Abstract
The yeast Gal4/UAS transcriptional activation system is a powerful tool for regulating gene expression in Drosophila and has been increasing in popularity for developmental studies in zebrafish. It is also useful for studying the basis of de novo transcriptional silencing. Fluorescent reporter genes under the control of multiple tandem copies of the upstream activator sequence (UAS) often show evidence of variegated expression and DNA methylation in transgenic zebrafish embryos. To characterize this systematically, we monitored the progression of transcriptional silencing of UAS-regulated transgenes that differ in their integration sites and in the repetitive nature of the UAS. Transgenic larvae were examined in three generations for tissue-specific expression of a green fluorescent protein (GFP) reporter and DNA methylation at the UAS. Single insertions containing four distinct upstream activator sequences were far less susceptible to methylation than insertions containing fourteen copies of the same UAS. In addition, transgenes that integrated in or adjacent to transposon sequence exhibited silencing regardless of the number of UAS sites included in the transgene. Placement of promoter-driven Gal4 upstream of UAS-regulated responder genes in a single bicistronic construct also appeared to accelerate silencing and methylation. The results demonstrate the utility of the zebrafish for efficient tracking of gene silencing mechanisms across several generations, as well as provide useful guidelines for optimal Gal4-regulated gene expression in organisms subject to DNA methylation.
Keywords: Gal4, DNA methylation, transgene silencing
Introduction
Aberrant regulation of gene expression by epigenetic processes results in diverse developmental disorders and is a feature of tumorigenic cells (Jiang et al., 2004; Robertson, 2005; Sharma et al., 2010). Methylation of DNA and histones can lead to transcriptional repression, but the cues that cause specific genomic regions to be modified in this manner are not fully understood (Campos and Reinberg, 2009; Goll and Bestor, 2005).
One type of sequence that is prone to silencing by methylation is repetitive DNA. Endogenous regions carrying multiple blocks of similar sequences >1 kilobases (kb) in length, such as those found at centromeres or the D4Z4 and NBL2 microsatellite repeats of the human genome, are known to accumulate repressive chromatin marks including DNA methylation (Kondo et al., 2000; Miller et al., 1974; Ponzetto-Zimmerman and Wolgemuth, 1984). Such silencing of repetitive sequences can have functional consequences. For example, silencing of the D4Z4 repeats was recently shown to repress expression of a polymorphic allele of a gene that would otherwise trigger the human disease Facioscapulohumeral muscular dystrophy (Lemmers et al., 2010).
Studies using transgenic constructs newly introduced into the genome support the idea that the repetitive nature of a DNA sequence is a strong cue for silencing. In mouse and plants, transgenes that integrate into the genome as high copy number concatemeric arrays typically show decreased expression (Davis and MacDonald, 1988; Linn et al., 1990; Mittelsten Scheid et al., 1991; Robertson et al., 1995; Sharpe et al., 1993). The link between silencing and repetitive DNA was elegantly demonstrated by Garrick et al. (Garrick et al., 1998) who established a mouse line carrying approximately 100 repeats of an erythroid-specific LacZ transgene flanked by loxP sites. Initially, animals showed very low expression of LacZ in less than 1% of cells and a high accumulation of DNA methylation. However, when embryos were injected with Cre recombinase, the resultant mice carried a single copy of the transgene, which showed less methylation and, correspondingly, a more than 1000-fold increase in the number of cells expressing LacZ (Garrick et al., 1998).
Short tandem repeats with unit lengths of less than 100 base pairs (bp) are also widespread in eukaryotic genomes (Boby et al., 2005), and there is some evidence for their silencing. Short repeats that contain CpG dinucleotides, such as those associated with Fragile X syndrome and other trinucleotide expansion diseases, accumulate methylation that is correlated with reduced gene expression (Oberle et al., 1991). Short tandem repeats may also be involved in the epigenetic regulation of imprinted genes, as they are often enriched in the surrounding DNA (Hutter et al., 2006). However, the potential for short tandem repeats to accumulate epigenetic marks associated with silencing has not been explored in depth.
The Gal4/UAS regulatory system serves as a useful model for monitoring DNA methylation and transcriptional silencing of a short tandem repeat. In yeast, the Gal4 transcription factor binds to upstream activating sequences (UAS) to direct transcription of genes necessary for metabolism of galactose (Giniger et al., 1985). Each UAS is 17 base pairs long, roughly palindromic, and in the form of CGG-N11-CCG. The CpG dinuleotides are essential for Gal4 binding (Marmorstein et al., 1992) and serve as a target for methylation (Goll et al., 2009). The Gal4/UAS system was first adapted to zebrafish by Scheer and Campos-Ortega (Scheer and Campos-Ortega, 1999), who assayed reporter expression under the control of 5 UAS copies (5X UAS). It was difficult to obtain high levels of expression from these constructs, most likely because they were integrated as large concatemers of multiple transgenes, which made them susceptible to silencing. To compensate for the low expression, Köster and Fraser (Köster and Fraser, 2001) used the potent Gal4-VP16 fusion protein for transcriptional activation and modified constructs designed for over expression screens in Drosophila that contained fourteen tandem copies of a synthetically generated upstream activating sequence (14X UAS) (Rorth, 1996). While this approach resulted in robust expression, a high level of toxicity was observed and stable transgenic lines were not generated. Since this initial work, new technologies such as Tol2 transposition have become available that allow integration of transgenes as single copies, thereby eliminating the problems associated with insertions containing complex concatemeric arrays (Kawakami et al., 2000). High levels of gene expression are obtained in transient embryo injection assays when Gal4-VP16 binds to the 14X UAS to promote transcription of the gene encoding green fluorescent protein (GFP) (Köster and Fraser, 2001). However, when stably integrated into the genome as single copy sequence, the same 14X UAS is prone to CpG methylation. Transgenic embryos show variegated GFP expression that correlates with increased DNA methylation, and silenced transgenes can be reactivated in larvae with hypomethylated genomes (Feng et al., 2010; Goll et al., 2009). Strikingly, while there is minimal silencing in the first generation, it is exacerbated upon propagation through later generations (Goll et al., 2009). Therefore, using the Gal4/UAS system, one can monitor the progression of methylation of short repeats and probe the cues that cause their silencing.
Silencing of UAS-regulated transgenes can be a technical challenge for the zebrafish field. This especially applies to studies of developmental processes that require all cells of a given population to express the UAS-regulated transgene, such as in genetic ablation of a specific cell type. The presence of DNA methylation machinery in fish and the associated variegation or silencing of gene expression is an impediment to creating the repertoire of powerful Gal4-based tools currently available for the Drosophila community.
Some efforts have been made toward optimizing the Gal4/UAS system for zebrafish. Using a luciferase-based assay in cultured zebrafish fibroblasts, Distel et al. demonstrated that expression from UAS constructs increased linearly from 1 to 5 UAS copies until leveling off, indicating that fewer than 14 copies of the UAS can provide an effective substrate for Gal4-VP16 in zebrafish cells and in transgenic animals (Distel et al., 2009). In other work, stable transgenic lines carrying fluorescent reporter genes driven by 5 copies of the UAS were shown to produce strong labeling (Asakawa et al., 2008; Collins et al., 2010). However, these studies did not directly address the susceptibility of UAS variants to DNA methylation and transcriptional silencing over multiple generations.
We set out to test systematically how UAS sites with different copy number and sequence diversity behave in vivo, by monitoring reporter expression in transgenic animals for three generations and correlating it with methylation at the UAS repeats. Four distinct Gal4 binding sites were placed in tandem and expression from this non-repeating construct (4Xnr UAS) was compared to the 14X UAS commonly used for many studies in zebrafish (for example: (Campbell et al., 2007; Davison et al., 2007; Douglass et al., 2008; Köster and Fraser, 2001; Pisharath and Parsons, 2009; Scott et al., 2007). We show that the 4Xnr UAS drives high levels of reporter expression and is significantly less susceptible to methylation than the 14X UAS. In addition, we find that silencing and methylation are enhanced when promoter-driven Gal4 is placed upstream of UAS-regulated responder genes in a bicistronic construct. Our findings suggest strategies for effective Gal4-regulated gene expression in transgenic zebrafish. Moreover, the results support the hypothesis that sequence or structural cues embedded in short tandem repeats attract DNA methylation and demonstrate the utility of the zebrafish for elucidating the specific nature of these cues in a live organism.
