Abstract
A physical connection between each pair of homologous chromosomes is crucial for reductional chromosome segregation during the first meiotic division and therefore for successful meiosis. Connection is provided by recombination (crossing over) initiated by programmed DNA double-strand breaks (DSBs). Although the topoisomerase-like protein Spo11 makes DSBs and is evolutionarily conserved, how Spo11 (Rec12 in fission yeast) is regulated to form DSBs at the proper time and place is poorly understood. Several additional (accessory) proteins for DSB formation have been inferred in different species from yeast to mice. Here, we show that Rec24 is a bona fide accessory protein in Schizosaccharomyces pombe. Rec24 is required genome-wide for crossing-over and is recruited to meiotic chromosomes during prophase in a Rec12-independent manner forming foci on linear elements (LinEs), structurally related to the synaptonemal complex of other eukaryotes. Stabilization of Rec24 on LinEs depends on another accessory protein, Rec7, with which Rec24 forms complexes in vivo. We propose that Rec24 marks LinE-associated recombination sites, that stabilization of its binding by Rec7 facilitates the loading or activation of Rec12, and that only stabilized complexes containing Rec24 and Rec7 promote DSB formation. Based on the recent report of Rec24 and Rec7 conservation, interaction between Rec24 and Rec7 might be widely conserved in DSB formation.
Key words: Meiotic recombination, Spo11-accessory proteins, Recombination-initiation complex, Fission yeast
Introduction
Meiosis, a special cell division by which diploid organisms halve their chromosome number prior to sexual reproduction, is essential to maintain the ploidy of individuals from generation to generation. One key feature of meiosis is the physical exchange of genetic material between each pair of homologous chromosomes, known as meiotic recombination. One type of exchange, crossing over, contributes both to the generation of genetic diversity and to the proper segregation of homologous chromosomes during the first meiotic division (Petronczki et al., 2003).
Meiotic recombination is initiated by programmed DNA double-strand breaks (DSBs), which are made by a conserved topoisomerase-like protein Spo11 (Rec12 in fission yeast) (Keeney, 2008). How Spo11 or Rec12 is loaded onto recombination sites and is activated to form DSBs is poorly understood, although additional (accessory) proteins essential for DSB formation and recombination are present in numerous species, including budding and fission yeasts, flies, worms, plants and mice (Liu et al., 2002; Libby et al., 2003; Reddy and Villeneuve, 2004; Reinholdt and Schimenti, 2005; Cromie and Smith, 2008; Keeney, 2008; De Muyt et al., 2009; Cole et al., 2010; Kumar et al., 2010). The picture is clearest in budding yeast where systematic physical interactions, chromosome localization and chromatin-association requirements of these accessory proteins have been studied (Jiao et al., 2003; Arora et al., 2004; Kee et al., 2004; Li et al., 2006; Maleki et al., 2007). These studies suggest that, rather than forming a single holoenzyme, accessory proteins are organized into different sub-complexes with different genetic dependencies for protein–protein and protein–chromosome interactions: the conserved MRX complex [Mre11–Rad50–Xrs2 in budding yeast or MRN (Mre11–Rad50–Nbs1) in other species], the Rec114–Mei4–Mer2 complex, the Rec102–Rec104 complex, and Ski8–Spo11. Although the MRX complex is required for formation of DSBs in budding yeast (Cao et al., 1990), its homolog MRN is not required in other species, such as fission yeast, Coprinus, Tetrahymena, Drosophila and Arabidopsis (Puizina et al., 2004; Young et al., 2004; Mehrotra and McKim, 2006; Acharya et al., 2008; Lukaszewicz et al., 2010).
Based on studies in budding yeast, complex interactions of the Spo11 accessory proteins have emerged. Ski8 is a conserved cytoplasmic protein involved in RNA metabolism, which associates with chromosomes only during meiosis in a Spo11-dependent manner. It has been proposed that Ski8 functions as a bridge that connects Spo11 to the Rec102–Rec104 subcomplex (Arora et al., 2004). Ski8 is required for Spo11 binding to chromatin and, after binding Spo11, also for loading Rec102 (and presumably Rec104) onto chromatin (Arora et al., 2004; Kee et al., 2004). Rec102 and Rec104 are essential for Spo11 self-oligomerization and association with recombination hotspots (Prieler et al., 2005; Sasanuma et al., 2007). Rec114 might function independently, because it is not required for Spo11 oligomerization but is for recruitment of Spo11 to DSB hotspots (Prieler et al., 2005; Sasanuma et al., 2007). However, this function of Rec114 is not shared by Mei4 and Mer2, indicating that the Rec114–Mei4–Mer2 complex is not a functional unit (Prieler et al., 2005; Sasanuma et al., 2007). Indeed, Rec114 chromosome localization is independent of any other DSB protein (Li et al., 2006; Maleki et al., 2007). Mer2 has a crucial role in connecting DSB formation to meiotic progression (Henderson et al., 2006; Sasanuma et al., 2008; Wan et al., 2008). Mer2 is phosphorylated by S-phase CDK (cyclin-dependent kinase) and DDK (Dbf4-dependent kinase) activities, and phosphorylation of Mer2 is essential for its interaction with Rec114, association of Spo11 with hotspots, and DSB formation. In summary, the accessory proteins have several essential roles in DSB formation by Spo11.
In the fission yeast Schizosaccharomyces pombe, in addition to Rec12, seven proteins – Rec6, Rec7, Rec10, Rec14, Rec15, Rec24 and Mde2 – are essential for DSB formation based on genetic data and physical analysis of meiotic DSBs (Ponticelli and Smith, 1989; DeVeaux et al., 1992; Cervantes et al., 2000; Young et al., 2002; Ellermeier and Smith, 2005; Gregan et al., 2005; Martin-Castellanos et al., 2005). Except for Spo11 (mainly its domains involved in catalytic activity), there is little conservation of amino acid sequences between budding and fission yeast accessory proteins (Keeney, 2001; Keeney, 2008; Cromie and Smith, 2008), and only two of them show obvious homology. Budding yeast Ski8 and fission yeast Rec14 (26% amino acid identity) have a common structure containing WD repeats and a mitotic function (Evans et al., 1997). However, the role of Ski8 and Rec14 in recombination is not universally conserved (Jolivet et al., 2006). In addition, there is limited amino acid sequence homology (9% amino acid identity) between budding yeast Rec114 and fission yeast Rec7 (Malone et al., 1997; Molnar et al., 2001; Maleki et al., 2007; Steiner et al., 2010). The general lack of obvious conservation among accessory proteins is observed even in very closely related species of Saccharomyces, apparently reflecting the rapid evolution of proteins involved in DSB formation (Henderson et al., 2006; Maleki et al., 2007; Keeney, 2008).
Little is known about accessory proteins in species other than budding yeast, including fission yeast. Among the fission yeast accessory proteins, the localization of only Rec7 has been reported. During meiotic prophase, Rec7 forms nuclear foci that localize to linear elements (LinEs, related to the lateral elements of the synaptonemal complex of other eukaryotes), and the number of foci corresponds well with the average number of crossovers per meiosis (Molnar et al., 2001; Lorenz et al., 2006). Localization of Rec7 depends on Rec10 (a major linear element component), but is independent of Rec12, indicating that Rec7 is loaded onto LinEs before DSB formation (Lorenz et al., 2006). A recent study has started to reveal physical interactions among the accessory proteins and identified interaction domains between Rec7 and Rec24, and between Rec12 and Rec14 (Steiner et al., 2010). Interestingly, the association of Rec12 with the ura4A hotspot depends on Rec14 (Ludin et al., 2008).
Here, we confirm that Rec24 is indeed a novel accessory protein required for Rec12 activity, and we demonstrate that Rec24 functionally interacts with Rec7. We propose a model in which Rec24 marks potential recombination sites on LinEs and in which Rec7 stabilizes this association and, therefore, regulates the loading or activation of Rec12. Recently, Rec24 has been reported to be an ortholog of Mei4 in mice and budding yeast (Kumar et al., 2010), suggesting that the interactions we report here may be widely conserved among eukaryotes (Cole et al., 2010).
