Abstract
Cis-regulatory modules (CRMs) function by binding sequence specific transcription factors, but the relationship between in vivo physical binding and the regulatory capacity of factor-bound DNA elements remains uncertain. We investigate this relationship for the well-studied Twist factor in Drosophila melanogaster embryos by analyzing genome-wide factor occupancy and testing the functional significance of Twist occupied regions and motifs within regions. Twist ChIP-seq data efficiently identified previously studied Twist-dependent CRMs and robustly predicted new CRM activity in transgenesis, with newly identified Twist-occupied regions supporting diverse spatiotemporal patterns (>74% positive, n = 31). Some, but not all, candidate CRMs require Twist for proper expression in the embryo. The Twist motifs most favored in genome ChIP data (in vivo) differed from those most favored by Systematic Evolution of Ligands by EXponential enrichment (SELEX) (in vitro). Furthermore, the majority of ChIP-seq signals could be parsimoniously explained by a CABVTG motif located within 50 bp of the ChIP summit and, of these, CACATG was most prevalent. Mutagenesis experiments demonstrated that different Twist E-box motif types are not fully interchangeable, suggesting that the ChIP-derived consensus (CABVTG) includes sites having distinct regulatory outputs. Further analysis of position, frequency of occurrence, and sequence conservation revealed significant enrichment and conservation of CABVTG E-box motifs near Twist ChIP-seq signal summits, preferential conservation of ±150 bp surrounding Twist occupied summits, and enrichment of GA- and CA-repeat sequences near Twist occupied summits. Our results show that high resolution in vivo occupancy data can be used to drive efficient discovery and dissection of global and local cis-regulatory logic.
In animal genomes, cis-acting regulatory modules (CRMs) average ∼300–500 bp in size and typically contain one or more binding motif instances for several transcription factors (Davidson 2006). DNA binding motif instances can now be readily mapped in silico by similarity to a consensus binding motif that has been defined through in vitro methods, or they can be derived from careful functional dissection of a few well-studied CRMs. However, many transcription factors recognize short sequence motifs that occur so frequently in the genome that virtually all gene loci have one or more, raising questions about which of these sites is occupied in the cell and what regulatory impact that occupancy has. We also know that binding motifs in the best-studied CRMs are often clustered (e.g., Ip et al. 1992a; Small et al. 1992; Berman et al. 2002; Markstein et al. 2002), presumably to facilitate coordinated and cooperative interaction among factors and cofactors and to achieve specificity relative to isolated single motif occurrences. However, we do not yet understand the logic by which motif combinations specify the functional output of the vast majority of CRMs in the genome (e.g., Lusk and Eisen 2010), and efficient identification and analysis of many more CRMs are needed to uncover these principles.
Advances in identifying candidate CRMs are coming from whole-genome approaches in which either chromatin immunoprecipitation (ChIP) is employed to find the region of DNA bound by a given transcription factor in vivo (e.g., Zeitlinger et al. 2007; Zinzen et al. 2009), or high-throughput screening assays are utilized to identify promoter and CRM functions (e.g., Landolin et al. 2010; Nam et al. 2010), although the latter have not yet been widely applied. Global ChIP assays also allow one to define de novo or refine binding motifs used by a factor in vivo and to compare this with in vitro defined motifs. ChIP-seq is a particular form of genome-wide chromatin immunoprecipitation, which can produce high positional resolution of observable DNA binding in vivo (Johnson et al. 2007). In particular, the resolution of ChIP-seq data can be used to infer, within a given binding region, which specific motif occurrence is likely to account for the majority of the observed ChIP signal (Valouev et al. 2008). We refer to the motif instances most likely to drive observed binding as candidate “explanatory” sites, and we explore the value of making explanatory site models for all ChIP signals to guide detailed functional assays.
We apply ChIP-seq and ChIP-chip analyses to Twist, a key transcription factor in the dorsal-ventral (DV) patterning network of the Drosophila early embryo. Patterning the DV axis depends partly on Twist, a bHLH transcription factor present at high levels in ventral regions of the embryo (for review, see Chopra and Levine 2009; Reeves and Stathopoulos 2009). Many previous studies have contributed to the current picture of a developmental gene network that describes embryonic DV patterning, in which more than 50 genes and 30 CRMs have been linked (for review, see Stathopoulos and Levine 2005). Previous published ChIP-chip studies conducted using Twist antibodies have demonstrated that its occupancy can be detected in vivo (Sandmann et al. 2007; Zeitlinger et al. 2007). Our goals are to relate the global Twist occupancy pattern to functional CRM activity, as assayed by transgenesis, and to relate the local ChIP-seq profile to specific motif instances and combinations and their contribution to individual CRM activity.
Results
Comparison of ChIP-chip and ChIP-seq in the identification of CRMs
We performed ChIP-chip and ChIP-seq analysis on sheared chromatin isolated from Drosophila embryos from 1 to 3 h in age, using an antibody that is specific to Twist protein, and subsequently assessed the overlap between sets of regions identified by each approach (see Supplemental Fig. 1A–C and Methods). For ChIP-chip, we used a script to call peaks based on a minimum signal score, whereas for ChIP-seq, we used the ERANGE software suite to call peaks based on the number, orientation, and ratio of short sequence reads relative to a background control. The results from these methods were compared at several sensitivity thresholds to accommodate different numbers of peaks called by their informatics pipelines (Supplemental Fig. 1D). Given the substantial technical and computational differences between ChIP-chip and ChIP-seq, the fact that the vast majority of ChIP-seq signals overlap with some ChIP-chip regions lends mutual confidence, although a large number of ChIP-chip sites lacked support from ChIP-seq. Inspection of multiple ChIP-seq regions for which Twist activity was previously studied in detail showed that ChIP-seq regions are generally better resolved and provide superior guidance for experimental tests of function that are the central focus of this study (Supplemental Table 1).
Functional analysis of Twist-occupied regions
We quantified how frequently and strongly ChIP-seq regions function as CRMs at the same time and place in development as the ChIP assays. To first identify a set of known gold-standard Twist CRMs, we applied a conservative standard that allowed only CRMs having prior genetic and molecular evidence. Enhancers (i.e., CRMs supporting gene expression rather than acting as silencers) along the DV axis were categorized as three types: Type I (ventral regions), Type II (ventro-lateral regions), and Type III (dorsal-lateral and dorsal regions) (Supplemental Table 2B; for review, see Chopra and Levine 2009; Reeves and Stathopoulos 2009). Many enhancers of Types I and II require Twist for expression based on genetic and molecular genetic evidence, but not until recent ChIP-chip analyses was it thought that Twist might function to regulate Type III patterns (Zeitlinger et al. 2007). We observed very strong ChIP signals at sog and brk Type III CRMs but not at ind, dpp, zen, and tld (Supplemental Table 2B; Supplemental Fig. 2). When only Type I and II CRMs were considered, 11 of 15 were present in our medium confidence (MC) data set (see Methods). Known CRMs for the four not present (i.e., Ady43A, phm, E(spl), and wntD) had below-threshold or no Twist ChIP-seq signal. The threshold for calling peaks could, of course, be reduced in order to recapture some (e.g., wntD and phm), but at the expense of increasing the false positive rate. Taken at face value, this gold standard comparison suggests we miss ∼25% of true positives at the threshold selected.