Materials and methods
Zebrafish strains
All studies were performed with the Oregon AB strain of wild type zebrafish (Walker, 1999). Dual reporter transgenic lines were maintained by outcrossing to AB fish. The Tg(ptf1a:Gal4-VP16)jh16 driver line (Pisharath and Parsons, 2009), was used to evaluate expression from independently derived Tg(UAS:GFP) reporter lines. Embryos and larvae were reared at 27°C and scored at the indicated hours (hpf) and days (dpf) post fertilization.
Constructs
Gal4-VP16/UAS dual reporters
A Gal4-VP16-2A-mCherry construct was generated by overlap-extension PCR (Wurch et al., 1998) using the SAGVG and UAS-E1b:nfsB-mCherry plasmids (Davison et al., 2007) as templates. During translation of the viral 2A peptide, a peptide bond fails to form between Gly-Pro, resulting in equimolar amounts of Gal4-VP16 and mCherry from a single transcript (Donnelly et al., 2001; Provost et al., 2007), enabling the intensity of the fluorescent label to be used as a read-out of Gal4-VP16 expression. The entire fragment was inserted into the BamHI site of pT2KXIG in (Urasaki et al., 2006) just downstream of the EF1α promoter. Sequences containing 14, 9, 6, or 1 copies of the UAS (CGGAGTACTGTCCTCCG) along with the E1b minimal promoter were PCR amplified from SAGVG (Davison et al., 2007), and inserted upstream of the GFP coding sequence using BclII and MluI sites. The 4Xnr UAS (see below) was also tested. The EF1α:Gal4-VP16-2A-mCherry and UAS:GFP components are flanked by the Tol2 arms in the modified pT2KXIGΔ in plasmids.
4Xnr UAS synthesis
To create the non-repetitive 4X UAS, four unique upstream activation sequences were cloned in tandem. Two UAS sequences (UAS I: CGGATTAGAAGCCACCG, UAS II: CGGGTGACAGCCCTCCG) that exhibited high affinity for the Gal4 DNA binding domain in vitro (Kang et al., 1993) were derived from the UASG promoter of the yeast GAL1 and GAL10 genes (Giniger et al., 1985). A single G=>A mutation was introduced into the UAS II sequence to abolish a CpG dinucleotide not essential for Gal4 binding. The other two UAS were synthetic near-consensus sequences (CGGAAGACTCTCCTCCG, CGGAGTACTGTCCTCCG) previously found to drive robust expression of reporter genes (Giniger et al., 1985; Webster et al., 1988). The four UAS sequences were separated by 10 bp spacer sequences and the second and 4th UAS were placed in reverse orientation to minimize further the repetitive nature of the multicopy UAS (Supplemental Fig. 1).
Gal4FF/UAS bicistronic reporters
Gal4FF-2A-mCherry was generated by overlap-extension PCR using the Gal4-VP16 dual reporter vector and pT2KSAGFF (Asakawa et al., 2008) as templates. The Gal4FF-2A-mCherry fragment was cloned into pT2KXIG in as above with either 14X or 4Xnr UAS:GFP.
UAS:GFP
To produce UAS-regulated reporter plasmids to test with the Gal4 driver line Tg(ptf1a:Gal4-VP16)jh16, UAS:GFP components were excised from the dual reporter constructs in pT2KXIG in and subcloned into the BamHI site of a pBluescript (Stratagene) plasmid modified by the addition of Tol2 arms (gift from S. Fisher, U. Pennsylvania).
Production of transgenic lines
Plasmid DNA for transgenic constructs (50 ng/μL) and Tol2 transposase mRNA (50 ng/μL) were coinjected into 1-cell stage embryos, which were raised to adulthood. To identify transgenic founders, F0 adults that had been injected with the dual reporter constructs were mated to AB, whereas those injected with UAS:GFP constructs were mated to the Tg(ptf1a:Gal4-VP16)jh16 driver line. Progeny were screened and only fluorescent F1 larvae were used to establish stable transgenic lines.
Fluorescence intensity analysis
At 2 dpf, individual larvae (n=10) were sampled from Tg(ptf1a:Gal4-VP16)jh16 lines bearing either 14X UAS:GFP or 4Xnr UAS:GFP single copy insertions. Images were captured with identical settings on a Leica MZ16 dissecting microscope outfitted with a Leica DC500 camera. Fluorescent pixel intensities were quantified using MetaMorph Offline (v.7.6) and compared by one-way ANOVA followed by Tukey’s post-hoc comparison using JMP 8.0 software.
Analysis of transgene copy number
Total genomic DNA (10 μg) was extracted from fin clips of F1 adults, digested with EcoRI, electrophoresed on 1% agarose gels, and analyzed by Southern blotting (Southern, 1975). Membranes were probed with radiolabeled DNA corresponding to GFP sequence, generated by digesting pEGFP-1 plasmid (Clontech) with SacII and NotI.
Mapping transgenic insertion sites
The genomic positions of UAS-regulated transgenic insertions were mapped by linker-mediated PCR as in Davison et al. (2007), using total genomic DNA extracted from fin clips of F1 adults. Sequences flanking the Tol2 arms were used to BLAT search the UCSC genome browser (Zv8/danRer6 assembly) to map sites of insertion within the zebrafish genome.
DNA bisulfite sequencing
DNA bisulfite sequencing was performed on individual 3 dpf larvae, as described (Goll et al., 2009). Doubly transgenic adult fish carrying both the Tg(ptf1a:Gal4-VP16)jh16 driver and each UAS transgene of interest were mated with wild type adults and the resultant GFP-positive progeny (25%) scored for the level of GFP labeling. Larvae showing the highest and lowest GFP labeling from within the clutch were selected for analysis of DNA methylation patterns. 14X and 4Xnr sequence was amplified using primers TTTAAGATGAAATGTGTTTT and TCCATTATATACCCTCTAAA followed by GGGATTATATTAAGTTTAGGT and CCATTATATACCCTCTAAAA. EF1α sequence was amplified using primers GGTTGAATGTTTTGTTAAGA and CAAAAACATCTTCCCATTC followed by GGTTGAATGTTTTGTTAAGA and TAAAAACTTTACCCCCTCCATATA. In all DNA bisulfite sequencing experiments, CpG methylation patterns were determined for 2–3 individual larvae from each subgroup, with at least 8 cloned sequences examined per individual. Less than 1% of CpH dinucleotides (where H=A, C or T) were methylated in all samples. Statistical analyses of bisulfite data were performed using QUMA (Kumaki et al., 2008).
Results
Toxic effects of ubiquitous Gal4-VP16 expression
Our initial plan was to evaluate transcription from five different UAS copy-number variants (14X, 9X, 6X, 4X, 1X) in a bipartite construct that also contained Gal4-VP16 under the control of the EF1α promoter (Supplemental Fig. 1A). The EF1α promoter drives fairly ubiquitous expression in transgenic zebrafish larvae (Amsterdam et al., 1995; Linney et al., 1999). Inclusion of the viral 2A peptide followed by mCherry yields Gal4-VP16 and the red fluorescent protein in equimolar amounts (Provost et al., 2007). Thus, with this construct, we could monitor Gal4-VP16 protein production indirectly through mCherry labeling and confirm the extent of expression from the EF1α promoter.
We recovered several founder fish for each UAS construct whose F1 progeny displayed widespread mCherry and GFP labeling, suggesting that the bipartite vector functioned effectively when integrated into the genome (Supplemental Fig. 1B and data not shown). Although mCherry-positive transgenic F1 larvae appeared morphologically wild type at 24 hpf, those that were brightly fluorescent, regardless of UAS copy-number, developed defects by 5 dpf and did not survive. Larvae with the highest fluorescence labeling exhibited gross morphological abnormalities after a few days (Supplemental Fig. 1B), whereas lower expressing larvae lived longer but rarely survived to adulthood (two escapers developed into adults with prominent eye defects). As had been suggested previously (Köster and Fraser, 2001), these results provided additional evidence that ubiquitous expression of Gal4-VP16 is incompatible with normal development and precluded this approach for analyzing the efficacy of UAS variants.
