Abstract
MicroRNAs (miRNAs) add a previously unexpected layer to the post-transcriptional regulation of protein production. Although locked nucleic acids (LNAs) reveal the distribution of mature miRNAs by in situ hybridization (ISH) experiments in zebrafish and other organisms, high cost has restricted their use. Further, LNA probes designed to recognize mature miRNAs do not distinguish expression patterns of two miRNA genes that produce the same mature miRNA sequence. Riboprobes are substantially less expensive than LNAs, but have not been used to detect miRNA gene expression because they do not bind with high affinity to the short, 22-nucleotide-long mature miRNAs. To solve these problems, we capitalized on the fact that miRNAs are initially transcribed into long primary transcripts (pri-mRNAs). We show here that conventional digoxigenin-labeled riboprobes can bind to primary miRNA transcripts in zebrafish embryos. We tested intergenic and intronic miRNAs (miR-10d, miR-21, miR-27a, miR-126a, miR-126b, miR-138, miR-140, miR-144, miR-196a1, miR-196a2, miR-196a2b [miR-196c], miR-196b, miR-196b1b [miR-196d], miR-199, miR-214, miR-200, and miR-222) in whole mounts and some of these in histological sections. Results showed that pri-miRNA ISH provides an attractive and cost-effective tool to study miRNA expression by ISH. We use this method to show that miR-126a and miR-126b are transcribed in the caudal vasculature in the pattern of their neighboring gene ci116 or host gene egfl7, respectively, and that the chondrocyte miRNA mir-140 lies downstream of Sox9 in development of the craniofacial skeleton.
Introduction
Research in the last decade has shown that microRNAs (miRNAs) regulate embryological development and physiological function in zebrafish and other organisms and that disregulation of miRNAs can lead to disease.1–12 miRNAs are about 22 nucleotides (nt) long and generally act by binding to complementary sequences in target messenger RNAs, often in the 3′ untranslated region (3′UTR), which either suppresses translation or enhances transcript degradation.13 RNA polymerase II, which transcribes protein-coding genes, transcribes most miRNA genes into long primary transcripts (pri-miRNAs), but RNA polymerase III, which transcribes transfer RNA, transcribes other miRNA genes.14,15 In the nucleus, the enzyme Drosha processes pri-miRNA transcripts to shorter precursor-miRNAs (pre-miRNAs) and after export from the nucleus, the enzyme Dicer processes pre-miRNAs to mature forms (miRNAs).16,17 Mice lacking Dicer die before axis formation, suggesting that Dicer activity and mature miRNAs are crucial for patterning and morphogenesis in mammalian embryos.18 Zebrafish mutants lacking Dicer form an axis and several cell types but show disturbed morphogenesis of many organ systems,19–21 demonstrating the general importance of miRNAs for zebrafish development.20,22 As yet, we know the function of few individual miRNAs in zebrafish development.
The investigation of miRNA function in zebrafish can lead to insights into the mechanisms of human disease. For example, overexpression of miR-140 was found to lead to cleft lip and cleft palate in zebrafish,7 and subsequently, a single-nucleotide polymorphism located in pre-miRNA140 was found to be associated with nonsyndromic cleft palate in humans and to act by altering miR-140 processing.23
Due to two whole genome duplication events in early vertebrate phylogeny24,25 and to an additional genome duplication event in the teleost lineage,26–28 identical or very similar sequences of many mature miRNAs are encoded by two or more duplicated genes in zebrafish and other vertebrates (see miRNA collection at miRBase [http://miRbase.org]).29–31 Zebrafish, for example, has five different genes for mir-10 and five for mir-196 that are located in hox clusters.2,32,33 Because miRNAs act only in cells in which they are expressed, the careful description of a miRNA's expression pattern over time, when compared to the expression of protein-coding genes in those same cells, can give important clues to a miRNA's targets, and hence, to miRNA functions. It is not yet known whether primary transcripts of miRNAs accumulate to sufficiently high levels or have a sufficiently long lifetime to be detected by in situ hybridization (ISH) using techniques similar to those used to detect expression for protein coding genes.34,35
Despite the importance of knowing which tissues express which miRNA genes at what time, the short length of miRNAs poses a problem for detecting miRNAs by ISH in zebrafish embryo whole mounts. Because mature miRNAs are just 22 nts long and are often contained in cells at low levels, typical ISH riboprobes do not give a detectable signal in ISH experiments. To overcome this problem, various methods have been used to amplify the signal. One method uses modified nucleic acid probes to increase affinity and specificity; such probes include radioactively labeled synthetic RNA oligonucleotide probes, 2′-O-methyl RNA oligonucleotides, and locked nucleic acids (LNA).36–38 The most successful method so far uses LNA probes, in which the ribose of the nucleotide contains a bridge connecting its 2′ oxygen to its 4′ carbon that locks the ribose in the 3′-endo conformation, thereby increasing base stacking forces and backbone preorganization.37,39 This modification significantly increases the specificity and sensitivity of binding to mature miRNAs. Hybridization to LNAs has revealed expression patterns for over 100 miRNAs in zebrafish, medaka, and chicken embryos.40–43 LNA probes are so specific that they can distinguish single-nucleotide differences in mature miRNAs.37,44–47
Unfortunately, LNAs and similar probes are too expensive for most laboratories to use routinely for the study of a large number of miRNAs. In addition, because most LNA probes are designed against mature miRNAs, they do not distinguish identical miRNAs that arise from different miRNA genes, for example, miR-196a1 and miR-196a2 in zebrafish. To develop a cost-effective method to detect miRNA gene expression in zebrafish embryos by ISH and to distinguish between genes that encode the same mature miRNA sequence, we developed the use of digoxigenin-labeled riboprobes designed to bind to miRNA primary transcripts. Results showed that digoxigenin-labeled probes directed against pri-miRNAs permit the efficient observation of miRNA expression for both intergenic and intronic miRNAs; further, we show that the method works in both whole mounts and histological sections. In addition, pri-miRNA ISH (PriMiSH) can distinguish among miRNA genes that produce the same mature miRNA sequence. We used this method to test the hypothesis that the transcription factor Sox9 is upstream to miR-140 in a developmental regulatory hierarchy. We conclude that ISH to pri-miRNA provides an important alternative for investigating miRNA expression with several advantages over current methodologies.
