB, Karplus PA. Structure-based insights into the catalytic power and conformational dexterity of peroxiredoxins. Antioxid Redox Signal. doi: 10.1089/ars.2010.3624. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizohata E, Sakai H, Fusatomi E, Terada T, Murayama K, Shirouzu M, Yokoyama S. Crystal structure of an archaeal peroxiredoxin from the aerobic hyperthermophilic crenarchaeon Aeropyrum pernix K1. J Mol Biol. 2005;354:317–29. doi: 10.1016/j.jmb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 19.Rouhier N, Jacquot JP. The plant multigenic family of thiol peroxidases. Free Radic Biol Med. 2005;38:1413–21. doi: 10.1016/j.freeradbiomed.2004.07.037. [DOI] [PubMed] [Google Scholar]
- 20.Sarma GN, Nickel C, Rahlfs S, Fischer M, Becker K, Karplus PA. Crystal structure of a novel Plasmodium falciparum 1-Cys peroxiredoxin. J Mol Biol. 2005;346:1021–34. doi: 10.1016/j.jmb.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 21.Lee TH, Kim SU, Yu SL, Kim SH, Park do S, Moon HB, Dho SH, Kwon KS, Kwon HJ, Han YH, et al. Peroxiredoxin II is essential for sustaining life span of erythrocytes in mice. Blood. 2003;101:5033–8. doi: 10.1182/blood-2002-08-2548. [DOI] [PubMed] [Google Scholar]
- 22.Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–5. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- 23.Bork P, Bairoch A. Go hunting in sequence databases but watch out for the traps. Trends Genet. 1996;12:425–7. doi: 10.1016/0168-9525(96)60040-7. [DOI] [PubMed] [Google Scholar]
- 24.Karp PD. What we do not know about sequence analysis and sequence databases. Bioinformatics. 1998;14:753–4. doi: 10.1093/bioinformatics/14.9.753. [DOI] [PubMed] [Google Scholar]
- 25.Andorf C, Dobbs D, Honavar V. Exploring inconsistencies in genome-wide protein function annotations: a machine learning approach. BMC Bioinformatics. 2007;8:284. doi: 10.1186/1471-2105-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naumoff DG, Xu Y, Stalon V, Glansdorff N, Labedan B. The difficulty of annotating genes: the case of putrescine carbamoyltransferase. Microbiology. 2004;150:3908–11. doi: 10.1099/mic.0.27640-0. [DOI] [PubMed] [Google Scholar]
- 27.Jones CE, Brown AL, Baumann U. Estimating the annotation error rate of curated GO database sequence annotations. BMC Bioinformatics. 2007;8:170. doi: 10.1186/1471-2105-8-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kyrpides NC, Ouzounis CA. Whole-genome sequence annotation: ‘Going wrong with confidence’. Mol Microbiol. 1999;32:886–7. doi: 10.1046/j.1365-2958.1999.01380.x. [DOI] [PubMed] [Google Scholar]
- 29.Pallen M, Wren B, Parkhill J. ‘Going wrong with confidence’: misleading sequence analyses of CiaB and clpX. Mol Microbiol. 1999;34:195. doi: 10.1046/j.1365-2958.1999.01561.x. [DOI] [PubMed] [Google Scholar]
- 30.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol. 2009;5:e1000605. doi: 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cammer SA, Hoffman BT, Speir JA, Canady MA, Nelson MR, Knutson S, Gallina M, Baxter SM, Fetrow JS. Structure-based active site profiles for genome analysis and functional family subclassification. J Mol Biol. 2003;334:387–401. doi: 10.1016/j.jmb.2003.09.062. [DOI] [PubMed] [Google Scholar]
- 32.Baxter SM, Rosenblum JS, Knutson S, Nelson MR, Montimurro JS, Di Gennaro JA, Speir JA, Burbaum JJ, Fetrow JS. Synergistic computational and experimental proteomics approaches for more accurate detection of active serine hydrolases in yeast. Mol Cell Proteomics. 2004;3:209–25. doi: 10.1074/mcp.M300082-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Huff RG, Bayram E, Tan H, Knutson ST, Knaggs MH, Richon AB, Santago P, 2nd, Fetrow JS. Chemical and structural diversity in cyclooxygenase protein active sites. Chem Biodivers. 2005;2:1533–52. doi: 10.1002/cbdv.200590125. [DOI] [PubMed] [Google Scholar]
- 34.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fetrow JS. Active site profiling to identify protein functional sites in sequences and structures using the Deacon Active Site Profiler (DASP) Curr Protoc Bioinformatics. 2006;Chapter 8(Unit 8–10) doi: 10.1002/0471250953.bi0810s14. [DOI] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonnet GH, Cohen MA, Benner SA. Exhaustive matching of the entire protein sequence database. Science. 1992;256:1443–5. doi: 10.1126/science.1604319. [DOI] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeong W, Cha MK, Kim IH. Thioredoxin-dependent hydroperoxide peroxidase activity of bacterioferritin comigratory protein (BCP) as a new member of the thiol-specific antioxidant protein (TSA)/Alkyl hydroperoxide peroxidase C (AhpC) family. J Biol Chem. 2000;275:2924–30. doi: 10.1074/jbc.275.4.2924. [DOI] [PubMed] [Google Scholar]