Transgenerational silencing of Gal4FF dual reporters
To reduce the toxic effects that were observed when constructs expressing ubiquitous Gal4-VP16 were stably integrated into the genome, the Gal4-VP16 coding sequence was replaced with sequence encoding Gal4FF. Gal4FF consists of the DNA binding component of the Gal4 protein fused to two phenylalanine-bearing motifs from the VP16 transcriptional activator (Asakawa et al., 2008). Robust activation of UAS-regulated transgenes and minimal toxicity had been reported for Gal4FF in zebrafish (Asakawa et al., 2008). We focused on constructing two modified dual reporter constructs, one with GFP under the control of the commonly used 14X UAS and another containing 4 different upstream activation sequences that, individually, are known to function as Gal4 binding sites (Giniger et al., 1985; Kang et al., 1993; Webster et al., 1988). In addition to decreasing the number of copies, the repetitive nature of the UAS regulatory region was further reduced by including distinct rather than identical copies of the UAS (~50% identity, refer to Materials and methods and Supplemental Fig. 2). This synthetic construct is referred to as the 4 copy, non-repetitive UAS or 4Xnr UAS.
We recovered one 14X UAS and three different 4Xnr UAS transgenic lines, all of which showed widespread expression of mCherry and GFP in the F1 generation (Fig. 1A and data not shown). Unlike Gal4-VP16, the Gal4FF modified construct did not interfere with viability. F1 transgenic larvae were successfully raised to adulthood and their progeny analyzed.
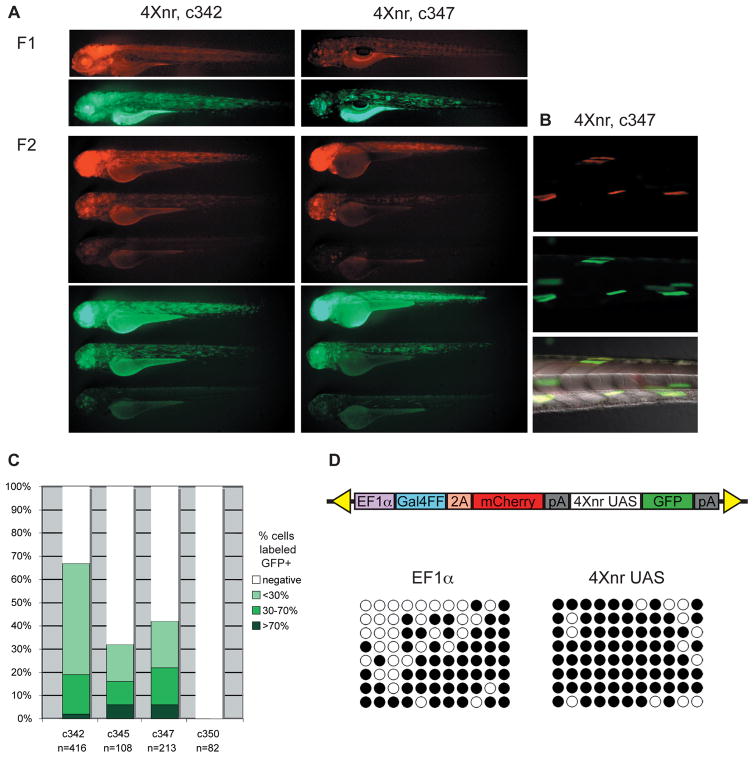
Figure 1. Silencing across Gal4FF bicistronic transgenic insertions.
(A) Lateral views of GFP and mCherry labeling in 3 dpf larvae from two transgenic lines in which GFP is regulated by the 4Xnr UAS. GFP expression recapitulates the pattern of mCherry in independently derived F1 larvae, which can be variable in their fluorescence. Representative sibling larvae in the F2 generation show widespread, highly mosaic, or largely absent mCherry and GFP labeling. (B) Colocalization of variegated mCherry and GFP fluorescence in muscle fibers of a c347 F2 larva. (C) Comparison of approximate number of GFP labeled cells in F2 larvae from lines carrying the 4Xnr UAS (c342, c345, c347) and the 14X UAS (c350). (D) Schematic of Gal4FF bipartite reporter construct and analysis of CpG methylation in c347 F2 larva from DNA bisulfite sequencing. Methylation at eleven CpGs within the EF1α promoter and the 4Xnr UAS are indicated on the horizontal axis, with black circles indicating methylated CpGs and open circles representing unmethylated CpGs. Patterns from eight different representative clones from one larva are shown on the vertical axis.
In contrast to the robust fluorescence observed in F1 larvae, F2 larvae showed significantly fewer cells with GFP labeling (Figs. 1A–C). Neither expression of GFP nor mCherry was detected in F2 larvae carrying the 14X UAS. In F2 larvae where GFP was under control of the 4Xnr UAS, only a subset of cells was labeled with GFP. Moreover, mCherry labeling showed a variegated pattern that colocalized with GFP labeled cells (Fig. 1A, B). Variegation or loss of mCherry-positive cells indicated that the Gal4FF protein was not being expressed ubiquitously, as would be expected under the regulation of the EF1α promoter.
To explore the reasons for the reduction in mCherry labeling, we examined the methylation status of the variegating transgenes by DNA bisulfite sequencing. We found substantial methylation of not only the multicopy UAS (average of 84% CpG methylation), but also of the EF1α promoter sequence (average of 64% CpG methylation) in F2 larvae (n=2 larvae assayed; Fig. 1D and data not shown). Methylation of the EF1α and the correlated transcriptional silencing of Gal4FF prevented the comparative analysis of UAS variants. We therefore turned to a binary approach, assaying UAS variants as independent transgenes introduced into Gal4 driver lines by mating of adult fish.
Increased variegated expression from 14X UAS transgenic insertions
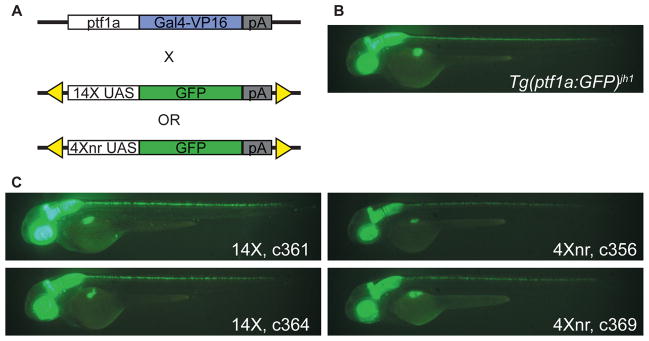
To compare the expression of GFP under control of the 14X or 4Xnr UAS, we generated new transgenic lines from separate UAS:GFP Tol2 constructs and identified carriers using the established Gal4 driver line Tg(ptf1a:Gal4-VP16)jh16 (Pisharath and Parsons, 2009 and Fig. 2A). At 2 dpf, the pancreas specific transcription factor 1a (ptf1a) gene is expressed in the retina, hindbrain, spinal cord, and pancreas primordium (Lin et al., 2004). Regulatory elements contained within 150 kb of genomic DNA in a bacterial artificial chromosome drive expression in the same tissues with high fidelity (Park et al., 2008). Six independent founders carrying 14X UAS:GFP and 8 carrying 4Xnr UAS:GFP were identified by mating adults raised from injected embryos to Tg(ptf1a:Gal4-VP16)jh16 fish and their fluorescent F1 progeny were raised to adulthood. Although the expected tissue-specific pattern of fluorescence was recovered (compare Figs. 2B and C), considerable variability in GFP labeling was observed between F1 larvae from different founders (Supplemental Fig. 3).
Figure 2. Assay of UAS constructs in binary transgenic system.
(A) Experimental scheme using the Tg(ptf1a:Gal4-VP16)jh16 driver line for transcriptional activation of 14X and 4Xnr UAS:GFP reporter transgenes. (B) Pattern of tissue-specific fluorescence in Tg(ptf1a:GFP)jh1/+ larvae at 2 dpf (Pisharath et al., 2007). (C) In the presence of the ptf1a driver, larvae from independently derived lines that carry either 14X UAS:GFP or 4Xnr UAS:GFP transgenes show a similar pattern of GFP labeling.
Variability in expression in the progeny from independently derived founders could be due to differences in the number of Tol2 insertions or reflect position effects associated with sites of integration. To address this issue, we examined genomic DNA isolated from tail fin clips of F1 adults by Southern blotting (Supplemental Fig. 4). We focused on two 14X UAS (c361 and c364) and two 4Xnr UAS carriers (c356 and c369) that had shown the complete ptf1a pattern of GFP labeling in F1 larvae (Fig. 2C), as well as one 4Xnr UAS F1 that had exhibited considerably fewer expressing cells in all GFP positive tissues (c368, refer to Fig. 4). The 14X UAS F1 c364 had one insertion, whereas the other, c361, contained more than 10 insertions. The high expressing 4Xnr UAS F1 individuals contained a single transgene insertion, while the c368 F1 carried two insertions. These data confirmed previous observations (Goll et al., 2009) that variegation in expression is not correlated with lower transgene copy number.
