Abstract
Gemcitabine and other cytidine antimetabolites require metabolic activation by phosphorylation. Deoxycytidine kinase (DCK) and cytidine monophosphate kinase (CMPK) catalyze these reactions. We have applied a genotype-to-phenotype strategy to study DCK and CMPK pharmacogenomics. Specifically, we resequenced DCK and CMPK using 240 DNA samples, 60 each from African-American, Caucasian-American, Han Chinese-American and Mexican-American subjects. We observed 28 DCK polymorphisms and 28 polymorphisms in CMPK, 33 of which were novel. Expression in COS-1 cells showed that variant allozyme enzyme activities ranged from 32 to 105% of the wild type (WT) for DCK, and from 78 to 112% of WT for CMPK – with no significant differences in apparent Km values for either enzyme except for a DCK Val24/Ser122 double variant allozyme. Relative levels of DCK and CMPK immunoreactive protein in the COS-1 cells paralleled relative levels of enzyme activity and were significantly correlated for DCK (Rp = 0.89, P = 0.0004) but not for CMPK (Rp = 0.82, P = 0.095). The results of an analysis of DCK and CMPK structural models were compatible with the observed functional consequences of sequence alterations in variant allozymes. We also confirmed that the CMPK protein expressed in COS-1 cells and in a rabbit reticulocyte lysate was 196 rather than 228 amino acids in length. In summary, we determined common sequence variation in DCK and CMPK and systematically evaluated its functional implications. These gene sequence differences may contribute to variation in the metabolic activation of gemcitabine and other cytidine antimetabolites.
Introduction
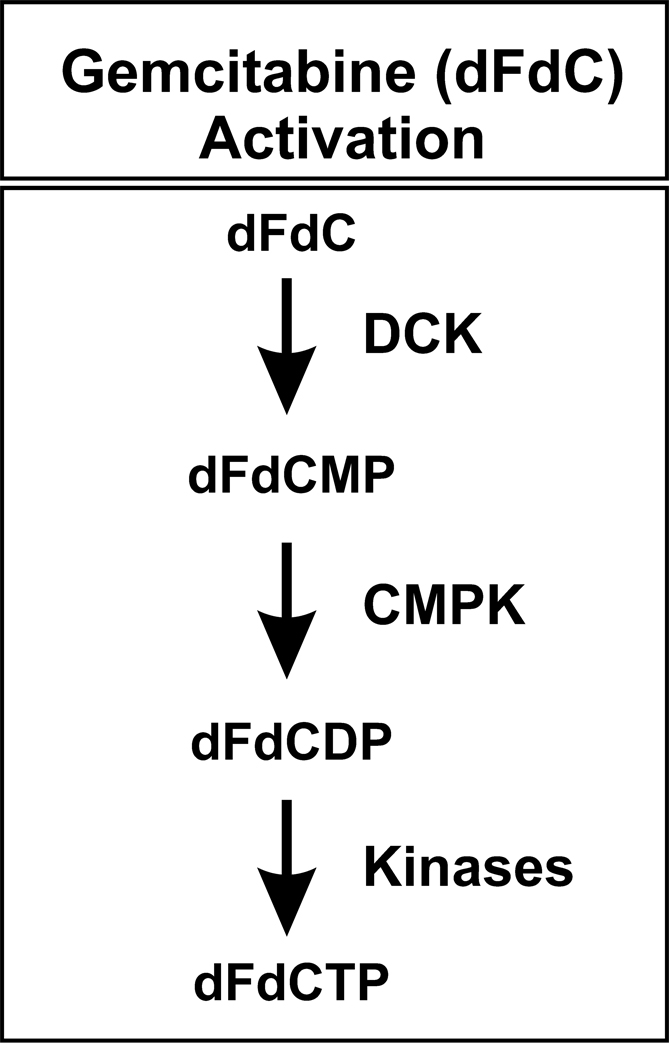
Gemcitabine (2′-deoxy-2′,2′-difluorocytidine; dFdC) is an antineoplastic deoxycytidine analog that has proven activity against a wide spectrum of solid tumors, including pancreatic, lung and breast cancer (Carmichael, 1998; Ulrich et al., 2003; Giovannetti et al., 2006). Gemcitabine and other cytidine analog antimetabolites such as cytosine arabinoside (AraC) require cellular uptake and subsequent phosphorylation for activation (Mini et al., 2006). These drugs are phosphorylated by deoxycytidine kinase (DCK EC2.7.1.74) to form a monophosphate, which is then converted to the diphosphate (dFdCDP) by cytidine monophosphate kinase (CMPK EC2.7.4.14) (Mini et al., 2006; Nakano et al., 2007), as illustrated in Figure 1. The phosphorylation of gemcitabine by DCK is the rate-limiting step in metabolic activation (Kroep et al., 2002; Mini et al., 2006). CMPK also plays an important role in the activation of many antineoplastic and antiviral deoxycytidine analogs (Cheng, 2001; Galmarini et al., 2001; Galmarini et al., 2002; Hsu et al., 2005). The cytotoxic effects of gemcitabine occur as a result of a series of actions of the diphosphate (dFdCDP) and triphosphate (dFdCTP) metabolites. dFdCDP is an inhibitor of ribonucleotide reductase, while dFdCTP is incorporated into DNA. Both of these actions result in the inhibition of DNA synthesis (Plunkett et al., 1995; Galmarini et al., 2001; Pauwels et al., 2006), so the antitumor effects of this drug are dependent on intracellular metabolism – metabolism that includes phosphorylation as a critical step (Maring et al., 2005).
Figure 1. Gemcitabine activation.
DCK, deoxycytidine kinase; CMPK, deoxycytidylate kinase; dFdC, gemcitabine; dFdCMP, gemcitabine monophosphate; dFdCDP, gemcitabine diphosphate; and dFdCTP, gemcitabine triphosphate.
We set out to study the pharmacogenomics of gemcitabine phosphorylation by applying a genotype-to-phenotype research strategy to genes encoding the two proteins that catalyze the intracellular phosphorylation of gemcitabine and structurally-related pyrimidine antagonists such as cytosine arabinoside (Maring et al., 2005). Specifically, we resequenced DCK and CMPK using 240 DNA samples from four different ethnic groups, i.e., 60 samples each from African-American (AA), Caucasian-American (CA), Han Chinese-American (HCA) and Mexican-American (MA) subjects. Functional genomic studies were then performed using expression constructs for all nonsynonymous coding single nucleotide polymorphisms (cSNPs) that were observed in these two genes, as well as combinations of cSNPs that were inferred from haplotype analyses. Since the CMPK gene has two different potential ATG translation initiation codons, there has been confusion with regard to which of those codons is utilized in vivo (Maring et al., 2005). Therefore, to make it possible to perform our functional genomic studies, we also verified which of these translation initiation codons is utilized in vivo. Finally, we took advantage of the crystal structures of DCK and CMPK to evaluate the potential effect of these naturally occurring alterations in encoded amino acid sequence on protein structure and to determine whether our results were plausible based on the known structures of these proteins. These results represent a step toward future translational studies of the role of these two genes in cytotoxic cytidine analog pharmacogenomics.
Materials and Methods
DNA Samples
DNA samples from 60 CA, 60 AA, 60 HCA, and 60 MA subjects (sample sets HD100CAU, HD100AA, HD100CHI, HD100MEX) were obtained from the Coriell Cell Repository (Camden, NJ). The present study was reviewed and approved by the Mayo Clinic Institutional Review Board.
DCK and CMPK Gene Resequencing
Each of the 240 DNA samples studied was used to perform PCR amplifications of all DCK and CMPK exons, splice junctions and a portion of the 5′-FRs for each gene. This study was powered to make it possible to reliably detect variant alleles with minor allele frequency (MAF) of 2% in the 120 alleles required for each of the 4 ethnic groups studies. For CMPK, a region identified by rVISTA (Loots et al., 2002) located upstream of the site of transcription initiation contained a cluster of putative transcription factor binding sites and showed high sequence homology to orthologous sequences in the dog and mouse genomes. This region was also resequenced. Primer sequences are listed in the Supplementary Material (Tables 1 and 2). Amplification reactions were performed with FastStart Taq DNA polymerase, and amplicons were sequenced on both strands in the Mayo Molecular Biology Core Facility with an ABI 3730×1 DNA sequencer using BigDye™ dye terminator sequencing chemistry (Perkin-Elmer). To exclude PCR-related artifacts, independent amplifications were performed for any SNP observed in only a single DNA sample or for any sample with an ambiguous chromatogram. The sequencing chromatograms were analyzed using Mutation Surveyor version 2.41, PolyPhred 3.0 (Maring et al., 2005) and Consed 8.0 (Maring et al., 2005). GenBank accession numbers for the DCK reference sequences used in these experiments were NT_006216.16 and NM_000788.1, and for CMPK were NT_032977.7 and NM_016308.1.
