Abstract
Rice expressing the Pi-ta gene is resistant to strains of the rice blast fungus, Magnaporthe grisea, expressing AVR-Pita in a gene-for-gene relationship. Pi-ta encodes a putative cytoplasmic receptor with a centrally localized nucleotide-binding site and leucine-rich domain (LRD) at the C-terminus. AVR-Pita is predicted to encode a metalloprotease with an N-terminal secretory signal and pro-protein sequences. AVR-Pita176 lacks the secretory and pro-protein sequences. We report here that transient expression of AVR-Pita176 inside plant cells results in a Pi-ta-dependent resistance response. AVR-Pita176 protein is shown to bind specifically to the LRD of the Pi-ta protein, both in the yeast two-hybrid system and in an in vitro binding assay. Single amino acid substitutions in the Pi-ta LRD or in the AVR-Pita176 protease motif that result in loss of resistance in the plant also disrupt the physical interaction, both in yeast and in vitro. These data suggest that the AVR-Pita176 protein binds directly to the Pi-ta LRD region inside the plant cell to initiate a Pi-ta-mediated defense response.
Keywords: ligand/Magnaporthe grisea/receptor/rice/signal recognition
Introduction
Plants have evolved sophisticated multi-faceted defense mechanisms against pathogens. These defense responses include a rapid, localized cell death termed the hypersensitive response (HR), production of antimicrobial compounds, lignin formation, an oxidative burst and increased expression of pathogenesis-related genes (Cutt and Klessig, 1992; Goodman and Novarcky, 1994; Levine et al., 1994; Mehdy, 1994). The defense responses are often activated by the action of a host resistance (R) gene and a pathogen avirulence (AVR) gene as proposed by the gene-for-gene hypothesis (Flor, 1971). One possible explanation for the molecular basis of gene-for-gene interactions is a ligand and receptor model where the R gene product acts as a receptor that recognizes a ligand, or elicitor, produced directly or indirectly by the pathogen’s AVR gene. This interaction then results in defense response activation. Structures of cloned R genes seem to support the ligand–receptor model because the majority of plant R genes contain leucine-rich repeat (LRR) domains (Bent, 1996; Hammond-Kosack and Jones, 1997). LRR proteins from animals and fungi mediate specific protein–protein interactions or ligand binding (Braun et al., 1991; Kobe and Deisenhofer, 1993; Jones and Jones, 1996). LRR-containing plant R genes fall into two classes, those with extra-cytoplasmic LRRs and those with cytoplasmic LRRs. The largest R gene class encodes cytoplasmic proteins with centrally localized nucleotide-binding sites (NBS) and LRRs near the C-terminus (Jones and Jones, 1996; Leister et al., 1996; Vos et al., 1998). Although the presence of the LRR domains in R proteins is consistent with their proposed receptor function, direct binding between an LRR-containing R protein and an AVR ligand has not yet been demonstrated.
In contrast to the similarities among cloned plant R genes, sequence analysis of cloned AVR genes from viral, bacterial and fungal pathogens revealed few similarities and few clues to their functions for the pathogen (for reviews see Dangl, 1994; Joosten et al., 1994; Rohe et al., 1995). A number of AVR genes have been shown to encode specific elicitor proteins, which induce HR either when infiltrated into the apoplastic space outside plant cells (de Wit, 1995; Knogge, 1996) or when expressed inside plant cells (Culver and Dawson, 1991; Gopalan et al., 1996; Leister et al., 1996; Van den Ackerveken et al., 1996; Bonas and Van den Ackerveken, 1997). The diversity among AVR genes is consistent with theories of evolution of gene-for-gene resistance as host plants acquired the ability specifically to recognize random molecules produced by pathogens.
Molecular characterization of several matched pairs of plant R genes and pathogen AVR genes made it possible to test the gene-for-gene receptor–ligand model. A direct interaction has been demonstrated between the tomato bacterial speck resistance gene product, Pto, and the corresponding avrPto avirulence gene product of Pseudomonas syringae pv. tomato (Scofield et al., 1996;Tang et al., 1996). Pto, a serine/threonine kinase, is a unique R gene product that lacks an LRR domain (Martin et al., 1993). Physical interaction between Pto and avrPto proteins was discovered initially with the yeast two-hybrid system (Scofield et al., 1996; Tang et al., 1996), and several lines of evidence suggest that the interaction is specific and essential for activation of the host defense response. First, the bacterial speck-susceptible cultivar Alisa Craig encodes an active Pto kinase that failed to bind avrPto in the yeast two-hybrid system (Tang et al., 1996; Jia et al., 1997). Secondly, a single amino acid residue, Thr204 of the Pto kinase, determines the recognition specificity for avrPto in both the yeast two-hybrid system and when Pto is transiently expressed in plant cells using Agrobacterium-based DNA transfer (Frederick et al., 1998). Thirdly, C-terminal deletions of the avrPto protein that interact with the Pto protein in the yeast two-hybrid system also elicited HR and disease resistance in a Pto-dependent manner in plants (Tang et al., 1996).
The interaction between rice (Oryza sativa) and the fungal pathogen Magnaporthe grisea (Hebert) Barr (anamorph Pyricularia grisea Sacc.) is a well documented gene-for-gene system (Silue et al., 1992; Valent, 1997). The rice blast disease caused by M.grisea is one of the most devastating plant diseases worldwide (Valent and Chumley, 1991). This hemibiotropic pathogen penetrates directly through the plant cuticle and outer cell wall into epidermal cells of the host (Howard et al., 1991). The fungus grows intracellularly, filling individual plant cells with fungal mycelium before moving on to the next cell. It is unknown whether the intracellular hyphae of this pathogen are enveloped with plant plasma membrane, as is the case for haustoria produced by biotrophic fungi, or if the pathogen breaches the plasma membrane. Thus it is also unknown how M.grisea might deliver AVR-encoded proteins into the cytoplasm of plant cells.
Intense research with the M.grisea system has led to the isolation of the matched pair of resistance gene Pi-ta and avirulence gene AVR-Pita, previously referred to as AVR2-YAMO (Pi-ta, Bryan et al., 2000; AVR-Pita, Orbach et al., 2000). Pi-ta is a single copy gene encoding a putative cytoplasmic protein of 928 amino acids with a centrally localized NBS domain and a leucine-rich domain (LRD) at the C-terminus. Pi-ta contains 16.4% leucine within this LRD region (amino acids 586–928, http:/www.expasy.ch/), which approximately corresponds to the LRR domain of the RPM1 protein (Grant et al., 1995; Bryan et al., 2000). Although Pi-ta is most similar to R proteins of the NBS-LRR class, the Pi-ta LRD has areas of LXXLXXL motifs, but it does not contain LRRs fitting any previously reported consensus (Jones and Jones, 1996). AVR-Pita is hypothesized to be a neutral zinc metalloprotease, although direct biochemical evidence is lacking (Orbach et al., 2000).
We report here evidence that AVR-Pita functions as an elicitor molecule that directly binds the Pi-ta protein and triggers a signal transduction cascade leading to resistance. First, we demonstrate that AVR-Pita176, a putative mature form of the protease containing the C-terminal 176 amino acids, is a genotype-specific elicitor, i.e. biolistic transient expression of AVR-Pita176 inside cells of resistant Pi-ta rice results in reduced activity of β-glucuronidase (GUS), an indicator of HR (Mindrinos et al., 1994; Gopalan et al., 1996). Next, we demonstrate a specific physical interaction between AVR-Pita176 and the Pi-ta LRD region in the yeast two-hybrid system. Finally, we corroborate our two-hybrid interaction by showing that recombinant Pi-ta LRD protein binds specifically to the AVR-Pita176 protein in far-western analysis. We suggest that AVR-Pita is translocated from the fungus into the cytoplasm of the plant cell where it binds to the Pi-ta LRD region to initiate a Pi-ta-mediated defense response.