Next, we tested 31 new candidate Twist CRMs drawn from the entire ChIP-seq set in a standard reporter gene assay (see Supplemental Table 2A). Of the 31 test regions, 23 (74%) supported expression; 21 supported expression in a classic dorso-ventral pattern or a subregion thereof, and 2 supported distinct patterns (i.e., ubiquitous or purely anterior-posterior) (Supplemental Fig. 3). The 23 new CRMs were distributed throughout the ChIP-seq signal range (Supplemental Fig. 2, “Positive signal”). Peaks near genes Cyp310a1, Traf4, mirror (mirr), and Mef2 were clearly defined by the ChIP-seq data, while the equivalent ChIP-chip data in these regions was much broader and, in some cases, gave multiple peaks, making the location of a candidate CRM ambiguous (see Fig. 1A–D). While Twist ChIP-seq data led to a high recovery rate of CRM detection, surprisingly, only ∼25% of the associated genes including Cyp310a1, Asph, and emc (i.e., 3 of 12 assayed) actually required Twist to support expression in embryos. For instance, mirr, Traf4, and Mef2 expression was unaffected in twist mutants, even though their Twist-ChIP-seq signals were equally prominent and numerous (data not shown; see Discussion).
Figure 1.
In vivo Twist occupancy supported by Twist ChIP-seq identifies functional CRMs. Representative examples of newly identified enhancers (brown boxes) and those previously identified (pink boxes) are shown for Cyp310a1 (A), mirr (B), Traf4 (C), and Mef2 (D). Upper left panels show ChIP-chip data and lower left panels show ChIP-seq data for Twist-IP and control samples. In upper right panels, lateral views of whole mount in situ hybridizations of the endogenous genes of stage 5–8 embryos are shown. In lower right panels, lateral views of whole mount in situ hybridizations of similar staged embryos containing either cherry (for Traf4, mirr, and Cyp310a1 enhancers) or lacZ (for Mef2 5′ enhancer) reporter constructs.
Twist recognition motifs in vivo and in vitro
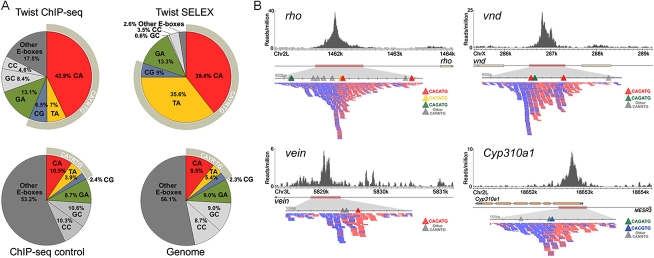
Twist belongs to a large bHLH family of DNA-binding factors that recognize a core DNA consensus, CANNTG, called an E-box (for review, see Massari and Murre 2000). Prior work using in vitro and in vivo approaches highlighted a subfamily preferred by Twist, led by CATATG (i.e., TA E-box). We asked which, if any, of the 10 possible E-box recognition motifs (counting reverse complements as the same motif) are selectively concentrated within 50 bp of called ChIP-seq signal summits (Fig. 2A). We found that CA and GA core E-boxes were most prominent, while GC, TA, and CG were relatively minor (Fig. 2A, “Twist ChIP-seq”). Compared with regions sampled from ChIP-seq control data or from the entire non-repeat genome, only CA, TA, CG, and GA core E-boxes were statistically enriched in Twist-occupied regions (Fig. 2A, colored slices). When larger radii from the ChIP signal summits were interrogated, the number of E-boxes of all types increased, and the specific enrichment trend was less apparent (i.e., enrichment of CA, TA, CG, and GA core E-boxes). In contrast, when ChIP-chip regions were similarly examined (Supplemental Figs. 5, 6), no specific enrichment of any motif was detected at any radius from the called Twist peaks. Overall, the enrichment and resolution results suggest that the ChIP-seq data could be used to model individual binding domains and causal motif instances in them (see below).
Figure 2.
A comparison of Twist in vivo and in vitro binding preferences. (A) The frequency of E-boxes associated with HC twist peaks (±50 bp), SELEX-bound sequences, ChIP-seq enriched control regions (±50 bp of summits), and the non-repeat dm3 genome was calculated. (B) Twist ChIP-seq data in the vicinity of CRMs shown to support expression of the genes rho (Ip et al. 1992b), vnd (Stathopoulos et al. 2002), vein (Markstein et al. 2004), and Cyp310a1 (this work). The directionality within ChIP-seq sequencing reads points to the position of the “explanatory” site. Blue and red ticks symbolize individual sequencing reads acquired, which match either the Watson or Crick strand.
Previously published foot-printing data and small-scale SELEX had found that the in vitro Twist protein binding consensus is CAYRTG (i.e., core E-box residues YR = TA, CG, and CA) (Ip et al. 1992b; Zinzen et al. 2006). To test how Twist in vivo binding results relate to in vitro preferences, we determined E-box frequencies in high-throughput Twist SELEX data, and compared them with our ChIP-seq data (see Supplemental Text). For the most part, the same E-boxes were highlighted, except that the TA-core E-box motif, which was the most highly bound by Twist in vitro (35.6% occupancy by SELEX), was less enriched in vivo (7% by ChIP-seq versus 5.3% frequency in the genome). A simple explanation is that there are real differences between the in vivo and in vitro binding conditions that affect Twist motif preference. Among alternative explanations, one or more species of bHLH heterodimers might be acting in vivo, while only homodimers were assayed in vitro (see Discussion).
Motif composition of Twist ChIP-seq regions
We examined the positions of all E-box motifs within Twist-ChIP-seq regions (Fig. 2B). The ChIP-seq protocol used here is a standard Illumina platform one that retains information about whether a sequenced fragment end originated from the Watson (red) or Crick (blue) strand (Fig. 2B; Valouev et al. 2008). With appropriate data preprocessing to account for fragment length (for review, see Pepke et al. 2009, see Methods), the summit location within each peak region can be identified computationally. Inspection of known Twist CRMs showed that this agrees well with, on average, 1–2 dominant binding motif instances within ±50 bp (e.g., see Fig. 2B). A subset of previously known Twist-bound regions consists of multiple peaks aggregated together, and these are typically associated with multiple Twist motifs (e.g., see Fig. 2B, vnd).
We mapped and visualized the position of each motif instance relative to its peak summit and calculated the cumulative frequency for each motif type as a function of distance from the peak (Fig. 3). Within the top ranked ∼1000 peaks the concentration of CAYRTG motifs was stronger than in lower ranked peaks, with CACATG sites, rather than CACGTG and CATATG, being most prominent near peak summits (Fig. 3B, top). Several criteria, including manual inspection of peaks throughout the ranking and the presence of previously studied Twist-dependent CRMs, led us to define a high confidence (HC) threshold of 513 regions (FDR 1%; see Methods and Supplemental Text); however we also found that binding motif centrality extends to ∼1000 sites in the genome, and for most analyses we use this more inclusive set of ∼1000 medium confidence (MC) calls (FDR 17%).