DCK and CMPK Expression Constructs and Transient Expression
The wild type (WT) DNA open reading frame (ORF) sequences for both genes were cloned into the eukaryotic expression vector pCR3.1 (Invitrogen, Carlsbad, CA). These expression constructs were then used to perform site-directed mutagenesis with the QuikChange kit (Stratagene, La Jolla, CA). Circular PCR was used to create variant allozyme constructs. The sequences of primers used to perform site-directed mutagenesis are also listed in the Supplementary Material (Tables 1 and 2). The sequences of all inserts were confirmed by sequencing both DNA strands. COS-1 cells were then transfected with expression constructs encoding WT and variant DCK and CMPK allozymes, as well as “empty” vector that lacked an insert as a control, using the TransFast reagent (Promega, Madison, WI) at a charge ratio of 1:2. Specifically, 7 µg of construct DNA was cotransfected with 7 µg of pSV-β-galactosidase DNA (Promega) to correct for possible variation in transfection efficiency. The coefficient of variation for the cotransfected β-galactosidase averaged 8.2%. After 48 h, the cells were harvested in 50 mM Tris-HCl (pH 7.6), 100 mM KCl, 1 mM MgCl2 and 2 mM dithiothreitol for DCK, or with 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2 and 10 mM dithiothreitol for cells transfected with CMPK constructs. The cells were then homogenized with a Polytron homogenizer (Brinkmann Instruments, Westbury, NY); homogenates were centrifuged at 100,000 × g for 1 h; and supernatant preparations were stored at −80°C for use in the functional genomic studies.
Enzyme Assays and Substrate Kinetics
DCK catalyzes the formation of 2′,2′-difluorodeoxycytidine 5′-monophosphate (gemcitabine monophosphate) from gemcitabine. DCK activity was measured using a modification of the assay of Usova and Eriksson (Usova and Eriksson, 2002) with gemcitabine (Eli Lilly, Indianapolis, IN) as a substrate. Our assay measured the product of the reaction using the extraction procedure and HPLC separation conditions described by Ruiz van Haperen et al. (Ruiz van Haperen et al., 1994). CMPK catalyzes the formation of 2′,2′-difluorodeoxycytidine 5′-diphosphate (gemcitabine diphosphate) from gemcitabine monophosphate. A modification of the assay described by van Rompay et al. (Van Rompay et al., 1999) was used to measure CMPK activity. This assay measured the depletion of gemcitabine monophosphate (Eli Lilly) during the reaction and employed the extraction procedure and HPLC separation described by Ruiz van Haperen et al. (Ruiz van Haperen et al., 1994).
Western Blot Analysis
Quantitative Western blot analyses were performed with recombinant DCK and CMPK allozymes after correction for transfection efficiency on the basis of the cotransfected β-galactosidase activity measured using the Promega β-Galactosidase Enzyme Assay System. For DCK, these assays used rabbit polyclonal antibody generated by Cocalico Biologicals that was directed against the C-terminal 59–80 amino acids of DCK. A rabbit polyclonal antibody to purified recombinant CMPK was kindly provided by Dr. Yung-Chi Cheng, Yale University School of Medicine, New Haven, CT. To perform quantitative Western blot analysis, COS-1 cell cytosol was loaded on 12% Tris-HCl acrylamide gels on the basis of the cotransfected β-galactosidase activity, and proteins were separated by SDS-PAGE prior to transfer to PVDF membranes (BioRad). Immunoreactive proteins were detected using the ECL Western Blotting System (Amersham Pharmacia, Piscataway, NJ). The IPLab Gel H (Biosystemetica, Plymouth, UK) system and the NIH image program (http//rsb.info.nih.gov/nih-image) were used to quantify immunoreactive proteins.
Rabbit Reticulocyte Lysate (RRL) Studies
Radioactively-labeled WT and variant CMPK allozyme proteins were synthesized using the TNT® RRL system (Promega, Madison, WI) as described by Wang et al. (Wang et al., 2003). CMPK has two potential in-frame ATG translation initiation codons, resulting in an ORF encoding either a 228 amino acid protein or a 196 amino acid protein (Van Rompay et al., 1999).
Data Analysis
Values for π, θ and Tajima’s D were calculated as described by Tajima (Tajima, 1989). D′ values, a measure of linkage disequilibrium that is independent of allele frequency, were calculated as described by Hartl and Clark (Hartl and Clark, 2000) and Hendrick (Hendrick, 2000). Haplotype analysis was performed as described by Schaid et al. (Schaid et al., 2002) using the E-M algorithm. Mean protein and Km values were compared using Students t-test. To construct models of the DCK and CMPK variants, the altered amino acids were computationally substituted within the 1.9 Å resolution crystal structure of the human DCK dimer bound to deoxycytidine and ADP (PDB accession code 1P60) (Sabini et al., 2003) and the 2.1 Å resolution crystal structure of the human CMPK monomer (PDB accession code 1TEV) (Segura-Peña et al., 2004) using the interactive graphics program O (Jones, 1991). Molecular figures were prepared with Molscript (Kraulis, 1991) and Raster3D (Merritt and Bacon, 1997).
Results
DCK and CMPK Gene Resequencing
The areas of DCK and CMPK that we resequenced included exons, splice junctions, a portion of the 5′-FR and a 1 kb region located approximately 2.4 kb upstream of the site of CMPK transcription initiation. This area was amplified because it displayed high cross-species homology with mouse and dog sequences and was predicted by rVISTA (Loots et al., 2002) to possibly be involved in transcription regulation. These areas of both genes were resequenced using 240 DNA samples from 4 ethnic groups. The polymorphisms observed in DCK and CMPK are listed in Table 1. For DCK, 28 polymorphisms, including 26 SNPs and 2 indels, were observed in the 480 alleles resequenced. Two upstream variants, 5’-FR (−248) and 5’-FR (−201), were located in potential regulatory elements identified by rVISTA within conserved DNA sequence. Further studies will be needed to determine whether these variants influence binding of transcription factors and therefore regulate transcription. The 3 nonsynonymous cSNPs observed in DCK resulted in the following amino acid substitutions: Ile24Val, Ala119Gly and Pro122Ser (Table 1). Two of these polymorphisms, Ile24Val and Ala119Gly, were observed only in samples from AA subjects, while Pro122Ser occurred in both the AA and HCA populations. Sixteen of the 28 DCK SNPs were “novel” (Table 1). For CMPK, all exons, intron sequence flanking exons, and approximately 5 kb of the 5′-FR were resequenced. Once again, 28 polymorphisms, including 26 SNPs and 2 indels, were observed. Three nonsynonymous cSNPs that resulted in the following amino acid alterations were observed: Gln48His, Glu75Lys and Asn83Ser (Table 1). The Gln48His and Asn83Ser polymorphisms were observed in all four populations studied. The Gln48His polymorphism had a minor allele frequency (MAF) of 15% in the HCA samples, but 5% or less in three other ethnic groups. The Asn83Ser polymorphism occurred with low frequency (0.8%) in all four ethnic groups, and the Glu75Lys polymorphism was observed only in MA subjects. Seventeen of the 28 CMPK SNPs were novel (Table 1). All of the SNPs that we identified were deposited in the NIH database PharmGKB.
Table 1.
Human DCK and CMPK polymorphisms.