Results
Optimization of a transient expression system in rice using biolistic bombardment
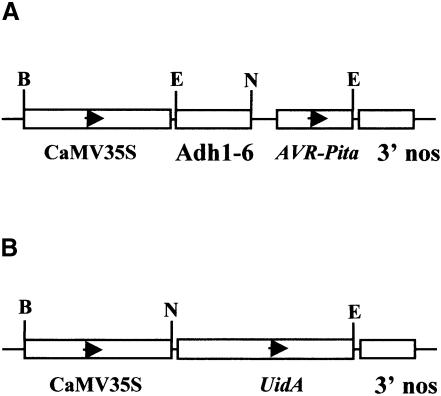
Transient gene expression systems have been used previously to investigate elicitor activity of avirulence proteins in dicot plants (Mindrinos et al., 1994). The assay is based on association of resistance with HR, and on reduced expression of a GUS reporter gene as an indicator of HR (Dixon et al., 1994; Mindrinos et al., 1994). We modified the transient expression system to analyze the function of the AVR-Pita protein in intact rice seedlings. We co-introduced two plasmids by particle bombardment, one expressing a GUS reporter gene and the other expressing AVR-Pita polypeptides. Resistant and susceptible seedlings were positioned side-by-side in the same Petri dish for bombardment. This was important since the efficiency of transformation varies both between bombardments and between different areas of the target tissue within a single bombardment (data not shown). To ensure efficient expression of AVR-Pita, we engineered a plasmid that contained a 493 bp Adh1-6 intron behind the cauliflower mosaic virus 35S (CaMV 35S) promoter (Figure 1A; Freeling and Bennet, 1985). This construct previously has been shown to mediate high levels of expression in rice (Freeling and Bennet, 1985; S.A.McAdams, unpublished data). We also used the CaMV 35S promoter to express the GUS reporter gene uidA (Figure 1B). These constructs were assayed in two resistant Pi-ta rice cultivars (Yashiro-mochi and YT14), and two susceptible pi-ta cultivars (Nipponbare and YT16). Equal GUS activity was observed repeatedly in all four cultivars in the absence of AVR-Pita constructs (data not shown).
Fig. 1. Physical maps of the transgenes used for transient expression analysis. (A) AVR-Pita expression vector. The maize Adh1-6 intron was inserted after the CaMV 35S promoter resulting in a fusion promoter for expression of constructs encoding AVR-Pita223, AVR-Pita176 and AVR-Pita166. (B) GUS expression vector. Plasmid pML63 contained the uidA gene, encoding GUS, under control of the CaMV 35S promoter. The restriction endonucleases used for generating constructs are shown (B, BamHI; E, EcoRI; N, NcoI). Both expression cassettes contain bacterial 3′ nos terminator sequences.
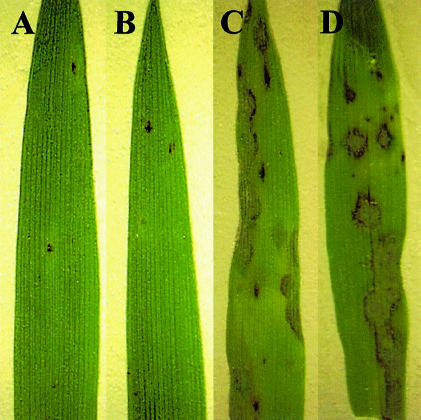
Pi-ta-containing rice plants undergo HR when they are inoculated with an avirulent M.grisea strain containing AVR-Pita. To ensure that Pi-ta specificity is maintained in seedlings used for transient expression analysis, we inoculated seedlings grown to the two-leaf stage on agar medium with an avirulent pathogen. Figure 2A and B shows sparse HR flecking in resistant Pi-ta cultivars, and Figure 2C and D shows typical leaf blast symptoms in susceptible pi-ta cultivars. Thus, we conclude that intact two-leaf rice seedlings are suitable for transient gene expression analysis.
Fig. 2. Genotype-specific HR in rice seedlings induced by M.grisea carrying AVR-Pita. Sparse HR flecking is seen in Pi-ta-containing rice seedlings (A) Yashiro-mochi and (B) YT14, as expected. In contrast, typical symptoms of rice blast disease are seen in susceptible rice seedlings (C) Nipponbare and (D) YT16. Representative leaves are shown from rice seedlings germinated in plant nutrient medium and infected with avirulent M.grisea strain 4360-R-62 (see Materials and methods for details). Shown at 4 days after inoculation.
AVR-Pita176 is an elicitor that acts inside the plant cell
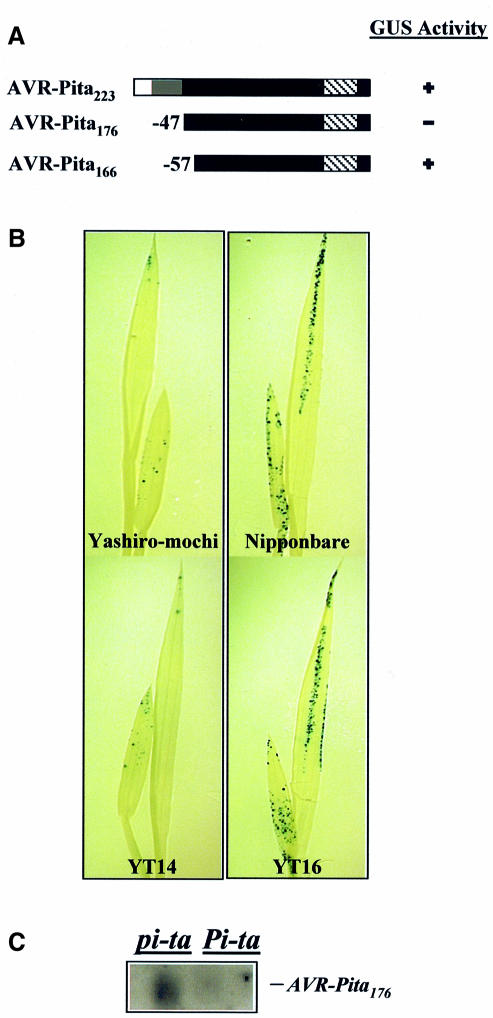
AVR-Pita is predicted to encode a 223 amino acid protein with similarity to fungal zinc metalloproteases. Specifically, the homology occurs in the C-terminal 176 amino acids of the proteins, a region corresponding to the mature processed version of known metalloproteases (Tatsumi et al., 1994; Orbach et al., 2000). Due to uncertainty as to the active form of the AVR-Pita protein in vivo, we produced constructs for expressing the full-length protein (AVR-Pita223), the putative mature protease (AVR-Pita176) and a protein with 10 amino acids deleted from the N-terminus of AVR-Pita176 (AVR-Pita166) (Figure 3A). Plasmids expressing these AVR-Pita constructs and plasmids expressing the GUS reporter were co-introduced into leaves of Pi-ta and pi-ta rice seedlings. GUS activity was then assayed histochemically 2 days after bombardment. No reduction of GUS activity was observed in any of the rice cultivars when AVR-Pita223 was co-expressed with the GUS reporter (Figure 3A). However, we repeatedly observed reduced GUS activity in leaves of resistant Pi-ta varieties but not in leaves of susceptible pi-ta varieties when AVR-Pita176 and the GUS reporter were co-expressed (Figure 3A and B). Co-expression of the GUS reporter and AVR-Pita166 did not result in reduced GUS activity on either susceptible or resistant rice varieties (Figure 3A). Thus, the putative mature AVR-Pita176 protein, but not AVR-Pita223 or AVR-Pita166, induces HR when produced inside cells of Pi-ta-containing rice.
Fig. 3. AVR-Pita176 is an elicitor. (A) AVR-Pita polypeptides tested in the transient assay. The white region indicates the putative secretory signal sequence, the gray region indicates the putative pro-protein domain and the hatched region indicates the putative protease motif. The black region indicates the putative mature protein. The number of amino acids missing from the N-terminus is indicated. GUS activity is indicated by ‘+’, whereas decreased GUS activity is indicated by ‘–’. (B) Representative rice seedlings showing GUS activity. Two-leaf Pi-ta (Yashiro-mochi and YT14) and pi-ta (Nipponbare and YT16) seedlings were co-bombarded with 35S/Adh1-6::AVR-Pita176 and 35S::uidA. Leaves were assayed histochemically for GUS activity and cleared in 70% ethanol to visualize GUS staining. (C) RNA gel blot analysis of AVR-Pita expression in the transient assay. YT14 (Pi-ta) and YT16 (pi-ta) were co-bombarded with the 35S/Adh1-6::AVR-Pita176 and 35S::uidA plasmids. Leaf tissue was harvested 2 days after bombardment. Poly(A)+ mRNA was then extracted, blotted to Hybond-N and hybridized with a radiolabeled AVR-Pita176 probe. The AVR-Pita transcript is indicated. Similar loading was verified before blotting by visualizing mRNA in the gel stained with ethidium bromide.
