Abstract
How genetic variations in apoptosis pathway interact with environmental factors to contribute to esophageal adenocarcinoma (EA) risk has not been comprehensively investigated. We conducted a case-only analysis in 335 Caucasian EA patients that were genotyped for 242 single nucleotide polymorphisms (SNPs) in 43 apoptotic genes. Gene–environment interactions were assessed using a two-step approach. First, random forest algorithm was used to screen for the potential interacting markers. Next, we used case-only logistic regression model to estimate the effects of gene–environment interactions on EA risk. Four SNPs (PERP rs648802; PIK3CA rs4855094, rs7644468 and TNFRSF1A rs4149579) had significant interaction with gastroesophageal reflux disease (GERD). The presence of variant alleles in TP53BP1 rs560191, CASP7 rs7907519 or BCL2 rs12454712 enhanced the risk of smoking by 2.08–2.58 times [interaction odds ratio (ORi) = 2.08–2.58, adjusted P-value (Padj) = 0.02–0.04]. Compared with patients carrying ≤1 risk genotype, the risk of GERD on EA was increased in persons with two (ORi = 1.89, Padj = 0.016) or ≥3 (ORi = 4.30, Padj < 0.0001) risk genotypes. Compared with cases with ≤1 risk genotype, smoking-associated EA risk increased by 3.15 times when ≥2 risk genotypes were present (ORi = 3.15, Padj < 0.0001). In conclusion, interactions among apoptotic SNPs and GERD or smoking play an important role in EA development.
Introduction
Cumulative evidence has indicated that symptomatic gastroesophageal reflux disease (GERD), obesity, smoking and male gender are four major risk factors for developing esophageal adenocarcinoma (EA) (1,2). Although GERD is the strongest individual risk factor and subjects who have the most frequent reflux symptom carry >4-fold EA risk, most of them follow an indolent course for the entire life (3). On the other hand, nearly 50% of EA patients do not experience GERD-associated symptoms (2,4,5). Therefore, challenges arise when considering the benefit and cost of endoscopic surveillance for EA among subjects with these risk factors, especially GERD (6). One of the explanations for this phenomenon stems from the complex interactions between lifestyle/environmental exposure and genetic factors. Since most EA cases are sporadic, genetic influences are more probably to be polymorphisms in multiple genes instead of single gene mutation. Evidence from pathway-based studies suggests that genetic variance may modulate the susceptibility of developing EA (7–9). Incorporating one’s genetic background in the clinical setting could be a better strategy for risk assessment and cancer surveillance, and possibly provide a better understanding about how complex factors contribute to EA risk jointly. However, fewer studies have assessed interactions using a more comprehensive approach.
Apoptosis, or programmed cell death, is essential for normal tissues to regulate cell number and to eliminate unwanted or aging cells as an organism develops. Mutations and single nucleotide polymorphisms (SNPs) in apoptotic pathway genes that alter the ability of the cell to undergo apoptosis may induce cancers by allowing transformed cells to keep accumulating rather than dying (10). Our recent studies indicated that genetic polymorphisms in the apoptosis pathway, by themselves or through interaction with environmental factors, play an important role in the carcinogenesis of EA (7,11). In a case–control study covering 1330 functional/tagging SNPs categorized into14 cancer-related pathways, two apoptotic SNPs (Caspase 7 rs312707 and Caspase 9 rs4661636) were significantly associated with EA risk. Moreover, apoptosis pathway was found to be the most import one in pathway-based analysis (11). However, it is possible that other SNPs further down the significance list of individual effects exert their importance through gene–environment interaction. Here, we conducted a pathway-based case-only study to further explore the interactions between 242 apoptosis-related SNPs and those well-known EA risk factors. We applied a two-step approach to identify gene–environment interaction markers in EA cases. Our results showed that gene–environment interactions play important roles in the development of EA.
Materials and methods
Study population, interview and DNA preparation
Details of patient recruitment were described in our previous paper (11). In brief, they were Caucasian, >18 years old and histologically confirmed to have EA at Massachusetts General Hospital between 1999 and 2005 and at Dana Farber Cancer Institute between 2004 and 2005. All of them had a tumor center located at or above the gastroesophageal junction and had at least two-thirds of the bulk tumor located in the esophagus. Patients with secondary or recurrent cancers were excluded. The recruitment rate was 86%. Participants were interviewed using a modified questionnaire (12) immediately after enrollment to collect information of their demographic characteristics and smoking history. Smoking status was defined based on whether the patient smoked 1 year prior to diagnosis. Lifetime GERD-related symptoms were collected based on the questions described before (13). The presence of GERD was defined as having heartburn or regurgitation symptoms at least once per month for more than six continuous months in one’s lifetime (7).