- 41.Rouhier N, Gelhaye E, Gualberto JM, Jordy MN, De Fay E, Hirasawa M, Duplessis S, Lemaire SD, Frey P, Martin F, et al. Poplar peroxiredoxin Q. A thioredoxin-linked chloroplast antioxidant functional in pathogen defense. Plant Physiol. 2004;134:1027–38. doi: 10.1104/pp.103.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang G, Olczak AA, Walton JP, Maier RJ. Contribution of the Helicobacter pylori thiol peroxidase bacterioferritin comigratory protein to oxidative stress resistance and host colonization. Infect Immun. 2005;73:378–84. doi: 10.1128/IAI.73.1.378-384.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shenkin PS, Farid H, Fetrow JS. Prediction and evaluation of side-chain conformations for protein backbone structures. Proteins. 1996;26:323–52. doi: 10.1002/(SICI)1097-0134(199611)26:3<323::AID-PROT8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 44.Bailey TL, Gribskov M. Methods and statistics for combining motif match scores. J Comput Biol. 1998;5:211–21. doi: 10.1089/cmb.1998.5.211. [DOI] [PubMed] [Google Scholar]
- 45.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trivelli X, Krimm I, Ebel C, Verdoucq L, Prouzet-Mauleon V, Chartier Y, Tsan P, Lauquin G, Meyer Y, Lancelin JM. Characterization of the yeast peroxiredoxin Ahp1 in its reduced active and overoxidized inactive forms using NMR. Biochemistry. 2003;42:14139–49. doi: 10.1021/bi035551r. [DOI] [PubMed] [Google Scholar]
- 47.Cha MK, Kim HK, Kim IH. Thioredoxin-linked “thiol peroxidase” from periplasmic space of Escherichia coli. J Biol Chem. 1995;270:28635–41. doi: 10.1074/jbc.270.48.28635. [DOI] [PubMed] [Google Scholar]
- 48.Van der Pol B. Trichomonas vaginalis infection: the most prevalent nonviral sexually transmitted infection receives the least public health attention. Clin Infect Dis. 2007;44:23–5. doi: 10.1086/509934. [DOI] [PubMed] [Google Scholar]
- 49.Parsonage D, Youngblood DS, Sarma GN, Wood ZA, Karplus PA, Poole LB. Analysis of the link between enzymatic activity and oligomeric state in AhpC, a bacterial peroxiredoxin. Biochemistry. 2005;44:10583–92. doi: 10.1021/bi050448i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peskin AV, Low FM, Paton LN, Maghzal GJ, Hampton MB, Winterbourn CC. The high reactivity of peroxiredoxin 2 with H2O2 is not reflected in its reaction with other oxidants and thiol reagents. J Biol Chem. 2007;282:11885–92. doi: 10.1074/jbc.M700339200. [DOI] [PubMed] [Google Scholar]
- 51.Liao SJ, Yang CY, Chin KH, Wang AH, Chou SH. Insights into the alkyl peroxide reduction pathway of Xanthomonas campestris bacterioferritin comigratory protein from the trapped intermediate-ligand complex structures. J Mol Biol. 2009;390:951–66. doi: 10.1016/j.jmb.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 52.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to doughnuts: redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 53.Yuan Y, Knaggs MH, Poole LB, Fetrow JS, Salsbury FR., Jr Conformational and oligomeric effects on the cysteine pK(a) of tryparedoxin peroxidase. J Biomol Struct Dyn. 2010;28:51–70. doi: 10.1080/07391102.2010.10507343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim SJ, Woo JR, Hwang YS, Jeong DG, Shin DH, Kim K, Ryu SE. The tetrameric structure of Haemophilus influenza hybrid Prx5 reveals interactions between electron donor and acceptor proteins. J Biol Chem. 2003;278:10790–8. doi: 10.1074/jbc.M209553200. [DOI] [PubMed] [Google Scholar]
- 55.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–3. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 56.Poole LB. Bacterial defenses against oxidants: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–54. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 57.Krauth-Siegel RL, Comini MA. Redox control in trypanosomatids, parasitic protozoa with trypanothione-based thiol metabolism. Biochim Biophys Acta. 2008;1780:1236–48. doi: 10.1016/j.bbagen.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 58.Reynolds CM, Meyer J, Poole LB. An NADH-dependent bacterial thioredoxin reductase-like protein in conjunction with a glutaredoxin homologue form a unique peroxiredoxin (AhpC) reducing system in Clostridium pasteurianum. Biochemistry. 2002;41:1990–2001. doi: 10.1021/bi011802p. [DOI] [PubMed] [Google Scholar]