We tested the specificity of the HR induction in the transient assay using virulent avr-pita alleles with single amino acid substitutions that abolish AVR-Pita avirulence function in whole-plant infection assays. Changing the putative protease motif active site residue (amino acid 177 in AVR-Pita223) from glutamic acid to aspartic acid (E177D) eliminated avirulence activity in standard fungal infection assays. Co-expression of virulent avr-pita176E177D with the GUS reporter gene failed to reduce GUS activity in either resistant Pi-ta or susceptible pi-ta rice cultivars (data not shown). Likewise, changing methionine to tryptophan at amino acid residue 178 (M178W) eliminated avirulence in infection assays as well as activity in the transient assay (data not shown). Thus, AVR-Pita mutations that abolish avirulence in fungal infection assays also abolish the ability to induce HR upon transient expression in Pi-ta-containing rice cells.
To confirm that AVR-Pita176 was expressed in the bombarded seedlings, we extracted poly(A)+ mRNA from leaves 2 days after co-bombardment and performed RNA gel blot analysis using an AVR-Pita176 probe. Transcripts of AVR-Pita176 were detected in susceptible pi-ta seedlings, but not in resistant Pi-ta seedlings (Figure 3C). These data demonstrate that the AVR-Pita176 gene is expressed in susceptible seedlings, and that this expression has no effect on GUS activity. The absence of AVR-Pita176 transcripts in Pi-ta-containing plant cells is consistent with the occurrence of HR-associated cell death in transformed cells. Thus, all data are consistent with the conclusion that the AVR-Pita176 protein specifically induces a defense response when introduced into plant cells expressing Pi-ta.
AVR-Pita176 binds specifically to the Pi-ta LRD region in the yeast two-hybrid system
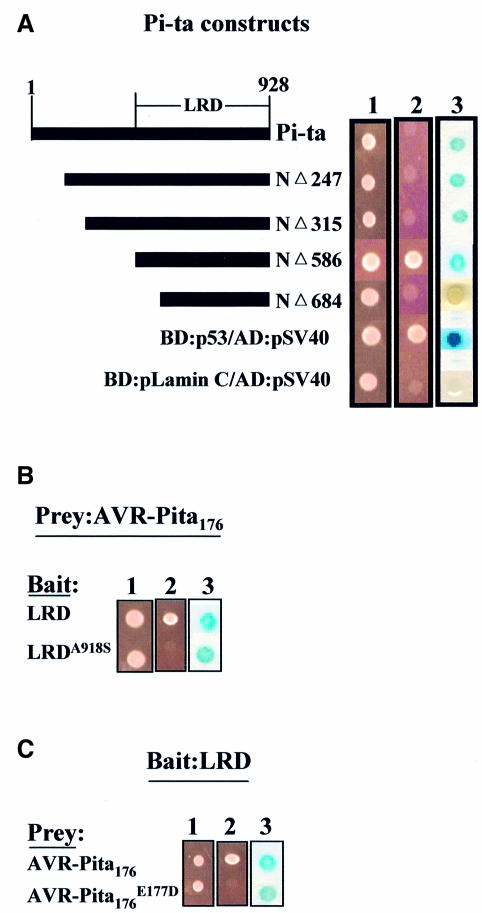
Since the Pi-ta protein confers recognition specificity in blast resistance and AVR-Pita176 is an elicitor of HR in a Pi-ta-dependent manner, we examined whether the Pi-ta protein interacts with AVR-Pita176 using the yeast two-hybrid system (Fields and Song, 1989). We constructed a series of N-terminal deletions of the Pi-ta protein to examine the requirement for different protein domains for any physical interaction (Figure 4A). These Pi-ta constructs were expressed as N-terminal fusions to the GAL4 DNA-binding domain (BD). The AVR-Pita176 protein was expressed as an N-terminal fusion to the transcriptional activation domain (AD). Physical interaction can be detected by activation of two reporter genes, HIS3 conferring histidine prototrophy and lacZ, when BD and AD fusions are expressed in the same yeast cell. Expression of either the BD-Pi-ta construct or the AD-AVR-Pita176 construct alone did not activate either reporter gene (data not shown). Positive and negative controls provided with Stratagene’s two-hybrid kit gave the expected results (Figure 4A).
Fig. 4. AVR-Pita176 interacts specifically with the Pi-ta LRD in the yeast two-hybrid system. (A) Mapping the interaction domain of the Pi-ta protein using the yeast two-hybrid system. The diagram (left) depicts the Pi-ta protein, the LRD region and a series of deletion constructs of Pi-ta. (1) YRG2 yeast cells expressing both bait and prey fusions were grown on yeast synthetic minimal (SD, Stratagene) liquid medium with omission of leucine and tryptophan (SD-LT), and (2) on yeast SD medium with omission of leucine, histidine and tryptophan for examination of HIS3 reporter gene (–LHT). (3) Yeast cells expressing both bait and prey fusions were grown on yeast SD-LT plates and assayed for lacZ activity as described in Materials and methods. Color development is shown after 24 h. AVR-Pita176 was cloned as an AD fusion (in pAD-GAL4) and each Pi-ta deletion construct was cloned as a BD fusion (in pBD-GAL4 Cam). The positive control was YRG2 cells containing p53 and pSV40 fusion constructs, which express proteins that interact in vivo, and the negative control was YRG2 cells containing pLamin C and pSV40 fusion constructs, which express proteins that do not interact in vivo (Stratagene). (B and C) Specificity of interaction of AVR-Pita176 with LRD polypeptide. Single amino acid substitutions that inactivate either LRD or AVR-Pita 176 in vivo eliminated growth in the absence of histidine and slowed color development with the lacZ reporter (referred to as an impaired physical interaction). LRD refers to the NΔ586 deletion of the Pi-ta protein from (A) above. LRDA918S refers to LRD containing S substituted for A at position 918 of the Pi-ta protein. avr-pita176E177D refers to the putative processed polypeptide that no longer confers avirulence due to an E to D substitution at residue 177 of AVR-Pita223. Similar expression of each BD fusion protein in yeast was verified by western blots.
Only one of the four deletion constructs tested in the two-hybrid assay activated both the HIS3 and lacZ reporter genes, demonstrating a physical interaction (Figure 4A). The active construct, deletion NΔ586, encodes the 341 amino acid C-terminal portion of the Pi-ta protein that we refer to as LRD. Interestingly, the reciprocal combination of LRD in AD fusion and AVR-Pita176 in BD fusion demonstrated an ‘impaired interaction’ in which the histidine biosynthetic gene was not activated, and the lacZ reporter gene was activated, but pigment accumulation was slowed in comparison with activation by the LRD in BD fusion (data not shown). This could result from steric constraints imposed by DNA AD and BD fusions in this particular orientation. Low levels of lacZ activity also indicated weak interactions between the full-length Pi-ta protein and AVR-Pita176, and between the remaining three Pi-ta polypeptides and AVR-Pita176 (Figure 4A).
To determine the specificity of the physical interaction between the Pi-ta LRD and AVR-Pita176, we constructed a BD fusion with the LRD region encoded by a susceptible pi-ta allele and tested its ability to interact with AD-AVR-Pita176. Because all susceptible Pi-ta LRDs have a single amino acid substitution, serine for alanine at residue 918 (Bryan et al, 2000), we designated susceptible Pi-ta LRD as LRDA918S. The impaired interaction of weak lacZ activity was observed between LRDA918S and AVR-Pita176 (Figure 4B), suggesting that substituting serine for Ala918 alters the binding specificity. As a further test to correlate the Pi-ta gene-for-gene interaction with physical interaction of the Pi-ta LRD and AVR-Pita176 proteins, we co-expressed the resistant LRD in BD fusion with a virulent form of AVR-Pita176 (avr-pitaE177D) in AD fusion. Similar to results with LRDA918S–AVR-Pita176 interaction, co-expression of these fusions gave weak lacZ activity, but did not activate the histidine reporter gene (Figure 4C). Finally, physical interaction of LRD with AVR-Pita223 was also examined, and this combination failed to activate either reporter gene (data not shown). Thus, the specific physical interaction between the LRD and AVR-Pita176 suggests that AVR-Pita176 serves directly as a fungal signal molecule and that Pi-ta serves as a cognate receptor molecule.