Blood samples were collected at the time of recruitment and DNA was extracted using the Puregene DNA Isolation Kit (Gentra Systems/Qiagen, Valencia, CA). This study was approved by the Human Subjects Committees of Massachusetts General Hospital, Dana Farber Cancer Institute and the Harvard School of Public Health (Boston, MA). All subjects signed the informed consent prior to study participation.
SNP selection, pathway categorization and genotyping
The criteria for SNP selection in the GoldenGate assay was described previously (11). The candidate SNPs selected in the apoptosis pathway were common SNPs with minor allele frequency (MAF) ≥ 5% in the HapMap Caucasian population (CEU). They were either missense/exonic SNPs, SNPs within untranslated regions and 2 kb 5′ of the gene or tagSNPs for genes with little functional information available. The common non-synonymous SNPs were selected using SNP500Cancer Project and International HapMap Project. Potential functional non-synonymous SNPs were selected from the Predicted Impact of Coding SNPs database (14) and FASTSNP (15). SNPs on the Illumina Cancer Panel were chosen with priority. TagSNPs were selected using the r2-based Tagger program (16) with pairwise r2 ≥ 0.80 and MAF ≥ 5% among Caucasians. Apoptosis pathway genes were chosen based on the literature reports and information in the following database, the National Center for Biotechnology Information, Genetic Association Database (17), National Institute for Environmental Health Science GeneSNPs (18) and Kyoto Encyclopedia of Genes and Genomes (19). More frequently cited genes were given greater priority.
Genotyping was performed on the Illumina GoldenGate assay at the Broad Institute (Cambridge, MA) by laboratory personnel blinded to the subjects’ clinical information. Forty-eight duplicate samples were randomly selected for quality control. The concordance rate of the replicate samples for all assays was >99%. As described previously (11), 1330 SNPs with MAF ≥ 0.01 and passed Hardy–Weinberg equilibrium (P ≥ 0.0001) in the control subjects were successfully genotyped. Among them, 242 SNPs (43 genes) in apoptosis pathway (supplementary Table S1 is available at Carcinogenesis Online) were used for interaction analysis in the case-only model.
Statistical analysis
We applied a two-step approach to identify the SNPs, which may have interactions with environmental factors (20). First, we used a random forest algorithm (21), with one individual environmental factor (GERD, smoking or body mass index [(BMI) at age 18] as the outcome and apoptotic SNPs as the predictors, to impute the missing values (<4% in each variable) and then obtain the mean decrease of accuracy (MDA) for each SNP. The MDA measures the degree of loss if the corresponding SNP was ‘removed’ from the model and could be regarded as an ‘importance’ score for the SNP. The top important SNPs were selected based on the turning point of MDA in random forest importance plots. The turning points in the MDA plots were defined as the cut points to select important markers for subsequent interaction analyses.
In the second step, we used the ‘important SNPs’ to perform case-only logistic regression analysis using dominant genetic model (mutant carriers versus wild-type homozygotes). SNPs with adjusted P-value (Padj) < 0.05 after adjustment for covariates and multiple comparison using false discovery rate were considered to be significant (22). To estimate cumulative risk effects of the significant SNPs, we combined the number of risk genotypes representing the degree of ‘genetic risk score’, which was included in the case-only logistic models to fit with individual environmental risk factors, respectively. Interaction odds ratios (ORi) and 95% confidence intervals (95% CIs) were calculated as an estimate of relative risk. All data analyses were performed using the SAS statistical package, version 9.l (SAS Institute, Cary, NC). Random forest analysis was performed using rpart package in R software, version 2.9.1.