Results
Rec24 is not required for formation of LinEs
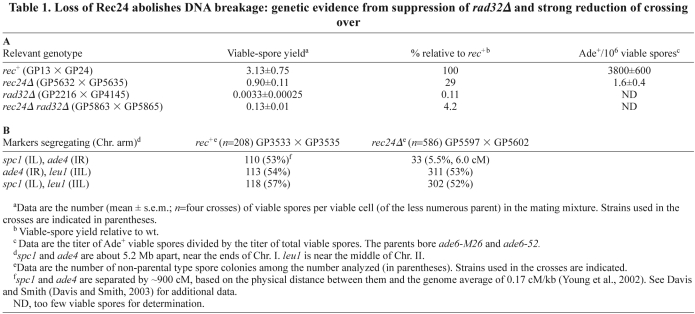
In a functional screening, we identified three genes, rec24, rec25 and rec27, that are required for meiotic recombination (Martin-Castellanos et al., 2005). Meiotic recombination in rec25Δ and rec27Δ mutants is reduced by factors of ~2 to 140, depending on the interval assayed, but is abolished in rec24Δ mutants in the few intervals tested. The strong reduction of recombination in rec24Δ mutants is nearly indistinguishable from that in mutants lacking Rec12 or its accessory proteins (Davis and Smith, 2001; Cromie and Smith, 2008) (see below).
The recombination defect in rec25Δ and rec27Δ mutants might be explained by the absence of LinE formation in these mutants (Davis et al., 2008). However, formation of LinE occurs in rec12Δ and previously tested accessory protein mutants (Molnar et al., 2003; Lorenz et al., 2004; Lorenz et al., 2006; Davis et al., 2008). Therefore, we addressed whether Rec24 has a role in LinE formation, using the major LinE component Rec10 as a marker (Lorenz et al., 2004). LinE formation was analyzed by Rec10 immunostaining of chromosome spreads prepared at different times (3 and 3.5 hours) during prophase in pat1-114 synchronous meiosis. The rec12Δ mutant was used as a negative control, and the rec8Δ mutant was used as an additional control because LinE formation is impaired in cohesin mutants (Molnar et al., 1995; Lorenz et al., 2004; Davis et al., 2008). Morphologically similar LinEs were observed with similar frequencies in wild-type, rec24Δ and rec12Δ mutants at the two time points analyzed (Fig. 1 shows data for 3 hours after meiotic induction; comparable unpublished data were obtained at 3.5 hours after meiotic induction). Therefore, as reported for Rec12 accessory proteins, Rec24 is not required for LinE formation, suggesting that Rec24 functions after these structures are formed.
Fig. 1.
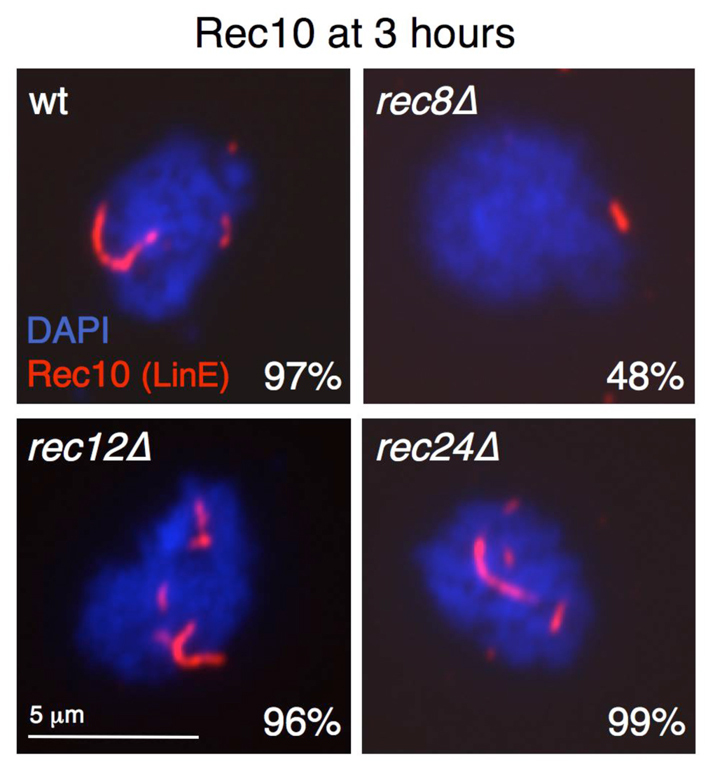
LinEs are formed normally in rec24Δ mutants. Diploid pat1-114 cells with the indicated deletions (strains CMC7, CMC40, CMC36 and CMC15) were induced for meiosis, and after 3 hours (prophase), cells were collected for nuclear spread preparation. Spreads were stained with DAPI (DNA; blue) and anti-Rec10 antibodies (red), and photographed under a fluorescence microscope. The fraction of nuclei with Rec10 structures (LinEs) is indicated (n=100 nuclei). Similar results were obtained with chromosome spreads prepared at 3.5 hours after induction and in an independent experiment (unpublished data).
Rec24 is essential for DSB formation and crossing over
To explore further a possible role for Rec24 in DSB formation, we tested the genetic interaction between rec24Δ and rad32Δ. Rad32 (the ortholog of Mre11 in other species) is part of the evolutionarily conserved MRN (Mre11–Rad50–Nbs1) nuclease complex that is required for DSB repair (Tavassoli et al., 1995; Young et al., 2004; Milman et al., 2009; Rothenberg et al., 2009). rad32Δ mutants exhibit a very low viable-spore yield (~10−3 of the wild-type yield) that is partially suppressed when DSB formation is abolished (Young et al., 2004; Ellermeier and Smith, 2005). Therefore, we analyzed whether rec24Δ can suppress the rad32Δ mutant phenotype. The viable-spore yield of rad32Δ rec24Δ double mutants was ~40 times higher than that of rad32Δ single mutants (Table 1A). This suppression is similar to that reported for rec12Δ and mutants lacking Rec6, another putative Rec12 accessory protein (Young et al., 2004; Ellermeier and Smith, 2005). A similar strong suppression of rad32Δ is not observed for rec8Δ or rec11Δ mutants, which lack sister chromatid cohesion and retain low levels of DSBs (Ellermeier and Smith, 2005). These genetic data are consistent with the lack of detectable DSBs by physical analysis at the two hotspots tested (mbs1 and ade6-3049) (Martin-Castellanos et al., 2005), suggesting that Rec24 is part of a complex with Rec12 and essential for meiotic DSB formation throughout the genome.
Table 1.
Loss of Rec24 abolishes DNA breakage: genetic evidence from suppression of rad32Δ and strong reduction of crossing over
As expected from this conclusion, crossing over was strongly reduced in rec24Δ mutants. Using the sty1 and ade4 markers located near each end of Chromosome I, the longest in S. pombe, we observed only 5.5% recombinants, equivalent to 6.0 cM (Table 1B). These markers segregate at random in the wild type and are calculated to be ~900 cM apart based on their physical distance (5.2 Mb) and the average rate of recombination across the S. pombe genome, 0.17 cM/kb (Young et al., 2002). Each marker segregated at random with leu1, a marker on Chromosome II, in both the wild type and rec24Δ, as expected for physically unlinked markers. The reduction of crossing over by a factor of ~150 in rec24Δ is similar to that in rec12Δ (Davis and Smith, 2003; Martin-Castellanos et al., 2005). Collectively, these data indicate that Rec24 is strongly required, and perhaps essential, for meiotic DSB formation.
Rec24 localizes to LinEs during meiotic prophase
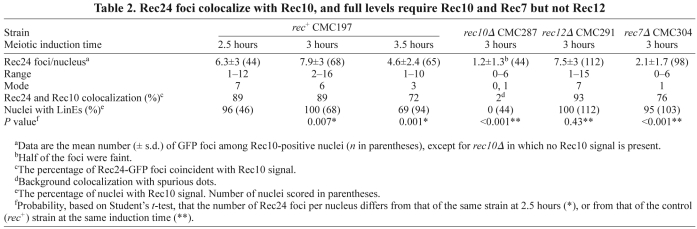
Because localization of Rec12 accessory proteins has been little studied in fission yeast, we determined the localization of Rec24 during meiotic prophase using a GFP-tagged version. Rec24–GFP fusion protein retains most of its activity, as shown by the production of normal asci (unpublished data) and nearly wild-type levels of recombination (supplementary material Table S1). In addition, Rec24–GFP protein is meiosis specific, as expected from the meiotic induction of rec24 transcripts (Mata et al., 2002) (supplementary material Fig. S1). Chromosome spreads were prepared during prophase of synchronous meiosis and double stained with anti-GFP and anti-Rec10 antibodies to visualize LinEs. Rec24–GFP showed specific chromosome localization during prophase with a dotted signal that colocalized with Rec10 (Fig. 2 and Table 2). Rec24–GFP appeared after LinE formation was initiated. At 1.5 hours after meiotic induction, only 16% of nuclei showed a weak Rec10 signal (mainly two dots per nucleus), and none of the Rec10-positive or Rec10-negative nuclei were positive for Rec24–GFP; at 2 hours the proportion of Rec10-positive nuclei rose to 80% (with more dots per nucleus and poorly elongated structures), none of the Rec10-negative nuclei showed Rec24–GFP signal, and only 37% of the Rec10-positive nuclei showed very few Rec24–GFP foci. These few foci were equally frequent in dotted or poorly elongated Rec10 signals. Rec24–GFP foci increased concomitantly with LinE development: focus number substantially increased at 2.5 hours (mean 6.3±3 foci per nucleus) (Table 2), when most of the nuclei showed clearly elongated Rec10 signals of different length (LinEs), and it was maximal at 3 hours after meiotic induction (mean 7.9±3 foci per nucleus) (Table 2), when all the nuclei showed LinEs (morphologically similar to those at 2.5 hours). There was no clear correlation between the length of a LinE and the presence of Rec24–GFP foci at either 2.5 or 3 hours. In both cases, foci were found on shorter and longer LinEs, and in single nuclei most of the LinEs, independently of their length, often contained Rec24–GFP foci (see Fig. 2 for examples of chromosome spreads at 3 hours). Rec24–GFP foci decreased in number (mean 4.6±2.4 foci per nucleus) (Table 2) and intensity at 3.5 hours after meiotic induction, when the proportion of nuclei with LinEs dropped, indicating that they were disassembling. At the time of maximal chromosome localization, strings of Rec24–GFP foci covering LinEs were often observed (Figs 2, 3 and 4). These foci frequently acquired a smeared and elongated shape (Figs 2, 3 and 4). These cytological data show that Rec24 localizes to LinEs during meiotic prophase, when formation of DSBs is known to take place (Cervantes et al., 2000).