| DCK | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene Location |
Nucleotide | Sequence Change |
Amino Acid Change |
MAF | HapMap and/or dbSNP |
|||
| AA | CA | HCA | MA | |||||
| 5'-FR | −822 | G>A | 0.000 | 0.025 | 0.000 | 0.008 | ||
| 5'-FR | −698 to −697 | TA deletion | 0.208 | 0.033 | 0.208 | 0.108 | ||
| 5'-FR | −520 | G>A | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 5'-FR | −289 | T>A | 0.000 | 0.000 | 0.017 | 0.000 | ||
| 5'-FR | −248 | G>C | 0.033 | 0.000 | 0.000 | 0.000 | ||
| 5'-FR | −201 | C>T | 0.000 | 0.033 | 0.208 | 0.100 | rs2306744 | |
| 5'-UTR | −79 | C>G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 5'-UTR | −52 | G>A | 0.033 | 0.000 | 0.000 | 0.000 | * | |
| Exon 1 | 70 | A>G | Ile24Val | 0.033 | 0.000 | 0.000 | 0.000 | * |
| IVS 1 | 37 | G>C | 0.008 | 0.025 | 0.000 | 0.000 | rs9997790 | |
| IVS 2 | 81 | T>G | 0.000 | 0.008 | 0.000 | 0.000 | ||
| IVS 2 | 102 | C>T | 0.042 | 0.000 | 0.000 | 0.000 | * | |
| IVS 2 | 114 | A>G | 0.458 | 0.975 | 0.933 | 0.992 | rs6446988 | |
| IVS 2 | 179 | C>T | 0.008 | 0.000 | 0.000 | 0.000 | ||
| IVS 2 | 186 | A>T | 0.017 | 0.000 | 0.000 | 0.000 | ||
| Exon 3 | 273 | A>G | 0.000 | 0.000 | 0.000 | 0.008 | ||
| Exon 3 | 300 | C>T | 0.008 | 0.092 | 0.000 | 0.042 | rs11544786 | |
| Exon 3 | 356 | C>G | Ala119Gly | 0.033 | 0.000 | 0.000 | 0.000 | * |
| Exon 3 | 364 | C>T | Pro122Ser | 0.083 | 0.000 | 0.067 | 0.000 | * |
| IVS 3 | 53 | G>A | 0.008 | 0.042 | 0.000 | 0.025 | * | |
| IVS 3 | −113 | C>T | 0.000 | 0.000 | 0.025 | 0.000 | ||
| Exon 4 | 513 | A>G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| IVS 5 | 33 | A>G | 0.000 | 0.000 | 0.000 | 0.008 | ||
| IVS 6 | 41 | A>T | 0.450 | 0.975 | 0.933 | 0.992 | rs1486271 | |
| IVS 6 | 137 | A>T | 0.000 | 0.042 | 0.000 | 0.025 | ||
| IVS 6 | 157 | T>G | 0.042 | 0.000 | 0.000 | 0.000 | ||
| 3’-UTR | 948 | C>T | 0.442 | 0.967 | 0.933 | 0.992 | rs4643786 | |
| 3’-UTR | 968 to 969 | C insertion | 0.033 | 0.000 | 0.000 | 0.000 | ||
| CMPK | ||||||||
|
Gene Location |
Nucleotide |
Sequence Change |
Amino Acid Change |
MAF |
HapMap and/or dbSNP |
|||
| AA | CA | HCA | MA | |||||
| 5'-FR | −3331 | C>G | 0.133 | 0.392 | 0.217 | 0.375 | rs12126139 | |
| 5'-FR | −3258 | A>C | 0.008 | 0.008 | 0.000 | 0.008 | ||
| 5'-FR | −3234 | T>C | 0.142 | 0.000 | 0.000 | 0.000 | rs3125637 | |
| 5'-FR | −3175 | G>C | 0.085 | 0.392 | 0.217 | 0.375 | rs12118514 | |
| 5'-FR | −3174 to −3173 | AG deletion | 0.233 | 0.000 | 0.000 | 0.000 | ||
| 5'-FR | −3138 | C>T | 0.000 | 0.000 | 0.067 | 0.000 | ||
| 5'-FR | −3076 | C>T | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 5'-FR | −3010 | G>T | 0.200 | 0.383 | 0.033 | 0.108 | rs9725517 | |
| 5'-FR | −2916 | G>A | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 5'-FR | −2835 | A>T | 0.042 | 0.000 | 0.000 | 0.000 | rs9436435 | |
| 5'-FR | −432 to -431 | GA deletion | 0.042 | 0.000 | 0.000 | 0.000 | ||
| 5'-FR | −299 | G>T | 0.017 | 0.000 | 0.000 | 0.000 | rs12097707 | |
| 5'-FR | −278 | G>A | 0.250 | 0.367 | 0.033 | 0.117 | ||
| 5'-FR | −229 | C>T | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 5'-FR | −132 | T>C | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 5'-UTR | −106 | G>C | 0.008 | 0.008 | 0.008 | 0.008 | ||
| 5'-UTR | −75 | C>G | 0.233 | 0.408 | 0.508 | 0.425 | rs7543016 | |
| IVS 1 | 15 | G>A | 0.000 | 0.000 | 0.000 | 0.017 | ||
| IVS 1 | −128 | A>G | 0.333 | 0.000 | 0.000 | 0.000 | rs12079592 | |
| IVS 1 | −3 | C>T | 0.017 | 0.000 | 0.000 | 0.000 | ||
| Exon 2 | 144 | G>T | Gln48His | 0.025 | 0.050 | 0.150 | 0.042 | rs35687416 |
| Exon 3 | 223 | G>A | Glu75Lys | 0.000 | 0.000 | 0.000 | 0.058 | |
| Exon 3 | 248 | A>G | Asn83Ser | 0.008 | 0.008 | 0.008 | 0.008 | |
| Exon 4 | 387 | A>G | 0.200 | 0.000 | 0.000 | 0.000 | rs6656779 | |
| IVS 4 | 95 | T>A | 0.000 | 0.000 | 0.000 | 0.008 | ||
| IVS 4 | 121 | C>T | 0.008 | 0.008 | 0.008 | 0.008 | ||
| IVS 4 | 153 | A>G | 0.000 | 0.000 | 0.000 | 0.008 | ||
| 3'-UTR | 605 | A>G | 0.150 | 0.000 | 0.000 | 0.008 | rs3125648 | |
Locations in the gene, nucleotide sequence alterations, amino acid sequence alterations, and minor allele frequencies (MAFs) of polymorphisms in the four populations studied are listed. If a polymorphism has been deposited in a public database (dbSNP or HapMap), an rs number is indicated. All other polymorphisms listed are unique to this study.
indicates polymorphisms also reported recently by Lamba et al. (2007). Polymorphisms within exons have been boxed. IVS is intervening sequence (intron). The numbering scheme for nucleotide positions is based on assignment of the (+1) position to the “A” in the translation initiation codon, with nucleotides 5′ to that position assigned negative, and those 3′ within the cDNA assigned positive numbers. Nucleotide positions in introns are numbered from the nearest splice site, with (+1) as the first nucleotide at the 5′-end, and (−1) as the first nucleotide at the 3′-end of the intron.
AA, African-American; CA, Caucasian-American; HCA, Han Chinese-American; MA, Mexican-American; FR, flanking region; UTR, untranslated region.
We also determined “nucleotide diversity”, a quantitative measure of genetic variation, adjusted for allele frequency, in all four ethnic groups by calculating θ, a population mutation measure that is theoretically equal to the neutral mutation variable, and π, the average heterozygosity per site (Maring et al., 2005). Tajima’s D values were also estimated as a test of the neutral mutation hypothesis (Table 2). For both genes, as anticipated, samples from AA subjects displayed the greatest nucleotide diversity. Although negative values for Tajima D indicate a departure from neutrality, none of the values listed in Table 2 was statistically significant (Table 2).
Table 2.
DCK and CMPK genetic diversity and neutrality test values.
| π, × 104 | θ, × 104 | Tajima's D | P Values | |
|---|---|---|---|---|
| DCK | ||||
| AA | 8.75 ± 5.2 | 10.8 ± 3.6 | −0.52 | 0.62 |
| CA | 2.51 ± 2.1 | 6.99 ± 2.6 | −1.63 | 0.10 |
| HCA | 4.42 ± 3.1 | 6.99 ± 2.6 | −0.93 | 0.36 |
| MA | 2.27 ± 1.9 | 7.01 ± 2.6 | −1.73 | 0.08 |
| CMPK | ||||
| AA | 12.8 ± 7.3 | 13.9 ± 4.4 | −0.23 | 0.83 |
| CA | 9.66 ± 5.8 | 8.99 ± 3.2 | 0.20 | 0.85 |
| HCA | 6.49 ± 4.2 | 6.92 ± 2.7 | −0.15 | 0.88 |
| MA | 8.09 ± 5.0 | 10.4 ± 3.6 | −0.60 | 0.57 |
Estimates of two measures of nucleotide diversity, π, θ are listed, as well as Tajima’s D, a test of the “neutral” mutation hypothesis.
The P values refer to Tajima’s D.
Haplotype and Linkage Disequilibrium Analysis
We also performed population-specific linkage disequilibrium and haplotype analysis for both genes. Haplotype designations were based on the encoded amino acid sequence of the allozyme, with the WT sequence designated as *1. For example, for DCK, the *2, *3 and *4 designations were used for sequences that encoded Val24, Gly119 and Ser122, respectively (Table 3A). Alleles encoding DCK Val24, Gly119 and Ser122, as well as Val24/Ser122 and Val24/Gly119/Ser122 variant allozymes were also present. Therefore, we designated these multiple amino acid variant haplotypes as *5 and *6, respectively. *1A was observed with high frequency in all four populations, while the *4 haplotype was observed in only AA and HCA subjects. The *2, *3, *5 and *6 haplotypes were observed only in AA DNA (Table 3A). For CMPK, the *2, *3 and *4 designations were used for sequences that encoded His48, Lys75 and Ser83, respectively. Combinations of these cSNPs were not observed or inferred in any of the DNA samples. CMPK haplotypes *1B, *1C, *1H, *1L and *2A were observed in all populations, while the *3 haplotype was seen in only MA subjects (Table 3B). We also calculated population-specific linkage disequilibrium based on pairwise D′ values for all DCK and CMPK SNPs (data not shown).
Table 3.