Materials and Methods
Animals
Wild-type fish were from an ABC/TU hybrid stock and sox9 mutants were as described.48 All experiments involving animals used protocols approved by the University of Oregon IACUC.
Nomenclature
miRNA nomenclature appears to be in a state of flux. MiRBase (www.mirbase.org/) and Ensembl (www.ensembl.org/index.html) use the form dre-mir-140 to indicate the Danio rerio gene encoding miR-140. The human symbol was MIRN140, until recently, when it changed to MIR-140, according to the Entrez Gene database (www.ncbi.nlm.nih.gov/gene). ZFIN (http://zfin.org) gives the official zebrafish gene name as mir-140. The mature miRNA we call miR-140 and the primary transcript we call pri-miR-140.
Cloning
Polymerase chain reaction (PCR) primers for intergenic miRNA primary transcripts were designed to span 300–700 nt of genomic DNA centered approximately on the mature miRNA sequence as given for the zebrafish genome at Ensembl (Zv8). PCR primers for intronic miRNA primary transcripts were also centered on the miRNA and were designed to include sequence from the intron but to exclude sequence from flanking exons. Because fewer than 200 nts separate the polycistronic sequences for miR-200a and miR-200b, primers for these miRNAs encompassed sequences encoding both miR-200a and miR-200b. To generate PCR products containing partial miRNA primary transcripts or protein-coding mRNAs, we used 1 day postfertilization zebrafish whole-embryo cDNA that was reverse transcribed with oligo-dT primer. The amplification of a band of the predicted size from cDNA confirmed that primers were within the primary transcript. Cloning of miRNA primary transcripts and protein coding genes used the following primers: mir-10d + 1719 TTTATTTAGCCCTTATGTTTTATTAGTCG and mir-10d2288 TAGAGAAGTGCATGCATTTTAGTCCTGTT; mir-21 + 777 TGAAAACGCGGCCACTATGAGAAAG and mir-21-1186 TGGCGACGCTAAAATAAGACAACAATACA; mir-27a + 272 AAGTTGAAGAAACAAAGCGTATGT and mir-27a566 TGTCCTTTTTCCGGTTTTGCTCT; mir-126a + 677 GTAATCAACCTCACAACAATCGCCTCAC and mir-126a1509 ACATGCCCTTTATTTCCTCCAGATTTTA; mir-126b + 399 TATGATGGCGGTGTATGAGAATGTGT and mir-126b1192 GTGTTTGCTTTGCTTGGACTTTTT; mir-138 + 604 TCAGCAGATGGTGGGTTATGGAGGAA and mir-138-1137 ATGTGACACTAAAGACTGGAGCAATGG; mir-140 + 507 GCAAGTCAAACCCTGTAGCATCCCGTT and mir-140-1386 GCGAGCCGATAGAGCGATTGTTT; mir-144 + 560 CGGGACGACTGAAGACGCACAT and mir-144-1338 TCTTATTTCATTTGGCAGCAGCAGTTA; mir-196a1 + 523 ATTAAATGAACGCTAGCGGCTGTATGATG and mir-196a1-1014 TTTTGCTAGCGCTTTGTCTTTGTAACCA; mir-196a2 + 1349 GCAGACAGGAGAGCGGCAAGAA and mir-196a2-1891 AGCAGGCAAGGCAAGATTATGGTA; mir-196b + 756 GTATCTCTTTGCCCCGCTGTGG and mir-196b1292 TGGAAAAACGATGGGAAAGTATTG; mir-196a2b(mir-196c) + 467 TATGCTACCTGGTGCCGTGAAG and mir-196a2b(mir-196c)-1325 CCGCTGATAATGGAAGACAACC; mir-196b1b(mir-196d) + 1016 ATTGCTTTAGATTATGCGCGGGTATTT and mir-196b1b(mir-196d)-1339 CAAGCTATGTCAAGGCGTGTCTGTCT; mir-200ab +452 GCATTATTACTTTGAGACTTTGTGTT and mir-200ab888 ACGAGCCCTGATGTGGTTTTT; mir-222 + 1003 GTTCGGGACGTCTGGAGTGGA and mir-222-2064 GGGCTTCAGGCTTTCTTCATTAGTT; wwp2ex + 395 AGTCGTCTCGGGGCTCGTGTG and wwp2ex-1160 ACCCCCTCTGCCGTGTACTTCATCT; wwp2int + 1562 GGTCTGTGTTGTTTTTGGAGGTCGTT and wwp2int-2519 ACAAGCAAAGGTAAAAGCAAGAGTCATAA; ci116(LOC557793) + 1217 TGAGGATTTGGCCGGATGGTGTTA and ci116(LOC557793)-1933 TGAGCGAGGAGCCCGTAGAGGAGT; egfl7 + 43 GCGCGGTGCTCTTCATCAG and egfl7-687 AGCCTCCTTACACTCGTCCACATC.