Fig. 2.
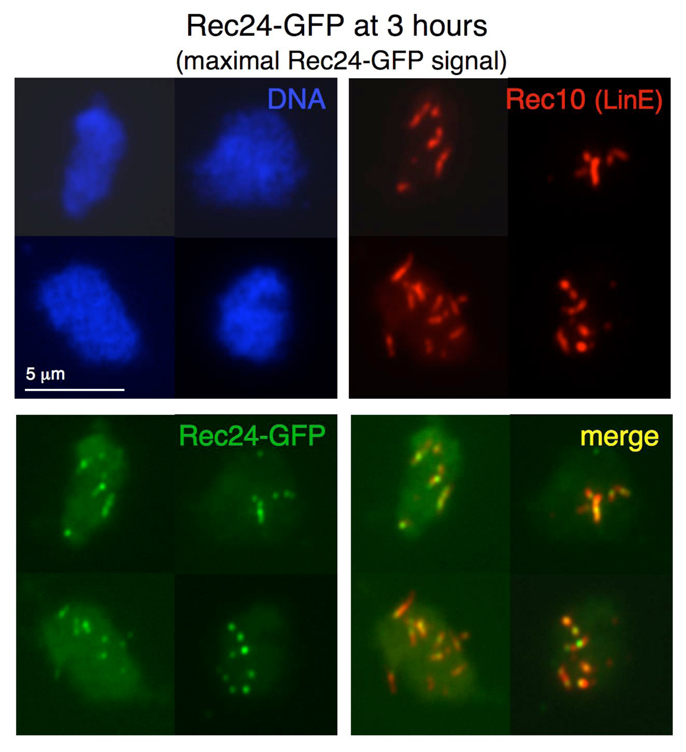
Rec24–GFP localizes to LinEs. Diploid pat1-114 rec24-GFP cells (strains CMC197) were induced for meiosis and collected during prophase for nuclear spread preparation. Spreads were stained with DAPI (DNA; blue), anti-GFP antibodies (Rec24–GFP; green) and anti-Rec10 antibodies (red) and photographed under a fluorescence microscope. Rec24–GFP signal was maximal at the time shown (3 hours). Similar results were obtained in two independent experiments. Rec24–GFP signal quantification at 2.5, 3 and 3.5 hours after meiotic induction for one time course is shown in Table 2.
Table 2.
Rec24 foci colocalize with Rec10, and full levels require Rec10 and Rec7 but not Rec12
Fig. 3.
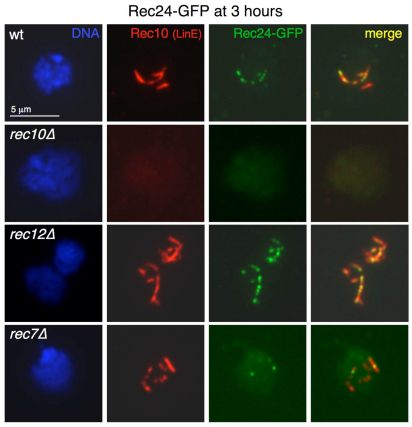
Genetic requirements for Rec24–GFP chromosome recruitment. Diploid pat1-114 rec24-GFP cells with the indicated deletions (strains CMC197, CMC287, CMC291 and CMC304) were induced for meiosis and collected 3 hours later (prophase) for nuclear spread preparation. Spreads were stained with DAPI (DNA; blue), anti-GFP antibodies (Rec24–GFP; green) and anti-Rec10 antibodies (red) and photographed under a fluorescence microscope. Rec24–GFP signal quantification is shown in Table 2. Rec24–GFP chromosome localization depends on Rec10 (second row) but not on Rec12 (third row). Similar results were obtained with 2.5 hour chromosome spreads of the same meiotic induction (unpublished data). Efficient Rec24–GFP chromosome localization depends on Rec7 (bottom row; see also Fig. 4).
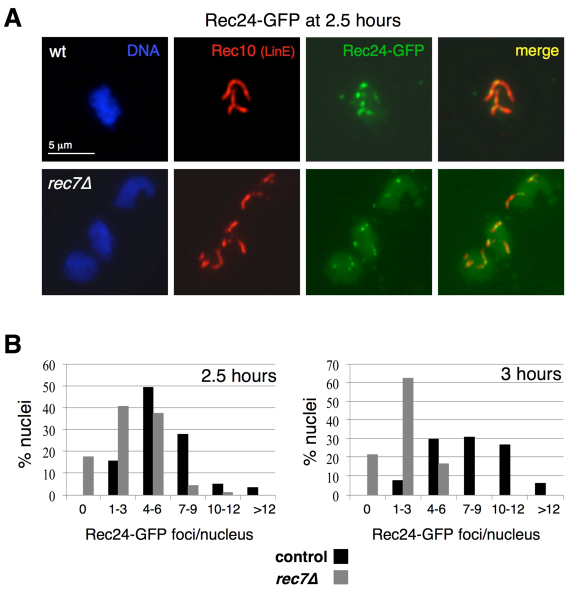
Fig. 4.
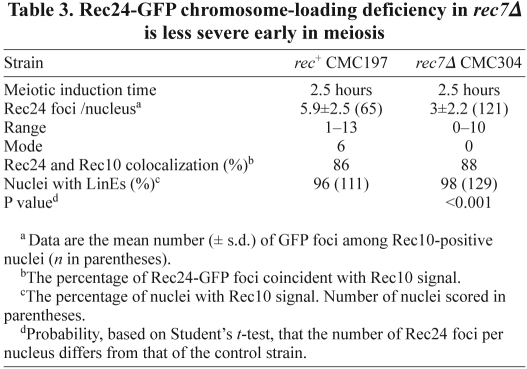
Maintenance of Rec24–GFP on LinEs requires Rec7. (A) Stains of spreads prepared 2.5 hours after the induction in Fig. 3. Spreads were stained with DAPI (DNA; blue), anti-GFP antibodies (Rec24–GFP; green) and anti-Rec10 antibodies (red) and photographed under a fluorescence microscope. Rec24–GFP signal quantification is shown in Table 3. (B) Distribution of the number of Rec24–GFP foci at 2.5 and 3 hours after meiotic induction in the wild type and rec7Δ mutants. Data from quantifications shown in Tables 2 and 3 are the percentage of Rec10-positive nuclei with the indicated number of Rec24–GFP foci. The number of nuclei analyzed were 65 and 121 at 2.5 hours and 68 and 98 at 3 hours, for wild-type and rec7Δ strains, respectively.
Localization of Rec24 requires formation of LinEs but not DSBs
We determined the requirements for localization of Rec24, because it was possible that Rec24 loading onto chromosomes was dependent on LinE formation and independent of DSB formation, as reported for Rec7 (Lorenz et al., 2006). The formation of LinEs in rec24Δ mutants supports this hypothesis (Fig. 1). Therefore, we studied the chromosome loading of Rec24–GFP in rec10Δ and rec12Δ mutants, in which no LinEs and no detectable DSBs are formed, respectively (Cervantes et al., 2000; Young et al., 2002; Molnar et al., 2003; Davis et al., 2008). First, we analyzed Rec24–GFP localization at 3 hours after meiotic induction, the time of maximal loading. As shown in Fig. 3, Rec24–GFP loading onto chromosomes was strongly impaired in rec10Δ but not in rec12Δ mutants. The number of Rec24–GFP foci in the rec12Δ mutant (mean 7.5±3 foci per nucleus) was similar to that in the wild type (mean 7.9±3 foci per nucleus), whereas most of the Rec24–GFP signal was lost in the rec10Δ mutant (mean 1.2±1.3 foci per nucleus) (see Table 2 for quantification). Similar data were obtained at an earlier time point, 2.5 hours after induction (unpublished data). The lack of Rec24–GFP from the meiotic chromosomes in the rec10Δ mutant is not explained by the absence of the protein, because it was expressed normally during the time course as shown by western blot (supplementary material Fig. S1). Therefore, Rec24–GFP chromosome localization is independent of DSB formation and requires formation of LinEs.