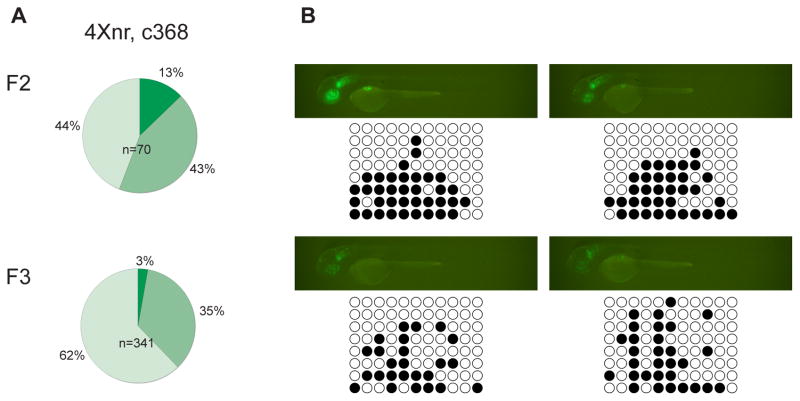
Figure 4. Correlation between variegated expression and CpG methylation.
(A) In the c368 line, variegation in GFP labeling was observed in F1 larvae (not shown) and was more frequently observed in F2 and F3 larvae compared to other 4Xnr UAS lines (compare with Fig. 3B). Few c368 GFPhigh larvae were found in either generation. (B) Fluorescence images and representative DNA bisulfite sequencing data for individual GFPmed and GFPlow c368 F2 larvae at 2 dpf. The upper larvae possess both the c368a and c368b transgenic insertions (Table 2), whereas the bottom two larvae have only the c368a transgene. GFP-labeled individuals carrying only the c368b insertion were not detected.
Integration sites were determined by linker-mediated PCR where possible. The single transgenes in c356, c364 and c369 were all found to be located in or near predicted genes (Table 1). The c368 genome contains two insertions, with one transgene situated in an intron and the second within a DNA2-2 DR repetitive element. Transgenic insertions from several other independently isolated F1 fish that had exhibited mosaic GFP labeling as larvae were mapped and positioned either directly in or immediately adjacent to repetitive elements (refer to Supplemental Fig. 3 and Table 2).
Table 1.
Insertion locations of transgenic lines with robust GFP expression
| UAS | Line | Linkage group | Insertion position | Insertion orientationa | Ensembl gene reference |
|---|---|---|---|---|---|
| 14X | c361 | multiple transgenes | ---- | ||
| 14X | c364 | 8: 27.029 Mb | Intron in novel protein similar to solute carrier family 26, member 5 (Slc26a5) gene | same transcriptional orientation | ENSDARG00000076957 |
| 4Xnr | c356 | 3: 20.455 Mb | 175 bp 5′ to casc3 | same transcriptional orientation | ENSDARG00000029911 |
| 4Xnr | c369 | 7: 18.123 Mb | Intron in coro1b | opposite transcriptional orientation | ENSDARG00000008660 |
Orientation of GFP in transgene relative to transcriptional orientation of nearest gene
Table 2.
Insertion locations of transgenic lines with variegated GFP expression
| UAS | Line | Linkage group | Insertion position | Insertion orientationa | Ensembl gene reference |
|---|---|---|---|---|---|
| 14X | c360a | ---- | DNA-8-9_DR repetitive element | opposite transcriptional orientation | ---- |
| 14X | c360b | ---- | TE-X-5_DR repetitive element | same transcriptional orientation | ---- |
| OR | |||||
| Kolobok-N7_DR repetitive element | opposite transcriptional orientation | ||||
| 14X | c362 | ---- | Tc1-4_DR repetitive element | same transcriptional orientation | ---- |
| 4Xnr | c357 | 17: 12.881 Mb | Intron in LOC571485 | opposite transcriptional orientation | ENSDARG00000073866 |
| Flanked by Tc1N1_DR repetitive elements | same transcriptional orientation | ||||
| 4Xnr | c367 | 9: 9.695 Mb | Intron in nrp2b | same transcriptional orientation | ENSDARG00000038446 |
| 13 bp dowstream from Polinton-1N1_DR repetitive element | opposite transcriptional orientation | ||||
| 4Xnr | c368a | Zv8_NA1912:32 Kb | Intron in PLA2G4C | opposite transcriptional orientation | ENSDARG00000036713 |
| 165 bp upstream from DNA-TTAA-2_DR repetitive element | same transcriptional orientation | ||||
| 4Xnr | c368b | ---- | DNA2-2 DR repetitive element | same transcriptional orientation | ---- |
| 4Xnr | c370 | ---- | DNA-8-13_DR repetitive element | opposite transcriptional orientation | ---- |
Orientation of GFP in transgene relative to transcriptional orientation of nearest gene or repetitive element
Doubly transgenic F1 fish bearing the Tg(ptf1a:Gal4-VP16)jh16 driver and 4Xnr UAS:GFP (c356, c369, c368) or 14X UAS:GFP (c361, c364) variants were outcrossed to the AB wild type strain to produce F2 progeny and to establish stable transgenic lines. We compared the intensity of GFP labeling between F2 larvae carrying single insertions of the 4Xnr UAS or 14X UAS. Larvae bearing one 4Xnr UAS transgene showed a modest reduction (20–30%) in mean fluorescence intensity levels (refer to Methods) compared to larvae with a single 14X UAS:GFP insertion (Supplemental Fig. 5).
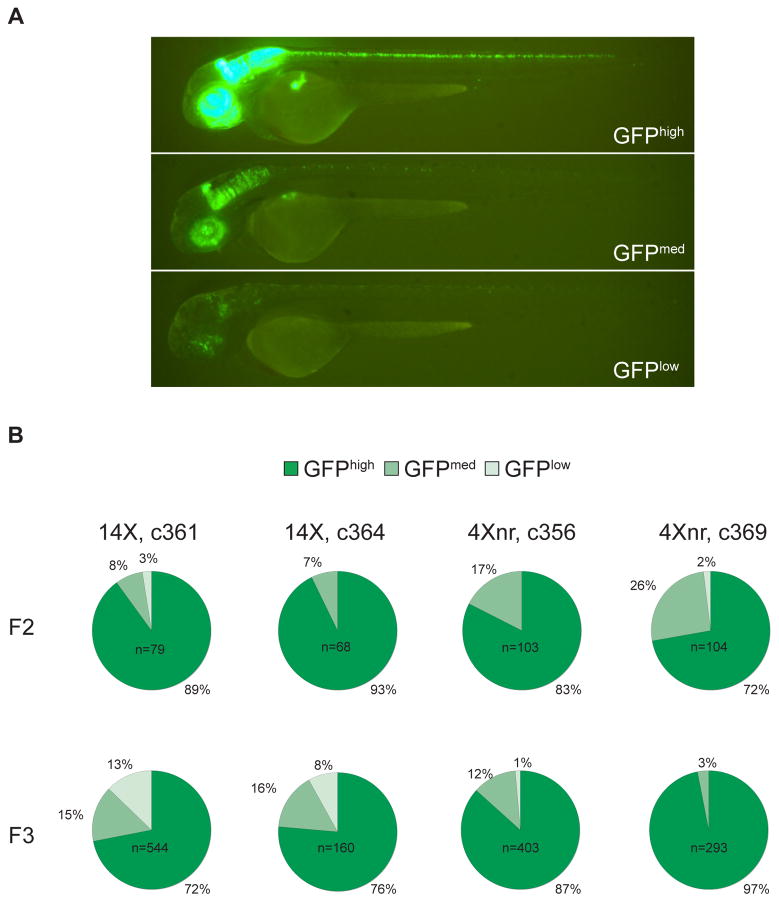
To assess the level of variegation in GFP expression, we devised a scoring strategy based on the proportion of fluorescence labeling observed at 2 dpf: Larvae exhibiting greater than 60% of the complete Tg(ptf1a:Gal4-VP16)jh16 pattern of labeling were scored as GFPhigh, larvae with 40–60% of the full pattern of GFP labeling were scored as GFPmed, and those with less than 40% were scored as GFPlow (refer to Fig. 3A). In the F2 generation, some variability in GFP labeling was observed between siblings derived from the same F1 parent, with a fraction of larvae in every line showing moderate levels of mosaicism (GFPmed) (Fig. 3B). Further analyses were performed on the F3 generation using the same conditions and scoring system applied to the F2 larvae. Whereas almost all F3 larvae from c356 and c369 4Xnr UAS lines were scored as GFPmed or GFPhigh, about 10% of those from 14X UAS lines showed significant mosaicism (GFPlow) (Fig. 3B). These data suggest that larvae bearing the 14X UAS:GFP are more prone to transcriptional silencing than those with the 4Xnr UAS:GFP transgene.