In situ hybridization
PCR products for riboprobes were cloned into pCR4-TOPO (Invitrogen) vector and ISH was as described.7 Antisense LNA probe for miR-196a with the sequence 5′Dig/CCCAACAACATGAAACTACCTA/3′Dig was ordered from Exiqon (LNA modification is not disclosed by Exiqon; http://Exiqon.com) and ISH was according to the manufacturer. Antisense LNA probe for miR-140 with sequences 5′Dig/CtACcATaGGgTAaAAcCAcTG-3′ (lowercase nucleotides represent LNA nts) was ordered from IDT (www.idtdna.com) and the ISH protocol was the same as for miR-196a LNA.
Embryos were fixed in 4% paraformaldehyde (PFA) overnight and dehydrated in a sequential methanol gradient for long-term storage. Before hybridization with riboprobes for pri-miRNAs or for protein coding genes, embryos were rehydrated and equilibrated in hybridization buffer for 4 h at 68°C. Hybridizations were performed at 68°C overnight. ISH using LNA probes was similarly equilibrated in hybridization buffer for 4 h and hybridized overnight at 42°C–45°C (21°C–25°C lower than the calculated Tm of LNA probes, as indicated by LNA probe manufacturers). After hybridization, embryos were washed in saline-sodium citrate (SSC) buffer and equilibrated in blocking buffer for 4 h at room temperature and then incubated with anti-digoxigenin-AP, Fab fragment at 4°C overnight. After antibody incubation, embryos were washed in phosphate-buffered saline (PBS) containing 0.3% Tween-20 for at least 2 h and then were incubated in 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium chloride (NBT). Expression of protein coding genes can sometimes appear in as little as 30 min, but pri-miRNA or LNA probe for miRNAs may require staining for up to 3 days. After ISH, zebrafish embryos were mounted in 100% glycerol and images were captured using a compound microscope. Although PriMiSH utilizes a lower hybridization temperature than riboprobe ISH for messenger RNAs, the rest of the method and solutions are those commonly used for regular riboprobes in zebrafish whole-mount ISH.34,35
For ISH to miRNA primary transcripts on histological sections, fish were fixed in 4% paraformaldehyde in PBS overnight at 4°C, then rinsed in PBS buffer, and embedded in the embedding medium (agar, agarose, and sucrose [9:10:50]). Cryostat sections were cut at 6 μm and transferred to Fisherbrand Superfrost/Plus microscope slides and air-dried overnight at room temperature. Hybridization was carried out overnight at 68°C for PriMiSH probes or at 42°C–45°C for LNA probes. After hybridization, sections were washed three times with 1 × SSC, 50% formamide, and 0.1% Tween-20 at 68°C. Digoxigenin-conjugated antibody was used to amplify the signal. Sections with antibody solution (1:5000 dilution of anti-dig in blocking solution) were incubated at 4°C over night and observed using NBT (3.5 μL/mL) and BCIP (2.6 μL/mL) incubated on sections at 37°C overnight. After staining, sections were washed twice in ddH2O, dehydrated quickly through a graded ethanol series to 100% ethanol, and cleared in xylene (or Histoclear) and coverslipped in Permount for imaging.
Results
pri-miRNA probe design
Most miRNA genes reside in intergenic regions (between two protein coding genes, like the zebrafish mir-10 family), but up to 25% of mammalian miRNA genes and at least 7% of zebrafish miRNA genes lie in introns.49 Intergenic miRNA genes usually use their own transcriptional machinery independent of neighboring protein-coding genes.49 Intronic miRNA genes are often encoded in the same direction as their host genes and are usually spliced out from the host gene's intron during maturation of the host gene transcript. Some intronic miRNA genes, however, are transcribed in the direction opposite to that of their host genes; such pseudo-intronic miRNA genes are similar to the intergenic miRNA category because they use their own transcriptional machinery and can be transcribed independent of their host genes.49 To construct riboprobes for pri-miRNAs, we used genomic DNA sequence as reference to design PCR primers to amplify a product of about 300 nt to 700 nt centered on the mature miRNA sequence. These primers amplified sequences corresponding to the presumed pri-miRNA using cDNA template extracted from 1-day-old intact zebrafish embryos. These pri-miRNA fragments were cloned and used to synthesize antisense digoxigenin-labeled probes. We used 17 PCR primer pairs to amplify pri-miRNA fragments from embryonic cDNAs, 14 of which produced probes that readily detected signal in ISH experiments on zebrafish embryos.