Figure 3.
Motif composition of Twist ChIP-seq regions shows preferential concentration of specific E-boxes near summits. (A) Locations of CAYRTG = CACATG CATATG and CACGTG E-box instances located within ±250 bp of the ChIP-seq peak (ERANGE-shifted called signal summit; see Methods) (y axis), plotted as a function of signal intensity rank from highest (1) to lowest (2000) (x axis). 1099 MC ChIP-seq data set is shown with a dashed line. CACATG is the most prevalent E-box motif in Twist ChIP regions and it shows the strongest central concentration. (B) Direct (top panel) and cumulative (bottom panel) motif density plots. In the MC data set, 65% of CACATG motifs and 50% of CAGATG occur within ±50 bp of Twist peaks. (C) CAGATG occurs more frequently in Twist ChIP-seq regions and is more centrally localized than (D). (D) CATATG is the motif most prominent in SELEX data (see text). (E) Other E-boxes (defined here as CANNTG motifs where NN is neither CA, GA, nor TA) display a more uniform distribution (B,E), though the other CABVTG E-boxes not pictured here (CG, GC, and CC) provide a minor central enrichment (see Supplemental Fig. 8). The number and distribution of explanatory E-boxes changes with ChIP-seq signal strength, suggesting that more E-boxes create a more robust Twist ChIP signal (A; Supplemental Fig. 7).
The accumulation of motif instances as a function of distance from the summit, over the entire set of Twist ChIP-seq regions, was analyzed (Fig. 3B, bottom). Using the K-S test, the P-value for CACATG distribution was defined as <2.2 × 10−16 (D = +0.44), meaning that the observed enrichment of CACATG near the peak summit is non-random and highly significant. It suggests that the CA-containing E-box drives in vivo binding at the majority of sites we called. Five other E-boxes also are enriched near summits, though they are less frequent in comparison to CACATG (Fig. 3B, top; Supplemental Fig. 8; Supplemental Table 3). In addition, the highest ranking peaks are associated with 2 or more matches to E-boxes; in particular the CACATG site is prominent (see Supplemental Fig.9).
CACATG and CATATG motifs are not functionally synonymous
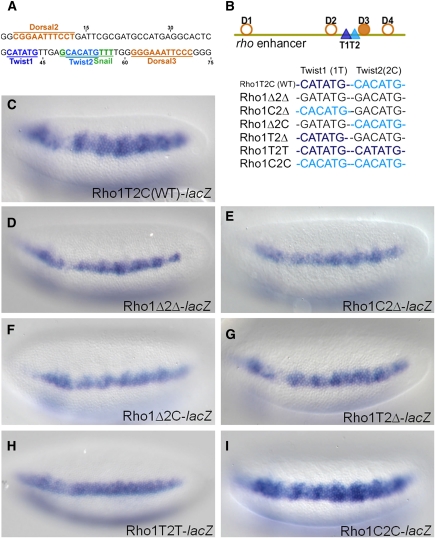
For many ChIP regions, detailed inspection of the primary data displayed in browser format confirms a single explanatory motif (e.g., vein CRM, Fig. 2B; Supplemental Fig. 10). However, some CRMs contain two or more closely spaced sites matching the CABVTG consensus, leading us to ask how closely positioned E-boxes interact. The rho early embryonic enhancer is such a case, with a highly directional single peak with two E-boxes sites (CATATG, T1, and CACATG, T2) separated by only 5 bp (Fig. 4A). We tested whether a series of enhancer constructs support expression in the lateral domain of the embryo, comparing the wild type CRM with Twist motif mutants.
Figure 4.
Mutagenesis of Twist binding sites at the ChIP-seq peak summit of rho enhancer. (A) The 75 bp sequence from the rho minimal enhancer which contains binding sites for Twist as well as for the transcription factors Dorsal and Snail. E-box sequences CATATG (T1, dark blue) and CACATG (T2, light blue) are separated by 5 bp, and Dorsal binding sites (orange) are positioned upstream and downstream of Twist sites. A Snail site that overlaps with T2 E-box is shown in green. (B) A diagram of the minimal 299 bp rho enhancer showing the relative positions of sites for Twist (dark and light blue triangles) and Dorsal (orange circles and filled circles, showing non-canonical and canonical sites, respectively). Lower schematic shows color-coded representations of the WT or mutant Twist binding sites present in various reporter constructs. Single nucleotide mutations were introduced into either T1 or T2 to eliminate binding (black: CATATG>GATATG or CACATG>GACATG) or to convert one site to the other (light blue: CATATG>CACATG or dark blue: CACATG>CATATG). (C) In situ staining of the wild type construct, minimal rho enhancer attached to the evep.lacZ reporter. (D) The Rho1Δ2Δ double mutant containing point mutations in both of the E-boxes, T1 and T2, supports reporter gene expression that is significantly weakened and more narrow compared to wild type (C). (E–G) Single mutations support expression that is weaker than wild type (C), more similar to the double mutant (D). (H) When a CATATG E-box is present in both the T1 and T2 positions, this change severely affects the expression domain of the reporter gene, reducing it to levels comparable to those observed in the double mutant Rho1Δ2Δ embryos (D). (I) When a CACATG E-box is present in both the T1 and T2 positions, the expression supported is comparable to the wild type (C).
Within the rho enhancer sequence, we introduced single-nucleotide changes to sites T1 and T2 (CANNTG→GANNTG). These subtle changes abrogated expression, such that instead of supporting expression in a wide domain (∼6–8 cells), the mutant enhancer supports expression in a more narrow domain (∼3–4 cells) (cf. Fig. 4D,C); this result is comparable to what others have found previously with more severe changes to the T1 and T2 E-box sequence (5 or more changes per site; Ip et al. 1992c). We also found that mutation of either site alone supported reporter gene expression, but neither was as severe as eliminating both (cf. Fig. 4E,F,G and 4C,D). This suggested that Twist binding to both T1 and T2 sites contributes to rho expression.
We then asked whether CA and TA E-boxes are interchangeable. When T1 and T2 are both CACATG (i.e., T1 site TA-core was converted into CA-core), reporter expression was comparable to wild type (Fig. 4I). In contrast, replacement of both sites by the CATATG was not sufficient to support expression over the full spatial domain (Fig. 4H); in fact, expression was comparable to the T2 mutant (Fig. 4G). This suggests that the CA E-box can function in both positions, while the TA E-box can function in T1 but not T2.