Efficient chromosomal localization of Rec24 requires the Rec12 accessory protein Rec7
Because Rec24 exhibits a localization pattern and genetic requirements for chromosome loading that are similar to those of Rec7, we explored the relationship between the two proteins. Thus, we analyzed localization of Rec24–GFP in the absence of Rec7 at 3 hours after meiotic induction. The loading of Rec24–GFP was dramatically reduced in rec7Δ mutants (mean 2.1±1.7 foci per nucleus) compared with the wild type (mean 7.9±3 foci per nucleus) (Fig. 3 and Table 2), although Rec24–GFP protein was expressed normally in rec7Δ meiosis (supplementary material Fig. S1). In addition, the foci remaining in rec7Δ were less intense and more rounded than the smeared and elongated shape of wild-type foci. However, some Rec24–GFP was still specifically present on LinEs. These data suggest that Rec7 is required for the loading or the stabilization of Rec24–GFP on linear elements.
To distinguish between these two possibilities, we analyzed the chromosome recruitment of Rec24–GFP at an earlier time point. The defect in Rec24–GFP loading in the rec7Δ mutant was less severe at 2.5 hours than at 3 hours after meiotic induction (Fig. 4). Nuclei with several Rec24–GFP foci aligned onto LinEs were frequently observed (Fig. 4A). Some elongated foci were also visible, although foci tended to be more rounded and less intense than wild-type foci. In addition, the mean and the range of foci were greater (mean 3±2.2 foci per nucleus in a range from 0–10 foci, Table 3) than the distribution observed at 3 hours (2.1±1.7 foci per nucleus in a range from 0–6 foci, Table 2). Moreover, the difference in foci number compared with the wild-type control was reduced at 2.5 hours (mean 5.9±2.5 foci per nucleus) (Table 3). When depicted as a distribution of the number of foci per nucleus (Fig. 4B), the difference between the wild-type and rec7Δ distributions was more evident at 3 hours than at 2.5 hours. Although the percentage of nuclei with a larger number of foci increased with time in the wild-type control, indicating increased loading, the percentage of nuclei with a larger number of foci decreased in the rec7Δ mutant. Thus, at 2.5 hours, the percentage of nuclei with more than four foci was 85% in the control and 42% in rec7Δ mutant; however, at 3 hours, the percentage increased in the control to 93% and decreased in rec7Δ to 16%, suggesting that some of the signal observed at 2.5 hours in rec7Δ was lost at the later time point. Therefore, Rec7 seems to be required for the stabilization of Rec24 on meiotic chromosomes.
Table 3.
Rec24-GFP chromosome-loading deficiency in rec7Δ is less severe early in meiosis
Rec7 is required for efficient chromatin association of Rec24
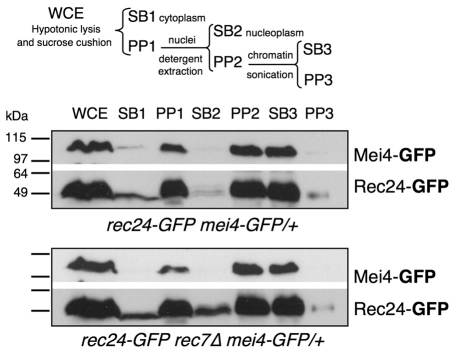
We studied in more detail the role of Rec7 by cellular fractionation to follow the subcellular distribution of Rec24–GFP in the wild type and rec7Δ mutants. Cells collected during meiotic prophase at 3 hours after meiotic induction were treated with zymolyase for cell wall permeabilization, subjected to a hypotonic lysis (whole cell extract, WCE), and separated into cytoplasmic (SB1) and nuclear fractions (PP1) by centrifugation through a sucrose cushion. The nuclear fraction was washed with detergent (1% Triton X-100) for solubilization of nucleoplasm proteins, and centrifuged to collect the soluble fraction (SB2) and the nuclear-insoluble fraction (PP2). This nuclear fraction resistant to detergent extraction was gently sonicated for further solubilization and separation of the soluble fraction (SB3) from the highly insoluble fraction (PP3). To follow the behavior of a chromatin-binding protein during the fractionation, we introduced a heterozygous mei4–GFP allele into the experimental diploid strains. Mei4 is a highly induced, meiosis-specific transcription factor that is essential for the induction of genes required for DSB formation and entry into meiosis I (Abe and Shimoda, 2000; Gregan et al., 2005). At 3 hours after meiotic induction, Mei4–GFP was specifically detected in the nuclear fractions PP1 and PP2, but was solubilized by sonication (SB3) (Fig. 5).
Fig. 5.
Rec24–GFP is less efficiently bound to the nuclear fraction in rec7Δ mutants. Diploids pat1-114 rec24-GFP and pat1-114 rec24-GFP rec7Δ carrying mei4-GFP in heterozygosis (strains CMC417 and CMC416) were induced for meiosis and collected after 3 hours for cellular fractionation. Scheme of the fractionation is shown in the upper part of the figure. The distribution of Rec24–GFP and Mei4–GFP was followed by western blot with anti-GFP antibodies. Mei4–GFP illustrates the distribution of a meiosis-specific transcription factor. Each lane contains equal extract equivalents (5% of the WCE).
Rec24–GFP showed a behavior similar to that of Mei4–GFP in the control strain, although a small fraction of Rec24–GFP was detected in SB1 (cytoplasmic fraction) (Fig. 5, upper panels). However, whereas Mei4–GFP did not change its localization in the rec7Δ mutant and was present only in the pellets PP1 and PP2, Rec24–GFP protein increased in SB1 and more importantly in SB2 (nucleoplasm fraction) (Fig. 5, lower panels). These data, along with the cytological data described above, support the idea that Rec7 is required for efficient association of Rec24–GFP with the nuclear fraction. This fraction contains the transcription factor Mei4–GFP and is highly resistant to detergent extraction.
Rec24 is essential for Rec7 recruitment to chromosomes
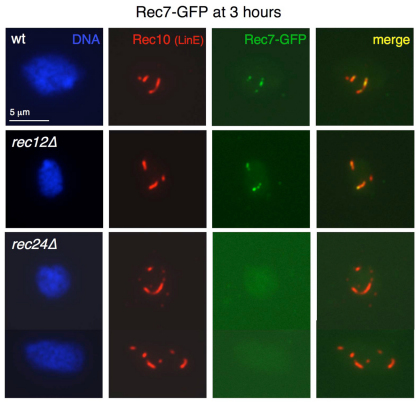
Our data suggest that Rec24 is loaded onto meiotic chromosomes before Rec7 and that the interaction with Rec7 stabilizes this loading. Therefore, we analyzed the chromosome association of Rec7–GFP in rec24Δ mutants. Chromosome spreads prepared at different times (2.5 and 3 hours) during prophase in rec7–GFP synchronous meiosis were double stained with anti-Rec10 antibodies to visualize LinEs and anti-GFP antibodies for detection of Rec7–GFP foci (Lorenz et al., 2006). In addition to rec24Δ, rec12Δ mutants were used as a control, because Rec7–GFP loading is independent of DSB formation (Lorenz et al., 2006). No substantial loading of Rec7–GFP was detected in rec24Δ mutants at 3 hours after meiotic induction (Fig. 6), and few nuclei (~14%, n=72) with a single GFP focus, in most cases very weak, that localized (~7%) or not (~7%) to LinEs were observed. However, foci were present in the wild type and rec12Δ mutants (see supplementary material Fig. S2A for distribution of Rec7–GFP foci in the wild-type strain). Similarly, no loading was observed at an earlier time point, 2.5 hours after meiotic induction (unpublished data). Thus, Rec24 but not Rec12 is crucial for loading of Rec7 onto meiotic chromosomes. This result, plus the ability of Rec24 to load in the absence of Rec7, is consistent with the idea that Rec7 is loaded after Rec24.
Fig. 6.