Intergenic pri-miRNAs
Several genes encoding miR-196 sequences reside between protein-coding genes for vertebrate Hox cluster paralogy groups 9 and 10.2,50 The human genome has three paralogous MIR-196 genes (MIR-196B, MIR-196A1, and MIR-196A2) that are located in the HOXA, HOXB, and HOXC clusters, respectively, and that originated in the vertebrate genome duplication event. Due to the teleost genome duplication, zebrafish has five intergenic copies of mir-196; as in humans, these genes are located between hox9 and hox10 genes in the hoxaa, hoxab, hoxba, hoxca, and hoxcb clusters, respectively.2,33,50,51 Because mir-196a1 and mir-196a2 encode the same miR-196a sequence, these five genes produce just four mature miR-196 molecules. To learn whether riboprobes can recognize intergenic pri-miRNAs and if riboprobes can distinguish between two transcripts encoding the same mature miRNAs, we designed PCR primers for mir-196a1 and mir-196a2 that cover 517 nt and 566 nt of genomic DNA, respectively, with the sequence encoding the mature miRNA roughly in the middle (Fig. 1A). Reverse transcription–PCR on embryo RNA amplified bands of the predicted sizes, thus verifying that primer sequences are located within the pri-miR-196a transcripts.
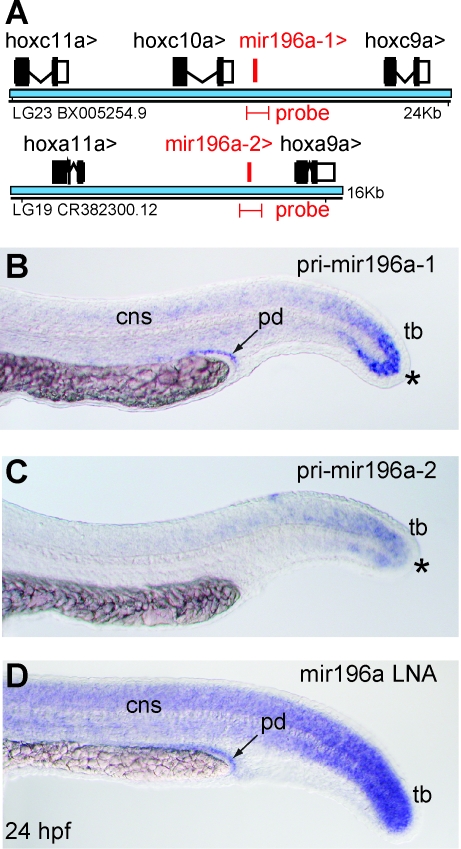
FIG. 1.
Expression of intergenic mir-196a paralogs observed by digoxigenin-labeled pri-miRNA probes. (A) The intergenic genes mir-196a1 and mir-196a2 lie between hoxc9a and hoxc10a (LG23) and between hoxa9a and hoxa11a (LG19), respectively. Riboprobes designed to cover mir-196a1 or mir-196a2 (517 nt and 566 nt in length, respectively) are marked in red. (B, C) Whole-mount in situ hybridization with digoxigenin-labeled fragments of primary transcripts showed expression of mir-196a1 in the tailbud, central nervous system, and pronephric duct and of mir-196a2 in the tailbud. (D) Expression of mature miR-196a labeled with LNA probe revealed expression in the neural tube, tailbud, and pronephric duct. cns, central nervous system; LNAs, locked nucleic acids; miRNA, microRNA; spd, pronephric duct; tb, tailbud; *note expression difference in the tail bud.
At 24 hour postfertilization, mir-196a1 and mir-196a2 riboprobes revealed overlapping but distinct expression patterns (Fig. 1B, C). The miR-196a1 probe stained the pronephric duct, posterior central nervous system (CNS), and the tail bud (Fig. 1B, asterisk). Although miR-196a2 did not stain the pronephric duct, it did stain the caudal CNS and the tail bud (Fig. 1C, asterisk). An LNA probe designed to recognize the mature miRNA product common to both miR-196a genes stained the pronephric duct, the tail bud, the CNS, and ventral posterior trunk (Fig. 1D), which resembles the combination of the expression patterns detected for mir-196a1 and mir-196a2 using riboprobes. The expression of mir-196a genes is similar to the expression of their neighboring hox9 and hox10 protein-coding genes.52 One possible explanation for the similar expression patterns of these miRNA genes and their flanking protein coding genes is that they share regulatory elements.