Human DCK and CMPK haplotypes.
| (A) DCK Haplotypes | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequency | 5'-FR (−698) |
5'-FR (−289) |
5'-FR (−201) |
Exon1 (70) |
IVS1 (37) |
IVS2 (102) |
IVS2 (114) |
Exon3 (300) |
Exon3 (356) |
Exon3 (364) |
IVS3 (53) |
IVS3 (−113) |
IVS6 (41) |
IVS6 (137) |
IVS6 (157) |
3'-UTR (948) |
3'-UTR (968) |
|||||
| AA | CA | HCA | MA | |||||||||||||||||||
| o | *1A | 0.424 | 0.803 | 0.667 | 0.808 | I | T | C | A | G | C | G | C | C | C | G | C | T | A | T | T | D |
| o | *1B | 0.168 | 0.008a | D | T | C | A | G | C | A | C | C | C | G | C | A | A | T | C | D | ||
| o | *1C | 0.150 | I | T | C | A | G | C | A | C | C | C | G | C | A | A | T | C | D | |||
| i | *1D | 0.025 | I | T | C | A | G | C | A | C | C | C | G | C | A | A | G | C | D | |||
| i | *1E | 0.023 | D | T | C | A | G | T | A | C | C | C | G | C | A | A | T | C | D | |||
| o | *1F | 0.072 | I | T | C | A | G | C | G | T | C | C | G | C | T | A | T | T | D | |||
| i | *1G | 0.037 | 0.025 | I | T | C | A | G | C | G | C | C | C | A | C | T | T | T | T | D | ||
| o | *1H | 0.017a | 0.207 | 0.092 | D | T | T | A | G | C | G | C | C | C | G | C | T | A | T | T | D | |
| o | *1I | 0.024 | I | T | C | A | G | C | G | C | C | C | G | T | T | A | T | T | D | |||
| o | *1J | 0.042 | I | T | C | A | G | C | G | T | C | C | G | C | T | A | T | T | D | |||
| i | *2 | 0.017 | I | T | C | G | G | C | A | C | C | C | G | C | A | A | T | C | D | |||
| i | *3A | 0.008 | D | T | C | A | G | C | A | C | G | C | G | C | A | A | T | C | I | |||
| i | *3B | 0.008 | D | T | C | A | G | C | G | C | G | C | G | C | A | A | T | C | I | |||
| i | *3C | 0.008 | I | T | C | A | G | C | A | C | G | C | G | C | A | A | G | C | I | |||
| i | *4A | 0.048 | 0.059 | I | T | C | A | G | C | A | C | C | T | G | C | A | A | T | C | D | ||
| i | *4B | 0.008 | I | T | C | A | G | C | A | C | C | T | G | C | T | A | T | C | D | |||
| i | *4C | 0.008 | I | T | C | A | C | C | A | C | C | T | G | C | A | A | T | C | D | |||
| i | *4D | 0.002 | I | T | C | A | G | T | A | C | C | T | G | C | A | A | T | C | D | |||
| i | *4E | 0.008 | I | A | C | A | G | C | A | C | C | T | G | C | A | A | T | C | D | |||
| i | *5 | 0.008 | I | T | C | G | G | C | A | C | C | T | G | C | A | A | G | C | D | |||
| i | *6 | 0.008 | I | T | C | G | G | C | A | C | G | T | G | C | A | A | T | C | I | |||
| (B) CMPK Haplotypes | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequency | 5'-FR (−3331) |
5'-FR (−3258) |
5'-FR (− 3234) |
5'-FR (−3175) |
5'-FR (−3174) |
5'-FR (−3138) |
5'-FR (−3010) |
5'-FR (−2835) |
5'-FR (−432) |
5'-FR (−278) |
5'-UTR (−106) |
5'-UTR (−75) |
IVS1 (−128) |
Exon2 (144) |
Exon3 (223) |
Exon3 (248) |
Exon4 (387) |
IVS4 (95) |
IVS4 (121) |
3'-UTR (605) |
|||||
| AA | CA | HCA | MA | ||||||||||||||||||||||
| o | *1A | 0.182 | C | A | T | G | D | C | G | A | I | G | G | C | G | G | G | A | G | T | C | A | |||
| o | *1B | 0.136 | 0.367 | 0.033a | 0.083 | C | A | T | G | I | C | T | A | I | A | G | C | A | G | G | A | A | T | C | A |
| o | *1C | 0.124 | 0.192 | 0.440 | 0.458 | C | A | T | G | I | C | G | A | I | G | G | C | A | G | G | A | A | T | C | A |
| o | *1D | 0.087 | 0.008 | C | A | T | G | I | C | G | A | I | A | G | C | A | G | G | A | A | T | C | A | ||
| i | *1E | 0.050 | C | A | C | G | I | C | G | A | I | G | G | C | A | G | G | A | A | T | C | G | |||
| i | *1F | 0.050 | C | A | C | G | I | C | G | A | I | G | G | C | G | G | G | A | A | T | C | A | |||
| o | *1G | 0.044 | C | A | T | G | I | C | G | A | I | G | G | C | A | G | G | A | A | T | C | G | |||
| o | *1H | 0.041a | 0.333 | 0.059 | 0.275 | G | A | T | C | I | C | G | A | I | G | G | G | A | G | G | A | A | T | C | A |
| i | *1I | 0.039 | C | A | T | G | I | C | T | A | I | G | G | G | A | G | G | A | A | T | C | A | |||
| i | *1J | 0.031 | G | A | T | G | I | C | G | A | D | G | G | G | A | G | G | A | A | T | C | A | |||
| i | *1K | 0.023 | C | A | C | G | I | C | G | T | I | G | G | C | A | G | G | A | A | T | C | G | |||
| o | *1L | 0.011a | 0.008a | 0.283 | 0.025a | C | A | T | G | I | C | G | A | I | G | G | G | A | G | G | A | A | T | C | A |
| i | *1M | 0.025 | G | A | T | C | I | T | G | A | I | G | G | G | A | G | G | A | A | T | C | A | |||
| o | *2A | 0.017a | 0.042a | 0.091 | 0.025 | G | A | T | C | I | C | G | A | I | G | G | G | A | T | G | A | A | T | C | A |
| i | *2B | 0.008 | 0.008 | 0.008 | G | C | T | C | I | C | G | A | I | G | G | G | A | T | G | A | A | T | C | A | |
| o | *2C | 0.041 | G | A | T | C | I | T | G | A | I | G | G | G | A | T | G | A | A | T | C | A | |||
| i | *2D | 0.018 | C | A | T | G | I | C | G | A | I | G | G | C | A | T | G | A | A | T | C | A | |||
| i | *2E | 0.008 | C | A | T | G | I | C | T | A | I | A | G | G | A | T | G | A | A | T | C | A | |||
| o | *3A | 0.050 | G | A | T | C | I | C | G | A | I | G | G | G | A | G | A | A | A | T | C | A | |||
| i | *3B | 0.008 | G | A | T | C | I | C | G | A | I | G | G | G | A | G | A | A | A | A | C | A | |||
| i | *4A | 0.008 | G | A | T | G | I | C | G | A | D | G | C | G | A | G | G | G | A | T | T | A | |||
| i | *4B | 0.008 | 0.008 | C | A | T | G | I | C | G | A | I | G | C | C | A | G | G | G | A | T | T | A | ||
| i | *4C | 0.008 | C | A | T | G | I | C | G | A | I | G | C | G | A | G | G | G | A | T | T | A | |||
Variant nucleotides compared with the “reference sequence” (i.e., the most common sequence in African-American subjects) are highlighted as white on black. Initial haplotype designations (*1, *2, etc.) were made on the basis of amino acids that vary, with the WT allozyme designated *1. Subsequent assignments/letter designations were made within ethnic groups based on descending allele frequencies, starting with haplotypes present in AA subjects. The *2, *3 and *4 designations for DCK were used for sequences that encoded Val24, Gly119 and Ser122, respectively. The combination of Val24/Ser122, and of Val24/Gly119/Ser122 were inferred, and these combinations were designated as *5 and *6,respectively. The symbols I and D in the 5'-FR (−698 to −697) and 3'-UTR (968 to 969) represent indels. The CMPK *2, *3, and *4 designations were used for sequences that encoded His48, Lys75 and Ser83, respectively. The symbols I and D in the 5'-FR (−3174 to −3173) and (−432 to−431) represent indels. o=observed haplotype; i=inferred haplotype with frequency ≥ 2%. Some inferred haplotypes with frequencies lower than 2% were included in the table because they included an amino acid change.
Indicates inferred data.