Rec7–GFP does not localize to LinEs in rec24Δ mutants. Diploid pat1-114 rec7-GFP cells with the indicated deletions (strains CMC271, CMC272 and CMC273) were induced for meiosis and collected in prophase for nuclear spread preparation. Spreads were stained with DAPI (DNA; blue), anti-GFP antibodies (Rec7–GFP; green) and anti-Rec10 antibodies (red) and photographed under a fluorescence microscope. Stains of chromosome spreads at 3 hours after meiotic induction are shown. No substantial loading was detected in 257 rec24Δ nuclei. Similar results were obtained with 2.5 hour spreads and in an independent experiment (unpublished data).
Rec24 and Rec7 physically interact during meiotic prophase
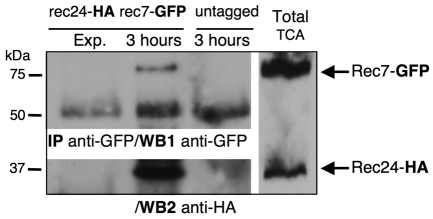
Given the above results, which point to a functional relationship between Rec24 and Rec7, we next addressed a possible physical interaction between these proteins, using functional Rec24–HA and Rec7–GFP tagged versions (Molnar et al., 2001) (supplementary material Table S1). Exponentially growing cells and cells at 3 hours after meiotic induction were collected, and native protein extracts were immunoprecipitated with anti-GFP antibodies (Fig. 7). Rec7–GFP in the immunoprecipitate was tested by anti-GFP western blot. A band corresponding to Rec7–GFP was detected in the double-tagged cells, but not in extracts from exponentially growing or untagged control cells at the same meiotic time point (Fig. 7, WB1). The same blot was then assayed for Rec24–HA with anti-HA antibody, and a specific band was detected only during meiotic prophase in extracts from the double-tagged cells (Fig. 7, WB2). Thus, Rec24 and Rec7 physically interact during meiotic prophase.
Fig. 7.
Rec24–HA and Rec7–GFP physically interact during prophase. Diploid pat1-114 and pat1-114 rec7-GFP rec24-HA cells (strains CMC7 and CMC302) were induced for meiosis and collected in prophase after 3 hours. Double-tagged cells were also collected before meiotic induction as a control for the specificity of the immunoprecipitation. Native protein extracts were immunoprecipitated with anti-GFP polyclonal antibodies and analyzed by western blot using first anti-GFP monoclonal antibodies (WB1, anti-GFP). The lower bands correspond to heavy chains of the antibodies used for immunoprecipitation. The same blot was then analyzed with anti-HA monoclonal antibodies (WB2, anti-HA). Total TCA extract of the double-tagged strain at 3 hours after meiotic induction was loaded in the same gel to show position of Rec7–GFP and Rec24–HA proteins (right lane).
Discussion
Although the role of Spo11 and Rec12 in DSB formation is evolutionarily conserved, the regulation of its DSB-forming activity is poorly understood. In addition to Spo11 (or Rec12) several proteins essential for DSB formation and recombination are present in numerous species, including budding and fission yeasts, flies, worms, plants and mice (Liu et al., 2002; Libby et al., 2003; Reddy and Villeneuve, 2004; Reinholdt and Schimenti, 2005; Cromie and Smith, 2008; Keeney, 2008; De Muyt et al., 2009; Cole et al., 2010; Kumar et al., 2010); however, these accessory proteins show very limited amino acid sequence similarity (see Introduction). Therefore, to better understand the regulation of Spo11 and Rec12 activity, we need to understand how these accessory proteins function and relate to each other, most readily done in the two experimentally tractable yeasts. Functional relationships might help to illuminate common regulatory mechanisms in other species, despite the limited amino acid conservation of the proteins.
Rec24 is a bona fide Rec12 accessory protein
In fission yeast, seven proteins can be considered as Rec12 accessory proteins based on genetic data and physical analysis of DSBs. Rec6, Rec7, Rec10, Rec14, Rec15, Rec24 and Mde2 are all essential for meiotic DSB formation and recombination (Ponticelli and Smith, 1989; DeVeaux et al., 1992; Cervantes et al., 2000; Young et al., 2002; Ellermeier and Smith, 2005; Gregan et al., 2005; Martin-Castellanos et al., 2005) (Table 1). In addition, LinEs have been analyzed in rec6, rec7, rec14 and rec15 mutants and, in agreement with the idea that formation of LinEs is independent of DSB formation, these structures are formed in all of these mutants (Molnar et al., 2003; Lorenz et al., 2004; Lorenz et al., 2006). However, very little is known about the accessory proteins (e.g. their localization) or the functional and physical relationships among them. An exception to this is Rec7, the only Rec12 accessory protein whose meiotic localization has been reported (Molnar et al., 2001; Lorenz et al., 2006).
Here, we confirm that Rec24 is indeed a Rec12 accessory protein. Rec24 is essential for DSB formation, as assayed by physical analysis at several DSB hotspots, and recombination (Martin-Castellanos et al., 2005) (Table 1). Moreover, rec24Δ mutants suppress the low viable-spore yield of rad32Δ to the same extent as rec6Δ, rec10Δ and rec12Δ, indicating that indeed Rec24 is required for most, and probably all, DSB formation (Table 1) (Young et al., 2004; Ellermeier and Smith, 2005). By contrast, cohesin mutants rec8Δ and rec11Δ do not suppress rad32Δ, probably because rec8Δ and rec11Δ retain residual DSB formation (Ellermeier and Smith, 2005). Also, unlike Rec8 and Rec11, Rec24 is not required for LinE formation, as reported for other Rec12 accessory proteins (Molnar et al., 2003; Lorenz et al., 2004; Lorenz et al., 2006) (Fig. 1). Thus, the accessory proteins appear to be essential for meiotic recombination and DSB formation throughout the genome, whereas cohesins stimulate recombination in a region-specific fashion (Ellermeier and Smith, 2005).
Similarly to the accessory protein Rec7, Rec24–GFP localizes to LinEs during meiotic prophase in a spotted and oval pattern with extensive colocalization with Rec10 (LinEs) (Figs 2, 3 and 4; Tables 2 and 3). Rec24–GFP appears after LinE formation is initiated and disappears when LinEs start to disassemble (unpublished data and Table 2). In addition, the genetic requirements for localization of Rec24–GFP are similar to those for Rec7 (Lorenz et al., 2006). Rec24–GFP focus formation requires LinE formation, because foci are absent in rec10Δ mutants, but is independent of DSB formation, because normal foci are found in rec12Δ mutants (Fig. 3). Therefore, Rec7 and Rec24 might be recruited to the same location on LinEs before DSB formation. Supporting this idea, both proteins form complexes in vivo (Fig. 7). This interaction has also been recently reported, using both two-hybrid assays and co-immunoprecipation experiments, as shown here (Steiner et al., 2010). However, our mean number of Rec24–GFP foci at maximal chromosome loading is smaller than the mean foci number reported for Rec7–GFP (Lorenz et al., 2006). This might be due to the different experimental conditions: pat1-114 synchronous meioses studied here and wild-type asynchronous meioses studied previously (Lorenz et al., 2006). Indeed, Rec7–GFP chromosome loading in our synchronous meioses is also less than reported previously (supplementary material Fig. S2A).
The interaction between Rec24 and Rec7 requires a phenylalanine residue on Rec7 (F325), whose substitution for alanine disrupts the two-hybrid interaction and confers a recombination defect in vivo (Steiner et al., 2010). Interestingly, this residue is not at the N-terminal part of the protein, which was previously reported to show some conservation with budding yeast Rec114 (Malone et al., 1997), but at the C-terminal region in a domain very modestly conserved in Ascomycetes. Moreover, a wider conservation of Rec7 has been uncovered very recently using a phylogenomic-oriented bioinformatics approach to search for proteins involved in DSB formation (Kumar et al., 2010). This conservation includes the crucial phenylalanine residue that is essential for the interaction between Rec24 and Rec7.
Although we have seen that Rec24–GFP (Figs 2, 3 and 4) and Rec7–GFP (Fig. 6) (Lorenz et al., 2006) localize to LinEs, these proteins did not appear in a purification of proteins associated with LinEs using the structural component Rec10 as a tool (Spirek et al., 2010). This suggests that the association of these proteins with LinEs is not very stable. Similarly, other known LinE-associated proteins, Mek1 and Rad51, were not co-purified with Rec10 (Spirek et al., 2010), indicating that purification conditions and perhaps timing, or both, are crucial to identify these interactions.