Membrane-immobilized AVR-Pita176 binds to Pi-ta and its LRD
To confirm our two-hybrid findings and test whether AVR-Pita176 is also capable of binding to the full-length Pi-ta protein, we next used bacterially produced recombinant proteins for in vitro binding experiments. We used a modified far-western (Chen and Evans, 1995) procedure to conduct these protein–protein interaction experiments because bacterially produced Pi-ta protein was soluble and bacterially produced AVR-Pita proteins were insoluble. For our studies, purified AVR-Pita proteins were refolded on a nitrocellulose membrane, and incubated with total Escherichia coli extracts containing soluble Pi-ta protein.
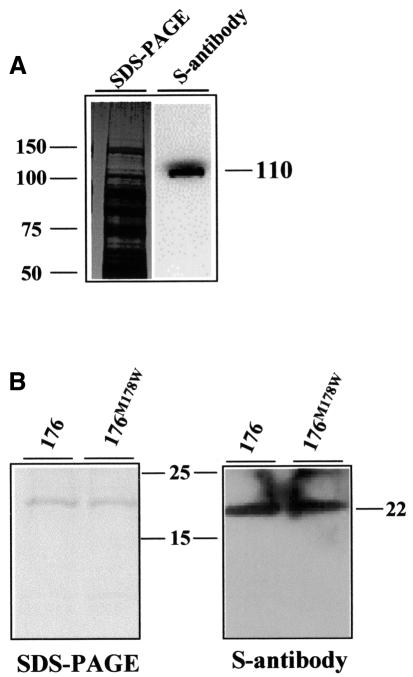
To facilitate detection and purification, the Pi-ta protein was engineered with an N-terminal S tag, a synthetic peptide composed of 15 amino acids (Novagen). We then expressed the S-Pi-ta fusion protein in E.coli and recovered it from the soluble fraction of total proteins. Figure 5A shows that the Pi-ta protein was highly expressed. The recombinant Pi-ta protein has an estimated molecular mass of ∼110 kDa as detected by S-antibody. Similarly, AVR-Pita176 was engineered to contain an N-terminal His tag to facilitate purification. The recombinant protein was found at high levels within inclusion bodies when expressed in E.coli. The corresponding virulent mutant protein avr-pita176M178W was also expressed and purified from inclusion bodies for use as a control. These recombinant AVR-Pita and avr-pita proteins had an estimated molecular mass of 22 kDa (Figure 5B).
Fig. 5. The Pi-ta protein binds to AVR-Pita176 and its mutant in far-western analysis. (A) Expression of the Pi-ta protein in E.coli. A total bacterial soluble extract expressing S-tagged Pi-ta protein was subjected to SDS–PAGE, and proteins were electroblotted onto PVDF membrane and detected by chemiluminescent visualization using monoclonal S-antibody to detect the S tag (right). The duplicate Coomassie Blue-stained gel is shown on the left. Molecular weights (kDa) were estimated using Perfect S protein markers (Novagen). (B) Binding of the Pi-ta protein to membrane-blotted AVR-Pita176 proteins. Purified AVR-Pita176 and avr-pita176M178W proteins were subjected to SDS–PAGE, and stained by Coomassie Blue (left). A duplicate SDS–PAGE gel was electroblotted onto a nitrocellulose membrane (right), and the soluble extract expressing S-tagged-Pi-ta protein was added for binding. Bound Pi-ta protein was visualized using the monoclonal S-antibody as shown in (A). Perfect S protein markers were used.
For far-western analysis, equal amounts of the purified AVR-Pita proteins were separated on a gradient SDS–polyacrylamide gel and immobilized on a nitrocellulose membrane for refolding. Immobilized, refolded AVR-Pita was then incubated with soluble extracts from E.coli transformants expressing S-tagged Pi-ta proteins. S-antibody was then used to detect Pi-ta protein bound to AVR-Pita on the membrane. Figure 5B shows that the Pi-ta protein bound to both AVR-Pita176 and avr-pita176M178W. Thus, AVR-Pita176 binds to the full-length Pi-ta protein, but the in vivo specificity is not retained under these conditions. Binding of full-length Pi-ta protein to avr-pita176M178W in this instance does not correlate with ability to trigger the Pi-ta-mediated resistance in vivo.
To confirm the yeast two-hybrid interaction between LRD and AVR-Pita176, we engineered plasmids expressing an N-terminal S tag fusion to both LRD and LRDA918S polypeptides. Each fusion construct expressed a polypeptide with an estimated molecular mass of 42 kDa in the soluble fraction of E.coli (Figure 6A). Figure 6B shows that the LRD protein bound specifically to AVR-Pita176, but not to avr-pita176M178W. Consistent with the results obtained from the yeast two-hybrid system, the susceptible LRDA918S protein did not bind to AVR-Pita176 under the same conditions (Figure 6C). These results in vitro confirm the interaction specificity of the Pi-ta LRD and AVR-Pita176 previously demonstrated in the yeast two-hybrid assay.
Fig. 6. AVR-Pita176 binds specifically to the Pi-ta LRD region in far-western analysis. (A) Expression of Pi-ta LRD polypeptides in E.coli. Total bacterial soluble extracts expressing S-tagged LRD and LRDA918S were subjected to SDS–PAGE, and then electroblotted onto PVDF membrane. LRD and LRDA918S were detected by chemiluminescent visualization using monoclonal S-antibody (right). The duplicate Coomassie Blue-stained SDS–PAGE gel is shown (left). The molecular weight (kDa) was estimated using the perfect S protein markers (Novagen). (B) Binding of the LRD polypeptide to membrane-bound AVR-Pita176. Purified AVR-Pita proteins were subjected to SDS–PAGE, and stained by Coomassie Blue (left). The duplicate SDS–PAGE gel was electroblotted onto a nitrocellulose membrane, and total soluble extract expressing S-tagged LRD was added for binding (see Materials and methods for details). The bound LRD was visualized by monoclonal S-antibody to detect S tag (right). Protein sizes (kDa) were determined as in (A). (C) In contrast, LRDA918S polypeptide failed to bind the membrane-blotted AVR-Pita176 proteins. Purified AVR-Pita proteins were subjected to SDS–PAGE, and stained by Coomassie Blue under identical conditions as in (B). The duplicate SDS–PAGE gel was electroblotted onto a nitrocellulose membrane, and the total soluble extract expressing S-tagged LRDA918S polypeptide was added for binding. Monoclonal S-antibody was added to detect bound LRDA918S (right). Protein sizes (kDa) were determined as in (A).
Discussion
Transient expression of the predicted mature AVR-Pita176 protease inside plant cells by particle bombardment transformation results in HR as measured by decreased GUS activity. HR occurs specifically on Pi-ta rice plants, not pi-ta plants. This result suggests that the AVR-Pita176 protein is a race-specific elicitor that triggers Pi-ta-mediated resistance to the rice blast fungus. We used the yeast two-hybrid system and in vitro binding assays to demonstrate that the C-terminal leucine-rich region of Pi-ta protein functions as an elicitor-binding domain. Our data suggest that the Pi-ta protein is an intracellular receptor that binds directly to the AVR-Pita176 protein inside the plant cell to initiate the defense response. These results may provide general insight into the way that the NBS class of plant R genes is able to detect the products of AVR genes.
AVR-Pita176 is an elicitor recognized inside the host plant cell
Our transient expression assay uses intact 7-day-old rice seedlings that retain Pi-ta-specific responses to infection with AVR-Pita-containing fungus. Co-bombardment of resistant and susceptible seedlings within a single Petri dish reduced variability that occurs between bombardments, and multiple repetitions compensate for variation between different areas of the target plant during each bombardment. Results described in this report validate the transient expression assay as a robust and specific assay for inducers of Pi-ta-mediated rice defense responses.
We demonstrated that the putative processed protease AVR-Pita176, but not the intact AVR-Pita223 protein, triggered the Pi-ta-dependent HR when produced inside rice cells by transient expression. This interaction is very specific. Deletion of an additional 10 amino acids from the N-terminus of AVR-Pita176 eliminated activity in this assay. The deletion removes a cysteine residue at position 53 that may be required for the protease function (Tatsumi et al., 1994). Indeed, M.grisea strains expressing an avr-pitaC53G allele substituting glycine for cysteine at residue 53 no longer exhibit full avirulence in standard infection assays (G.T.Bryan and B.Valent, unpublished). In addition, two separate mutations in the AVR-Pita protease motif that eliminate avirulence in fungal infection assays also eliminate elicitor activity in the transient expression assay. Thus, we have seen excellent correspondence between the structure and function of various AVR-Pita proteins, whether they are delivered via the normal route of fungal infection or through direct delivery of this single fungal gene into the plant cell by biolistic transformation.