Figure 3. Transgenerational analysis of UAS-regulated GFP expression.
(A) Scoring of representative c364 transgenic larvae carrying the ptf1a driver and 14X UAS:GFP reporter transgenes as GFPhigh, GFPmed, or GFPlow, based on the extent of GFP labeling within the ptf1a expression domain. (B) Transgenerational analysis of GFP fluorescence in independently derived lines. All F2 larvae were obtained from matings between single F1 adults and AB fish. After F2 larvae were evaluated for their GFP labeling, GFPmed and GFPhigh individuals were raised, mated to AB, and their progeny scored in the F3 generation. Only GFP positive F3 larvae were included in the pie charts, which represent the cumulative data from at least five different GFPmed and GFPhigh F2 parents for each line.
An exception to the more consistent labeling observed with 4Xnr UAS:GFP transgenes was the c368 line, which showed variegated expression as early as in the F1 generation. A greater proportion of F2 larvae were also GFPmed or GFPlow compared to other 4Xnr UAS:GFP lines (Fig. 4A). The fraction of GFPlow larvae increased to over 60% in the F3 generation and only 3% of larvae were scored as GFPhigh (Fig. 4A). Although the c368 F1 adult contained two transgenes (Table 2), all F2 larvae with GFP labeled cells carried the insertion located within the intron of the PLA2G4C gene (c368a), suggesting that the other transgene (c368b) that mapped within a transposable element was fully silenced. Because variegated expression was prominent earlier in c368, we examined the extent of UAS methylation by DNA bisulfite sequencing. Correlated with the increased variegation of GFP labeling, we found that the 4Xnr UAS was methylated in c368 F2 larvae (on average 35% of CpG dinucleotides, n=4 larvae; Fig. 4B). Partial methylation of the transgene within the PLA2G4C intron could be due to its proximity to a DNA-TTAA-2 repetitive element (Table 2). The analysis of the c368 line shows that the site of integration can still exert a strong effect on reporter gene expression even under the control of the superior 4Xnr UAS construct.
Significant accumulation of methylation at the 14X UAS compared to the 4Xnr UAS
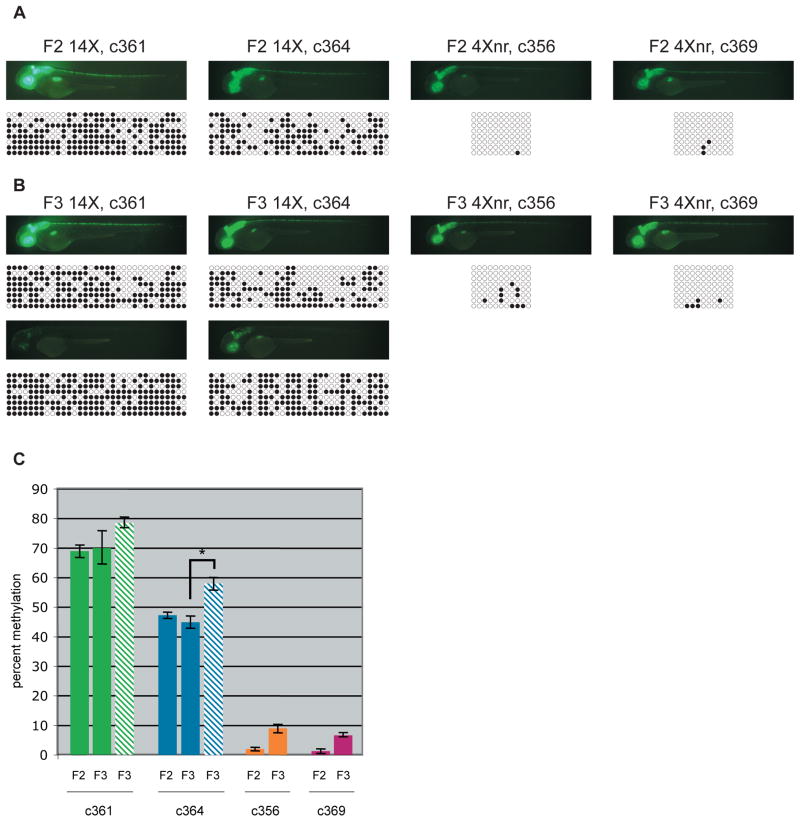
To monitor methylation status across generations, we compared 14X UAS and 4Xnr UAS lines that had shown robust GFP expression as F1s. Sodium bisulfite sequencing was performed on genomic DNA from individual F2 and F3 larvae and the difference in CpG dinucleotide methylation between 14X and 4Xnr UAS transgenes was striking (Fig. 5A). While F2 larvae derived from 14X lines showed significant methylation at the UAS (on average 69% for c361 and 47% c364), little was detected in genomic DNA from 4Xnr UAS lines (Figs. 5A, C). In the F3 generation, methylation was again prevalent at the 14X UAS. Moreover, in the c364 line with one 14X UAS insertion, GFPlow individuals had statistically significant increases in methylation (p<0.01) compared to GFPhigh siblings (Figs. 5B, C). In contrast, methylation levels at 4Xnr UAS sequences were consistently below 10% in c356 and c369 F3 larvae (Figs. 5B, C). Together, these results support a strong correlation between reduced transgene expression and UAS methylation and, in the appropriate genomic context, the greater resistance of 4Xnr UAS-containing transgenes to CpG methylation and silencing.
Figure 5. Reduced CpG methylation at the 4Xnr UAS.
(A) Fluorescence images and corresponding DNA bisulfite sequencing data for representative 2 dpf F2 larvae. Methylation at the 33 CpGs in the 14X UAS or the 11 CpGs in the 4Xnr UAS promoter are indicated on the horizontal axis, with black circles indicating methylated CpGs and open circles unmethylated CpGs. Patterns from eight different clones are shown on the vertical axis. (B) Fluorescence images and corresponding DNA bisulfite sequencing data for GFPhigh (top) and GFPlow (bottom) F3 larvae from 14X UAS lines and GFPhigh larvae from 4Xnr UAS lines. (C) Quantification of DNA bisulfite methylation data. Solid bars and striped bars indicate the average percentage of methylation of 14X UAS GFPhigh and GFPlow individuals, respectively. Percent methylation corresponds to the number of methylated CpG residues divided by the total number of CpG residues. Error bars represent standard error of the mean, P-values were calculated using the Fisher’s exact test and Mann-Whitney U test, with *p<0.01.
Discussion
The Gal4/UAS system of yeast is a powerful method for regulating gene expression in heterologous systems (Fischer et al., 1988; Ma et al., 1988; Ornitz et al., 1991; Webster et al., 1988), and has been used effectively in zebrafish for tissue-specific enhancer and gene traps (Davison et al., 2007; Scott et al., 2007), to label and track subsets of differentiating cells (Aramaki and Hatta, 2006; Distel et al., 2009; Hatta et al., 2006), for selective killing of specific cell types (Davison et al., 2007; Pisharath and Parsons, 2009; Zhao et al., 2009) and to modulate or detect neuronal activity (Asakawa et al., 2008; Douglass et al., 2008; Wyart et al., 2009). However, the transcriptional silencing of UAS transgenes has been a persistent problem that is often anecdotally reported, but less well documented. In some applications, the resultant mosaicism in gene expression can offer a technical advantage (Scott et al., 2007; Wyart et al., 2009), but often it is a hindrance. Silencing makes it difficult to maintain transgenic lines over multiple generations and can complicate the interpretation of results in experiments where every cell in a given population must express the gene of interest. The purpose of this study was to perform a systematic analysis of UAS-regulated transgene silencing across several zebrafish generations and to optimize reagents for long-term transgenic approaches.