Functional relationship between Rec24 and Rec7
Our data reveal a functional relationship between Rec24 and Rec7. Apart from the physical interaction between these proteins, we have shown that Rec7 is required to maintain the chromosome localization of Rec24. First, in the rec7Δ mutant, Rec24–GFP is largely lost from chromosomes later in prophase, although it seems to be substantially more abundant at earlier time points (Figs 3, 4; Tables 2 and 3). Second, the abundance of Rec24–GFP in the nucleus depends on Rec7. In rec7Δ mutants the amount of Rec24–GFP in the cytoplasm increases; in addition, it is solubilized more easily from the nuclei, suggesting that Rec7 is required for efficient interaction of Rec24 with meiotic chromosomes (Fig. 5). The dependencies for chromosome recruitment reported among accessory proteins in budding yeast has led to a proposed organization of accessory proteins into different subcomplexes (see Introduction). Our report is the first of this kind of regulation for any Rec12 accessory protein in fission yeast, or in any other species. In addition, analysis of Rec24–GFP localization at different time points during meiotic prophase has helped us to establish a dependency, not only for chromosome recruitment, but also for maintenance. Binding stabilization of accessory proteins (or subcomplexes) to meiotic chromosomes might be a point for additional regulation of DSB formation.
As recently proposed (Steiner et al., 2010), Rec24 and Rec7 might form a complex whose binding to chromosomes is, based on our results, stabilized by Rec7. Another possibility is that Rec7 is loaded onto chromosomes after Rec24 and not as a preformed complex. In both scenarios, we would expect loss of Rec7 binding in the absence of Rec24, as we observed. Rec7–GFP chromosome recruitment is dependent on Rec24 at different time points during prophase (Fig. 6). However, we favor the latter hypothesis because Rec24 forms a substantial number of foci on LinEs in the absence of Rec7, but not vice versa, and Rec24–GFP chromosome loading precedes Rec7–GFP chromosome loading (supplementary material Fig. S2). Therefore, Rec24 might mark loading sites where Rec7 is recruited. Only stabilized complexes containing Rec24 and Rec7 might then load or activate Rec12 to form DSBs, which is an essential step in successful meiosis.
Recently, the wide conservation of Rec24 and Rec7 in other eukaryotes has been reported using a bioinformatics approach (Cole et al., 2010; Kumar et al., 2010). Rec24 shows low homology with budding yeast Mei4 (7% amino acid identity), and budding yeast Mei4 and mouse Mei4 show only 8% amino acid identity. However, they share evolutionarily conserved short signature sequence motifs (SSMs) distributed similarly along the primary protein sequences. As we describe here for Rec24 in fission yeast and as previously reported for budding yeast Mei4 (Li et al., 2006; Maleki et al., 2007), mouse Mei4 localizes on meiotic chromosomes in a Spo11-independent manner, is required for DSB formation, and physically interacts, as assayed by immunoprecipitation and two-hybrid analysis, with Rec114 (the fission yeast Rec7 ortholog) (Kumar et al., 2010). Therefore, the functional relationship between Rec24 and Rec7 that we report here in fission yeast indicates that the interplay between these proteins might have a central role in the control of DSB formation in many species.
Materials and Methods
Yeast strains and manipulation
Strains used in this study are listed in supplementary material Table S2. Cells were grown and manipulated as described (Moreno et al., 1991). Cells were grown in yeast extract medium with supplements (YES) or Edinburgh minimal medium (EMM) with supplements at 32°C or 25°C (for the pat1-114 temperature-sensitive mutants). Genetic crosses were done on supplemented SPA agar (SPA; Table 1) or on supplemented malt extract plates (MEA-4S) at 25°C.
Viable-spore yield and recombinant frequencies
Viable-spore yields and recombinant frequencies in Table 1 were determined as described (Young et al., 2004). Functionality of epitope-tagged proteins was determined by ade6-M26 × ade6-3049 intragenic recombination (supplementary material Table S1). To eliminate possible remaining vegetative cells after glucuronidase treatment, spore preparations were incubated for 30 minutes at 55°C. Appropriate dilutions of spores were plated on YE minus supplements and incubated for 4 days at 32°C. Ade+ colony frequencies were calculated as the number of white colonies per 104 viable spores. Rec7–GFP recombination efficiency was also checked, since its functionality has been questioned (Molnar et al., 2001; Lorenz et al., 2006).
Epitope tagging and deletion construction
C-terminal Rec24 GFP and HA tagging and GFP tagging of Mei4, was carried out using a PCR-based method (Bahler et al., 1998). Oligonucleotides with 80 nucleotides of homology to regions flanking the stop codon were used to amplify GFP and kanMX6, or HA and kanMX6 from plasmids pFA6a-GFP-kanMX6 and pFA6a-3HA-kanMX6, respectively, in the case of Rec24 tagging, and GFP and hphMX6 from plasmid pFA6a-GFP-hphMX6 (pFA6a-kanMX6 derivative by hph replacement) in the case of Mei4 tagging; these were oligo rec24-HA1 5′-AGTTGTCTAAGAGTGCCTACTCTGCTCCATCCATCCTTTTGATTGGCCTCAAAGATATGCTATTTCCCGAAGACATTTCCCGGATCCCCGGGTTAATTAA-3′ and oligo rec24-HA2 5′-TAATATCAAAGTTCATCGAATGGATATGATGATCAACACTTTCACCACAACGTTCTTTGTACATTGAATTTCCAGAAAAAGAATTCGAGCTCGTTTAAAC-3′, oligo mei4-HA1 5′-AACTTAATCATGAGTCAAGTGGCTACTCAAGTGCGCCATTAATGCCTTCTAATCGAGCGTTTATCAATGACTTCTCTCTTCGGATCCCCGGGTTAATTAA-3′ and mei4-HA2 5′-ACAATATAGATGATGAACACTCTGTTCTTTCGTCGAAAAAAGGGATGTTGAATGCATGGCCTTTGATCTAGTTTTTTGATGAATTCGAGCTCGTTTAAAC-3′ (bold face indicates S. pombe genome sequence). In the case of Rec24 tagging, the PCR products were used to transform h+ ade6-M216 leu1-32 (strain S778) or h− pat1-114 ade6-M210 leu1-32 (strain CMC94) to G418-resistance using the lithium acetate protocol (Bahler et al., 1998). Correct integration was checked by PCR and sequencing, and the required strains were obtained by subsequent meiotic crosses. This method of epitope-tagging displaces the endogenous 3′UTR and therefore might alter the expression pattern (Rabitsch et al., 2004). However, rec24–GFP and rec24-HA fusions are quite functional, as shown by the production of asci with wild-type appearance and high levels of recombination (unpublished data and supplementary material Table S1), indicating that appropriate protein levels are produced. Diploid pat1-114 leu1-32 strains were generated by protoplast fusion and selection for complementation of the ade6-M210 and ade6-M216 alleles (Sipiczki and Ferenczy, 1977). PCR products for Mei4 tagging were used to transform diploids with rec24–GFP (strain CMC197) and rec24–GFP rec7Δ (strain CMC304) to hygromycin resistance, thereby generating strains CMC417 and CMC416, respectively. Heterozygous integration (+/mei4–GFP) was checked by PCR and correct GFP fusion by sequencing. mei4–GFP in strain CMC416 carries a mutation just after the stop codon at the AscI restriction site (G to A) and another one at the beginning of GFP (A118 to G). These mutations do not affect Mei4–GFP expression or subcellular localization (Fig. 5; compare Mei4–GFP in strains CMC417 and CMC416). Mei4–GFP functionality was checked by additional transformation of h90 (strain CMC3; the same as wild-type strain 968), sequencing and testing for normal sporulation (unpublished data).
A complete rec7 deletion was generated by the same PCR-based method using oligos to amplify kanMX6 from plasmid pFA6a-kanMX6 and transformation of h− ade6-M216 leu1-32 (strain CMC323) to G418 resistance. These were oligo rec7-D1 5′-CACCGTGACGGAACGTACTGCGTTATCAGATAAATGAGGTTCAGTCTAACGCGTCTTCTCATATTCAAACATAAACAAACCGTACGCTGCAGGTCGAC-3′ and oligo rec7-D2 5′-AAATAACCATTCCGATCCTGATTTCGTTCCATTTTTACTACTTTTTTAAATTTGGGATTAAGTTGAATTGGTCTGTCTTCATCGATGAATTCGAGCTCG-3′ (bold face indicates S. pombe genome sequence). Correct deletion was checked by PCR and the required strains were obtained by subsequent meiotic crosses and protoplast fusion.
Synchronous meiosis
Meiotic time-course experiments using pat1-114 leu1-32 diploid cells were done as described previously (Davis et al., 2008). The high temperature (34°C) used to inactivate the thermosensitive allele pat1-114 does not seem to strongly affect recombination rates as shown by Li and Smith (Li and Smith, 1997) and Hartsuiker and co-workers (Hartsuiker et al., 2009). Cells collected during synchronous meiosis were fixed with 70% ethanol and kept at 4°C until flow cytometry processing, as previously described (Sazer and Sherwood, 1990). A Becton Dickinson FACSort flow cytometer and CellQuest software were used for acquisition and analysis of data. In all experiments, the bulk of DNA replication occurred between 1.5 and 2 hours after meiotic induction.