The cytoplasmic site of action of AVR-Pita176 as determined by transient expression is consistent with the structural similarity of Pi-ta to members of the cytoplasmic NBS-LRR receptor class of R genes. These results are also consistent with a lack of elicitor activity in extensive experiments when recombinant AVR-Pita176 protein was applied to Pi-ta rice plants by vacuum infiltration or spray inoculation, techniques that would place these proteins in the apoplastic spaces between plant cells (G.T.Bryan and B.Valent, unpublished).
Other fungal AVR genes characterized so far are from pathogens that colonize the intercellular (apoplastic) spaces of their host plants, Avr9 and Avr4 from the tomato pathogen, Cladosporium fulvum (de Wit, 1995; Knogge, 1996), and NIP1 from the barley pathogen, Rhynchosporium secalis. Avr9 and Avr4 encode extracellular peptides that correspond to the products of R genes of the extracellular LRR class. A number of bacterial pathogens have AVR proteins corresponding to members of the NBS-LRR class of R genes and these appear to function inside plant cells. For example, it is lethal to express AvrB in RPM1 Arabidopsis plants by either transient or stable transformation (Gopalan et al., 1996). Transient expression of AvrBs3 in pepper cells using Agrobacterium-mediated gene delivery resulted in hypersensitive cell death specifically on Bs3 plants (Van den Ackerveken et al., 1996). Our finding that AVR-Pita176 acts inside the plant cell raises the interesting question of how M.grisea delivers AVR proteins into healthy plant cells. In some bacterial strains, AVR proteins are transferred into plant cells by a type III secretion system (Galan and Collmer, 1999). Perhaps hemibiotrophic and biotrophic fungi also employ specialized secretion systems for delivering proteins to the cytoplasm of their host cell without killing the cell. Details of the fungal–plant interface, such as whether M.grisea intracellular hyphae have breached the plasma membrane, need to be defined as a prelude to understanding how this fungus delivers proteins into the plant cytoplasm.
Ligand–receptor model in plant disease resistance
The ligand–receptor model suggests that a direct interaction between products of an R gene and an AVR gene is a key event in triggering resistance. So far, direct physical interaction between a cloned R gene product and an AVR gene product has only been demonstrated for Pto–avrPto (Scofield et al., 1996; Tang et al., 1996). Interaction between Pto and avrPto has a functional implication in bacterial speck resistance. However, Pto is unique among other cloned race-specific R gene products in lacking an obvious protein interaction domain. Initiation of Pto-mediated resistance does appear to require Prf, an NBS-LRR R gene, (Salmeron et al., 1996) in a probable downstream role in the Pto-mediated signal transduction pathway (Rathjen et al., 1999). Thus, in the Pto system, an NBS-LRR protein appears involved in a manner other than as a direct receptor for the AVR gene product. Although AvrB seems to be recognized inside the plant cells, a direct interaction between AvrB and RPM1 gene products has not been detected. Similarly, in the interaction between tomato and its fungal pathogen C.fulvum, the tomato Cf9 confers resistance to C.fulvum expressing Avr9. However, Avr9 bound to the membrane of both resistant Cf9 and susceptible cf9 tomato plants, suggesting that another protein may be the receptor (Kooman-Gersmann et al., 1996, 1998).
We demonstrate that the resistant Pi-ta LRD specifically interacts with AVR-Pita176 both in the two-hybrid assay and in the in vitro binding studies. The full-length Pi-ta protein binds to AVR-Pita176 protein that is bound to a membrane, but it shows minimal interaction, at best, in the yeast two-hybrid system. Pi-ta, with an estimated mol. wt of 105 kDa, and the GAL4 BD would form a large fusion protein that may not be suitable for the yeast two-hybrid system (Fields and Song, 1989). The lack of specificity in the binding of full-length Pi-ta to membrane-bound avr-pita176M178W remains to be explained, and may be a property of this particular mutant protein. Overall, the high degree of specificity maintained suggests that the physical interaction has functional implications in rice blast resistance. Therefore, this system provides direct biochemical evidence for the ligand–receptor model of gene-for-gene interactions.
LRRs in plant proteins have been implicated indirectly in binding pathogen-derived signal molecules that mediate recognition specificity. Wang et al. (1998) suggest that the LRR domain of Xa21D is sufficient for pathogen recognition. Recent work on the M locus of flax suggests that alterations in the LRR domain play a significant role in the evolution of rust resistance genes and generation of novel recognition specificities (Anderson et al., 1997). Similarly, differences in the LRR domains of the Cf gene products are suggested to be responsible for ligand binding specificity (Dixon et al., 1996).
Pi-ta encodes a protein with the same structural organization as the cytoplasmic NBS-LRR R genes, and it shows significant similarity to these R genes in its characteristic NBS domain. Pi-ta has an LRD that is similar in size and location to the LRR from the Arabidopsis RPM1 gene (Grant et al., 1995). The Pi-ta LRD has LXXLXXL repeats that may represent residual LRRs partially lost through evolution. Although the structure of the LRD of the Pi-ta protein differs from LRRs of other NBS-LRR proteins, the function of this LRD region from Pi-ta is likely to be similar to that of other LRRs. Thus, our data may also support the model that the LRR is the ligand-binding domain.
AVR-Pita176 may be a zinc metalloprotease
Results described here support the hypothesis that AVR-Pita encodes a pre-pro-protein that is processed to a 176 amino acid active form. AVR-Pita176, and not the intact 223 amino acid protein, is active as an HR elicitor when delivered into the plant cytoplasm. AVR-Pita176, and not AVR-Pita223, binds specifically to the LRD from resistant Pi-ta as determined by expression of the HIS3 reporter gene in the two-hybrid assay. Confirming the two-hybrid analysis, AVR-Pita176 that has been refolded on a membrane binds specifically to the LRD from resistant Pi-ta. For these in vitro studies, binding required addition of zinc during refolding of the AVR-Pita176 protein on the membrane, and during incubation of the membrane with Pi-ta and LRD extracts, as would be expected if AVR-Pita176 were a zinc metalloprotease. Mutant avr-pita proteins with conservative amino acid substitutions in the putative protease motif served as controls in our experiments. The avr-pita protein with the putative active site glutamic acid replaced with aspartic acid (E177D) failed to confer avirulence in whole-plant infection assays, failed as an HR elicitor in the transient expression assay and failed to interact with the Pi-ta LRD in the two-hybrid system. The avr-pita protein with tryptophan replacing methionine at residue 178 (M178W) in the putative protease motif failed to confer avirulence in infection assays, failed as an HR elicitor in the transient expression assay and failed to interact with the Pi-ta LRD in the in vitro binding studies. Our results are consistent with the hypothesis that AVR-Pita176 is a metalloprotease. Biochemical demonstration of enzymatic activity will ultimately answer this question.
How does the physical interaction between Pi-ta LRD and AVR-Pita176 trigger the Pi-ta-mediated defense response? One straightforward possibility is that the binding of Pi-ta to AVR-Pita176 leads to a conformational change that allows Pi-ta to interact with downstream proteins in the signal transduction pathway. Another possibility is that AVR-Pita176 may cleave the Pi-ta protein to an active form. Alternatively, the susceptible allele of Pi-ta might be inactive because it is degraded by AVR-Pita176. Pi-ta may be a HR inhibitor and degrading Pi-ta might activate other proteins to trigger the defense response. Arabidopsis RPM1 is the dicot R gene most closely related to Pi-ta, and RPM1 protein appears to be degraded with the onset of the HR and resistance response (Boyes et al., 1998). Again, demonstration of enzymatic activity and substrate specificity of AVR-Pita, as well as crystal structures of the AVR-Pita and Pi-ta proteins will provide insight into the triggering mechanism.
