Abstract
Summary: Easyfig is a Python application for creating linear comparison figures of multiple genomic loci with an easy-to-use graphical user interface. BLAST comparisons between multiple genomic regions, ranging from single genes to whole prokaryote chromosomes, can be generated, visualized and interactively coloured, enabling a rapid transition between analysis and the preparation of publication quality figures.
Availability: Easyfig is freely available (under a GPL license) for download (for Mac OS X, Unix and Microsoft Windows) from the SourceForge web site: http://easyfig.sourceforge.net/.
Contact: s.beatson@uq.edu.au
1 INTRODUCTION
Comparative genomics involves the comparison of sequenced genomes, particularly for the identification of insertions, deletions and variation in syntenic regions. Visualizing alignments between specific regions of multiple genomes is a critical step in identifying genotypic differences that underlie phenotypic changes between strains or species. For example, comparisons between related prokaryote genomes can highlight mobile elements such as integrons, prophage or pathogenicity islands. Preparation of clear and accurate images based on these genomic comparisons is typically accomplished ad hoc by tedious manual compilation (e.g. Thomson et al., 2004; Venturini et al., 2010) or using screen-snapshots from analysis tools (e.g. Jackson et al., 2010; Kozak et al., 2010). Artemis comparison tool (ACT; Carver et al., 2005) and Mauve (Darling et al., 2010) are both examples of excellent comparative genome analysis tools that are widely used to generate figures for publication, but are not designed for this purpose and generally lose clarity when displaying several regions at once. Recently, an elegant visualization tool was developed (Guy et al., 2010); however, its dependence on R makes it difficult for users unfamiliar with scripting languages.
Here, we describe Easyfig, a Python application for plotting comparison figures of multiple genomes or genomic regions from annotation files (e.g. GenBank and EMBL) and tabular comparison files [e.g. BLAST (Altschul et al., 1990)]. Easyfig has been designed to enable any biologist to visualize comparisons between multiple genomes or genomic regions and produce clear, publication quality images quickly and easily.
2 IMPLEMENTATION
Easyfig is a Python application that uses the Tkinter windows system. It is available as an executable file, or as a Python script. As such it is platform independent and can be used in a Microsoft Windows, Linux or Mac OS X environment. No Unix or scripting knowledge is required so that it is easily accessible to the average biologist with little or no bioinformatics or computing experience. The graphical user interface (GUI) permits images to be drawn with minimal user input, yet allows highly customizable figures to be generated for closer analysis or publication.
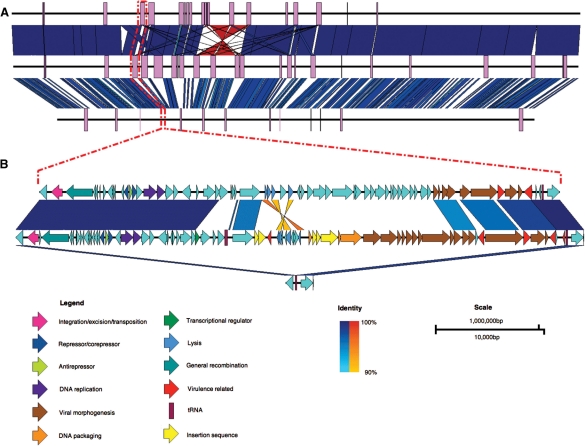
Easyfig accepts multiple sequences with or without annotation in standard formats (i.e. GenBank and EMBL). The input DNA sequence is rendered to scale as a solid black line centered vertically. Easyfig can handle a variety of loci lengths, from full prokaryote genomes (Fig. 1A) or small eukaryote chromosomes (5–10 Mb), down to individual loci or genes (Fig. 1B). The relative orientation of each region (forward/reverse) can be specified so that input sequences can be ‘flipped’ if required. By default, Easyfig will produce an image showing only gene features, but other features such as tRNAs, coding sequences (CDS), misc_features or a user-specified feature, can be added. Features can be displayed as rectangles, directional arrows, arrows representing frame and direction or a pointer to the start of the feature. Features can be coloured via the GUI or if the annotation file already has colour information, such as those that can be assigned using Artemis (Rutherford et al., 2000), each feature will be individually coloured according to the input file (i.e. using the feature qualifier : /colour=). Introns or pseudogenes with insertions in them are represented by dashed brackets joining each of the coding regions. The pixel height and width of features are customizable. Genomic regions can also be aligned left, right, centered or directly perpendicular to their best BLAST hit. A ‘zoom’ feature enables subregions of large sequence files to be specified via the GUI and examined in more detail. The figure can also show custom graphs displaying guanine-cytosine content, read coverage (calculated by Easyfig from an assembly file in .ace format), or a user-defined graph.