Results
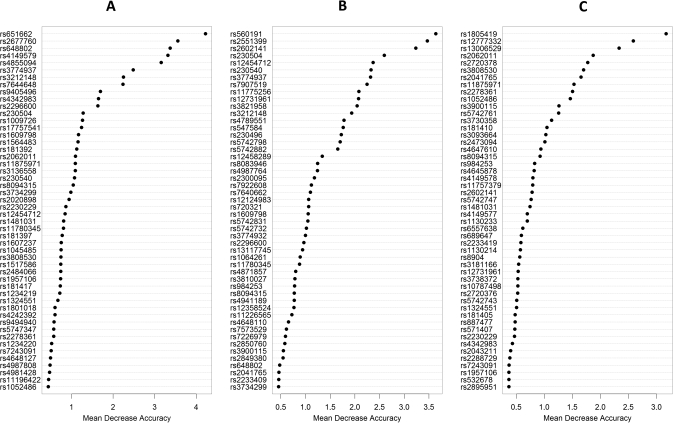
The mean age of the EA cases was 62.9 years and 295 (88%) were males. About half of them had history of GERD symptoms and 80.3% were ever smokers. Other demographic characteristics were shown in Table I. In random forest analysis, MDA scores were obtained by SNP–GERD, SNP-smoking and SNP–BMI models, respectively. To minimize random bias, 100 000 trees were constructed in each RF model, allowing each marker to have ∼100% of probability to be tested for 500 times. Based on MDA plot curves, the most important SNPs that had interaction with GERD (22 SNPs), smoking (17 SNPs) and BMI (15 SNPs) were selected for subsequent case-only logistic regression analysis (Figure 1).
Table I.
Characteristics of 335 EA cases
| Characteristics | EA cases |
| Age (years, mean ± SD) | 62.9 ± 11.8 |
| Gender, male (%) | 295 (88.0%) |
| GERDa, yes (%) | 173 (50.6%) |
| Smoking, yes (%) | 269 (80.3%) |
| Smoking (pack-years), mean ± SD, medium (minimum to maximum) | 30.5 ± 31.0, 24.4 (0–212.0) |
| BMI at age 18, (kg/m2, mean ± SD) | 23.6 ± 3.8 |
| BMI ≥ 25 at age 18 (%) | 104 (31.0%) |
| Stage | |
| I–IIA | 98 (29.3%) |
| IIB–IV | 237 (70.7%) |
7 (2%) missing and imputed.
Fig. 1.
Random forest importance plots for interaction between apoptotic SNPs and GERD (A), smoking (B) or body mass index (C).
In case-only logistic regression analysis, we found that five SNPs (rs651662, rs648802, rs4855094, rs7644468 and rs4149579) in three genes (PERP, PIK3CA and TNFRSF1A) had significant interaction with GERD; these interactions remained significant even after adjusting for covariates and false discovery rate (Table II). Two SNPs (rs651662 and rs648802) were in the gene PERP and were in high linkage disequilibrium (r2 = 0.99). Another two significant SNPs in PIK3CA (rs4855094 and rs7644468) were not tagged to each other. Subjects carrying the variant allele of rs651662, rs648802, rs4855094 or rs7644468 significantly enhanced the risk of GERD to develop EA compared with subjects carrying homozygous wild genotypes. On the contrary, the risk of GERD was significantly reduced when one carries the wide-type allele of rs4149579 (ORi = 0.42, 95% CI = 0.22–0.81, Padj = 0.04) (Table II). Since rs651662 and rs648802 were in linkage disequilibrium (r2 = 0.99) and the later was a non-synonymous SNP, whereas the former located in intron, only rs648802 was used to combine with the other three SNPs to calculate the joint effect. There was a significant cumulative risk when those four SNPs were considered jointly. The risk of GERD on EA was increased when one has two (ORi = 1.89, 95% CI = 1.13–3.16) or ≥3 (ORi = 4.30, 95% CI = 2.25–8.21) risk genotypes comparing with those with ≤1 risk genotype (Table II).
Table II.