Cytology
Preparation of chromosome spreads was done as described previously (Davis et al., 2008), with 10 mM DTT in the buffer for spheroplast formation, not 10 μM (as incorrectly stated by Davis and colleagues) and stored at −20°C until use. Immunostaining was performed as described (Lorenz et al., 2004). Polyclonal anti-Rec10 (1:200 dilution; a gift from Ramsey McFarlane, North West Cancer Research Fund Institute, Bangor, UK) or monoclonal anti-GFP (1:800 dilution, JL-8 Living Colors, Clontech) antibodies were used as primary antibodies. Mouse Alexa Fluor 488 (1:1000 dilution, A11001 Molecular Probes), rabbit Alexa Fluor 568 (1:1000 dilution, A11011 Molecular Probes) or rabbit Alexa Fluor 488 (1:1000 dilution, A11008 Molecular Probes) antibodies were used as secondary antibodies. Slides were mounted in PBS containing 50% glycerol, p-phenylenediamine (1 mg/ml) and DAPI (2 μg/ml). Immunofluorescence was detected with a Nikon Eclipse 90i microscope equipped with a 100×/1.4 Oil Plan APO VC lens, a Hamamatsu ORCA-ER camera and MetaMorph software.
Cellular fractionation
Cellular fractionation was done as described previously in budding yeast to follow the behavior of accessory proteins (Kee et al., 2004). 5×108 cells at 3 hours after meiotic induction were collected and resuspended in 2 ml of PEM-sorbitol (100 mM PIPES, pH 6.9, 1 mM EGTA, 1 mM MgSO4, 1.2 M Sorbitol) containing 10 mM DTT, 2 mg/ml 20T Zymolyase (Seikagaku Corporation), 2 mM PMSF (Sigma) and 2× protease inhibitors (Complete Protease Inhibitor Cocktail, EDTA-free, Roche), and incubated 30 minutes at 32°C. The subsequent steps were done at 0–4°C with ice-cold solutions in Eppendorf-type tubes and a microcentrifuge. The resulting spheroplasts were centrifuged at 1000 g for 1 minute, resuspended in 750 μl MES (0.1 M MES, pH 6.4, 1 mM EDTA, 0.5 mM MgCl2) containing 2 mM PMSF and 2× protease inhibitors, and incubated on ice for 10 minutes for hypotonic shock. 350 μl of this whole cell extract (WCE) were conserved (mixed with 4× Laemmli buffer, boiled and frozen; Laemmli buffer: 80 mM Tris-HCl pH 6.8, 5 mM EDTA, 5 mM DTT, 2% SDS, 7.5% glycerol, 0.002% Bromophenol Blue) and 500 μl loaded on a 500 μl 30% sucrose cushion in MES. After centrifugation for 10 minutes at 16,000 g, the supernatant (S1) was conserved, and the pellet (PP1) resuspended in 200 μl of Extraction Buffer (50 mM HEPES-NaOH pH 7.5, 100 mM KCl, 2.5 mM MgCl2, 1% Triton X-100) containing 2 mM PMSF and 2× protease inhibitors. 70 μl PP1 were conserved, and the rest incubated on ice for 10 minutes and centrifuged at 16,000 g for 10 minutes. The supernatant SB2 was conserved and the pellet PP2 resuspended in an equal volume of Extraction Buffer (140 μl), split into two, and conserved. One aliquot of the PP2 was sonicated using a Bioruptor (Diagenode) for 2.5 minutes at level L, 5 seconds ON/OFF, centrifuged at 16,000 g for 10 minutes at 20°C, and the supernatant SB3 collected. The pellet PP3 was resuspended in an equal volume of 1× Laemmli buffer (70 μl). Equal extract equivalents (5% of WCE) of each fraction were analyzed by western blot.
Immunoprecipitation assays and western blot
100 ml of exponentially growing cells (OD=1) and 100 ml of cells at 3 hours after meiotic induction (OD=1) were collected, washed once in sterile water, and stored at −20°C until use. Cell extracts were prepared in a 6770 Freezer/Mill (SPEX SamplePrep) always under frozen conditions. After 5 minutes of cooling, the frozen cell pellets were broken at level 12 for 10 minutes, with 1 minute ON/OFF cycles. The cell powder was resuspended in 1 ml ice-cold Lysis Buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate) with freshly added 2 mM PMSF (Sigma) and 4× protease inhibitors (Complete Protease Inhibitor Cocktail, EDTA-free, Roche). For a complete solubilization the powder suspension was gently sonicated using a Bioruptor (Diagenode) for 5 minutes at level H, 5 seconds ON/OFF cycles. 900 μl of WCEs were immunoprecipitated for 30 minutes at 4°C on a rotation wheel with 20 μl of antibody-precoated magnetic beads per sample (Dynabeads ProteinG, Invitrogen). The beads were coated for 45 minutes at room temperature in Lysis Buffer with 1 μl polyclonal rabbit anti-GFP antibody (A6455 Invitrogen) per immunoprecipitation. Using the Dynamag-Spin magnet (Invitrogen), the beads were collected from the WCE and washed with 1 ml of the following ice-cold buffers: twice with Lysis Buffer and twice with Washing Buffer (10 mM Tris-HCl, pH 8.0, 250 mM LiCl, 5 mM EDTA, 0.5% NP-40, 0.5% sodium deoxycholate). Elution of the immunoprecipitations was carried out by boiling in 20 μl of 2× Laemmli buffer. Total immunoprecipitates were loaded on a 12% SDS-PAGE mini-gel (BioRad) and transferred to nitrocellulose membrane (Hybond ECL, Amersham). The membrane was incubated first with monoclonal anti-GFP antibodies (1:3000 dilution, JL-8 Living Colors, Clontech) and goat anti-mouse conjugated to horseradish peroxidase (1:3000 dilution, A4416 Sigma) for GFP-tagged protein detection. After membrane washing, monoclonal mouse anti-HA (1:3000 dilution, 12CA5 Roche) and goat anti-mouse conjugated to horseradish peroxidase were used for HA-tagged protein detection. Immunoblots were developed with SuperSignal West DURA Extended kit (Pierce).
Assay of GFP-tagged proteins during synchronous meiosis using trichloroacetic acid (TCA) extracts was done as described (Davis et al., 2008). Monoclonal anti-GFP antibodies (1:3000 dilution, JL-8 Living Colors, Clontech) and monoclonal anti-actin antibodies (1:6000 dilution, clone C4, MP Biomedicals) for a loading control were used. GFP signal was detected with SuperSignal West DURA Extended Kit (Pierce) and actin signal with ECL Western Blotting Kit (Amersham, GE Healthcare).