Functional Genomic Studies
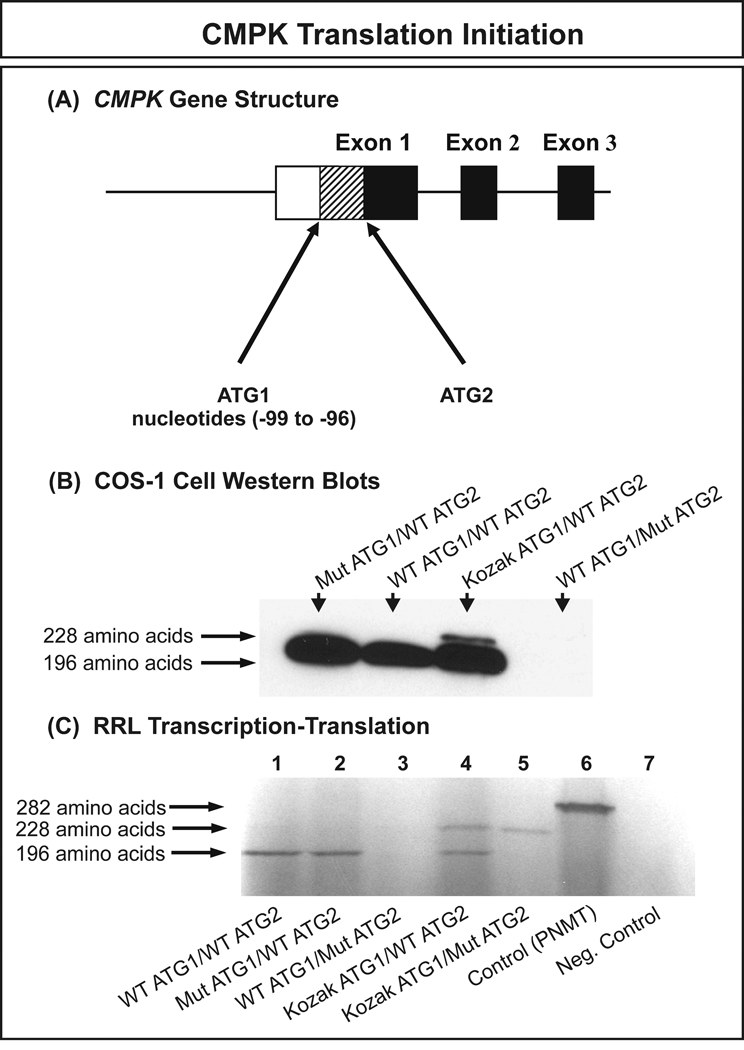
Expression constructs for the WT and variant allozymes for both DCK and CMPK were created to determine the effect of nonsynonymous SNPs on level of protein, level of enzyme activity and substrate kinetics. Since our haplotype analysis had shown that multiple variant DCK alleles were present that encoded Val24/Ser122 and Val24/Gly119/Ser122, we also created expression constructs for those allozymes. Although the DCK double variants, Gly119/Ser122 and Val24/Gly119 were not observed in our samples, we also created those variants to help us understand the effect on enzyme activity of combinations of the polymorphisms. However, before expressing the CMPK constructs, one issue required clarification. CMPK contains two in-frame ATGs at the 5′-end of the gene – creating confusion with regard to which might be used in vivo as the translation initiation codon. To address that issue before studying results obtained with our expression constructs, we created four CMPK “test” expression constructs to determine which ATG might be used to initiate translation. These additional constructs included one with a mutated initial ATG (Mut ATG1/WT ATG2); one with the second ATG mutated (WT ATG1/Mut ATG2); a construct with a perfect Kozak sequence (Kozak, 1986) for the initial ATG and the wild type second ATG (Kozak ATG1/WT ATG2); and a final construct with a perfect Kozak sequence surrounding the initial ATG and a mutated second ATG (Kozak ATG1/Mut ATG2) (Fig. 2). The sequences of the ATG in these constructs are listed in Table 4. The top panel in Fig. 2 shows this situation diagrammatically, with the location of the two inframe ATGs – separated by 99 nucleotides (33 putative codons) – indicated. When these constructs were expressed in COS-1 cells, the 196 amino acid sequence was always produced in the presence of WT ATG2 but the 228 amino acid sequence was only seen if a perfect Kozak sequence was created for the initial (Kozak ATG1/WT ATG2), putative, translational initiation codon (Fig. 2B). Identical results were observed during rabbit reticulocyte lysate expression, with the 196 amino acid recombinant protein encoded by only the ORF that began with naturally occurring “ATG2” being expressed (Fig. 2C). These results indicated clearly that the second inframe ATG (“ATG2”) was utilized as the translation initiation codon, so all subsequent data relate to the 196 amino acid CMPK protein.
Figure 2. CMPK translation initiation.
(A) Diagrammatic representation of the 5′-portion of the gene structure showing the locations of the two potential in-frame translation initiation ATGs. (B) COS-1 cell transfection with CMPK expression constructs. See text and Table 4 for a description of the constructs. (C) RRL in vitro transcription and translation. See text for a detailed explanation.
Table 4.
Sequence of the two potential CMPK translation initiation codons and results of studies performed to determine which codon might be functional in vivo (see also Fig. 2).
| ATG1 | ATG2 | COS-1 Cell Western Results |
RRL Results | |
|---|---|---|---|---|
| Ideal Kozak Sequence | ACCATGG | ACCATGG | ----- | ----- |
| WT ATG1/WT ATG2 | TGTATGC | CTCATGA | 196 amino acids | 196 amino acids |
| Mut ATG1/WT ATG2 | TGTTTGC | CTCATGA | 196 amino acids | 196 amino acids |
| Kozak ATG1/WT ATG2 | ACCATGG | CTCATGA | 228 and 196 amino acids | 228 and 196 amino acids |
| Kozak ATG1/Mut ATG2 | ACCATGG | CTCTTGA | ----- | 228 amino acids |
| WT ATG1/Mut ATG2 | TGTATGC | CTCTTGA | No protein | No protein |
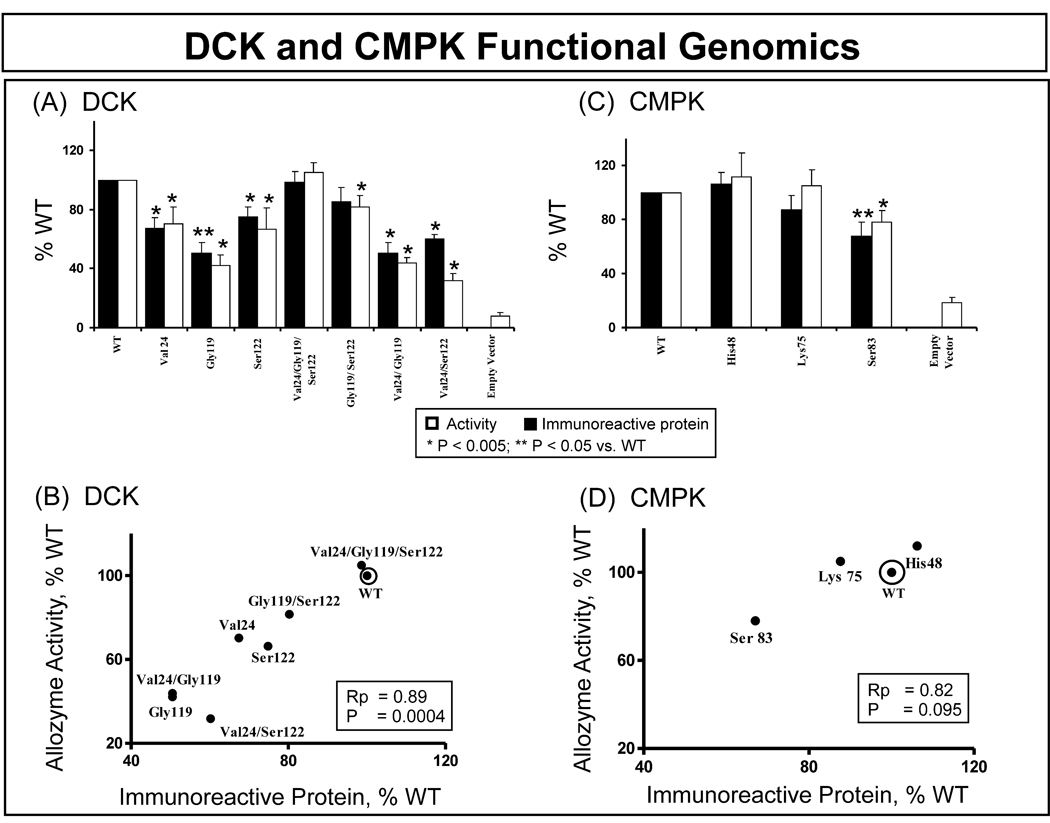
The DCK and CMPK expression constructs were transfected into COS-1 cells, and the cells were co-transfected with β-galactosidase to make it possible to correct for transfection efficiency. Immunoreactive protein levels were then measured by quantitative Western blot analysis, and levels of enzyme activity were measured in the same samples. Each transfection was performed in triplicate and was repeated three times. Levels of DCK and CMPK immunoreactive protein, as well as the enzyme activity for all of these transfections, are shown in Fig. 3. Because DCK and CMPK are both widely expressed, it was necessary to include an “empty vector” control in all transfections, and the results for those samples are shown in the figure for levels of enzyme activity. These empty vector control samples displayed an average 8.1% of WT construct values for DCK and 18.7% for CMPK. However, no evidence of endogenous immunoreactive protein for either DCK or CMPK was detected when the quantitative Western blot studies were performed.
Figure 3. DCK and CMPK allozyme functional genomics.