Case-only interaction between apoptotic SNPs and GERD on EA risk
| SNPs | GERD |
ORia (95% CI) | Padja | |
| No, n (%) | Yes, n (%) | |||
| PERP rs651662 | ||||
| GG | 68 (41.97) | 43 (24.86) | reference | |
| GA + AA | 94 (58.03) | 130 (75.14) | 2.13 (1.33–3.40) | 0.02 |
| PERP rs648802 | ||||
| CC | 67 (41.36) | 43 (24.86) | reference | |
| CG + GG | 95 (58.64) | 130 (75.14) | 2.09 (1.30–3.34) | 0.02 |
| PIK3CA rs4855094 | ||||
| GG | 151 (93.21) | 145 (83.82) | reference | |
| GA + AA | 11 (6.79) | 28 (16.18) | 2.76 (1.32–5.80) | 0.04 |
| PIK3CA rs7644648 | ||||
| AA | 123 (75.93) | 109 (63.00) | reference | |
| GA + GG | 39 (24.07) | 64 (37.00) | 1.89 (1.17–3.06) | 0.04 |
| TNFRSF1A rs4149579 | ||||
| GG | 130 (80.25) | 157 (90.75) | reference | |
| GA + AA | 32 (19.76) | 16 (9.25) | 0.42 (0.22–0.81) | 0.04 |
| bCumulative risks of rs648802, rs4855094, rs76044648 and rs4149579 | ||||
| ≤1 risk genotype | 65 (40.12) | 37 (21.39) | reference | |
| 2 risk genotypes | 75 (46.30) | 82 (47.40) | 1.89 (1.13–3.16) | 0.016 |
| ≥3 risk genotypes | 22 (13.58) | 54 (31.21) | 4.30 (2.25–8.20) | <0.0001 |
Adjusted for age, sex, BMI at age 18 (<25 versus ≥25) and smoking (+/−) (also false discovery rate for Padj).
Since rs651662 and rs648802 were in linkage disequilibrium, only the later was used to calculate the cumulative risks.
For the SNP-smoking interaction, 4 of the top 17 SNPs were identified to be significant in logistic regression models (Table III). The presence of variant alleles in TP53BP1 rs560191, rs2602141, CASP7 rs7907519 or BCL2 rs12454712 enhanced the risk of smoking by 2.08–2.58 times (ORi = 2.08–2.58, Padj = 0.02–0.04). TP53BP1 rs560191 and rs2602141 were tagged to each other (r2 = 0.99) and the former was non-synonymous; thus, we dropped the later in the cumulative risk analysis. Taking those three SNPs together, persons with ≥2 risk genotypes had 3.15 times of interaction effect with smoking (ORi = 3.15, 95% CI = 1.80–5.52, Padj < 0.0001) than those with ≤1 risk genotype (Table III). No SNPs were found to be significantly interacted with BMI in logistic regression analysis. The allele frequencies of the SNPs with significant interaction with environmental factors were shown in Supplementary Table S2, available at Carcinogenesis Online.
Table III.
Case-only interaction between apoptotic SNPs and smoking on EA risk
| SNPs | Smoking |
ORia (95% CI) | Padja | |
| No, n (%) | Yes, n (%) | |||
| TP53BP1 rs560191 | ||||
| CC | 44 (66.67) | 119 (44.24) | reference | |
| CG + GG | 22 (33.33) | 150 (45.76) | 2.58 (1.45–4.59) | 0.02 |
| TP53BP1 rs2602141 | ||||
| AA | 43 (65.15) | 118 (43.87) | reference | |
| AC + CC | 23 (34.85) | 151 (56.13) | 2.38 (1.35–4.19) | 0.02 |
| CASP7 rs7907519 | ||||
| CC | 30 (45.45) | 76 (28.25) | reference | |
| CA + AA | 36 (54.55) | 193 (71.75) | 2.14 (1.23–3.76) | 0.04 |
| BCL2 rs12454712 | ||||
| TT | 36 (54.55) | 99 (36.80) | reference | |
| TC + CC | 30 (45.45) | 170 (63.20) | 2.08 (1.20–3.63) | 0.04 |
| bCumulative risks of rs560191, rs7907519 and rs12454712 | ||||
| ≤1 risk genotype | 39 (59.09) | 85 (31.60) | reference | |
| ≥2 risk genotypes | 27 (40.91) | 184 (68.40) | 3.15 (1.80–5.52) | <0.0001 |
Adjusted for age, sex, BMI at age 18 (<25 versus ≥25) and GERD (+/−) (also false discovery rate for Padj).
Since rs560191 and rs2602141 were in linkage disequilibrium, only the former was used to calculate the cumulative risks.