Motif discovery in Twist ChIP-seq regions
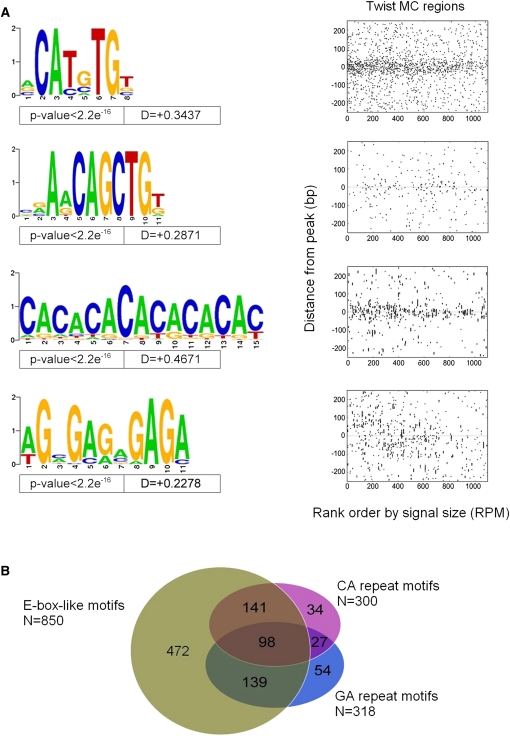
To uncover possible alternative Twist binding motifs or co-associated motifs for Twist-interacting factors, we used MEME, a motif discovery tool (Bailey et al. 2006), to search for statistically overrepresented motifs in and near Twist-occupied regions. As expected, prominent motifs found by MEME were E-box sequences (Fig. 5A) that overlap with CABVTG defined by our previous analyses (Fig. 3). In addition, MEME output highlighted residues flanking the E-box, such that a leading-A or lagging-T residue is preferred [e.g., CACATG-T (A-CATGTG) or A-CACATG (CATGTG-T)]. In contrast, a lagging A was very rare in Twist regions and in the genome at large (Fig. 5A). Other in vitro and in vivo bHLH binding studies support the idea that flanking bases may influence bHLH DNA binding (Grove et al. 2009; Cao et al. 2010).
Figure 5.
Motifs associated with Twist in vivo occupancy identified using MEME. MEME was run on the narrow 50 bp region surrounding each of the 1099 MC ChIP-seq peaks to identify all motifs that are enriched near the point of Twist occupancy. These motifs were mapped back to determine their spatial distribution relative to Twist peaks, and some motifs showing a non-uniform distribution near Twist peaks were selected. (A) Variations on CAYRTG and CAGCTG were returned, together specifying CABVTG (top two Weblogos). Note that a leading A residue or a lagging T residue is also suggested, which appears preferred by other non-Twist family DNA-binding bHLH factors (K Fisher-Aylor, S Kuntz, and A Kirilusha, unpubl. obs.; Grove et al. 2009). In addition, two simple repetitive sequences (CA and GA) are also significantly enriched at Twist-occupied sites (bottom two Weblogos). (B) Venn diagram illustrating the relationship between sets of peaks defined as having at least one occurrence of (i) either of the two E-box-like motifs; (ii) the CA-repeat-like sequence; or (iii) the GA-repeat-like sequence.
Several “simple” repeat sequences were significantly overrepresented in the Twist-occupied regions: the predominant one was a CA-repeat, and a similar GA-repeat sequence was also found (Fig. 5A). Of the 1099 peaks comprising the MC Twist ChIP-seq data set, 850 contain at least one match to either major E-box in the wide area around the peak (±250 bp), and 378 of these (or 44%) also contain at least one CA- or GA-repeat sequence (Fig. 5B). It is possible that the CA- and GA-repeats associated with Twist ChIP-seq peaks play some role in marking or phasing these regions as potentially “open chromatin”, as these same motifs were recently found associated with DNA occupied by Trithorax and Polycomb group/recruitment factors (see Schuettengruber and Cavalli 2009; and Discussion).
Interactions between Twist and other transcription factors might exist, yet not be identified by MEME for various reasons. We therefore tested additional motifs already known to bind transcription factors that pattern the DV axis in the early Drosophila embryo. Dorsal is a maternal transcription factor that functions cooperatively with Twist at some well-studied, closely-spaced sites (e.g., Ip et al. 1992c; Erives and Levine 2004), but the generality of this pattern across other Twist bound regions is not known. We found no significant global correlation between Dorsal motif occurrences and Twist peaks in our data. Among other factors (i.e., Su(H), Zelda, RGGNCAG/unknown, and Snail), only Snail exhibited significant motif co-enrichment in Twist ChIP regions, while Su(H) and RGGNCAG exhibited weak enrichment. The Snail result is neither surprising nor definitive because this factor can bind a sequence similar to that of Twist (Supplemental Fig. 12). Snail is thought to function as a repressor, at least in part, by competitively inhibiting binding of Twist (e.g., Ip et al. 1992b). Perhaps binding of both Twist and Snail to CRMs through the CA-core E-box plays a role that is more widespread than previously appreciated (see Discussion).
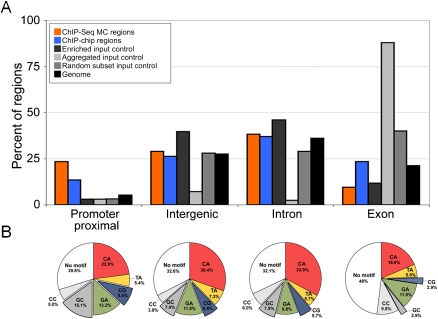
Twist-occupied regions were preferentially and significantly concentrated in proximal promoters (Fig. 6A), relative to several control samples, while intronic and intergenic classes were not enriched. Twist regions were slightly, but not significantly, depleted in exons. We tested whether the Twist regions near promoters were, more frequently than any others, lacking an explanatory E-box. This would be expected if many Twist promoter ChIP signals resulted from capture of indirect looping interactions from distant Twist-bound CRMS (e.g., Fullwood and Ruan 2009), rather than from primary motif binding, but it was not observed (Fig. 6B). We also asked if specific E-box motifs are selectively associated with any specific gene region class. Explanatory motifs at promoters showed higher CAGCTG and CACGTG E-box content, relative to intronic and intergenic groups, and a reduction in the dominant CACATG motif (Fig. 6B; Supplemental Fig. 13). These trends were not due to similar changes in the frequencies of GC, CG, or CA dinucleotides in promoters genome-wide (Supplemental Fig. 13). Exons also had distinctive signatures, presumably due to protein coding constraints.
Figure 6.