Supplementary Figure S1 (Supplementary Data are available online at www.liebertonline.com/zeb) shows riboprobe-generated expression patterns for pri-miR-10d in the caudal hindbrain and spinal cord, pri-mir-200ab in the olfactory primordium, pri-miR-138 and pri-miR-144 in the caudal vein of whole-mount embryos, and pri-miR-199 and pri-miR-214 in the mesenchyme surrounding the ceratohyal cartilage in histological sections.
These results show that for intergenic miRNA genes, normal digoxigenin-labeled riboprobes can help observe gene expression in zebrafish embryo whole mounts and can distinguish expression patterns of different miRNA paralogs that encode the same mature miRNA sequence, which LNA probes designed to recognize only the mature miRNA sequences fail to do.
Intronic pri-miRNAs
To learn if riboprobes can detect the expression of intronic miRNA genes, we investigated pri-miR-140 by ISH to digoxigenin-labeled riboprobe. Vertebrate mir-140 resides in an intron of the wwp2 gene and is transcribed in the same direction as its host gene (Fig. 2A).7 ISH for pri-miR-140-riboprobe amplified from a portion of intron-15 of wwp2 showed that pri-miR-140 riboprobe labeled expression of mir-140 in chondrocytes similar to an LNA probe (Fig. 2B, C).7,40 In addition, pri-miR-140 riboprobe mostly labeled expression of pri-miR-140 in the cell nucleus, whereas LNA probe labeled miR-140 in the cytoplasm (Fig. 2B, C, insets). This result is consistent with previous studies showing that pri-miRNAs reside in the nucleus, where they are processed, whereas mature miRNAs accumulate in the cytoplasm, where they exert their function. In contrast, overstaining embryos with a probe for intron-16 of wwp2, which does not contain a known miRNA gene, failed to show a tissue-specific ISH pattern (Fig. 2J, K), consistent with the rapid degradation of most introns.53
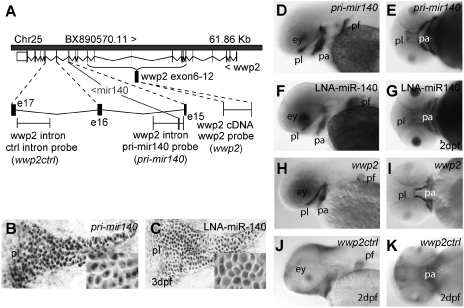
FIG. 2.
Expression of intronic pri-miR-140 and its host gene wwp2 in 2 dpf embryos. (A) Genomic organization of wwp2 and mir-140 on zebrafish LG25. Sequences of pri-miR-140 in intron-15 and wwp2 exon-6 to −12 that provided probes are delineated in red and green, respectively, and the location of probe for wwp2 intron-16, which lacks a miRNA gene, is shown in blue. (B, C) Expression of pri-miR-140 observed by riboprobe (B) and the mature form of miR-140 stained by LNA probe (C) in a histological section of a 3 dpf zebrafish palate. The inset in each panel shows expression at a cellular level, revealing that pri-miR-140 transcript accumulated in the nucleus and miR-140 accumulated in the cytoplasm. (D, E) Digoxigenin-labeled antisense probes against the mir-140 primary transcript showed strong expression in the palate portion of the neurocranium and in the pharyngeal arches and in skeletal tissues of the pectoral fin bud in lateral (D) and ventral (E) views. (F, G) LNA antisense probe to mature miR-140 showed that mature miR-140 accumulated in the palate in a pattern similar to that shown for the riboprobes, in lateral (F) and ventral (G) views. (H, I) The host gene wwp2 was expressed in a pattern similar to that of mir-140 (H, lateral view; I, ventral view). (J, K) Overstaining of riboprobe to wwp2 intron-16 showed background levels proportional to cell density rather than providing a tissue-specific signal. dpf, day postfertilization; ep, ethmoid plate; ey, eye; i15, i16, intron-15 and intron-16; pa, pharyngeal arch; pf, pectoral fin; pl, palate; tr, trabeculae.