The instability of M.grisea avirulence genes may enable the fungus to escape recognition by corresponding plant R genes (Valent et al., 1997). Thus, rapid evolution of novel resistance specificities would appear crucial for the host. We have found that the resistance specificity to M.grisea expressing AVR-Pita was altered by substitution of serine for alanine at residue 918 of the Pi-ta protein (Bryan et al., 2000). In the present study, we demonstrate that impaired binding to AVR-Pita176 occurs when Ala918 in the LRD region is replaced by serine in the pi-ta protein. In the tomato Pto kinase, T204 in subdomain VIII of the Pto kinase appears to determine the ability to interact with avrPto both in vitro and in vivo (Frederick et al., 1998). Thus, in these instances, it appears that the inability of R protein to bind the pathogen signal molecule accounts for the loss of resistance.
In the battle between the plant and pathogen, clearly AVR genes in the pathogen did not evolve to prevent the pathogen from infecting its host. It is likely that plants have evolved the ability to detect pathogen molecules. For M.grisea, AVR-Pita is an infection-specific pathogen molecule that is highly expressed during the later stages of the infection cycle (G.T.Bryan and B.Valent, unpublished). The timing of expression correlates well with the penetration process of M.grisea. An exact role for AVR-Pita in pathogenesis remains to be demonstrated. However, this system represents an example where rice plants containing Pi-ta produce a resistance gene product in which a single amino acid difference permits binding to a pathogen protein and activation of defense responses. Further elucidation of the molecular basis of these physical interactions might allow precise manipulation of recognition specificity of plant R proteins to produce durable resistance to important plant diseases.
Materials and methods
Standard methods were used for DNA isolation and restriction enzyme digests (Ausubel et al., 1987). Color photo images were obtained using a CCD (charged-coupled device) camera (Optionix DEI750) coupled to a Leica MZ12 microscope. Photographs were captured using Adobe Photoshop 5.
Fungal strains, transformation and infection assays
Fungal transformation and infection assays were performed as described previously (Valent et al., 1991; Sweigard et al., 1995). Conidial suspensions (2.5 × 105 spores/ml) from fungal strain 4360-R-62 that contains AVR-Pita were inoculated onto 7-day-old rice seedlings growing on medium [1/2× MS (Murashige and Skoog, 1962) salt, supplemented with 100 mg/l-casein hydrolysate, pH 5.8, 0.5% agarose]. After inoculation, plates were sealed with parafilm and kept on a laboratory bench at room temperature.
Transient expression of AVR-Pita in rice seedlings
Plasmids. pGEM9Z (Promega) vectors pML63 and pML142 were gifts from Mary Locke (DuPont Agricultural Products). pML142 has an Adh1-6 intron sequence behind the 35S promoter (Figure 1A). pML63 has a CaMV 35S promoter and bacterial 3′ nos terminator sequence for expressing GUS (Figure 1B). All AVR-Pita constructs were first cloned in the NcoI and EcoRI site of pML63. A 1434 bp fragment containing the 35S promoter of each resulting plasmid was replaced by an 1897 bp fragment containing the 35S/Adh1-6 promoter from pML142 in the BamHI–NcoI site. Sequences of all constructs were verified using an ABI automatic sequencer. Primer sequences and constructs (Table I) are available on request.
Table I. List of plasmids used in this study.
| Plasmids | Description | Source |
|---|---|---|
| pML63 | Expression cassette for GUS | M.Locke |
| pML142 | Expression cassette for AVR-Pita | M.Locke |
| pCB980 | AVR-Pita ORF in ClaI–BamHI site of pBSSK (+) | Orbach et al. (2000) |
| pCB1646 | 0.5 kb NdeI–NotI fragment encoding AVR-Pita176 in pET28a (+) | this study |
| pCB1650 | NΔ586: Pi-ta LRD in-frame fusion to EcoRI–SalI of GAL4 DNA activation domain of pAD-GAL4 | this study |
| pCB1657 | AVR-Pita176 in-frame fusion to EcoRI–SalI of GAL4 DNA-binding domain of pBD-GAL4 | this study |
| pCB1760 | Pi-ta LRDA918S in-frame fusion to EcoRI–SalI of pBD-GAL4 | this study |
| pCB1761 | AVR-Pita176 in-frame fusion to EcoRI–SalI of pAD-GAL4 | this study |
| pCB1896 | NΔ586: Pi-ta LRD in-frame fusion to EcoRI–SalI of pBD-GAL4 | this study |
| pCB1900 | Pi-ta LRD in-frame fusion to an S-tag in EcoRI–SalI of modified pET 29a (+) | this study |
| pCB1902 | AVR-Pita ORF in-frame fusion to EcoRI–SalI of pAD-GAL4 | this study |
| pCB1906 | 2.8 kb NcoI–HindIII of Pi-ta ORF in pSL1180 | Bryan et al. (2000) |
| pCB1917 | NΔ247: 2.1 kb EcoRI–HindIII of Pi-ta NΔ247 in pBD-GAL4 | this study |
| pCB1921 | NΔ315: 1.8 kb EcoRI–SalI of Pi-ta in pBD-GAL4 | this study |
| pCB1922 | NΔ684: 0.7 kb EcoRI–HindIII of Pi-ta N684 in pBD-GAL4 | this study |
| pCB1925 | AVR-Pita176 E177D in-frame fusion to EcoRI–SalI of pAD-GAL4 | this study |
| pCB1927 | 0.5 kb NdeI–NotI fragment encoding AVR-Pita176M177W in pET28 (+) | this study |
| pCB1941 | 0.5 kb NcoI–KpnI fragment encoding AVR-Pita166 in pML142 | this study |
| pCB1947 | 0.6 kb NcoI–KpnI fragment encoding AVR-Pita176 in pML142 | this study |
| pCB2012 | 0.6 kb NcoI–KpnI fragment encoding avr-pita176E177D in pML142 | this study |
| pCB2013 | 0.6 kb NcoI–KpnI fragment encoding avr-pita176M178W in pML142 | this study |
| pCB2074 | 2.8 kb EcoRI–SalI Pi-ta ORF in-frame fusion to pBD-GAL4 | this study |
| pCB2111 | 0.7 kb KpnI–SpeI fragment encoding AVR-Pita223 in pML142 | this study |
Plant materials, seedling preparation and bombardment. Two resistant Pi-ta rice cultivars, Yashiro-mochi and the doubled haploid rice line YT14, and two susceptible pi-ta cultivars, Nipponbare and doubled haploid line YT16, were used for leaf transient assays. YT14 and YT16 are related doubled haploid rice lines derived from a cross between Yashiro-mochi and the susceptible variety Tsuyuake. Seeds were germinated on agarose medium, 1/2× MS salt with casein hydrolysate, and grown for 1 week in an incubator at 25°C in a 12 h photoperiod with 100 µE–1 of cool white light. At the two-leaf stage, seedlings were removed from the agarose medium, labeled and placed in a Petri dish containing a pre-wetted filter paper. Biolistic bombardment was performed using a Bio-Rad PDS-1000/He apparatus and 1150 p.s.i. rupture disks. Gold particles, 0.6 µm in diameter, were prepared according to the instructions provided by the manufacturer (Bio-Rad). For each co-bombardment, 0.5 µg of gold particles were coated with 0.5 µg of 35S::uidA and 1 µg of AVR-Pita expression plasmid. After bombardment, seedlings were maintained at 25°C for 48 h in Petri dishes containing pre-wetted filter paper in the above seedling conditions. Histochemical GUS staining was done using 5-bromo-4-chloro-3-indolyl glucuronide (X-gluc) (Biosynth AG) as a substrate (Seki et al., 1991).
RNA isolation and northern blot analysis
RNA isolation and northern blot analysis were done as previously described (Jia and Martin, 1999). The DNA probe for AVR-Pita was a PCR product amplified from AVR-Pita cDNA pCB980 (Orbach et al., 2000) using primers YL29 (5′-GGCGGGCTCCATGGGAACGCTATTC-3′) and YL33 (5′-CCCCCATGGCACAATATTTATAACGT-3′). Hybridization was performed at 42°C using a standard procedure (Ausubel, 1987). Hybridized filters were washed in 1× SSC, 0.1% SDS at 59°C for 20 min.