Enrichment of Twist ChIP-seq summits and explanatory E-box motifs in different genic and intergenic locations. (A) Enrichment of Twist ChIP-seq and ChIP-chip summits at particular positions in the genome, relative to a genome random sample and several sequencing negative controls. The genome was segregated into four mutually exclusive categories: promoter proximal (relative to the set of promoters from S. Celniker, including 500 bp upstream), exonic, intronic, and intergenic (see Supplemental Methods). While the majority of Twist regions fall into intergenic and intronic regions, there is a significant overabundance of Twist peaks in promoters relative to the amount of promoters in the genome (24%, or 258 of the ChIP-seq peaks). Intergenic and intronic Twist occurrences are comparable to that expected from a random genomic sample (29%, or 319 intergenic, and 38%, or 420 intronic). The number of summits within exonic regions is relatively disenriched (9%, or 102). In order to assess these numbers compared to expected values, we also compared the same number of Twist ChIP-chip regions (largest by area), the input control DNA regions enriched over Twist, the aggregated input DNA, and a random sampling of sequenced reads mapping uniquely to the genome (see Supplemental Text). We also report the total amount of the genome falling into each of these categories. The aggregated control and, to a lesser degree, the random control reads draw attention to the fact that there are many sequenced reads falling into exons. The enriched control does not show the exon bias perhaps because a directionality requirement was used; there is a mild enrichment of these sequences in the gene flanking category relative to the random genomic sample but a significant depletion in the promoter proximal that is likely due to the fact that Twist peaks are enriched at promoters. (B) The frequency of explanatory E-box sequences as a function of position of Twist-bound peaks in the genome (i.e., promoter proximal, intergenic, intronic, and exonic position). The CA, CG, and GA core E-boxes show enrichment in promoter, intergenic, and intronic positions; the GC core E-box is specifically enriched in the promoter proximal position.
Evolutionary conservation of ChIP-seq regions and motifs
Preferential sequence conservation is a signature of many biologically-significant regulatory regions and sequence motif instances. On average, our Twist-occupied regions were more conserved over a sequence domain of ∼300 bp compared to random genomic background conservation (blue versus red trace, Fig. 7A). In the HC Twist ChIP-seq data set of 513 peaks, conservation was highest over the motif when regions were centered on the explanatory CABVTG instance, and conservation gradually dropped to background levels as a function of distance from the center (green versus blue trace, Fig. 7A). Slight preferential conservation is observed in the background control sequence when they are aligned using the same set of E-boxes (cyan versus red trace, Fig. 7A). This is consistent with E-boxes being targets of a large family of transcription factors that exhibit varying degrees of motif preference. Furthermore, this regional conservation was less prominent in lower ranked peaks, suggesting that the higher ranked peaks are more likely to be functional (see Supplemental Fig. 14).
Figure 7.
Conservation analysis of sequences defined by Twist binding. (A) Averaged conservation profiles using phastCons scores for ChIP-seq regions and random genome samples. The blue curve shows average conservation in ChIP-seq peak regions is significantly elevated ±150–200 bp from the ChIP-seq signal summit. The green curve shows the same data but with regions recentered over the nearest CABVTG binding motif within 150 bp of the original summit. For the random sample, 500 regions containing one of the motifs were selected with the region start point selected at random for the uncentered distribution. Here “midpoint” refers to the location in the center of the randomly determined region. The error bar shows two standard deviations of 30 trials of 500 samples each. A maximum over the motifs is manifest, though substantially smaller than within the ChIP-seq peak regions. (B) Histogram of phastCons scores for bp occurring within the 6 E-box binding motif candidates (gray) compared to that for bp within the ChIP-seq regions, but outside any of the E-box motifs (black). Bp in the motif sites are found to be statistically more conserved than bp outside of motifs (0.005 significance level). (C) Fraction of sites in various sequence patterns falling within the top decile of phastCons scores for a 150 bp radius surrounding ChIP-seq summits versus the chi squared statistic for distributions within 150 bp of the summit compared to those of region 250–500 bp from the summit. CACATG, CATATG, and GA repeat sequences exhibit significantly greater conservation in ChIP-seq regions compared to flanking sequence than other motifs (as shown by their clustering at high values of the chi squared statistic), though CATATG and GA repeats do not exhibit high absolute levels of conservation.
To assess conservation of E-box sites more quantitatively, we compared the distribution of phastCons scores for inferred Twist binding motifs in peak domains (±150 bp from the ChIP-seq summit) to those for other sequences in the same regions (Fig. 7B). E-box motifs were significantly more conserved than the rest of the domain, suggesting that they are more functionally relevant than the average sequence around them. This supports the view that E-boxes in proximity to detected peaks are not only “explanatory” for binding, but that many of these have some function in vivo. The function implied by conservation may or may not occur during the embryonic stage at which we have made our measurements, and it is even possible that some are conserved due to binding by a different bHLH factor during the life of the animal.
We examined the degree of conservation of individual E-boxes of interest relative to one another and to CA and GA repeats that were found to be prevalent in the ChIP-seq signals. We sought to distinguish those with functions associated specifically with the Twist-occupied CRMs by comparison to flanking sequence, by comparing the fraction of conserved (phastCons > 0.9) motif occurrences within ±150 bp of the ChIP-seq summit to those in flanking regions 250–500 bp away from the summit (Fig. 7C); the latter is assumed to be statistically equivalent to genomic background from data in Figure 6A. We find that CATATG, CACATG, and GA repeats stand out in terms of the change in conservation between peak and flanking sequences. In contrast, CAGATG, CACGTG, CACCTG, and CA repeats show minimal change between peak and non-peak sequences.
Discussion
This analysis of in vivo Twist occupancy in the developing Drosophila embryo provides general and specific insights into relationships of Twist DNA binding motifs and in vivo Twist occupancy with regulatory function. We found that the in vivo consensus binding motif, as derived from Twist ChIP-seq data, is CABVTG (Figs. 2 and 5). Within that subfamily of E-boxes, CACATG is most prevalent within tested CRMs and is occupied preferentially within ChIP-seq defined peaks in general (Supplemental Tables 1 and 2; Fig. 3). Our detailed analysis of the rho enhancer showed that within the Twist-subfamily of E-boxes, individual members are not always interchangeable, and this suggests that they can support different functions (Fig. 4). When we searched for other motifs in addition to the E-box sequence that are associated with Twist peaks, we found that two repeat sequences, in particular, are associated with Twist ChIP-seq peaks, CA- and GA- repeat sequences, and that A/T-rich sequences are generally depleted from the region around ChIP signals (Supplemental Fig. 11). E-boxes and the over-represented motifs, in particular CACATG, CATATG, and a GA-repeat, are more conserved within peaks than background, suggesting that they have significant functions, presumably in transcriptional regulation.
We investigated the relationship between Twist occupancy and CRM regulatory activity by conducting functional tests and through analyses of conservation. Because the numbers of Twist-occupied sites we detected (500–1100) is large compared to the number of known Twist-regulated genes, it was not a foregone conclusion that most occupied regions would have any regulatory function. Our observed 74% CRM activity rate (23 positive CRMs of 31 tested) is high, and it argues that ChIP occupancy is efficiently highlighting functional regulatory DNA segments (Supplemental Table 2A); this analysis also captured the majority of gold standard enhancers identified by a number of previous studies (Supplemental Table 2B). Results showing preferential conservation of the Twist-bound cohort provide additional support for the idea that many other candidate regions that we did not test directly for function will also turn out to be CRMs.
A natural question is why the remaining ∼25% did not score as active enhancers to support gene expression. Simple biological possibilities are that some Twist occupancy is not associated with any regulatory activity; that the module's regulatory activity is to silence or to insulate, rather than to enhance; that the module is bound but is not active at this time in development (for review, see Levine and Tjian 2003; Arnosti and Kulkarni 2005; Gurudatta and Corces 2009; Cao et al. 2010). There are precedents for all these possibilities, although not all have been explicitly shown for Twist. Technical explanations are that CRM activity might not have been successfully captured in a segment tested, or that the original ChIP region calls include an unrecognized class of false positives.