These experiments showed that riboprobe against pri-miR-140 labeled the same set of craniofacial skeletal structures as the LNA probe for mature miR-140, verifying the methodology for intronic miRNAs. In addition, riboprobe directed against exons of wwp2, the host gene for mir-140, showed that the host gene was co-expressed with the miRNA. This would be expected under the hypothesis that intronic miRNAs are associated with the transcription of their host genes and would thus be available to support the biological functions of their host gene.54
Duplicates of miRNA mir-126
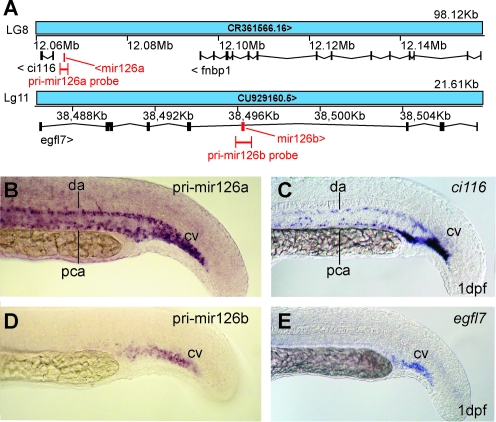
Zebrafish has two copies of mir-126. The mir-126a gene (alias dre-mir-126 in Ensembl and mir-126 in ZFIN) is intergenic between ci116 (LOC557793, the ortholog of which is called C9orf116 in human) and fnbp1 in linkage group 8 (LG8) (Fig. 3A). Zebrafish mir-126b is located in an intron of egfl7 in LG11 (Fig. 3A) in a position orthologous to the human gene. To determine if riboprobes can distinguish between zebrafish miRNA duplicates, we investigated mir-126a and mir-126b expression. Results showed that riboprobes directed against pri-miR-126a and pri-miR-126b showed overlapping expression patterns in the caudal vein and strong expression of pri-miR-126a in the dorsal aorta and posterior cardinal vein (Fig. 3B, D). To investigate functional relationships of these miRNAs to their nearby genes, we studied the expression patterns of ci116, which has not been described for any species, and egfl7, which is expressed in endothelial cells and regulates vascular tube formation in zebrafish.55 Results showed that ci116 and egfl7 are both expressed in the caudal vasculature in patterns virtually identical to the adjacent miRNAs. This result would be predicted by the hypothesis that miRNAs sometimes share regulatory features with their neighboring protein-coding genes.54
FIG. 3.
Duplicate mir-126 genes have expression patterns related to their genomic contexts. (A) Organization of zebrafish mir-126 paralogs in the genome. (B, C) Riboprobe detected mir-126a expression in the caudal vasculature (B), and expression of the nearest neighboring gene ci116 (LOC557793) had a similar expression pattern in the caudal vasculature (C). (D, E) Riboprobes detected expression of the intronic gene mir-126b in the caudal vein (D) and expression of the host gene egfl7 also in the caudal vein (E). cv, caudal vein; da, dorsal aorta; pca, posterior cardinal vein.
Regulation of mir-140 expression by Sox9 transcription factors
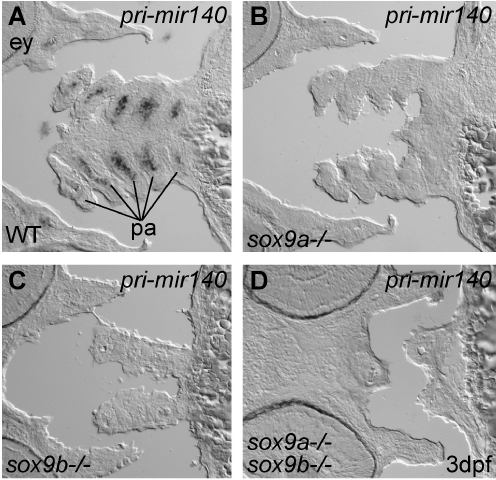
Mechanisms that regulate miRNA gene expression are not yet well understood. To investigate factors responsible for the expression of mir-140 in chondrogenic cells (see Fig. 2),7 we studied mir-140 expression in mutants lacking activity of Sox9, a master regulator of cartilage development.48,56 Riboprobe detection of pri-miR-140 showed that mutant zebrafish lacking activity of either sox9a only, sox9b only, or double mutants lacking activity of both sox9a and sox9b lacked mir-140 expression in craniofacial cartilages (Fig. 4). These results show, first, that mir-140 is downstream of Sox9 in the pathway of chondrogenesis and, second, that riboprobe detection of miRNA primary transcripts is sufficiently robust to detect miRNA gene expression on histological sections.
FIG. 4.
mir-140 is downstream of Sox9 transcription factors. Riboprobe detection of pri-miR-140 in histological sections of wild-type 3 dpf embryos (A), a sox9a mutant (B), a sox9b mutant (C), and a sox9a;sox9b double mutant (D). The lack of expression of mir-140 in sox9a and sox9b mutants shows that Sox9 acts upstream of mir-140. ey, eye; pa, pharyngeal arches; WT, wild type.
Discussion
miRNA primary transcripts are produced by either independent transcription (intergenic miRNAs) or from the splicing of introns from protein-coding genes (intronic miRNAs). Investigations reported here showed that the primary transcripts of both intronic and intergenic miRNAs accumulate in zebrafish embryos to levels sufficient for detection by ISH using riboprobes in both whole mounts and histological sections (also see Supplementary Fig. S1). This discovery will stimulate research on the mechanisms of zebrafish miRNA action for several reasons. First, the use of riboprobes for miRNA detection reduces the cost of detecting miRNA expression about 20-fold over the currently favored LNA probes. This dramatic improvement in economy will allow researchers to investigate many more miRNAs using far less resources. Second, riboprobes directed against the primary miRNA transcript allow one to distinguish expression patterns from different miRNA genes that encode the same mature miRNA. Because gene duplicates arising in the teleost genome duplication are often expressed in different tissues due to subfunctionalization,57 because miRNAs arising from the teleost genome duplication are likely to have identical sequences, and because miRNAs with identical sequences but encoded by different genes might be responsible for tissue-specific phenotypes, the riboprobe method we describe should be of high value for zebrafish miRNA research.