Fig. 1.
Comparison between the genomes of Escherichia coli O157:H7 str. EDL933 (top), E.coli O157:H7 str. Sakai (middle) and E.coli K12 str. MG1655 (bottom). (A) Whole-genome comparison with prophage regions shown as purple boxes. (B) A zoomed-in view showing that prophages have inserted at tRNA-Ser in the O157:H7 strains EDL933 and Sakai (prophages CP933M and Sp4, respectively), but not in K12. Dashed red lines indicate the site in the whole-genome sequences of the prophages and flanking genes shown in the bottom figure. Vertical blocks between sequences indicate regions of shared similarity shaded according to BLASTn (blue for matches in the same direction or orange for inverted matches). CDS in prophage Sp4 have been coloured according to Asadulghani et al. (2009) and functions of the CDS in CP933M have been inferred from BLAST hits and existing annotation.
BLAST comparisons (BLASTn, tBLASTx) between two or more loci can also be generated by the Easyfig interface, provided BLAST+ or legacy BLAST is available in a users path (details of how to set this up are included in the documentation for Easyfig). Alternatively, previously generated tabular comparison files can be loaded into Easyfig, including any pairwise alignment output (e.g MUMmer) that has been converted to BLAST hit table format. The Easyfig interface allows customization of the minimum expect values, lengths and identities of BLAST hit to be displayed in the final image. The hits are coloured on a gradient according to the BLAST identity value. Inverted matches can be shown using a different colour gradient. The colour scheme, gradient settings and height in pixels of the alignment can also be defined by the user.
If required, identity and scale legends will be embedded in the image along with specific annotations such as a scale bar and a colour gradient representing the identities of the BLAST hits.
Figures generated by Easyfig are saved in compressed bitmap (bmp) or vector graphics (svg) format at a user-defined resolution so that they can be easily annotated and manipulated in an image-editing program such as GIMP (www.gimp.org) if necessary.
In conclusion, Easyfig enables a variety of high-quality comparative genomic images to be generated locally using a simple GUI. A command-line version of Easyfig is also available, enabling it to be incorporated into analysis pipelines.
Funding: National Health and Medical Research Council of Australia (grant no. 511224); Australian Research Council Australian Research Fellowship (DP00881347 to S.A.B.).
Conflict of Interest: none declared.
REFERENCES
- Altschul S.F., et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asadulghani M., et al. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions and horizontal transfer of virulence determinants. PLoS Pathog. 2009;5:e1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T., et al. ACT: the artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Darling A.E., et al. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L., et al. genoPlotR: comparative gene and genome visualization in R. Bioinformatics. 2010;26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.P., et al. The genome sequence of Trypanosoma brucei gambiense causative agent of Chronic Human African Trypanosomiasis. PLoS Negl. Trop. Dis. 2010;4:e658. doi: 10.1371/journal.pntd.0000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak N.A., et al. Virulence factors encoded by Legionella longbeachae identified on the basis of the genomes sequence analysis of clinical isolate D-4968. J. Bacteriol. 2010;192:1030–1044. doi: 10.1128/JB.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–955. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Thomson N., et al. The role of prophage-like elements in the diversity of Salmonella enterica Serovars. J. Mol. Biol. 2004;339:279–300. doi: 10.1016/j.jmb.2004.03.058. [DOI] [PubMed] [Google Scholar]
- Venturini C., et al. Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 2010;24:1160–1166. doi: 10.1096/fj.09-144972. [DOI] [PubMed] [Google Scholar]