Although our ChIP data efficiently identified CRMs, we emphasize that there is a distinction between significant in vivo Twist occupancy, as indicated by the ChIP-seq data, versus significant regulatory dependence on Twist, which appears to be rarer. Lower levels of regulatory dependency are, at present, difficult to measure, and they might be common. At the extreme, Twist-binding at most CRMs could be entirely opportunistic, arising by protein-protein interactions with other already bound factors and cofactors and/or binding to an E-box that has been made accessible by other unrelated factors nearby.
Incongruity between in vivo and in vitro preferred motifs
Our findings suggest that the TA-core and CA-core E-boxes are similarly preferential for Twist binding in vitro, but in vivo the Twist ChIP-seq explanatory sites are enriched in CA-core E-boxes. If Twist protein sees CA and TA motifs similarly, then the in vivo preference might simply reflect general base composition. When we specifically tested for this, the magnitude of CA enrichment in Twist bound E-boxes was much larger than in the non-coding genome at large (Supplemental Fig. 13). Alternatively, bHLH proteins are known to form heterodimers in addition to homodimers, and an explanation for CA differences is that Twist binding detected in vivo is a combination of homo- and heterodimers (e.g., Murre et al. 1989). The enrichment of CA core E-boxes in vivo could reflect a particular Twist–bHLH heterodimer, since ChIP will, in principle, recover any Twist-containing complex. In particular, there is some genetic interaction data that suggests that Twist and Daughterless (Da), a bHLH ubiquitously expressed in the embryo, may interact to affect patterning in the early embryo (Jiang et al. 1992; Gonzalez-Crespo and Levine 1993; Stathopoulos and Levine 2002). Other data with forced heterodimers showed that Twist can partner with Da at later stages to influence somatic mesoderm specification (Castanon et al. 2001). When we examined overlap between our Twist ChIP-seq binding events and that of Da ChIP-chip data available (Li et al. 2008), using relaxed criteria for overlap, we found 30% of our high confidence sites have some evidence for Da binding at the same locus. When the explanatory E-box instances for these regions from our data were interrogated, we found no positive correlation with CA core E-boxes and Da, but we did find a positive correlation with GC core E-boxes and possible Twist/Da co-occupancy (data not shown). Since other bHLH factors in the embryo might also partner with Twist, the specific role, if any, of heterodimers in this system will be speculative until the full partnering repertoire for Twist is quantified and characterized. It is also possible that post-translational modifications and local conditions in the nucleus that differ from the in vitro conditions affect DNA binding preferences.
Our mutagenesis experiments with the rho CRM further demonstrate that the TA-core and CA-core E-boxes are not equivalent, at least in some instances. What could be different about CA- versus TA-core E-boxes? CACATG and CATATG E-boxes (e.g., T1 and T2; see Fig. 4) were first identified as Twist-binding sites within the rho early embryonic enhancer in 1991 by Ip et al. (1992c) using in vitro footprinting. They showed that the CA-core E-box (but not TA-core) can also be bound by the repressor Snail. It is therefore possible that the preference we see for CA core E-boxes near ChIP-seq peaks indicates that Twist/Snail combined sites have been favorably selected, and that this combination site has a distinct role in regulating the activity of many CRMs in the early embryo. In 2002, the CA-core E-box was also found to be overrepresented in a small group of CRMs that specifically support expression in ventro-lateral domains of the embryo (Stathopoulos et al. 2002), and since then others have studied cooperativity between Twist and Dorsal binding (e.g., Erives and Levine 2004; Zinzen et al. 2006; Crocker et al. 2008). It might follow that the CA-core E-box is generally required to support cooperative interactions with Dorsal or with other collaborating factors, although we did not detect Dorsal motifs in most Twist ChIP-seq defined regions.
We favor the view that in the majority of regions the Twist motif highlighted by ChIP-seq is the one most likely to contribute to regulating gene expression (or other unidentified functions), but we cannot dismiss contributions from other E-box sites present in the region. Our experiments with the rho enhancer illustrate this, as both E-boxes CACATG and CATATG, located five nucleotides apart, affect gene expression. Within Twist ChIP-seq peaks, we find that TA core E-boxes are less frequent overall and only weakly enriched under peaks of binding (±250 bp from the peak summit), and as a result they are not often “explanatory” (<±50 bp from the peak summit). Yet these accessory TA core E-boxes may also contribute to regulating gene expression, whether by binding Twist more transiently or by interacting with some other factor. Because the CA core E-box is also bound by Snail, the balance of activation/repression may require that a combination of CA and TA core E-boxes is optimal to support expression. Furthermore, while Twist bound to the explanatory sites may serve a major role in regulating gene expression and these accessory sites may provide less input, even marginal input may be crucial to support gene expression patterns in ways that matter for viability and selection, even though some of these may also be too subtle for our assays to detect.
Simple sequence motifs and chromatin status
Apart from the CA- and GA-repeat sequences, no motifs other than the E-boxes were found to co-cluster with Twist binding sites in a large fraction of Twist-bound regions even when a wider window around the peaks of detected binding was interrogated. This does not preclude that other factors function in important combinations with Twist, but it suggests that no single transcription factor motif is commonly used in the entire Twist-occupied set. Finding specific combinations will require focus on subsets of regions selected by other criteria, such as expression pattern of nearby genes, performance of CRMs in transgenic assays, or direct binding assays for known or suspected accessory factors.
We do not know the significance of CA- and GA-simple repeat motifs that are enriched in Twist binding regions, but their association in other studies with open chromatin regions is suggestive (Auerbach et al. 2009). We hypothesized that GAGA-binding factor (GAF) which binds to promoters (for review, see Lehmann 2004) might do so here in promoter proximal regions through recognition of the GA-repeats. However, we did not find an enrichment of GA-repeat sequences associated with promoter proximal Twist peaks; the GA-repeats were located in many different positions suggesting a broader role than regulation of promoters, such as making DNA regions accessible.
Depletion of A/T-rich sequences from peaks was striking and it proved to be non-specific, as it is associated with a multitude of ChIP-seq samples. Further analyses showed there is a similar depletion of A/T-rich sequences around ChIP-seq peaks for diverse factors and in multiple genomes, including worm, mouse, and human (Supplemental Fig. 15; K Fisher-Aylor and B Wold, unpubl. obs.). This depletion was also seen when “peaks” of reads were selected from matching control samples of input chromatin (cross-linked, sheared, and reverse cross-linked). The sonication step associated with ChIP-seq has recently been shown to enrich for promoter regions, DNase I hypersensitive sites, and other “open” chromatin regions (Auerbach et al. 2009), but in that work no specific sequence content biases were reported. The depletion of A/T rich runs might arise from a role these sequences have been suggested to play in nucleosome exclusion and positioning (e.g., Iyer and Struhl 1995; Peckham et al. 2007). Our observations of broad A/T depletion arose from a study of motif representation that happened to be A-rich (Supplemental Fig. 11), and it suggests that careful examination of background input chromatin is needed when evaluating the sequence composition of ChIP regions.