- 59.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–7. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 60.Koshkin A, Nunn CM, Djordjevic S, Ortiz de Montellano PR. The mechanism of Mycobacterium tuberculosis alkylhydroperoxidase AhpD as defined by mutagenesis, crystallography, and kinetics. J Biol Chem. 2003;278:29502–8. doi: 10.1074/jbc.M303747200. [DOI] [PubMed] [Google Scholar]
- 61.Jennings AJ, Edge CM, Sternberg MJ. An approach to improving multiple alignments of protein sequences using predicted secondary structure. Protein Eng. 2001;14:227–31. doi: 10.1093/protein/14.4.227. [DOI] [PubMed] [Google Scholar]
- 62.Teichert F, Minning J, Bastolla U, Porto M. High quality protein sequence alignment by combining structural profile prediction and profile alignment using SABER-TOOTH. BMC Bioinformatics. 2010;11:251. doi: 10.1186/1471-2105-11-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ouzounis C, Sander C, Scharf M, Schneider R. Prediction of protein structure by evaluation of sequence-structure fitness. Aligning sequences to contact profiles derived from three-dimensional structures. J Mol Biol. 1993;232:805–25. doi: 10.1006/jmbi.1993.1433. [DOI] [PubMed] [Google Scholar]
- 64.Rost B, Schneider R, Sander C. Protein fold recognition by prediction-based threading. J Mol Biol. 1997;270:471–80. doi: 10.1006/jmbi.1997.1101. [DOI] [PubMed] [Google Scholar]
- 65.Bernsel A, Viklund H, Falk J, Lindahl E, von Heijne G, Elofsson A. Prediction of membrane-protein topology from first principles. Proc Natl Acad Sci U S A. 2008;105:7177–81. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juretic D, Zucic D, Lucic B, Trinajstic N. Preference functions for prediction of membrane-buried helices in integral membrane proteins. Comput Chem. 1998;22:279–94. doi: 10.1016/s0097-8485(97)00070-3. [DOI] [PubMed] [Google Scholar]
- 67.Brenner SE, Berry A. A quantitative methodology for the de novo design of proteins. Protein Sci. 1994;3:1871–82. doi: 10.1002/pro.5560031026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31:3635–41. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su QJ, Lu L, Saxonov S, Brutlag DL. eBLOCKs: enumerating conserved protein blocks to achieve maximal sensitivity and specificity. Nucleic Acids Res. 2005;33:D178–82. doi: 10.1093/nar/gki060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wass MN, Sternberg MJ. ConFunc--functional annotation in the twilight zone. Bioinformatics. 2008;24:798–806. doi: 10.1093/bioinformatics/btn037. [DOI] [PubMed] [Google Scholar]
- 71.Yaffe MB, Leparc GG, Lai J, Obata T, Volinia S, Cantley LC. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat Biotechnol. 2001;19:348–53. doi: 10.1038/86737. [DOI] [PubMed] [Google Scholar]
- 72.Gribskov M, McLachlan AD, Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987;84:4355–8. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Claudel-Renard C, Chevalet C, Faraut T, Kahn D. Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 2003;31:6633–9. doi: 10.1093/nar/gkg847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bairoch A. PROSITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991;19 (Suppl):2241–5. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Attwood TK, Beck ME, Bleasby AJ, Parry-Smith DJ. PRINTS--a database of protein motif fingerprints. Nucleic Acids Res. 1994;22:3590–6. [PMC free article] [PubMed] [Google Scholar]
- 76.Henikoff S, Henikoff JG, Pietrokovski S. Blocks+: a non-redundant database of protein alignment blocks derived from multiple compilations. Bioinformatics. 1999;15:471–9. doi: 10.1093/bioinformatics/15.6.471. [DOI] [PubMed] [Google Scholar]
- 77.Kleywegt GJ. Recognition of spatial motifs in protein structures. J Mol Biol. 1999;285:1887–97. doi: 10.1006/jmbi.1998.2393. [DOI] [PubMed] [Google Scholar]
- 78.Soito L, Williamson C, Knutson ST, Fetrow JS, Poole LB, Nelson KJ. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1060. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pegg SC, Brown SD, Ojha S, Seffernick J, Meng EC, Morris JH, Chang PJ, Huang CC, Ferrin TE, Babbitt PC. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry. 2006;45:2545–55. doi: 10.1021/bi052101l. [DOI] [PubMed] [Google Scholar]
- 80.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–34. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.