Discussion
This is a comprehensive study covering 242 apoptotic SNPs to evaluate the gene–environment interaction in EA. Case-only study with random forest analysis has been proven to be a reliable method to investigate gene–environment interaction in complex diseases (20). The EA patients in this study were incident cases; therefore, the ORi estimated the interaction under the assumption that the genetic and environment factors were independent. In addition to identifying potential modifiers, we also demonstrated increased gene–GERD and gene–smoking interactions when those significant SNPs were considered jointly. Our results provide more evidence to how apoptotic genes modify the risk of GERD and smoking on EA risk. Dominant model of each SNP was used to calculate the ORi because it was more convenient and the sample size was limited in this study. We also used case–control model to test the interaction between smoking and the four significant SNPs found in this case-only analysis. The control subjects were described in our previous study (11). Three of them had very consistent results (rs560191, rs260214 and rs12454712; ORi = 2.11–2.28, P-value = 0.02–0.04). Although we did not find significant interaction between CASP7 rs7907519 and smoking in case–control model, the trend was consistent (ORi = 1.20, 95% CI, 0.58–2.48, P-value = 0.61). Because GERD information was missing in >50% of our control subjects (11), we cannot perform case–control analysis for genotype–reflux interaction.
Evading apoptosis, a characteristic of transformed cells, has been well documented in Barrett’s carcinogenesis (23,24). Gastric acid and bile acid are two principle components of refluxate that cause chronic esophageal inflammation and Barrett’s transformation. Direct exposure of an EA cell line to acid leads to suppression of apoptotic genes, which may occur via p53-dependent mechanism (25). TP53 tumor suppressor induces the intrinsic apoptotic pathway and activates many downstream genes, including PERP (TP53 apoptosis effecter related to PMP-22) (26). Previous study also suggests that there is a reciprocal regulation between p53 and the PI3K–AKT pathway (27). The PIK3-AKT signaling pathway regulates many normal cell functions, including cell proliferation, survival and growth. It is also known to be anti-apoptotic in many different cell types (28). Amplification of PIK3CA, which encodes the 110 kDa subunit of PI3K, has been found in many human cancers (29,30). Treatment with a PI3K inhibitor decreases proliferation and increases apoptosis; therefore, it has become a target for cancer therapy (28). Genetic polymorphisms of PIK3CA have been studied in the susceptibility of ovarian cancer (31). In addition to PERP and PIK3CA, we also found a significant interaction between the TNFRSF1A polymorphism and GERD. TNFRSF1A is one of the cognate receptors that binds to tumor necrosis factor superfamily ligands to activate the extrinsic apoptotic pathway (32). TNFRSF1A rs4149570 has been linked to the survival of early-stage non-small cell lung cancer (33). However, no studies investigated the association between PERP, PIK3CA or TNFRSF1A polymorphism and esophageal cancer risk.
Our data revealed that polymorphisms in CASP7, TP53BP1 and BCL2 modified the risk of smoking to develop EA. Long-term exposure to carcinogens in cigarette smoke such as polycyclic aromatic hydrocarbons leads to DNA damage, which accumulates if a cell evades cell cycle regulation and apoptotic mechanism. Caspase activation plays a primary role in the apoptotic cascade. A previous report has shown an inactivation mutation of CASP7 in esophageal cancer cells (34). Polymorphisms in CASP7 have been examined in several human cancers (35,36) but not in esophageal cancer until our recent report (11). In this study, we identified a significant interaction between CASP7 rs7907519 and smoking in EA cases. Such information adds more evidence to the importance of genes in caspase cascade in the susceptibility of EA. TP53BP1 binds to TP53 and plays a role in responses to DNA damage. A study in Japan revealed that TP53BP1 Asp353Glu (rs560191) and TP53 Arg72Pro polymorphism had significant interaction in lung cancer risk. However, no gene–smoking interaction was found (37). BCL2 protein is one of the products of BCL2 family and has anti-apoptotic function. It has been found to aberrantly expressed in Barrett’s carcinogenesis (38). Polymorphisms in BCL2 were reported in many human cancers such as ovarian cancer, leukemia and esophageal cancer (39–41). However, no study examined the association of TP53BP1 or BCL2 polymorphism and EA risk among Caucasians.