The conservation profile around explanatory Twist motifs implies CRMs of ∼300 bp
The genomes of Drosophilids are known to exhibit more conservation, in general, than many other animal species separated by what are thought to be an equivalent length of evolutionary distance. Thus, it has proven difficult to identify putative CRMs based on a simple search for increased local conservation of non-coding DNA sequence among Drosophilid genomes. Early comparative studies of enhancer regions in Drosophila species suggested that local increases in conservation of non-coding sequence imply regulatory function (Bergman et al. 2002). More recently, it has been suggested that this idea should be narrowed to conservation of specific binding sites only within CRMs or even just conservation of site number without strong primary sequence conservation (Sosinsky et al. 2007; Ho et al. 2009; Liberman and Stathopoulos 2009). Here we provide evidence to support both views: increased general conservation of sequence within putative CRMs relative to genomic background, as well as higher conservation of particular binding sites (Fig. 7). We asked if there is a genome-wide average conservation signature that would characterize candidate CRMs; ChIP-chip data previously detected a conservation preference but without clarity about the dimensions of regions under selective pressure (MacArthur et al. 2009). Our data suggests that sequences around these motif instances are preferentially conserved compared with genomic background in a window of ∼300 bp on average, a size that corresponds well with anecdotal samplings of individual CRMs. We also found evidence that the explanatory sites identified by Twist binding are preferentially conserved compared with their surroundings, arguing for their biological salience.
Methods
Fly stocks and general molecular biology
Drosophila melanogaster fly stocks were reared under standard conditions at 25°C. Transgenic flies were obtained using standard P-element transformation or by site-directed integration. Wild type refers to the background yw. P-element transformations were achieved in yw flies, while site-directed integration was carried out using D. mel stock containing attP insertion at position ZH-86Fb. Enhancer sequences were amplified from genomic DNA (primer sequences are available upon request) and cloned into eve.promoter-LacZ-attB or eve.promoter-cherry-attB vectors (Liberman and Stathopoulos 2009). Anti-sense riboprobes labeled with Digoxigenin-UTP (Roche) were used for in situ hybridization to detect transcripts.
Chromatin preparation, DNA isolation, amplification, hybridization, and sequencing
Chromatin was prepared as described previously (Sandmann et al. 2006) from 2 g of yw embryos of from 1 to 3 h in age. Rat anti-Twist antibody (gift of M. Levine, UC Berkeley) was used for both ChIP-chip and ChIP-seq experiments. For ChIP-chip, the resulting DNA library was labeled and hybridized to arrays by NimbleGen Systems, Inc.; 10 ng of immunoprecipitated (IP) DNA was amplified using the Whole Genome Amplification kit (Sigma) according to the manufacturer's instructions. The mock ChIP-chip sample used preimmune antibody, rather than anti-Twist. For ChIP-seq, 50 ng of IP material was used to prepare a library (Johnson et al. 2007), and DNA sequencing of samples was performed by the Illumina protocol at Caltech Genome Center. The ChIP-seq input control was processed equivalently to the Twist ChIP-seq sample, except that it was not immunoprecipitated (no antibody or bead processing). Each ChIP-seq library was sequenced to a total of 9 million reads.
SELEX
SELEX experiments using in vitro binding to a column were carried out as described (Ogawa and Biggin 2011). See the Supplemental Text for more details, including processing of SELEX data.
Bioinformatics
ChIP-chip and ChIP-seq data processing: Methods used to call ChIP-chip versus ChIP-seq peaks are described in detail within the Supplemental Text. In brief, we used the ERANGE software suite to call peaks based on the number, orientation, and ratio of short sequenced reads relative to a background control. We considered an alternate peak caller (MACS), overlap of ChIP-seq regions with ChIP-chip regions, and the inclusion of known Twist targets to determine the threshold for calling Twist occupied sites (i.e., ChIP-seq signals). We selected a high confidence (HC) set of 513 sites based on high inclusion in ChIP-chip regions (87%), MACS regions (72%), and validated Twist targets (75%). We also selected a medium confidence (MC) set of 1099 regions based on the similarity in motif organization around these peaks (E-box, Fig. 3A).
ChIP-seq summit refinement
After ChIP-seq enriched regions were identified by the ERANGE program, post-processing was performed to refine the summit location by utilizing directional tag information. For each peak region, plus and minus tags were simultaneously shifted toward the imputed fragment center by a trial amount, ranging from 0 to 100 bp. The shift that maximized area overlap of the plus and minus tag density profiles (i.e., a measure of “directionality”) was then implemented prior to calculating the location of the ChIP-seq tag count maximum (“summit”).
Explanatory site interval
The interval for designating “explanatory sites” near ChIP-seq summits was estimated utilizing count statistics for the CACATG motif, due to its being the most prevalent E-box in the set of Twist regions. Specifically, the motif occurrences within increasing radii around peak centers (binned by 5 bp) were compared to the number expected from a Poisson distribution with the mean equal to the genome average density of CACATG motifs. When the probability of the observed number of counts coming from the Poisson model fell below 0.001, the distribution was deemed indistinguishable from random fluctuations, and the boundary of the previous bin was set to be the cutoff for explanatory sites (±50 bp from the summit).
Conservation analysis
Conservation at each base pair was assessed using phastCons scores (Siepel et al. 2005). Genome-wide scores for the fifteen-way insect alignment including D. melanogaster, D. simulans, D. sechellia, D. yakuba, D. erecta, D. ananassae, D. pseudoobscura, D. persimilis, D. willistoni, D. virilis, D. mojavensis, D. grimshawi, A. gambiae, A. mellifera, and T. castaneum were downloaded from the UCSC genome gateway. Statistical analysis of the data is described in the Supplemental Methods.
Annotations
Precomputed annotation files for exons and introns were downloaded from the FlyBase website, release 5.27 (Tweedie et al. 2009). Here, exons and introns are mutually exclusive. 5′ UTRs data are from S. Celniker.
Acknowledgments
We thank the Caltech Jacobs Genome Facility members I. Antoshechkin and L. Schaeffer for library building and DNA sequencing, as well as D. Trout, B. King, and H. Amrhein for primary sequence data processing and visualization. We are grateful to A. Mortazavi and A. Kirilusha (Caltech Biology) for software and discussion of analysis; M. Biggin and S. Celniker (Lawrence Berkeley Lab) for sharing unpublished data; and M. Levine (University of California at Berkeley) for antibodies. K.I.F.-A. was funded by a NSF pre-doctoral fellowship, and S.P. was funded by The Gordon and Betty Moore Foundation. Work at Lawrence Berkeley National Laboratory was conducted under Department of Energy contract DE-AC02-05CH11231. This work was funded by the Functional Genomics Resource Center of the Caltech Beckman Institute, NIH grant R01GM077668 (A.S.), NIH grant U54HG004576 (B.J.W.), and the Bren Chair (B.J.W).