Two-hybrid plasmid construction, interactions and immunoblot analysis of bait protein in yeast
Mutant alleles encoding various regions of the Pi-ta protein and different AVR-Pita coding sequences were generated by PCR with Pfu DNA polymerase (Stratagene). Oligonucleotides containing 5′ EcoRI and 3′ SalI sites were used to amplify sequences from Pi-ta genomic clone pCB1641 and from cDNA clone pCB1906 (Bryan et al., 2000) for bait fusions. Similarly, oligonucleotides containing 5′ EcoRI and 3′ SalI were used to amplify AVR-Pita cDNA clone pCB980 (Orbach et al., 2000) for prey fusions. All amplified products were digested with EcoRI and SalI and cloned into the corresponding sites in two-hybrid bait pBD-GAL4 Cam phagemid vector and prey vector pAD-GAL4 phagemid (Stratagene). Each in-frame fusion was verified by sequencing. Primer sequences and constructs (Table I) are available on request.
Each BD and AD fusion presented in Figure 4 was co-transformed into yeast strain YRG2 using competent cells for examination of interaction (Stratagene) according to the manufacturer’s instruction. Briefly, transformed YRG2 cells were plated on synthetic minimal medium (SD) lacking leucine and tryptophan for expression of BD and AD fusions at 30°C for 3 days. The HIS3 reporter was examined by plating YGR2 cells on SD medium lacking histidine, leucine and tryptophan for 3 days. LacZ activity was examined by growing YRG2 cells containing both bait and prey fusions on SD lacking leucine and tryptophan at 30°C for 2 days. The yeast colonies were then blotted onto filter papers (VWR Scientific Products), and the filter papers were frozen rapidly in liquid nitrogen and air-dried. Each air-dried filter paper was then placed on top of a filter paper pre-wetted with Z buffer containing X-gal (98 ml of Z buffer, 0.27 ml of β-mercaptoethanol, 1.67 ml of X-gal stock solution/l). LacZ activities were recorded at 24 h after incubation at room temperature.
Levels of expression of all bait constructs in the GAL4 BD were determined by western blots. Briefly, total proteins were extracted and analyzed as described in the instructions with the two-hybrid kit (Clontech). The protein profile was visualized by Coomassie Blue staining. Equal amounts of proteins were electrophoresed on 10–15% gradient polyacrylamide gels and transferred to a nitrocellulose membrane (Novex) by electroblotting according to the manufacturer’s recommendations (Bio-Rad). GAL4 fusion proteins were detected by ECL (ECL kit, Pierce) using a monoclonal antibody to the GAL4 BD (Santa Cruz Biotechnology Inc.) according to the manufacturer’s instructions.
Expression of Pi-ta and LRD polypeptides in E.coli
A 2800 bp NcoI–HindIII DNA fragment encoding the open reading frame of the Pi-ta gene was isolated from pCB1906. The 1036 bp EcoRI–SalI DNA fragments encoding LRD and LRDA918S were isolated and purified from the two-hybrid bait vector. Each DNA fragment was then cloned into the corresponding site of a modified version of Novagen vector pET29a (+) (kindly provided by Dr Qun Zhu, DuPont CR&D) to introduce an N-terminal S tag (15 amino acids, Novagen). Bacterial host cells BL21 (DE3, Novagen) for expressing the fusion proteins were grown at 30°C with shaking at 250 r.p.m. Expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (1 mM final concentration) to the culture for 3 h. The bacterial pellets were suspended in buffer ST (50 mM Tris–HCl pH 8.0, 150 mM NaCl) with 1 mg/ml lysozyme (Sigma) and sonicated briefly on ice. Triton X-100 (Sigma) was added to 1% final concentration. Extracts were cleared by centrifugation at 13 000 r.p.m. for 10 min.
Modified far-western analysis
In vitro binding studies were performed with modification of the methods described by Chen and Evans (1995). Briefly, both AVR-Pita176 and virulent avr-pita176M178W were purified using an Ni-NTA column (Qiagen) and separated on 10–20% gradient SDS–polyacrylamide gels without previously boiling to denature, and blotted onto nitrocellulose membrane (Millipore). Non-specific binding to membranes was first blocked using 1% non-fat milk (Sigma), and then the AVR-Pita polypeptides were subjected to conditions for refolding. Membranes were first soaked for 30 min in 6 M urea. They were then soaked in 3 M urea for 15 min, in 1.5 M urea for 15 min, in 0.75 M urea (1 mM ZnCl2) for 15 min and subsequently in each half reduction of molarity of the solution for 15 min until the final solution was 0.05 M urea (1 mM ZnCl2). The final solution was replaced with blocking buffer (25 mM HEPES–KOH pH 7.7, 25 mM NaCl, 5 mM MgCl2, 1 mM ZnCl2 and 0.05% NP-40) containing total E.coli extracts expressing S-tagged-Pi-ta and LRD proteins. The membranes were rinsed briefly with 1× TBST (1× TBST = 10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.1% Tween-20). Immunodetection was done by a modification of methods described by Novagen. Briefly, membranes were incubated with a 1:5000 dilution of an S-antibody–horseradish peroxidase (HRP) conjugate (Novagen) in TBST for 15 min at room temperature, and membranes were washed thoroughly three times (1–2 min each time) with 25 ml of TBST at room temperature. The resulting image was visualized by detecting HRP chemiluminescence (ECL kit, Pierce) with X-ray film (Kodak). S protein perfect marker (Novagen) was used for molecular weight standards.
Acknowledgments
Acknowledgements
We thank Melissa Jia for proofreading and Todd Dezwaan, Qun Zhu, Greg Martin, Y.-Q.Gu, Guido Sessa, Adam Bogdanove, Zhixiong Xue, James Sweigard, Bob Larossa and anonymous reviewers for helpful comments on the manuscript. We gratefully acknowledge Qun Zhu and Allan Shapiro for advice on performing the far-western experiments, Mary Locke for plasmids pML63 and pML142, and Leonard Farrall, Kristina Faulk and Sue Morgan-Hoffman for technical assistance. We are indebted to DuPont sequencing and oligonucleotide synthesis laboratories for their assistance in designing primers and sequencing all constructs reported in this paper.
Note added in proof
Support for the ligand–receptor model to explain gene-for-gene specificity in plant disease resistance was obtained through transient expression of the Arabidopsis thaliana resistance gene RPS2 and its corresponding avirulence gene avrRpt2 in leaf mesophyll protoplasts, followed by co-immunoprecipitation of an in vivo complex containing the RPS2 and AvrRpt2 proteins.
Leister,R.T. and Katagiri,F. (2000) A resistance gene product of the nucleotide binding site–leucine rich repeats class can form a complex with bacterial avirulence proteins in vivo. Plant J., 22, 345–354.