Obesity not only promotes reflux symptoms but also contributes to the progression of EA by inhibiting apoptosis through a reduced adiponectin (anti-inflammatory adipokine from adipose tissue) and ghrelin levels (42). It is possible that there is an interaction between increased BMI and apoptotic SNPs; however, our data does not support this hypothesis. This might suggest that the effect of obesity on EA involves in mechanisms other than polymorphisms in apoptotic genes. It is also possible that there is a high-dimensional interaction involving multiple SNPs and BMI, which is not easy to investigate. Because we only included 40 (12%) female cases, the interaction with gender is difficult to examine in the present study. IL1B rs1143634 has been shown to have significant interaction with GERD on EA risk in our previous case–control study (7). However, this association was not found in the current case-only model. This is possibly because case-only and case–control designs differ in estimating interaction parameters. In case-only analysis, the ORi tests whether the influence of a risk factor (e.g. GERD) is different among two groups of patients dichotomized by the presence or absence of a risk genotype. In case–control study, interaction was tested by measuring the departure of multiplicative joint effects of genetic and environmental factors on disease risk (43,44).
This study has several limitations. First, we did not validate our findings in another group, and these findings need to be replicated. Second, the case number in this study is relatively small because EA is an uncommon disease among Caucasians. However, we have >80% power to identify a significant pairwise interaction if the MAF, relative risk of interaction are ∼20% and 2.5, respectively (45). Third, it is not clear how these polymorphisms affect the biological function of the genes. Finally, we did not provide data to prove that the gene and environmental factors in this study are independent although it was reasonable since EA is an uncommon disease. Violation of the independence assumption could make the multiplicative odds ratio of a case-only study very different from the ORi obtained from a case–control study (43,44).
In conclusion, our data suggest that the interaction among apoptotic SNPs and GERD or smoking play an important role in the carcinogenesis of EA. Replication in other populations and further mechanistic studies are needed to validate the results and elucidate how the genes interact with environmental factors to contribute to the cancer risk.
Funding
National Institutes of Health (R01CA109193, R03CA110822, R01CA074386 and ES00002); the Canadian Institute of Health Research operating grant; the Kevin Jackson Memorial Fund; Alan B. Brown Chair in Molecular Genomics; National Cancer Institute of Canada Dorothy Lamont Award; Posluns Family Fund Princess Margaret Hospital Foundation; the Flight Attendant Medical Research Institute Award No. 062459. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research Resources.
Supplementary Material
Acknowledgments
The authors thank the physicians and patients from the Massachusetts General Hospital Thoracic Oncology Center and the Dana Farber Cancer Institute GI Oncology Center for their cooperation and participation. The authors also thank researchers from Excellence for cancer research center, Department of Health, Executive Yuan, Taiwan, ROC (grant, DOH100-TD-C-111-002).
Conflict of Interest Statement: All authors disclosed no conflict of interest; the institutes, which provided the grant supports did not participate in the study design, the collection, analysis or interpretation of data.
Glossary
Abbreviations
- BMI
body mass index
- CI
confidence interval
- EA
esophageal adenocarcinoma
- GERD
gastroesophageal reflux disease
- MAF
minor allele frequency
- MDA
mean decrease of accuracy
- ORi
interaction odds ratio
- Padj
adjusted P-value
- SNP
single nucleotide polymorphism
References
- 1.Whiteman DC, et al. Combined effects of obesity, acid reflux and smoking on the risk of adenocarcinomas of the oesophagus. Gut. 2008;57:173–180. doi: 10.1136/gut.2007.131375. [DOI] [PubMed] [Google Scholar]