(A) and (C) show levels of allozyme activity and immunoreactive protein for DCK and CMPK, respectively, expressed as percentages of WT. Each bar represents the average of 9 independent transfections (mean ± SE). All values were corrected for transfection efficiency. (B) and (D) show correlations between average levels of immunoreactive protein and enzyme activity for DCK and CMPK, respectively.
Immunoreactive DCK protein levels showed significant differences from WT, specifically Val24 (67 ± 7%); Gly119 (51 ± 7%); Ser122 (75 ± 7%); Val24/Gly119 (50 ±7%); and the combination of Val24/Ser122 (60 ± 3%) as compared with WT (Fig. 3A). The “triple” variant (*6) which included Val24/Gly119/Ser122 (99 ± 7%) and the combination Gly119/Ser122 (85 ± 8%) did not show significant differences from the WT protein. DCK allozyme enzyme activity levels when compared with that for WT were Val24 (70 ± 11%), Gly119 (42 ± 7%), Ser122 (66 ± 15%), Val24/Gly119/Ser122 (105 ± 7%), Gly119/Ser122 (82 ± 8%), Val24/Gly119 (44 ± 4%) and Val24/Ser122 (32 ± 5%). These results for activity, with gemcitabine as a substrate, correlated significantly with corresponding protein levels (Rp = 0.89, P = 0.0004) (Fig. 3B). Recombinant CMPK protein levels did not differ significantly from those for WT for His48 (106 ± 9%) or Lys75 (88 ± 10%). However, the protein level for CMPK Ser83 was significantly lower than that for WT (68 ± 11%) (Fig. 3C). Once again, levels of protein and enzyme activity were very similar (Rp = 0.82) but, in this case, the correlation was not significant (P = 0.095).
Because of the possibility that some of the differences in levels of enzyme activity that we had observed might have been due to alterations in substrate kinetics as a result of changes in the encoded amino acids, substrate kinetic studies were also performed. For those experiments, gemcitabine substrate concentrations for the DCK studies ranged from 1.25 to 75 µM and gemcitabine monophosphate concentrations for the CMPK studies ranged from 50 to 2000 µM. The apparent Km values observed are listed in Table 5. Only the DCK Val24/Ser122 apparent Km value differed significantly from that for the WT of the enzyme being studied.
Table 5.
Recombinant DCK and CMPK allozyme substrate kinetics.
| DCK Allozyme | Apparent Km (µM) |
|---|---|
| WT | 14.6 ± 3.5 |
| Val24 | 13.1 ± 3.2 |
| Gly119 | 14.5 ± 2.5 |
| Ser122 | 14.1 ± 3.3 |
| Val24/Gly119/Ser122 | 14.8 ± 2.5 |
| Gly119/Ser122 | 9.5 ± 2.5 |
| Val24/Gly119 | 10.7 ± 1.1 |
| Val24/Ser122 | 6.3 ± 0.8* |
| CMPK Allozyme | Apparent Km (µM) |
| WT | 246 ± 61 |
| His48 | 208 ± 45 |
| Lys75 | 265 ± 67 |
| Ser83 | 236 ± 39 |
Gemcitabine substrate kinetics for WT and variant DCK allozymes and gemcitabine monophosphate substrate kinetics for CMPK were measured in the presence of 1 mM ATP. Values represent mean ± SEM (n ≥ 6).
Indicates a statistically significant difference from the WT allozyme (P < 0.025 by Student’s t-test).
DCK and CMPK Structural Models
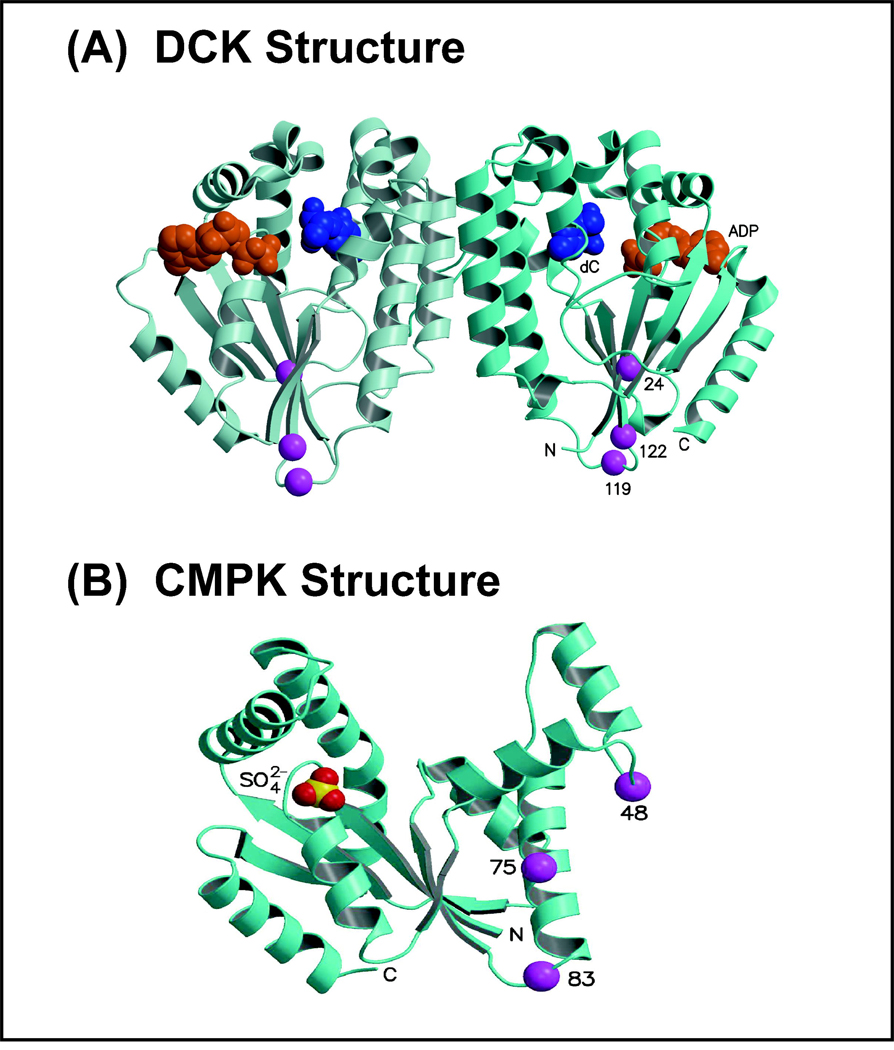
High resolution x-ray crystal structures of human DCK and CMPK (Sabini et al., 2003; Segura-Peña et al., 2004) were used as starting scaffolds to map variant amino acids encoded by the naturally occurring nonsynonymous cSNPs in DCK and CMPK. We then assessed the computationally substituted variant residues for compatibility with the wild-type native protein structures to determine whether our observations with recombinant protein were plausible within the context of the structures of these enzymes (Fig. 4). The DCK monomer structure has a central β-sheet surrounded by helices. Ile24 is located in a β-strand which is first in amino acid sequence, but physically in the middle of the β-sheet, and the Ile24 side chain is buried in a hydrophobic environment. Since the Val24 variant amino acid is smaller than Ile24, it could be accommodated structurally, but a small void resulting from the change in amino acid could result in a locally destabilizing conformational change. Ala119Gly is located in a loop near the N-terminus of a strand next to the Ile24 strand in the central β-sheet and far from the active site (~25 Å). The Ala119 carbonyl oxygen atom forms a hydrogen bond with the Arg20 side chain and tethers the Ala119 loop to the Ile24-containing strand. The Ala119 amide nitrogen atom also interacts with the carbonyl oxygen of Leu116 to stabilize the loop conformation. In addition, the Ala119 side chain is partially buried, since it packs against Pro122. The smaller Gly119 variant could be easily accommodated in the structure sterically, but Gly119 might introduce an increase in local flexibility due to its lack of a bulky side chain, altering the local conformation so that interactions between this loop and adjacent β strands would be destabilized, resulting in the striking decrease in enzyme protein that we observed (Fig. 3A). DCK Pro122Ser is located at the N-terminus of the strand next to the Ile24 strand in the central β-sheet, and distant from the active site (~22 Å). The Pro122 side chain is mostly buried in a hydrophobic environment formed by several residues, one of which is Ala119. In addition, the main chain Pro122 carbonyl oxygen atom forms a long hydrogen bond with the Lys22 main chain amide nitrogen (located in the adjacent Ile24 strand). The smaller Ser122 could be easily accommodated sterically, but it could introduce increased local flexibility as a result of loss of the rigid proline ring. This increased flexibility might alter the local conformation so that interactions between the Pro122 strand and the adjacent Ala119 loop and Ile24 strand would be destabilized. This analysis provides structural explanations for how the single DCK variants may result in the observed decreased protein quantity and activity. Unfortunately it is not possible to reliably model the specific altered conformations of the single variants, to provide a basis for a similar structural analysis of the double or triple variants.
Figure 4. Human DCK and CMPK structural models.