Our results showed that riboprobe detection of miRNA primary transcripts can be used to investigate mechanisms that regulate miRNA gene expression. Investigations of zebrafish sox9 mutants revealed that Sox9 is required for the expression of the mir-140 gene, suggesting that sox9a and sox9b are both upstream regulators of mir-140. Cartilages in sox9a mutants are fully formed and simply fail to express extracellular matrix genes, including collagen type II,58 raising the hypothesis that Sox9a might act directly on mir-140 to stimulate its transcription. In contrast, cartilages are substantially disrupted in the sox9b mutants,48 so it is possible that mir-140 may be substantially downstream of sox9b regulation.
Not all of the miRNA-directed riboprobes that we tested gave signals as strong as that of LNA probe. Three of the 17 pri-miRNA riboprobes that we examined, including those for mir-21, mir-27a, and mir-222, failed to show a strong and clean signal, although signals for these miRNA genes had been obtained using LNA probes.40 These results suggest that some pri-miRNAs may be exceptionally short-lived or expressed at levels below the detection limits of the riboprobe method. Nevertheless, for the majority of mir genes we tested, riboprobe detection of miRNA gene expression provides a convenient, inexpensive, and highly effective way to rapidly study miRNA expression for both intergenic and intronic miRNA genes in zebrafish embryo whole mounts and in histological sections.
Supplementary Material
Acknowledgments
The authors thank Ruth BreMiller for help with in situs and discussion, and NIH grants 1U01DE020076 and P01HD022486 for support.
Disclosure Statement
No competing financial interests exist.
References
- 1.Boehm M. Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 2.Hornstein E. Mansfield JH. Yekta S. Hu JK. Harfe BD. McManus MT, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 3.Chen JF. Mandel EM. Thomson JM. Wu Q. Callis TE. Hammond SM, et al. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J. Lee EJ. Schmittgen TD. Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes Cancer. 2006;45:103–106. doi: 10.1002/gcc.20264. [DOI] [PubMed] [Google Scholar]
- 5.Tuddenham L. Wheeler G. Ntounia-Fousara S. Waters J. Hajihosseini MK. Clark I, et al. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006;580:4214–4217. doi: 10.1016/j.febslet.2006.06.080. [DOI] [PubMed] [Google Scholar]
- 6.Begemann G. MicroRNAs and RNA interference in zebrafish development. Zebrafish. 2008;5:111–119. doi: 10.1089/zeb.2008.0528. [DOI] [PubMed] [Google Scholar]
- 7.Eberhart JK. He X. Swartz ME. Yan YL. Song H. Boling TC, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40:290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman RC. Farh KK. Burge CB. Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le MT. Teh C. Shyh-Chang N. Xie H. Zhou B. Korzh V, et al. MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 2009;23:862–876. doi: 10.1101/gad.1767609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y. Chao T. Li R. Liu W. Chen Y. Yan X, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med. 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z. Peng H. Chen J. Chen X. Han F. Xu X, et al. MicroRNA-21 protects from mesangial cell proliferation induced by diabetic nephropathy in db/db mice. FEBS Lett. 2009;583:2009–2014. doi: 10.1016/j.febslet.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Wang B. Majumder S. Nuovo G. Kutay H. Volinia S. Patel T, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rand TA. Petersen S. Du F. Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Lee Y. Kim M. Han J. Yeom KH. Lee S. Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez A. Griffiths-Jones S. Ashurst JL. Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutvagner G. Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 17.Lee Y. Ahn C. Han J. Choi H. Kim J. Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein E. Kim SY. Carmell MA. Murchison EP. Alcorn H. Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 19.Wienholds E. Koudijs MJ. van Eeden FJ. Cuppen E. Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 20.Giraldez AJ. Cinalli RM. Glasner ME. Enright AJ. Thomson JM. Baskerville S, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 21.Giraldez AJ. Mishima Y. Rihel J. Grocock RJ. Van Dongen S. Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 22.Kanellopoulou C. Muljo SA. Kung AL. Ganesan S. Drapkin R. Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L. Meng T. Jia Z. Zhu G. Shi B. Single nucleotide polymorphism associated with nonsyndromic cleft palate influences the processing of miR-140. Am J Med Genet A. 2010;152A:856–862. doi: 10.1002/ajmg.a.33236. [DOI] [PubMed] [Google Scholar]
- 24.Holland PW. Garcia-Fernandez J. Williams NA. Sidow A. Gene duplications and the origins of vertebrate development. Dev Suppl. 1994:125–133. [PubMed] [Google Scholar]
- 25.Dehal P. Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amores A. Force A. Yan Y-L. Joly L. Amemiya C. Fritz A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 27.Taylor JS. Braasch I. Frickey T. Meyer A. Van de Peer Y. Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jaillon O. Aury JM. Brunet F. Petit JL. Stange-Thomann N. Mauceli E, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 29.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S. Grocock RJ. van Dongen S. Bateman A. Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S. Saini HK. van Dongen S. Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woltering JM. Durston AJ. The zebrafish hoxDb cluster has been reduced to a single microRNA. Nat Genet. 2006;38:601–602. doi: 10.1038/ng0606-601. [DOI] [PubMed] [Google Scholar]