Supplementary Material
Acknowledgments
We thank Ramsay McFarlane for his constant generosity providing us with Rec10 antibody, Jürg Kohli for the rec7-GFP allele, and Jürg Kohli, Silvia Steiner and Katja Ludin for discussion and sharing data before publication. This research was supported by grants from the National Institutes of Health of the United States of America to G.R.S. (GM032194), from the Spanish Ministry of Science and Innovation (Consolider ingenio CSD2007-00015 and FEDER-BFU2008-01808) and Junta de Castilla y León (Grupo de Excelencia grant GR265) to S.M., and from the Spanish Ministry of Science and Innovation (FEDER-BFU2007-66366 and FEDER-BFU2010-14954), CSIC (PII-200820I125), and Junta de Castilla y León (SA038A07) to C.M.-C. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/124/8/1328/DC1
References
- Abe H., Shimoda C. (2000). Autoregulated expression of Schizosaccharomyces pombe meiosis-specific transcription factor Mei4 and a genome-wide search for its target genes. Genetics 154, 1497-1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya S. N., Many A. M., Schroeder A. P., Kennedy F. M., Savytskyy O. P., Grubb J. T., Vincent J. A., Friedle E. A., Celerin M., Maillet D. S., et al. (2008). Coprinus cinereus rad50 mutants reveal an essential structural role for Rad50 in axial element and synaptonemal complex formation, homolog pairing and meiotic recombination. Genetics 180, 1889-1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora C., Kee K., Maleki S., Keeney S. (2004). Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13, 549-559 [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943-951 [DOI] [PubMed] [Google Scholar]
- Cao L., Alani E., Kleckner N. (1990). A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell 61, 1089-1101 [DOI] [PubMed] [Google Scholar]
- Cervantes M. D., Farah J. A., Smith G. R. (2000). Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell 5, 883-888 [DOI] [PubMed] [Google Scholar]
- Cole F., Keeney S., Jasin M. (2010). Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 24, 1201-1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie G., Smith G. R. (2008). Meiotic recombination in Schizosaccharomyces pombe: a paradigm for the genetic and molecular analysis. In Genome Dynamics & Stability, Recombination and Meiosis, Vol. 3 (ed. Egel R., Lankenau D.), pp. 195-230 Berlin, Heidelberg: Springer-Verlag; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L., Smith G. R. (2001). Meiotic recombination and chromosome segregation in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 98, 8395-8402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L., Smith G. R. (2003). Nonrandom homolog segregation at meiosis I in Schizosaccharomyces pombe mutants lacking recombination. Genetics 163, 857-874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L., Rozalen A. E., Moreno S., Smith G. R., Martin-Castellanos C. (2008). Rec25 and Rec27, novel linear-element components, link cohesin to meiotic DNA breakage and recombination. Curr. Biol. 18, 849-854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A., Pereira L., Vezon D., Chelysheva L., Gendrot G., Chambon A., Laine-Choinard S., Pelletier G., Mercier R., Nogue F., et al. (2009). A high throughput genetic screen identifies new early meiotic recombination functions in Arabidopsis thaliana. PLoS Genet. 5, e1000654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux L. C., Hoagland N. A., Smith G. R. (1992). Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics 130, 251-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C., Smith G. R. (2005). Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA 102, 10952-10957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. H., Li Y. F., Fox M. E., Smith G. R. (1997). A WD repeat protein, Rec14, essential for meiotic recombination in Schizosaccharomyces pombe. Genetics 146, 1253-1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J., Rabitsch P. K., Sakem B., Csutak O., Latypov V., Lehmann E., Kohli J., Nasmyth K. (2005). Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr. Biol. 15, 1663-1669 [DOI] [PubMed] [Google Scholar]
- Hartsuiker E., Mizuno K., Molnar M., Kohli J., Ohta K., Carr A. M. (2009). Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol. Cell. Biol. 29, 1671-1681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson K. A., Kee K., Maleki S., Santini P. A., Keeney S. (2006). Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125, 1321-1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao K., Salem L., Malone R. (2003). Support for a meiotic recombination initiation complex: interactions among Rec102p, Rec104p, and Spo11p. Mol. Cell. Biol. 23, 5928-5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet S., Vezon D., Froger N., Mercier R. (2006). Non conservation of the meiotic function of the Ski8/Rec103 homolog in Arabidopsis. Genes Cells 11, 615-622 [DOI] [PubMed] [Google Scholar]
- Kee K., Protacio R. U., Arora C., Keeney S. (2004). Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J. 23, 1815-1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S. (2001). Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1-53 [DOI] [PubMed] [Google Scholar]
- Keeney S. (2008). Spo11 and the formation of DNA double-strand breaks in meiosis. In Genome Dynamics & Stability, Recombination and Meiosis, Vol. 2 (ed. Egel R., Lankenau D.), pp. 81-123 Berlin, Heidelberg: Springer-Verlag; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Bourbon H. M., de Massy B. (2010). Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24, 1266-1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hooker G. W., Roeder G. S. (2006). Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173, 1969-1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. F., Smith G. R. (1997). The Schizosaccharomyces pombe rec16 gene product regulates multiple meiotic events. Genetics 146, 57-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libby B. J., Reinholdt L. G., Schimenti J. C. (2003). Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc. Natl. Acad. Sci. USA 100, 15706-15711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jang J. K., Kato N., McKim K. S. (2002). mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162, 245-258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A., Wells J. L., Pryce D. W., Novatchkova M., Eisenhaber F., McFarlane R. J., Loidl J. (2004). S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 117, 3343-3351 [DOI] [PubMed] [Google Scholar]
- Lorenz A., Estreicher A., Kohli J., Loidl J. (2006). Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma 115, 330-340 [DOI] [PubMed] [Google Scholar]
- Ludin K., Mata J., Watt S., Lehmann E., Bahler J., Kohli J. (2008). Sites of strong Rec12/Spo11 binding in the fission yeast genome are associated with meiotic recombination and with centromeres. Chromosoma 117, 431-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukaszewicz A., Howard-Till R. A., Novatchkova M., Mochizuki K., Loidl J. (2010). MRE11 and COM1/SAE2 are required for double-strand break repair and efficient chromosome pairing during meiosis of the protist Tetrahymena. Chromosoma 119, 505-518 [DOI] [PubMed] [Google Scholar]
- Maleki S., Neale M. J., Arora C., Henderson K. A., Keeney S. (2007). Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma 116, 471-486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone R. E., Pittman D. L., Nau J. J. (1997). Examination of the intron in the meiosis-specific recombination gene REC114 in Saccharomyces. Mol. Gen. Genet. 255, 410-419 [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C., Blanco M., Rozalen A. E., Perez-Hidalgo L., Garcia A. I., Conde F., Mata J., Ellermeier C., Davis L., San-Segundo P., et al. (2005). A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr. Biol. 15, 2056-2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Lyne R., Burns G., Bahler J. (2002). The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 32, 143-147 [DOI] [PubMed] [Google Scholar]
- Mehrotra S., McKim K. S. (2006). Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2, e200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N., Higuchi E., Smith G. R. (2009). Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol. Cell. Biol. 29, 5998-6005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M., Bahler J., Sipiczki M., Kohli J. (1995). The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics 141, 61-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M., Parisi S., Kakihara Y., Nojima H., Yamamoto A., Hiraoka Y., Bozsik A., Sipiczki M., Kohli J. (2001). Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics 157, 519-532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M., Doll E., Yamamoto A., Hiraoka Y., Kohli J. (2003). Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J. Cell Sci. 116, 1719-1731 [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795-823 [DOI] [PubMed] [Google Scholar]
- Petronczki M., Siomos M. F., Nasmyth K. (2003). Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell 112, 423-440 [DOI] [PubMed] [Google Scholar]
- Ponticelli A. S., Smith G. R. (1989). Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics 123, 45-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieler S., Penkner A., Borde V., Klein F. (2005). The control of Spo11's interaction with meiotic recombination hotspots. Genes Dev. 19, 255-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J., Siroky J., Mokros P., Schweizer D., Riha K. (2004). Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16, 1968-1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabitsch K. P., Gregan J., Schleiffer A., Javerzat J. P., Eisenhaber F., Nasmyth K. (2004). Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 14, 287-301 [DOI] [PubMed] [Google Scholar]
- Reddy K. C., Villeneuve A. M. (2004). C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118, 439-452 [DOI] [PubMed] [Google Scholar]
- Reinholdt L. G., Schimenti J. C. (2005). Mei1 is epistatic to Dmc1 during mouse meiosis. Chromosoma 114, 127-134 [DOI] [PubMed] [Google Scholar]
- Rothenberg M., Kohli J., Ludin K. (2009). Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 5, e1000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H., Murakami H., Fukuda T., Shibata T., Nicolas A., Ohta K. (2007). Meiotic association between Spo11 regulated by Rec102, Rec104 and Rec114. Nucleic Acids Res. 35, 1119-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasanuma H., Hirota K., Fukuda T., Kakusho N., Kugou K., Kawasaki Y., Shibata T., Masai H., Ohta K. (2008). Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 22, 398-410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazer S., Sherwood S. W. (1990). Mitochondrial growth and DNA synthesis occur in the absence of nuclear DNA replication in fission yeast. J. Cell Sci. 97, 509-516 [DOI] [PubMed] [Google Scholar]
- Shiozaki K., Russell P. (1995). Cell-cycle control linked to extracellular environment by MAP kinase pathway in fission yeast. Nature 378, 739-743 [DOI] [PubMed] [Google Scholar]
- Sipiczki M., Ferenczy L. (1977). Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating-type. Mol. Gen. Genet. 151, 77-81 [DOI] [PubMed] [Google Scholar]
- Spirek M., Estreicher A., Csaszar E., Wells J., McFarlane R. J., Watts F. Z., Loidl J. (2010). SUMOylation is required for normal development of linear elements and wild-type meiotic recombination in Schizosaccharomyces pombe. Chromosoma 119, 59-72 [DOI] [PubMed] [Google Scholar]
- Steiner S., Kohli J., Ludin K. (2010). Functional interactions among members of the meiotic initiation complex in fission yeast. Curr. Genet. 56, 237-249 [DOI] [PubMed] [Google Scholar]
- Tavassoli M., Shayeghi M., Nasim A., Watts F. Z. (1995). Cloning and characterisation of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 23, 383-388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan L., Niu H., Futcher B., Zhang C., Shokat K. M., Boulton S. J., Hollingsworth N. M. (2008). Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 22, 386-397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. A., Schreckhise R. W., Steiner W. W., Smith G. R. (2002). Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9, 253-263 [DOI] [PubMed] [Google Scholar]
- Young J. A., Hyppa R. W., Smith G. R. (2004). Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics 167, 593-605 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.