(A) The two monomers in the DCK dimeric crystal structure are shown as ribbon structures in two shades of blue. Bound in the active site of each monomer are ADP (orange) and deoxycytidine (dC, blue), shown as space-filling structures. The Cα atoms of the three residues altered by SNPs (Ile24, Ala119, Pro122) are drawn as purple spheres. The N- and C- termini, active site ligands, and positions of the altered residues are labeled for the right monomer. (B) The human CMPK monomer is shown as a blue ribbon structure. Bound in the active site is a sulfate ion shown as space-filling spheres. The Cα atoms of the three residues altered by SNPs (Gln48, Glu75, and Asn83) are drawn as purple spheres.
All three CMPK variant residues are located far from the active site (Fig. 4B). However, Gln48His alters a highly exposed residue that forms a hydrogen bond with Arg74. His48 would be easily accommodated sterically, and it would allow the hydrogen bond to be retained. Glu75Lys affects an exposed residue that interacts only with a water molecule, so the Lys substitution would be accommodated easily. Asn83Ser also alters an exposed residue, and the Ser substitution would be easily accommodated. The structural predictions for these CMPK residues are compatible with the observed functional effects of all three SNPs, indicating modest to no consequences after variant substitution (Fig. 3C). Overall, the three DCK variant substitutions were less compatible with the native protein structure than were the three CMPK variants, results consistent with our experimental observations that the decreases in protein and activity levels for the DCK variants were more significant than for the CMPK variants.
Discussion
DCK and CMPK catalyze the phosphorylation of antineoplastic nucleoside analogs such as gemcitabine and Ara-C (Mini et al., 2006). DCK is thought to be the rate-limiting step for nucleoside analogue phosphorylation, but CMPK also plays an essential role in the activation of these drugs (Van Rompay et al., 1999). In addition, previous studies have reported that mutations in DCK (Owens et al., 1992; Flasshove et al., 1994; Stegmann et al., 1995; Galmarini et al., 2004), low levels of DCK enzyme activity (Bergman et al., 1999; Kroep et al., 2002), and increased levels of the active gemcitabine triphosphate metabolite (Sebastiani et al., 2006) might all be factors that contribute to variation in gemcitabine response.
In the present study, we set out to perform comprehensive studies of DCK and CMPK pharmacogenomics. We began by resequencing both genes using 240 DNA samples from four different ethnic groups. We observed a total of 28 SNPs in DCK, 16 of which were novel. Joerger et al (2006) had previously discovered 6 DCK SNPs in their resequencing study of a mixed ethnic cohort of healthy volunteers (−243 G>T, −135 G>C, 261 G>A, 364 C>T, 727 A>C, IVS6 +41T>A). We observed two of those SNPs, 364C>T and IVS6 +41T>A, but not the other four in our samples. Very recently, Lamba et al (2007) studied polymorphisms in DCK, using genomic DNA from the 30 CEPH (CA) and 30 YRI (African) trios (90 subjects of each ethnicity in groups of 3 that included related parent-child trios) that had been used in the HapMap Project. Those investigators resequenced DCK exons, all of the first intron plus an additional 300 bp of the 5′-FR beyond what we resequenced. In the studies reported here, we have extended DCK resequencing to include previously unreported ethnic populations (HCA and MA), as well as performed haplotype analysis, genetic diversity determination, neutrality testing, and linkage disequilibrium analysis for all four (AA, CA, HCA, and MA) ethnic groups.
To determine the effect of nonsynonymous SNPs on protein and activity levels, 8 DCK expression constructs were created, four of which included more than one SNP, three double variants (Val24/Gly119; Val24/Ser122; Gly119/ Ser122), as well as a triple variant (Val24/Gly119/Ser122). Levels of immunoreactive protein and allozyme activity were determined when these constructs were used to transfect COS-1 cells. The common DCK 24 A>G (Ile24Val) and DCK 122 C>T (Pro122Ser) polymorphisms resulted in moderate decreases in levels of both activity and protein. The correlation between levels of immunoreactive protein and allozyme activity for all DCK variant allozymes was highly significant (Fig. 3B). It was of interest that the DCK “compound variants” with two or three of the naturally occurring variant amino acids displayed less striking alterations in activity and protein levels than did those with only a single variant amino acid (Fig. 3A). The mechanism(s) responsible will have to be the subject of future studies, but these results clearly indicate that the functional effects of nonsynonymous SNPs cannot be assumed to be purely “additive”. Our substrate kinetics analyses indicated that, except for the compound variant Val24/Ser122, no variant DCK allozyme had a Km value significantly different from that of WT. Our results contrasted with those of Lamba et al (2007) who reported that not only were “single variant” DCK allozymes expressed at equivalent levels to WT but that Gly119 and Ser122 had significantly lower Km values than WT. However, no direct comparison of those results with our data can accurately be made, as mammalian COS-1 cells were employed in our studies for the expression of DCK allozymes to ensure appropriate post-translational modification of expressed proteins as well as the presence of mammalian protein degradation systems, as opposed to the bacteria employed by Lamba et al. In addition, we employed the pyrimidine analog gemcitabine (2’, 2’-difluorodeoxycytidine) as substrate in our deoxycytidine kinase kinetics studies, as opposed to the purine analog cladribine (2-chloro-2’-deoxyadenosine) employed by Lamba et al.
The CMPK cDNA presented an interesting challenge since it has two possible inframe translational start codons, one encoding a 228 and the other a 196 amino acid protein, assuming that the N-terminal methionine is retained (Liou et al., 2002). To determine which ATG was utilized in vitro, we mutated each ATG in turn. The results shown in Fig. 2 indicated that the 196 residue protein was that expressed in mammalian cells, results consistent with previous reports (Bucurenci et al., 1996; Liou et al., 2002). Therefore, this 196 amino acid protein was used in our recombinant variant allozyme studies. We identified 17 novel CMPK SNPs among the 28 observed. Functional genomic studies of variant allozymes after COS-1 cell transfection showed that protein levels for the CMPK His48 and Lys75 allozymes were similar to that for the WT allozyme. However, activity and protein levels for CMPK Ser83 were decreased to 78% and 67% of WT, respectively (p<0.05) (Fig. 3). Our apparent Km value of 250 µmol/L for recombinant WT CMPK with gemcitabine monophosphate as a substrate can be compared with a value of 450 µmol/L that Van Rompay et al. (1999) observed using the same substrate. Analysis of the CMPK structure was compatible with our observation that these CMPK variants failed to alter apparent Km values since they were far from the active site.
In summary, we have performed comprehensive pharmacogenomic studies of genes encoding two enzymes required for the metabolic activation of antineoplastic cytidine antimetabolites. We identified a series of novel SNPs for these two genes in four ethnic groups by resequencing these genes – followed by comprehensive functional genomic characterization of variant allozymes. These observations may now contribute to translational pharmacogenomic studies of gemcitabine, AraC and other cytidine antimetabolites.
Supplementary Material
Acknowledgements
We thank Mrs. Luanne Wussow for her assistance with the preparation of this manuscript and Dr. Yung-Chi Cheng (Yale University School of Medicine, Department of Pharmacology, New Haven, CT) for providing the CMPK antibody.
Supported in part by NIH grants R01 GM28157, R01 GM35720, R01 CA132780 and U01 GM61388 (The Pharmacogenetics Research Network) as well as a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award and a grant from the Commonwealth Foundation for Cancer Research.
The gene resequencing data described in this article have been deposited in the NIH-funded database PharmGKB with submission numbers PS207098 and PS207100.