References
- Anderson P.A., Lawrence,G.J., Morris,B.C., Ayliffe,M.A., Finnegan,E.J. and Ellis,J.G. (1997) Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell, 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Smith,J.A., Seidman,J.G. and Struhl,K. (eds) (1987) Protocols in Molecular Biology. John Wiley and Sons, New York, NY. [Google Scholar]
- Bent A.F. (1996) Plant disease resistance genes: function meets structure. Plant Cell, 8, 1757–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas U. and van den Ackerveken,G. (1997) Recognition of bacterial avirulence proteins occurs inside the plant cell: a general phenomenon in resistance to bacterial disease? Plant J., 12, 1–7. [DOI] [PubMed] [Google Scholar]
- Boyes D.C., Nam,J. and Dangl,J.L. (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc. Natl Acad. Sci. USA, 95, 15849–15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T., Schofield,P.R. and Sprengel,R. (1991) Amino-terminal leucine-rich repeats in gonadotropin receptors determine hormone selectivity. EMBO J., 10, 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan G.T., Wu,K.-S., Farrall,L., Jia,Y., Hershey,H.P., McAdams,S.A., Donaldson,G.K., Tarchini,R. and Valent,B. (2000) A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D.J. and Evans,R.M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Culver J.N. and Dawson,W.O. (1991) Tobacco mosaic virus elicitor coat protein genes produce a hypersensitive phenotype in transgenic Nicotiana sylvestris plants. Mol. Plant Microbe Interact., 4, 458–463. [Google Scholar]
- Cutt J.R. and Klessig,D.F. (1992) Pathogenesis-related proteins. In Boller,T. and Meins,F. (eds), Genes Involved in Plant Defense. Spring-Verlag, New York, NY, pp. 209–243. [Google Scholar]
- Dangl J.L. (1994) The enigmatic avirulence genes of phytopathogenic bacteria. Curr. Top. Microbiol. Immunol., 192, 99–118. [DOI] [PubMed] [Google Scholar]
- de Wit P.J.G.M. (1995) Fungal avirulence genes and plant resistance genes: unraveling the molecular basis of gene-for-gene interactions. Adv. Bot. Res., 21, 147–185. [Google Scholar]
- Dixon R.A., Harrison,M.J. and Lamb,C.J. (1994) Early events in the activation of plant defense responses. Annu. Rev. Phytopathol., 32, 479–501. [Google Scholar]
- Dixon M.S., Jones,D.A., Keddie,J.S., Thomas,C.M., Harrison,K. and Jones,J.D.G. (1996) The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell, 84, 451–459. [DOI] [PubMed] [Google Scholar]
- Fields S. and Song,O. (1989) A novel genetic system to detect protein–protein interactions. Nature, 340, 245–246. [DOI] [PubMed] [Google Scholar]
- Flor H.H. (1971) Current status of the gene-for-gene concept. Annu. Rev. Phytopathol., 9, 275–296. [Google Scholar]
- Freeling M. and Bennett,D.C. (1985) Maize Adh1. Annu. Rev. Genet., 19, 297–323. [DOI] [PubMed] [Google Scholar]
- Frederick R.D., Thilmony,R.L., Sessa,G. and Martin,G.B. (1998) Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol. Cell, 2, 241–245. [DOI] [PubMed] [Google Scholar]
- Galan J.E. and Collmer,A. (1999) Type III secretion machines: bacterial device for protein delivery into host cells. Science, 284, 1322–1328. [DOI] [PubMed] [Google Scholar]
- Goodman R.N. and Novarcky,A.J. (1994) The Hypersensitive Reaction in Plant to Pathogens: A Resistance Phenomenon. American Phytopathology Press, St Paul, MN. [Google Scholar]
- Gopalan S., Bauer,D.W., Alfano,J.R., Loniello,A., He,S. and Collmer,A. (1996) Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenecity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell, 8, 1095–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M.R., Godiard,L., Straube,E., Ashfield,T., Lewald,J., Sattler,A., Innes,R.W. and Dangl,J.L. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science, 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack K.E. and Jones,J.D.G. (1997) Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol., 48, 575–607. [DOI] [PubMed] [Google Scholar]
- Howard R.J., Ferrari,M., Roach,D.H. and Money,N.P. (1991) Penetration of hard substrates by a fungus employing enormous turgor pressures. Proc. Natl Acad. Sci. USA, 88, 11281–11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y. and Martin,G.B. (1999) Rapid transcript accumulation of pathogenesis-related genes during an incompatible interaction in bacterial speck disease resistant tomato plants. Plant Mol. Biol., 40, 455–465. [DOI] [PubMed] [Google Scholar]
- Jia Y., Loh,Y.-T., Zhou,J. and Martin,G.B. (1997) Alleles of Pto and Fen occur in bacterial speck-susceptible and fenthion-insensitive tomato lines and encode functional protein kinases. Plant Cell, 9, 61–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten M.H.A.J., Cozijnsen,A.J. and de Wit,P.J.G.M. (1994) Host resistance to a fungal tomato pathogen lost by a single base-pair change in an avirulence gene. Nature, 367, 384–386. [DOI] [PubMed] [Google Scholar]
- Jones D.A. and Jones,J.D.G. (1996) The roles of leucine rich repeats in plant defences. Adv. Bot. Res., 24, 89–167. [Google Scholar]
- Knogge W. (1996) Fungal infection of plants. Plant Cell, 8, 1711–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B. and Deisenhofer,J. (1993) Crystal structure of porcine ribonuclease inhibitor, a protein with leucine rich repeats. Nature, 366, 751–756. [DOI] [PubMed] [Google Scholar]
- Kooman-Gersmann M., Honee,G., Bonnema,G. and de Wit,P.J.G.M. (1996) A high-affinity binding site for the AVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell, 8, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooman-Gersmann M., Vogelsang,R., Vossen,P., van den Hooven,H.W., Mahe,E., Honee,G. and de Wit,P.J.G.M. (1998) Correlation between binding affinity and necrosis-inducing activity of mutant AVR9 peptide elicitors. Plant Physiol., 117, 607–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister R.T., Ausubel,F.M. and Katagiri,F. (1996) Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc. Natl Acad. Sci. USA, 93, 15497–15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A., Tenhaken,R., Dixon,R. and Lamb,C. (1994) H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell, 79, 583–593. [DOI] [PubMed] [Google Scholar]
- Mehdy M.C. (1994) Active oxygen species in plant defense against pathogens. Plant Physiol., 105, 467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G.B., Brommonschenkel,S., Chunwongse,J., Fray,A., Ganal,M.W., Spivey,R., Wu,T., Earle,E.D. and Tanksley,S.D. (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science, 262, 1432–1436. [DOI] [PubMed] [Google Scholar]
- Mindrinos M., Katagiri,F., Yu,G.L. and Ausubel,F.M. (1994) The A.thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell, 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Murashige T. and Skoog,F. (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant, 15, 473–479. [Google Scholar]
- Orbach M.J., Farrall,L., Sweigard,J.A., Chumley,F.G. and Valent,B. (2000) A telomeric avirulence gene determines efficacy for rice blast resistance gene Pi-ta. Plant Cell, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen J.P., Chang,J.H., Staskawicz,B.J. and Michelmore,R.W. (1999) Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J., 18, 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohe M., Gierlich,A., Hermann,H., Hahn,M., Schmidt,B., Rosahl,S. and Knogge,W. (1995) The race-specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J., 14, 4168–4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J.M., Oldroyd,G.E.D., Rommens,C.M.T., Scofield,S., Kim,H.S., Lavelle,D.T., Dahlbeck,D. and Staskawicz,B.J. (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell, 86, 123–133. [DOI] [PubMed] [Google Scholar]
- Scofield S.R., Tobias,C.M., Rathjen,J.P., Chang,J.H., Lavelle,D.T., Michelmore,R.W. and Staskawicz,B.J. (1996) Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science, 274, 2063–2065. [DOI] [PubMed] [Google Scholar]
- Seki M., Komeda,Y., Lida,A., Yamada,Y. and Morikawa,H. (1991) Transient expression of β-glucuronidase in Arabidopsis thaliana leaves and Brassica napus stems using a pneumatic particle gun. Plant Mol. Biol., 17, 259–263. [DOI] [PubMed] [Google Scholar]
- Silue D., Nottenghem,J.L. and Tharreau,D. (1992) Evidence for a gene-for-gene relationship in the Oryza sativa–Magnaporthe grisea pathosystem. Phytopathology, 82, 577–580. [Google Scholar]
- Sweigard J.A., Carroll,A.M., Kang,S., Farrall,L., Chumley,F.G. and Valent,B. (1995) Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus. Plant Cell, 7, 1221–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Frederick,R.D., Zhou,J., Halterman,D.A., Jia,Y. and Martin,G.B. (1996) Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science, 274, 2060–2063. [DOI] [PubMed] [Google Scholar]
- Tatsumi H., Ikegaya,K., Murakami,S., Kawabe,H. and Nakano,E. (1994) Elucidation of the thermal stability of the neutral proteinase II from Aspergillus oryzae. Biochim. Biophys. Acta, 1208, 179–185. [DOI] [PubMed] [Google Scholar]
- Valent B. (1997) The rice blast fungus, Magnaporthe grisea. Plant relationships. In G.C.Carroll and P.Tudzynoski (eds), The Mycota V Part B, Springer-Verlag, Heidelberg, Germany, pp. 37–54. [Google Scholar]
- Valent B. and Chumley,F.G. (1991) Molecular genetic analysis of the rice blast fungus, Magnaporthe grisea. Annu. Rev. Phytopathol., 29, 443–467. [DOI] [PubMed] [Google Scholar]
- Valent B., Farrall,L. and Chumley,F.G. (1991) Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics, 127, 87–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ackerveken G., Marois,E. and Bonas,U. (1996) Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell, 87, 1307–1316. [DOI] [PubMed] [Google Scholar]
- Vos P. et al. (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nature Biotechnol., 16, 1365–1369. [DOI] [PubMed] [Google Scholar]
- Wang G.-L. et al. (1998) Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subjected to adaptive evolution. Plant Cell, 10, 765–779. [DOI] [PMC free article] [PubMed] [Google Scholar]