- 2.Reid BJ, et al. Barrett's oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat. Rev. Cancer. 2010;10:87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holmes RS, et al. Epidemiology and pathogenesis of esophageal cancer. Semin. Radiat. Oncol. 2007;17:2–9. doi: 10.1016/j.semradonc.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Anderson LA, et al. The association between alcohol and reflux esophagitis, Barrett's esophagus, and esophageal adenocarcinoma. Gastroenterology. 2009;136:799–805. doi: 10.1053/j.gastro.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Lassen A, et al. Esophagitis: incidence and risk of esophageal adenocarcinoma–a population-based cohort study. Am. J. Gastroenterol. 2006;101:1193–1199. doi: 10.1111/j.1572-0241.2006.00550.x. [DOI] [PubMed] [Google Scholar]
- 6.Kahrilas PJ, et al. American Gastroenterological Association Institute technical review on the management of gastroesophageal reflux disease. Gastroenterology. 2008;135:1392–1413. doi: 10.1053/j.gastro.2008.08.044. 1413 e1391–e1395. [DOI] [PubMed] [Google Scholar]
- 7.Zhai R, et al. Interactions among genetic variants in apoptosis pathway genes, reflux symptoms, body mass index, and smoking indicate two distinct etiologic patterns of esophageal adenocarcinoma. J. Clin. Oncol. 2010;28:2445–2451. doi: 10.1200/JCO.2009.26.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tse D, et al. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control. 2008;19:1077–1083. doi: 10.1007/s10552-008-9171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doecke J, et al. Polymorphisms in MGMT and DNA repair genes and the risk of esophageal adenocarcinoma. Int. J. Cancer. 2008;123:174–180. doi: 10.1002/ijc.23410. [DOI] [PubMed] [Google Scholar]
- 10.Reed JC. Mechanisms of apoptosis. Am. J. Pathol. 2000;157:1415–1430. doi: 10.1016/S0002-9440(10)64779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu CY, et al. A Large-scale genetic association study of esophageal adenocarcinoma risk. Carcinogenesis. 2010;31:1259–1263. doi: 10.1093/carcin/bgq092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferris BG. Epidemiology Standardization Project (American Thoracic Society) Am. Rev. Respir. Dis. 1978;118:1–120. [PubMed] [Google Scholar]
- 13.Cheung WY, et al. Epidermal growth factor A61G gene polymorphism, gastroesophageal reflux disease and esophageal adenocarcinoma risk. Carcinogenesis. 2009;30:1363–1367. doi: 10.1093/carcin/bgp126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rudd MF, et al. The predicted impact of coding single nucleotide polymorphisms database. Cancer Epidemiol. Biomarkers Prev. 2005;14:2598–2604. doi: 10.1158/1055-9965.EPI-05-0469. [DOI] [PubMed] [Google Scholar]
- 15.Yuan HY, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke X, et al. Efficient selective screening of haplotype tag SNPs. Bioinformatics. 2003;19:287–288. doi: 10.1093/bioinformatics/19.2.287. [DOI] [PubMed] [Google Scholar]
- 17.Becker KG, et al. The genetic association database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 18.Chandanos E, et al. Endogenous estrogen exposure in relation to distribution of histological type and estrogen receptors in gastric adenocarcinoma. Gastric Cancer. 2008;11:168–174. doi: 10.1007/s10120-008-0475-6. [DOI] [PubMed] [Google Scholar]
- 19.Kyoto Encyclopedia of Genes and Genomes. Kyoto (JP): Kanehisa Laboratory, Kyoto University.; http://www.genome.jp/kegg/ (19 April 2010, date last accessed) [Google Scholar]
- 20.Maenner MJ, et al. Detecting gene-by-smoking interactions in a genome-wide association study of early-onset coronary heart disease using random forests. BMC Proc. 2009;3(suppl. 7):S88. doi: 10.1186/1753-6561-3-s7-s88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore JH, et al. Bioinformatics challenges for genome-wide association studies. Bioinformatics. 2010;26:445–455. doi: 10.1093/bioinformatics/btp713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, et al. Quantitative trait Loci analysis using the false discovery rate. Genetics. 2005;171:783–790. doi: 10.1534/genetics.104.036699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dvorakova K, et al. Apoptosis resistance in Barrett's esophagus: ex vivo bioassay of live stressed tissues. Am. J. Gastroenterol. 2005;100:424–431. doi: 10.1111/j.1572-0241.2005.40932.x. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald RC. Molecular basis of Barrett's oesophagus and oesophageal adenocarcinoma. Gut. 2006;55:1810–1820. doi: 10.1136/gut.2005.089144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan C, et al. In vitro acid exposure has a differential effect on apoptotic and proliferative pathways in a Barrett's adenocarcinoma cell line. Am. J. Gastroenterol. 2004;99:218–224. doi: 10.1111/j.1572-0241.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- 26.Attardi LD, et al. PERP, an apoptosis-associated target of p53, is a novel member of the PMP-22/gas3 family. Genes Dev. 2000;14:704–718. [PMC free article] [PubMed] [Google Scholar]