Abbreviations
- DCK
deoxycytidine kinase
- CMPK
cytidine monophosphate kinase
- dFdC
gemcitabine
- dFdCMP
gemcitabine monophosphate
- dFdCDP
gemcitabine diphosphate
- dFdCTP
gemcitabine triphosphate
- WT
wild type
- AA
African-American
- CA
Caucasian-American
- HCA
Han Chinese-American
- MA
Mexican-American
- SNP
single nucleotide polymorphism
- cSNP
coding single nucleotide polymorphism
- AraC
cytosine arabinoside
- ORF
open reading frame
- 5’-FR
5’-flanking region
- UTR
untranslated region
References
- Bergman AM, Pinedo HM, Jongsma AP, Brouwer M, Ruiz van Haperen VW, Veerman G, Leyva A, Eriksson S, Peters GJ. Decreased resistance to gemcitabine (2',2'-difluorodeoxycitidine) of cytosine arabinoside-resistant myeloblastic murine and rat leukemia cell lines: role of altered activity and substrate specificity of deoxycytidine kinase. Biochem Pharmacol. 1999;57:397–406. doi: 10.1016/s0006-2952(98)00318-9. [DOI] [PubMed] [Google Scholar]
- Bucurenci N, Sakamoto H, Briozzo P, Palibroda N, Serina L, Sarfati RS, Labesse G, Briand G, Danchin A, Barzu O, Gilles AM. CMP kinase from Escherichia coli is structurally related to other nucleoside monophosphate kinases. J Biol Chem. 1996;271:2856–2862. doi: 10.1074/jbc.271.5.2856. [DOI] [PubMed] [Google Scholar]
- Carmichael J. The role of gemcitabine in the treatment of other tumours. Br J Cancer. 1998;78 Suppl 3:21–25. doi: 10.1038/bjc.1998.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YC. Potential use of antiviral L(−)nucleoside analogues for the prevention or treatment of viral associated cancers. Cancer Lett. 2001;162 Suppl:S33–S37. doi: 10.1016/s0304-3835(00)00650-9. [DOI] [PubMed] [Google Scholar]
- Flasshove M, Strumberg D, Ayscue L, Mitchell BS, Tirier C, Heit W, Seeber S, Schütte J. Structural analysis of the deoxycytidine kinase gene in patients with acute myeloid leukemia and resistance to cytosine arabinoside. Leukemia. 1994;8:780–785. [PubMed] [Google Scholar]
- Galmarini CM, Clarke ML, Jordheim L, Santos CL, Cros E, Mackey JR, Dumontet C. Resistance to gemcitabine in a human follicular lymphoma cell line is due to partial deletion of the deoxycytidine kinase gene. BMC Pharmacol. 2004;4:8. doi: 10.1186/1471-2210-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues: mechanisms of drug resistance and reversal strategies. Leukemia. 2001;15:875–890. doi: 10.1038/sj.leu.2402114. [DOI] [PubMed] [Google Scholar]
- Galmarini CM, Mackey JR, Dumontet C. Nucleoside analogues and nucleobases in cancer treatment. Lancet Oncol. 2002;3:415–424. doi: 10.1016/s1470-2045(02)00788-x. [DOI] [PubMed] [Google Scholar]
- Giovannetti E, Mey V, Nannizzi S, Pasqualetti G, Del Tacca M, Danesi R. Pharmacogenetics of anticancer drug sensitivity in pancreatic cancer. Mol Cancer Ther. 2006;5:1387–1395. doi: 10.1158/1535-7163.MCT-06-0004. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Clark AG. Organization of genetic variation. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- Hendrick P. Genetics of Populations. Sudbury, Massachusetts: Jones and Bartlett Publishers; 2000. [Google Scholar]
- Hsu CH, Liou JY, Dutschman GE, Cheng YC. Phosphorylation of cytidine, deoxycytidine, and their analog monophosphates by human UMP/CMP kinase is differentially regulated by ATP and magnesium. Mol Pharmacol. 2005;67:806–814. doi: 10.1124/mol.104.006098. [DOI] [PubMed] [Google Scholar]
- Joerger M, Bosch TM, Doodeman VD, Beijnen JH, Smits PH, Schellens JH. Novel deoxycytidine kinase gene polymorphisms: a population screening study in Caucasian healthy volunteers. Eur J Clin Pharmacol. 2006;62:681–684. doi: 10.1007/s00228-006-0162-7. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Cryst. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Links. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. Cell. 1986 Jan 31;44(2):283-92. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Cryst. 1991;24:946–950. [Google Scholar]
- Kroep JR, Loves WJ, van der Wilt CL, Alvarez E, Talianidis I, Boven E, Braakhuis BJ, van Groeningen CJ, Pinedo HM, Peters GJ. Pretreatment deoxycytidine kinase levels predict in vivo gemcitabine sensitivity. Mol Cancer Ther. 2002;1:371–376. [PubMed] [Google Scholar]
- Lamba JK, Crews K, Pounds S, Schuetz EG, Gresham J, Gandhi V, Plunkett W, Rubnitz J, Ribeiro R. Pharmacogenetics of deoxycytidine kinase: identification and characterization of novel genetic variants. J Pharmacol Exp Ther. 2007;323:935–945. doi: 10.1124/jpet.107.128595. [DOI] [PubMed] [Google Scholar]
- Liou JY, Dutschman GE, Lam W, Jiang Z, Cheng YC. Characterization of human UMP/CMP kinase and its phosphorylation of D- and L-form deoxycytidine analogue monophosphates. Cancer Res. 2002;62:1624–1631. [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I, Pachter L, Dubchak I, Rubin EM. rVista for comparative sequence-based discovery of functional transcription factor binding sites. Genome Res. 2002;12:832–839. doi: 10.1101/gr.225502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maring JG, Groen HJ, Wachters FM, Uges DR, de Vries EG. Genetic factors influencing pyrimidine-antagonist chemotherapy. Pharmacogenomics J. 2005;5:226–243. doi: 10.1038/sj.tpj.6500320. [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ. Raster3D: Photorealistic molecular graphics. Meth Enzymol. 1997:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- Mini E, Nobili S, Caciagli B, Landini I, Mazzei T. Cellular pharmacology of gemcitabine. Ann Oncol. 2006;17 Suppl 5:v7–v12. doi: 10.1093/annonc/mdj941. [DOI] [PubMed] [Google Scholar]
- Nakano Y, Tanno S, Koizumi K, Nishikawa T, Nakamura K, Minoguchi M, Izawa T, Mizukami Y, Okumura T, Kohgo Y. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br J Cancer. 2007;96:457–463. doi: 10.1038/sj.bjc.6603559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens JK, Shewach DS, Ullman B, Mitchell BS. Resistance to 1-beta-D-arabinofuranosylcytosine in human T-lymphoblasts mediated by mutations within the deoxycytidine kinase gene. Cancer Res. 1992;52:2389–2393. [PubMed] [Google Scholar]
- Pauwels B, Korst AE, Pattyn GG, Lambrechts HA, Kamphuis JA, De Pooter CM, Peters GJ, Lardon F, Vermorken JB. The relation between deoxycytidine kinase activity and the radiosensitising effect of gemcitabine in eight different human tumour cell lines. BMC Cancer. 2006;6:142. doi: 10.1186/1471-2407-6-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plunkett W, Huang P, Xu YZ, Heinemann V, Grunewald R, Gandhi V. Gemcitabine: metabolism, mechanisms of action, and self-potentiation. Semin Oncol. 1995;22:3–10. [PubMed] [Google Scholar]
- Ruiz van Haperen VW, Veerman G, Boven E, Noordhuis P, Vermorken JB, Peters GJ. Schedule dependence of sensitivity to 2',2'-difluorodeoxycytidine (Gemcitabine) in relation to accumulation and retention of its triphosphate in solid tumour cell lines and solid tumours. Biochem Pharmacol. 1994;48:1327–1339. doi: 10.1016/0006-2952(94)90554-1. [DOI] [PubMed] [Google Scholar]
- Sabini E, Ort S, Monnerjahn C, Konrad M, Lavie A. Structure of human dCK suggests strategies to improve anticancer and antiviral therapy. Nat Struct Biol. 2003;19:513–519. doi: 10.1038/nsb942. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani V, Ricci F, Rubio-Viqueira B, Kulesza P, Yeo CJ, Hidalgo M, Klein A, Laheru D, Iacobuzio-Donahue CA. Immunohistochemical and genetic evaluation of deoxycytidine kinase in pancreatic cancer: relationship to molecular mechanisms of gemcitabine resistance and survival. Clin Cancer Res. 2006;12:2492–2497. doi: 10.1158/1078-0432.CCR-05-2655. [Erratum in: Clin Cancer Res. 2007 Jul 15;13(14):4313] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Peña D, Sekulic N, Ort S, Konrad M, A L. Substrate-induced conformational changes in human UMP/CMP kinase. J Biol Chem. 2004;279:33882–33889. doi: 10.1074/jbc.M401989200. [DOI] [PubMed] [Google Scholar]
- Stegmann AP, Honders MW, Hagemeijer A, Hoebee B, Willemze R, Landegent JE. In vitro-induced resistance to the deoxycytidine analogues cytarabine (AraC) and 5-aza-2'-deoxycytidine (DAC) in a rat model for acute myeloid leukemia is mediated by mutations in the deoxycytidine kinase (dck) gene. Ann Hematol. 1995;71:41–47. doi: 10.1007/BF01696231. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich CM, Robien K, McLeod HL. Cancer pharmacogenetics: polymorphisms, pathways and beyond. Nat Rev Cancer. 2003;3:912–920. doi: 10.1038/nrc1233. [DOI] [PubMed] [Google Scholar]
- Usova EV, Eriksson S. Identification of residues involved in the substrate specificity of human and murine dCK. Biochem Pharmacol. 2002;64:1559–1567. doi: 10.1016/s0006-2952(02)01389-8. [DOI] [PubMed] [Google Scholar]
- Van Rompay AR, Johansson M, Karlsson A. Phosphorylation of deoxycytidine analog monophosphates by UMP-CMP kinase: molecular characterization of the human enzyme. Mol Pharmacol. 1999;56:562–569. doi: 10.1124/mol.56.3.562. [DOI] [PubMed] [Google Scholar]
- Wang L, Sullivan W, Toft D, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics. 2003;13:555–564. doi: 10.1097/01.fpc.0000054124.14659.99. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.