Non-repetitive 4X UAS maintains expression and is less prone to methylation
Our work focused on the comparison of a newly constructed less repetitive 4 copy UAS to the widely used 14X UAS (Köster and Fraser, 2001). We reasoned that the repetitive nature of the 14X UAS likely triggers methylation and, by reducing repetitiveness through decreasing the number of UAS copies and their degree of sequence identity, we might be able to diminish silencing. In designing the 4Xnr UAS, we aimed to use functional variants that were as divergent as possible and had minimal CpG dinucleotides. Unfortunately, the six outermost bases of the UAS, including two CpG dinucleotides, could not be altered since they contact Gal4 directly and are necessary for efficient binding (Carey et al., 1989; Marmorstein et al., 1992). Nonetheless, differences within the 11 internal bases of the UAS led to an overall 50% divergence between UAS variants.
Despite having ten fewer Gal4 binding sites, 4Xnr UAS:GFP transgenic larvae demonstrated only a modest reduction in fluorescence intensity compared to those bearing 14X UAS:GFP. Most larvae showed the expected pattern of GFP labeling from the ptf1a driver, although a subset of both F2 and F3 larvae showed partial expression patterns (i.e., GFPmed). Irrespective of this difference, methylation of the UAS was negligible in both GFPhigh and GFPmed larvae from stable lines carrying single insertions of the 4Xnr UAS:GFP transgene. This suggests that other mechanisms besides methylation are influencing transgene expression. Variability in UAS-regulated gene expression has also been described in Drosophila (Skora and Spradling, 2010), an organism that lacks DNA methlyation, although its cause is unknown.
A small increase in methylation was detected at the 4Xnr UAS in F3 larvae compared to F2, but levels remained below 10% in both generations. Analyses of additional generations will be required to determine whether this increase is a significant trend that might ultimately lead to loss of gene expression from the 4Xnr UAS.
Silencing of 14X UAS-regulated gene expression in transgenic fish
In contrast to the 4Xnr UAS, and consistent with previous observations (Goll et al., 2009), F2 and F3 individuals carrying the ptf1a driver showed extensive CpG methylation at the 14X UAS. It was difficult to correlate precise levels of methylation with the extent of variegation in reporter expression, as even GFPhigh larvae showed significant methylation at the 14X UAS. Nonetheless, we did observe a statistically significant increase in the percent of methylation in GFPlow compared to GFPhigh larvae carrying a single transgenic insertion. In GFPhigh larvae, expression was attributed to retention of some unmethylated Gal4 binding sites in the multicopy UAS. We hypothesize that in cells resistant to Gal4 activation, increased methylation prevents access to all Gal4 binding sites. This suggests that a threshold level of methylation must be achieved in order for expression to be silenced, and that sub-threshold levels of methylation at the UAS may be a harbinger of silencing in future generations.
Differing patterns of methylation were detected among bisulfite clones from the same individual, indicating that methylation patterns varied from cell to cell in a single larva. Such variability could account for the mosaic expression observed in GFPlow individuals and suggests that methylation of the UAS is somewhat dynamic and stochastic in the early embryo.
Position effects and transgene silencing
It is well known that the local chromatin environment at their position of integration can influence expression of transgenes (refer to Wilson et al., 1990). For this reason, it is preferable to compare constructs inserted at the same genomic position using a targeted approach such as PhiC31 integrase or Cre recombinase-mediated cassette exchange; however, this technology is still under development for the zebrafish (Boniface et al., 2009; Lister, 2010; Lu et al., 2010).
Instead, we attempted to control for position effects by using bicistronic vectors expressing Gal4FF-2A-mCherry from a ubiquitous promoter and UAS:GFP. The rationale was that by monitoring Gal4 levels via coordinately produced mCherry, we could account for differences in expression between transgenic insertions. However, while the integrated bicistronic transgenes performed as expected in the F1 generation, F2 progeny exhibited markedly mosaic mCherry and GFP labeling, independent of UAS copy number. Variegated mCherry fluorescence suggested silencing of the EF1α promoter, and consistent with this, we found an accumulation of methylation at this promoter as well as at the UAS. Methylation of the 4Xnr UAS in the context of the bicistronic construct was unexpected and inconsistent with what was observed for transgenes just containing 4Xnr UAS:GFP, implying that silencing is triggered by some feature of the bicistronic transgene itself. It is possible that silencing initiates at the EF1α promoter and then spreads to the UAS. Alternatively, read-through transcription from the initially strong EF1α promoter past a weak polyA terminator may lead to low levels of UAS RNA synthesis, which, in turn, targets the corresponding UAS DNA repeats for silencing. RNA based mechanisms of silencing are widely used in plants and have also been described in mammals (Matzke et al., 2009; Morris et al., 2004; Wassenegger et al., 1994); but have not yet been documented in zebrafish. While the mechanism underlying the rapid methylation and silencing observed in the bicistronic construct is one worth pursuing, it significantly complicated the analyses of reporter expression.
An alternative strategy was to distribute 14X or 4Xnr UAS:GFP transgenes throughout the genome and generate multiple, independent reporter lines. By using the same Tg(ptf1a:Gal4-VP16)jh16 driver line to evaluate expression from UAS:GFP reporters, we abolished concerns of variable Gal4-VP16 expression.
A drawback to the binary approach is that it does not control for position effects resulting from differences between UAS:GFP transgene integration sites. Several 14X and 4Xnr transgenic insertions showed mosaic expression in larvae as early as in the first generation. Remarkably, F1 individuals that exhibited mosaic GFP labeling had transgenes inserted within or in close proximity to transposable elements, sequence elements that are enriched in DNA methylation in the zebrafish genome (Feng et al., 2010). Others have observed such variability between transgene integration sites. For example, Asakawa and Kawakami (Asakawa and Kawakami, 2009) tested approximately 75 different insertions to obtain an optimal UAS:TeTxLC:CFP zebrafish transgenic line. Fortunately, the ease of generating numerous insertions with Tol2-mediated transposition allows for the selection of high expressing integrants, which has become routine practice in the field. A systematic approach to map all integration sites and to correlate genomic position with expression, as in our study, would be valuable for obtaining a more comprehensive portrait of the genome-wide chromatin landscape.
Application of the Gal4/UAS system in transgenic zebrafish
Given the time and effort required to make transgenic lines, it is prudent to incorporate strategies that produce optimal gene expression and regulation. The rapid silencing of bicistronic Gal4/UAS vectors used in this study suggests that this type of construct is not ideal to ensure continued expression from transgenic insertions. Inclusion of strong polyA signals may eliminate the potential for read-through transcription from strong promoters and thereby improve the reliability of multicistronic constructs.
In contrast to Drosophila where the Gal4/UAS system was shown to be highly effective and rapidly adopted by the field, initial studies of the Gal4 transcriptional activator in zebrafish indicated that reporters under UAS control were only weakly induced (Scheer and Campos-Ortega, 1999). To obtain high expression levels, vectors were reengineered with the addition of the strong transcriptional activator VP16 and 14 copies of the UAS (Köster and Fraser, 2001). Robust expression was successfully achieved in transient assays, but a problem with this approach is the high toxicity of Gal4-VP16 thought to be due to “squelching” of factors necessary for normal gene regulation (Gill and Ptashne, 1988; Köster and Fraser, 2001). Our work provides additional evidence for toxicity from singly integrated, stable insertions and suggests that caution should be taken when using Gal4-VP16 in zebrafish, especially when widespread expression is required. Use of the attenuated Gal4FF driver appears to circumvent this problem (Asakawa et al., 2008). However, neither Gal4-VP16 nor Gal4FF retain sequences necessary for Gal80 modulation (Johnston et al., 1987; Ma and Ptashne, 1987; Suster et al., 2004), removing this added level of regulatory control. Thus, for some applications use of full-length Gal4 may be preferable.
The selection of an appropriate responder line is also critical. Reporter genes regulated by the 14 copy UAS are transcriptionally silenced, owing to CpG methylation of the repetitive UAS (Goll et al., 2009). As discussed above, the 4Xnr UAS generates high levels of gene expression and correspondingly low levels of methylation, properties that are maintained for at least 3 generations. However, as with any transgene, and as evidenced by the c368 line, several independent integration events should still be examined to identify those that reside in a favorable chromatin environment. Nevertheless, our results demonstrate the superiority of the 4Xnr UAS for producing and preserving transgenic lines that show a consistent response to Gal4 activation and, accordingly, reproducible patterns of reporter expression. We expect that this tool will be useful not only for generating UAS-regulated transgenes in zebrafish, but also for other organisms where DNA methylation is known to act on repetitive sequences, such as plants and mice.