- 33.He X. Eberhart JK. Postlethwait JH. MicroRNAs and micromanaging the skeleton in disease, development and evolution. J Cell Mol Med. 2009;13:606–618. doi: 10.1111/j.1582-4934.2009.00696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thisse C. Thisse B. Schilling TF. Postlethwait JH. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development. 1993;119:1203–1215. doi: 10.1242/dev.119.4.1203. [DOI] [PubMed] [Google Scholar]
- 35.Jowett T. Mancera M. Amores A. Yan Y-L. In situ hybridization to embryo whole mounts and tissue sections: mRNA detection and application to the developmental studies. In: Clark M, editor. In situ Hybridization. New York: Chapman and Hall; 1995. pp. 19–121. [Google Scholar]
- 36.Paul CP. Subcellular distribution of small interfering RNA: directed delivery through RNA polymerase III expression cassettes and localization by in situ hybridization. Methods Enzymol. 2005;392:125–145. doi: 10.1016/S0076-6879(04)92008-3. [DOI] [PubMed] [Google Scholar]
- 37.You Y. Moreira BG. Behlke MA. Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson RC. Deo M. Turner DL. Analysis of microRNA expression by in situ hybridization with RNA oligonucleotide probes. Methods. 2007;43:153–161. doi: 10.1016/j.ymeth.2007.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kloosterman WP. Wienholds E. de Bruijn E. Kauppinen S. Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 40.Wienholds E. Kloosterman WP. Miska E. Alvarez-Saavedra E. Berezikov E. de Bruijn E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 41.Ason B. Darnell DK. Wittbrodt B. Berezikov E. Kloosterman WP. Wittbrodt J, et al. Differences in vertebrate microRNA expression. Proc Natl Acad Sci U S A. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Darnell DK. Kaur S. Stanislaw S. Konieczka JH. Yatskievych TA. Antin PB. MicroRNA expression during chick embryo development. Dev Dyn. 2006;235:3156–3165. doi: 10.1002/dvdy.20956. [DOI] [PubMed] [Google Scholar]
- 43.Hicks JA. Tembhurne P. Liu HC. MicroRNA expression in chicken embryos. Poult Sci. 2008;87:2335–2343. doi: 10.3382/ps.2008-00114. [DOI] [PubMed] [Google Scholar]
- 44.Simeonov A. Nikiforov TT. Single nucleotide polymorphism genotyping using short, fluorescently labeled locked nucleic acid (LNA) probes and fluorescence polarization detection. Nucleic Acids Res. 2002;30:e91. doi: 10.1093/nar/gnf090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mouritzen P. Nielsen AT. Pfundheller HM. Choleva Y. Kongsbak L. Moller S. Single nucleotide polymorphism genotyping using locked nucleic acid (LNA) Expert Rev Mol Diagn. 2003;3:27–38. doi: 10.1586/14737159.3.1.27. [DOI] [PubMed] [Google Scholar]
- 46.Tolstrup N. Nielsen PS. Kolberg JG. Frankel AM. Vissing H. Kauppinen S. OligoDesign: optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. Nucleic Acids Res. 2003;31:3758–3762. doi: 10.1093/nar/gkg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson MP. Haupt LM. Griffiths LR. Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. Nucleic Acids Res. 2004;32:e55. doi: 10.1093/nar/gnh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan YL. Willoughby J. Liu D. Crump JG. Wilson C. Miller CT, et al. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132:1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 49.Li SC. Tang P. Lin WC. Intronic microRNA: discovery and biological implications. DNA Cell Biol. 2007;26:195–207. doi: 10.1089/dna.2006.0558. [DOI] [PubMed] [Google Scholar]
- 50.Mansfield JH. Harfe BD. Nissen R. Obenauer J. Srineel J. Chaudhuri A, et al. MicroRNA-responsive “sensor” transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 51.Tanzer A. Amemiya CT. Kim CB. Stadler PF. Evolution of microRNAs located within Hox gene clusters. J Exp Zoolog B Mol Dev Evol. 2005;304:75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- 52.Prince VE. Joly L. Ekker M. Ho RK. Zebrafish hox genes: genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- 53.Nott A. Meislin SH. Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–617. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lutter D. Marr C. Krumsiek J. Lang EW. Theis FJ. Intronic microRNAs support their host genes by mediating synergistic and antagonistic regulatory effects. BMC Genomics. 2010;11:224. doi: 10.1186/1471-2164-11-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker LH. Schmidt M. Jin SW. Gray AM. Beis D. Pham T, et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature. 2004;428:754–758. doi: 10.1038/nature02416. [DOI] [PubMed] [Google Scholar]
- 56.Mori-Akiyama Y. Akiyama H. Rowitch DH. de Crombrugghe B. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci U S A. 2003;100:9360–9365. doi: 10.1073/pnas.1631288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Force A. Lynch M. Pickett FB. Amores A. Yan YL. Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan YL. Miller CT. Nissen RM. Singer A. Liu D. Kirn A, et al. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129:5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.