Footnotes
[Supplemental material is available for this article. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE26285, and the sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA027330.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.104018.109.
References
- Arnosti DN, Kulkarni MM 2005. Transcriptional enhancers: Intelligent enhanceosomes or flexible billboards? J Cell Biochem 94: 890–898 [DOI] [PubMed] [Google Scholar]
- Auerbach RK, Euskirchen G, Rozowsky J, Lamarre-Vincent N, Moqtaderi Z, Lefrancois P, Struhl K, Gerstein M, Snyder M 2009. Mapping accessible chromatin regions using Sono-Seq. Proc Natl Acad Sci 106: 14926–14931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW 2006. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–373 (Web Server issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman CM, Pfeiffer BD, Rincon-Limas DE, Hoskins RA, Gnirke A, Mungall CJ, Wang AM, Kronmiller B, Pacleb J, Park S, et al. 2002. Assessing the impact of comparative genomic sequence data on the functional annotation of the Drosophila genome. Genome Biol 3: RESEARCH0086 doi: 10.1186/gb-2002-3-12-research0086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci 99: 757–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. 2010. Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev Cell 18: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon I, Von Stetina S, Kass J, Baylies MK 2001. Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 128: 3145–3159 [DOI] [PubMed] [Google Scholar]
- Chopra VS, Levine M 2009. Combinatorial patterning mechanisms in the Drosophila embryo. Brief Funct Genomics Proteomics 8: 243–249 [DOI] [PubMed] [Google Scholar]
- Crocker J, Tamori Y, Erives A 2008. Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol 6: e263 doi: 10.1371/journal.pbio.0060263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH 2006. The regulatory genome: Gene regulatory networks in development and evolution. Academic, Burlington, MA [Google Scholar]
- Erives A, Levine M 2004. Coordinate enhancers share common organizational features in the Drosophila genome. Proc Natl Acad Sci 101: 3851–3856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Ruan Y 2009. ChIP-based methods for the identification of long-range chromatin interactions. J Cell Biochem 107: 30–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Crespo S, Levine M 1993. Interactions between dorsal and helix-loop-helix proteins initiate the differentiation of the embryonic mesoderm and neuroectoderm in Drosophila. Genes Dev 7: 1703–1713 [DOI] [PubMed] [Google Scholar]
- Grove CA, De Masi F, Barrasa MI, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJ 2009. A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell 138: 314–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurudatta BV, Corces VG 2009. Chromatin insulators: Lessons from the fly. Brief Funct Genomics Proteomics 8: 276–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho MC, Johnsen H, Goetz SE, Schiller BJ, Bae E, Tran DA, Shur AS, Allen JM, Rau C, Bender W, et al. 2009. Functional evolution of cis-regulatory modules at a homeotic gene in Drosophila. PLoS Genet 5: e1000709 doi: 10.1371/journal.pgen.1000709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip YT, Levine M, Small SJ 1992a. The bicoid and dorsal morphogens use a similar strategy to make stripes in the Drosophila embryo. J Cell Sci Suppl 16: 33–38 [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Bier E, Levine M 1992b. The dorsal gradient morphogen regulates stripes of rhomboid expression in the presumptive neuroectoderm of the Drosophila embryo. Genes Dev 6: 1728–1739 [DOI] [PubMed] [Google Scholar]
- Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M 1992c. dorsal–twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev 6: 1518–1530 [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J 14: 2570–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Rushlow CA, Zhou Q, Small S, Levine M 1992. Individual dorsal morphogen binding sites mediate activation and repression in the Drosophila embryo. EMBO J 11: 3147–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science 316: 1497–1502 [DOI] [PubMed] [Google Scholar]
- Landolin JM, Johnson DS, Trinklein ND, Aldred SF, Medina C, Shulha H, Weng Z, Myers RM 2010. Sequence features that drive human promoter function and tissue specificity. Genome Res 20: 890–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M 2004. Anything else but GAGA: A nonhistone protein complex reshapes chromatin structure. Trends Genet 20: 15–22 [DOI] [PubMed] [Google Scholar]
- Levine M, Tjian R 2003. Transcription regulation and animal diversity. Nature 424: 147–151 [DOI] [PubMed] [Google Scholar]
- Li XY, MacArthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Luengo Hendriks CL, et al. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol 6: e27 doi: 10.1371/journal.pbio.0060027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LM, Stathopoulos A 2009. Design flexibility in cis-regulatory control of gene expression: Synthetic and comparative evidence. Dev Biol 327: 578–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk RW, Eisen MB 2010. Evolutionary mirages: Selection on binding site composition creates the illusion of conserved grammars in Drosophila enhancers. PLoS Genet 6: e1000829 doi: 10.1371/journal.pgen.1000829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, et al. 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol 10: R80 doi: 10.1186/gb-2009-10-7-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci 99: 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M 2004. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development 131: 2387–2394 [DOI] [PubMed] [Google Scholar]
- Massari ME, Murre C 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. 1989. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58: 537–544 [DOI] [PubMed] [Google Scholar]
- Nam J, Dong P, Tarpine R, Istrail S, Davidson EH 2010. Functional cis-regulatory genomics for systems biology. Proc Natl Acad Sci 107: 3930–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa N, Biggin MD 2011. High-throughput SELEX determination of DNA sequences bound by transcription factors in vitro. In Methods in molecular biology (ed. Deplanke B), Humana Press, Clifton, New Jersey: (in press). [DOI] [PubMed] [Google Scholar]
- Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z 2007. Nucleosome positioning signals in genomic DNA. Genome Res 17: 1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A 2009. Computation for ChIP-seq and RNA-seq studies. Nat Methods 6 (11 Suppl): S22–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves GT, Stathopoulos A 2009. Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol 1: a000836 doi: 10.1101/cshperspect.a000836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Jakobsen JS, Furlong EE 2006. ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat Protoc 1: 2839–2855 [DOI] [PubMed] [Google Scholar]
- Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE 2007. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev 21: 436–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G 2009. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136: 3531–3542 [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M 1992. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J 11: 4047–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky A, Honig B, Mann RS, Califano A 2007. Discovering transcriptional regulatory regions in Drosophila by a nonalignment method for phylogenetic footprinting. Proc Natl Acad Sci 104: 6305–6310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2002. Linear signaling in the Toll-Dorsal pathway of Drosophila: Activated Pelle kinase specifies all threshold outputs of gene expression while the bHLH protein Twist specifies a subset. Development 129: 3411–3419 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Levine M 2005. Genomic regulatory networks and animal development. Dev Cell 9: 449–462 [DOI] [PubMed] [Google Scholar]
- Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M 2002. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell 111: 687–701 [DOI] [PubMed] [Google Scholar]
- Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. 2009. FlyBase: Enhancing Drosophila gene ontology annotations. Nucleic Acids Res 37: D555–D559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A 2008. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods 5: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M 2007. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev 21: 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen R, Senger K, Levine M, Papatsenko D 2006. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol 16: 1358–1365 [DOI] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE 2009. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70 [DOI] [PubMed] [Google Scholar]