Conclusions
The zebrafish offers an expeditious system to compare silencing of different sequences in live animals and to follow their propagation through the germline. Comparison of the 4Xnr and 14X UAS transgenes validates the utility of this approach for studying the sequence cues that direct silencing. Because vertebrate genomes are widely methylated, there has been some debate about whether methylation is targeted to particular regions of the genome or if unmethylated regions of the genome are protected from methylation. The fact that 14 identical UAS sites become rapidly methylated, but four non-identical copies do not, supports the conclusion that repetitive sequences attract methylation. Additional work will be required to define the relative importance of repeat number and percent sequence identity, as both variables were altered in the 4Xnr UAS construct. It will be interesting to probe the exact features that make short tandem repeats attract methylation. Ideally, once targeted integration methods become routine for the zebrafish genome, UAS variants should be compared within the same chromosomal context. Given the preponderance of short variable tandem repeats in the vertebrate genome, understanding the cues that trigger their silencing may provide important insights into how these repeated sequences influence the expression of nearby genes.
Supplementary Material
Supplemental Figure 1. Dual reporter system to monitor Gal4-VP16 expression and UAS-driven transcription simultaneously
(A) Schematic diagram of the bicistronic construct used to generate transgenic lines for testing UAS copy number variants. Under the control of the EF1α promoter, Gal4-VP16 and mCherry are produced in equimolar amounts due to incorporation of the viral 2A peptide sequence. UAS copy number variants and the E1b minimal promoter were inserted into a multiple cloning site (MCS) upstream of GFP. (B) Transgenic F1 larvae generated with different UAS copy number variants show widespread, mCherry (left) and GFP (right) fluorescence at 2 dpf, but develop defects and do not survive to adulthood.
Supplemental Figure 2. Comparison of multi-copy UAS constructs
The 14X UAS is comprised of 14 identical copies of a near consensus synthetic UAS (Webster et al., 1988) with various spacer sequences interspersed (Rorth, 1996). The 4Xnr UAS contains two UAS derived from the yeast UASG promoter and two distinct consensus synthetic UAS (Giniger et al., 1985; Webster et al., 1988).
Supplemental Figure 3. Variability in GFP labeling in offspring from independent transgenic founders.
F1 larvae carrying the Tg(ptf1a:Gal4-VP16)jh16 driver line and either 14X or 4Xnr UAS transgenes. Some individuals display robust GFP labeling in the ptf1a expression domain (retina, hindbrain, spinal cord, pancreas primordium) at 2 dpf, while others exhibit fluorescence in only a subset of cells in the expected pattern.
Supplemental Figure 4. Analysis of transgene insertion number
Genomic DNA derived from fin clips of individual F1 adults was digested with EcoRI. The Southern blot was probed using radiolabeled GFP sequence. Single transgenic insertions were detected for lines c364, c356, and c369. Two insertions were detected in c368, whereas c361 contained greater than 10.
Supplemental Figure 5. Fluorescence intensity quantification.
Transgenic F3 larvae bearing single insertions were imaged using identical camera and microscope settings (n=10 larvae for each line). Mean pixel intensity in the green channel was calculated for GFP-positive tissues. Fluorescence intensity in the 4Xnr UAS lines was 70–80% of the level achieved with the 14X UAS. Error bars represent standard error of the mean.
Acknowledgments
We are grateful to Lucilla Facchin and Tagide deCarvalho for assistance with statistics, Koichi Kawakami and Shannon Fisher for Tol2 reagents, and Steve Leach and Michael Parsons for plasmids, transgenic zebrafish lines and for valuable discussions. MGG was supported by the Damon Runyon Cancer Research Foundation as a Damon Runyon Fellow (DRG#1945-07) and was a Carnegie Collaborative Fellow with Allan Spradling. This study was funded by a grant from the National Institute of Child Health and Human Development to MEH (R01HD058530).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amsterdam A, Lin S, Hopkins N. The Aequorea victoria green fluorescent protein can be used as a reporter in live zebrafish embryos. Dev Biol. 1995;171:123–9. doi: 10.1006/dbio.1995.1265. [DOI] [PubMed] [Google Scholar]
- Aramaki S, Hatta K. Visualizing neurons one-by-one in vivo: optical dissection and reconstruction of neural networks with reversible fluorescent proteins. Dev Dyn. 2006;235:2192–9. doi: 10.1002/dvdy.20826. [DOI] [PubMed] [Google Scholar]
- Asakawa K, Kawakami K. The Tol2-mediated Gal4-UAS method for gene and enhancer trapping in zebrafish. Methods. 2009;49:275–81. doi: 10.1016/j.ymeth.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Suster ML, Mizusawa K, Nagayoshi S, Kotani T, Urasaki A, Kishimoto Y, Hibi M, Kawakami K. Genetic dissection of neural circuits by Tol2 transposon-mediated Gal4 gene and enhancer trapping in zebrafish. Proc Natl Acad Sci U S A. 2008;105:1255–60. doi: 10.1073/pnas.0704963105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boby T, Patch AM, Aves SJ. TRbase: a database relating tandem repeats to disease genes for the human genome. Bioinformatics. 2005;21:811–6. doi: 10.1093/bioinformatics/bti059. [DOI] [PubMed] [Google Scholar]
- Boniface EJ, Lu J, Victoroff T, Zhu M, Chen W. FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis. 2009;47:484–91. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DS, Stringham SA, Timm A, Xiao T, Law MY, Baier H, Nonet ML, Chien CB. Slit1a inhibits retinal ganglion cell arborization and synaptogenesis via Robo2-dependent and -independent pathways. Neuron. 2007;55:231–45. doi: 10.1016/j.neuron.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–32. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- Collins RT, Linker C, Lewis J. MAZe: a tool for mosaic analysis of gene function in zebrafish. Nat Methods. 2010;7:219–23. doi: 10.1038/nmeth.1423. [DOI] [PubMed] [Google Scholar]
- Davis BP, MacDonald RJ. Limited transcription of rat elastase I transgene repeats in transgenic mice. Genes Dev. 1988;2:13–22. doi: 10.1101/gad.2.1.13. [DOI] [PubMed] [Google Scholar]
- Davison JM, Akitake CM, Goll MG, Rhee JM, Gosse N, Baier H, Halpern ME, Leach SD, Parsons MJ. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev Biol. 2007;304:811–24. doi: 10.1016/j.ydbio.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel M, Wullimann MF, Koster RW. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc Natl Acad Sci U S A. 2009;106:13365–70. doi: 10.1073/pnas.0903060106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein 'cleavage' mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal 'skip'. J Gen Virol. 2001;82:1013–25. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Douglass AD, Kraves S, Deisseroth K, Schier AF, Engert F. Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr Biol. 2008;18:1133–7. doi: 10.1016/j.cub.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–94. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988;332:853–6. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–9. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–4. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- Giniger E, Varnum SM, Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985;40:767–74. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Goll MG, Anderson R, Stainier DY, Spradling AC, Halpern ME. Transcriptional silencing and reactivation in transgenic zebrafish. Genetics. 2009;182:747–55. doi: 10.1534/genetics.109.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat Protoc. 2006;1:960–7. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- Hutter B, Helms V, Paulsen M. Tandem repeats in the CpG islands of imprinted genes. Genomics. 2006;88:323–32. doi: 10.1016/j.ygeno.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Bressler J, Beaudet AL. Epigenetics and human disease. Annu Rev Genomics Hum Genet. 2004;5:479–510. doi: 10.1146/annurev.genom.5.061903.180014. [DOI] [PubMed] [Google Scholar]
- Johnston SA, Salmeron JM, Jr, Dincher SS. Interaction of positive and negative regulatory proteins in the galactose regulon of yeast. Cell. 1987;50:143–6. doi: 10.1016/0092-8674(87)90671-4. [DOI] [PubMed] [Google Scholar]
- Kang T, Martins T, Sadowski I. Wild type GAL4 binds cooperatively to the GAL1-10 UASG in vitro. J Biol Chem. 1993;268:9629–35. [PubMed] [Google Scholar]
- Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci U S A. 2000;97:11403–8. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Bobek MP, Kuick R, Lamb B, Zhu X, Narayan A, Bourc'his D, Viegas-Pequignot E, Ehrlich M, Hanash SM. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum Mol Genet. 2000;9:597–604. doi: 10.1093/hmg/9.4.597. [DOI] [PubMed] [Google Scholar]
- Köster RW, Fraser SE. Tracing transgene expression in living zebrafish embryos. Dev Biol. 2001;233:329–46. doi: 10.1006/dbio.2001.0242. [DOI] [PubMed] [Google Scholar]