- 27.Trotman LC, et al. PTEN and p53: who will get the upper hand? Cancer Cell. 2003;3:97–99. doi: 10.1016/s1535-6108(03)00022-9. [DOI] [PubMed] [Google Scholar]
- 28.Luo J, et al. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 29.Shayesteh L, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat. Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- 30.Ma YY, et al. PIK3CA as an oncogene in cervical cancer. Oncogene. 2000;19:2739–2744. doi: 10.1038/sj.onc.1203597. [DOI] [PubMed] [Google Scholar]
- 31.Quaye L, et al. Tagging single-nucleotide polymorphisms in candidate oncogenes and susceptibility to ovarian cancer. Br. J. Cancer. 2009;100:993–1001. doi: 10.1038/sj.bjc.6604947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aggarwal BB. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 2003;3:745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 33.Lee EB, et al. Polymorphisms in apoptosis-related genes and survival of patients with early-stage non-small-cell lung cancer. Ann. Surg. Oncol. 2010;17:2608–2618. doi: 10.1245/s10434-010-1082-4. [DOI] [PubMed] [Google Scholar]
- 34.Soung YH, et al. Inactivating mutations of CASPASE-7 gene in human cancers. Oncogene. 2003;22:8048–8052. doi: 10.1038/sj.onc.1206727. [DOI] [PubMed] [Google Scholar]
- 35.Yoo SS, et al. Polymorphisms in the CASPASE genes and survival in patients with early-stage non-small-cell lung cancer. J. Clin. Oncol. 2009;27:5823–5829. doi: 10.1200/JCO.2009.23.1738. [DOI] [PubMed] [Google Scholar]
- 36.Xu HL, et al. Polymorphisms and haplotypes in the caspase-3, caspase-7, and caspase-8 genes and risk for endometrial cancer: a population-based, case-control study in a Chinese population. Cancer Epidemiol. Biomarkers Prev. 2009;18:2114–2122. doi: 10.1158/1055-9965.EPI-09-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiyohara C, et al. Cigarette smoking, TP53 Arg72Pro, TP53BP1 Asp353Glu and the risk of lung cancer in a Japanese population. Oncol. Rep. 2010;23:1361–1368. doi: 10.3892/or_00000772. [DOI] [PubMed] [Google Scholar]
- 38.Katada N, et al. Apoptosis is inhibited early in the dysplasia-carcinoma sequence of Barrett esophagus. Arch. Surg. 1997;132:728–733. doi: 10.1001/archsurg.1997.01430310042007. [DOI] [PubMed] [Google Scholar]
- 39.Heubner M, et al. Association of the AA genotype of the BCL2 (-938C>A) promoter polymorphism with better survival in ovarian cancer. Int. J. Biol. Markers. 2009;24:223–229. doi: 10.1177/172460080902400402. [DOI] [PubMed] [Google Scholar]
- 40.Kim DH, et al. Genetic variants in the candidate genes of the apoptosis pathway and susceptibility to chronic myeloid leukemia. Blood. 2009;113:2517–2525. doi: 10.1182/blood-2008-07-169110. [DOI] [PubMed] [Google Scholar]
- 41.Jain M, et al. Role of BCL2 (ala43thr), CCND1 (G870A) and FAS (A-670G) polymorphisms in modulating the risk of developing esophageal cancer. Cancer Detect. Prev. 2007;31:225–232. doi: 10.1016/j.cdp.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 42.Konturek PC, et al. Effect of adiponectin and ghrelin on apoptosis of Barrett adenocarcinoma cell line. Dig. Dis. Sci. 2008;53:597–605. doi: 10.1007/s10620-007-9922-1. [DOI] [PubMed] [Google Scholar]
- 43.Albert PS, et al. Limitations of the case-only design for identifying gene-environment interactions. Am. J. Epidemiol. 2001;154:687–693. doi: 10.1093/aje/154.8.687. [DOI] [PubMed] [Google Scholar]
- 44.Piegorsch WW, et al. Non-hierarchical logistic models and case-only designs for assessing susceptibility in population-based case-control studies. Stat. Med. 1994;13:153–162. doi: 10.1002/sim.4780130206. [DOI] [PubMed] [Google Scholar]
- 45.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat. Med. 2002;21:35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.