- Kumaki Y, Oda M, Okano M. QUMA: quantification tool for methylation analysis. Nucleic Acids Res. 2008;36:W170–5. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers RJ, van der Vliet PJ, Klooster R, Sacconi S, Camano P, Dauwerse JG, Snider L, Straasheijm KR, van Ommen GJ, Padberg GW, Miller DG, Tapscott SJ, Tawil R, Frants RR, van der Maarel SM. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–3. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Biankin AV, Horb ME, Ghosh B, Prasad NB, Yee NS, Pack MA, Leach SD. Differential requirement for ptf1a in endocrine and exocrine lineages of developing zebrafish pancreas. Dev Biol. 2004;270:474–86. doi: 10.1016/j.ydbio.2004.02.023. [DOI] [PubMed] [Google Scholar]
- Linn F, Heidmann I, Saedler H, Meyer P. Epigenetic changes in the expression of the maize A1 gene in Petunia hybrida: role of numbers of integrated gene copies and state of methylation. Mol Gen Genet. 1990;222:329–36. doi: 10.1007/BF00633837. [DOI] [PubMed] [Google Scholar]
- Linney E, Hardison NL, Lonze BE, Lyons S, DiNapoli L. Transgene expression in zebrafish: A comparison of retroviral-vector and DNA-injection approaches. Dev Biol. 1999;213:207–16. doi: 10.1006/dbio.1999.9376. [DOI] [PubMed] [Google Scholar]
- Lister JA. Transgene excision in zebrafish using the phiC31 integrase. Genesis. 2010;48:137–43. doi: 10.1002/dvg.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Maddison LA, Chen W. PhiC31 integrase induces efficient site-specific excision in zebrafish. Transgenic Res. 2010 doi: 10.1007/s11248-010-9394-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Przibilla E, Hu J, Bogorad L, Ptashne M. Yeast activators stimulate plant gene expression. Nature. 1988;334:631–3. doi: 10.1038/334631a0. [DOI] [PubMed] [Google Scholar]
- Ma J, Ptashne M. The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell. 1987;50:137–42. doi: 10.1016/0092-8674(87)90670-2. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–14. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- Matzke M, Kanno T, Daxinger L, Huettel B, Matzke AJ. RNA-mediated chromatin-based silencing in plants. Curr Opin Cell Biol. 2009;21:367–76. doi: 10.1016/j.ceb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- Miller OJ, Schnedl W, Allen J, Erlanger BF. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974;251:636–7. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- Mittelsten Scheid O, Paszkowski J, Potrykus I. Reversible inactivation of a transgene in Arabidopsis thaliana. Mol Gen Genet. 1991;228:104–12. doi: 10.1007/BF00282454. [DOI] [PubMed] [Google Scholar]
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small Interfering RNA-Induced Transcriptional Gene Silencing in Human Cells. Science. 2004;305:1289–92. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- Oberle I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Moreadith RW, Leder P. Binary system for regulating transgene expression in mice: targeting int-2 gene expression with yeast GAL4/UAS control elements. Proc Natl Acad Sci U S A. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Davison JM, Rhee J, Hruban RH, Maitra A, Leach SD. Oncogenic KRAS induces progenitor cell expansion and malignant transformation in zebrafish exocrine pancreas. Gastroenterology. 2008;134:2080–90. doi: 10.1053/j.gastro.2008.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisharath H, Parsons MJ. Nitroreductase-mediated cell ablation in transgenic zebrafish embryos. Methods Mol Biol. 2009;546:133–43. doi: 10.1007/978-1-60327-977-2_9. [DOI] [PubMed] [Google Scholar]
- Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech Dev. 2007;124:218–29. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetto-Zimmerman C, Wolgemuth DJ. Methylation of satellite sequences in mouse spermatogenic and somatic DNAs. Nucleic Acids Res. 1984;12:2807–22. doi: 10.1093/nar/12.6.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost E, Rhee J, Leach SD. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–9. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- Robertson G, Garrick D, Wu W, Kearns M, Martin D, Whitelaw E. Position-dependent variegation of globin transgene expression in mice. Proc Natl Acad Sci U S A. 1995;92:5371–5. doi: 10.1073/pnas.92.12.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Rorth P. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc Natl Acad Sci U S A. 1996;93:12418–22. doi: 10.1073/pnas.93.22.12418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev. 1999;80:153–8. doi: 10.1016/s0925-4773(98)00209-3. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–6. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe JA, Wells DJ, Whitelaw E, Vyas P, Higgs DR, Wood WG. Analysis of the human alpha-globin gene cluster in transgenic mice. Proc Natl Acad Sci U S A. 1993;90:11262–6. doi: 10.1073/pnas.90.23.11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skora AD, Spradling AC. Epigenetic stability increases extensively during Drosophila follicle stem cell differentiation. Proc Natl Acad Sci U S A. 2010;107:7389–94. doi: 10.1073/pnas.1003180107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–17. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Suster ML, Seugnet L, Bate M, Sokolowski MB. Refining GAL4-driven transgene expression in Drosophila with a GAL80 enhancer-trap. Genesis. 2004;39:240–5. doi: 10.1002/gene.20051. [DOI] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–49. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker C. Haploid Screens and Gamma-Ray Mutagenesis. In: Detrich HW, Westerfield M, Zon LI, editors. The Zebrafish, Genetics and Genomics. Vol. 60. Academic Press; San Diego: 1999. pp. 44–68. [Google Scholar]
- Wassenegger M, Heimes S, Riedel L, Sanger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–76. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- Webster N, Jin JR, Green S, Hollis M, Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988;52:169–78. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- Wilson C, Bellen HJ, Gehring WJ. Position effects on eukaryotic gene expression. Annu Rev Cell Biol. 1990;6:679–714. doi: 10.1146/annurev.cb.06.110190.003335. [DOI] [PubMed] [Google Scholar]
- Wurch T, Lestienne F, Pauwels PJ. A modified overlap extension PCR method to create chimeric genes in the absence of restriction enzymes. Biotechnology Techniques. 1998;12:653–657. [Google Scholar]
- Wyart C, Del Bene F, Warp E, Scott EK, Trauner D, Baier H, Isacoff EY. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461:407–10. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XF, Ellingsen S, Fjose A. Labelling and targeted ablation of specific bipolar cell types in the zebrafish retina. BMC Neurosci. 2009;10:107. doi: 10.1186/1471-2202-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Dual reporter system to monitor Gal4-VP16 expression and UAS-driven transcription simultaneously
(A) Schematic diagram of the bicistronic construct used to generate transgenic lines for testing UAS copy number variants. Under the control of the EF1α promoter, Gal4-VP16 and mCherry are produced in equimolar amounts due to incorporation of the viral 2A peptide sequence. UAS copy number variants and the E1b minimal promoter were inserted into a multiple cloning site (MCS) upstream of GFP. (B) Transgenic F1 larvae generated with different UAS copy number variants show widespread, mCherry (left) and GFP (right) fluorescence at 2 dpf, but develop defects and do not survive to adulthood.
Supplemental Figure 2. Comparison of multi-copy UAS constructs
The 14X UAS is comprised of 14 identical copies of a near consensus synthetic UAS (Webster et al., 1988) with various spacer sequences interspersed (Rorth, 1996). The 4Xnr UAS contains two UAS derived from the yeast UASG promoter and two distinct consensus synthetic UAS (Giniger et al., 1985; Webster et al., 1988).
Supplemental Figure 3. Variability in GFP labeling in offspring from independent transgenic founders.
F1 larvae carrying the Tg(ptf1a:Gal4-VP16)jh16 driver line and either 14X or 4Xnr UAS transgenes. Some individuals display robust GFP labeling in the ptf1a expression domain (retina, hindbrain, spinal cord, pancreas primordium) at 2 dpf, while others exhibit fluorescence in only a subset of cells in the expected pattern.
Supplemental Figure 4. Analysis of transgene insertion number
Genomic DNA derived from fin clips of individual F1 adults was digested with EcoRI. The Southern blot was probed using radiolabeled GFP sequence. Single transgenic insertions were detected for lines c364, c356, and c369. Two insertions were detected in c368, whereas c361 contained greater than 10.
Supplemental Figure 5. Fluorescence intensity quantification.
Transgenic F3 larvae bearing single insertions were imaged using identical camera and microscope settings (n=10 larvae for each line). Mean pixel intensity in the green channel was calculated for GFP-positive tissues. Fluorescence intensity in the 4Xnr UAS lines was 70–80% of the level achieved with the 14X UAS. Error bars represent standard error of the mean.