Abstract
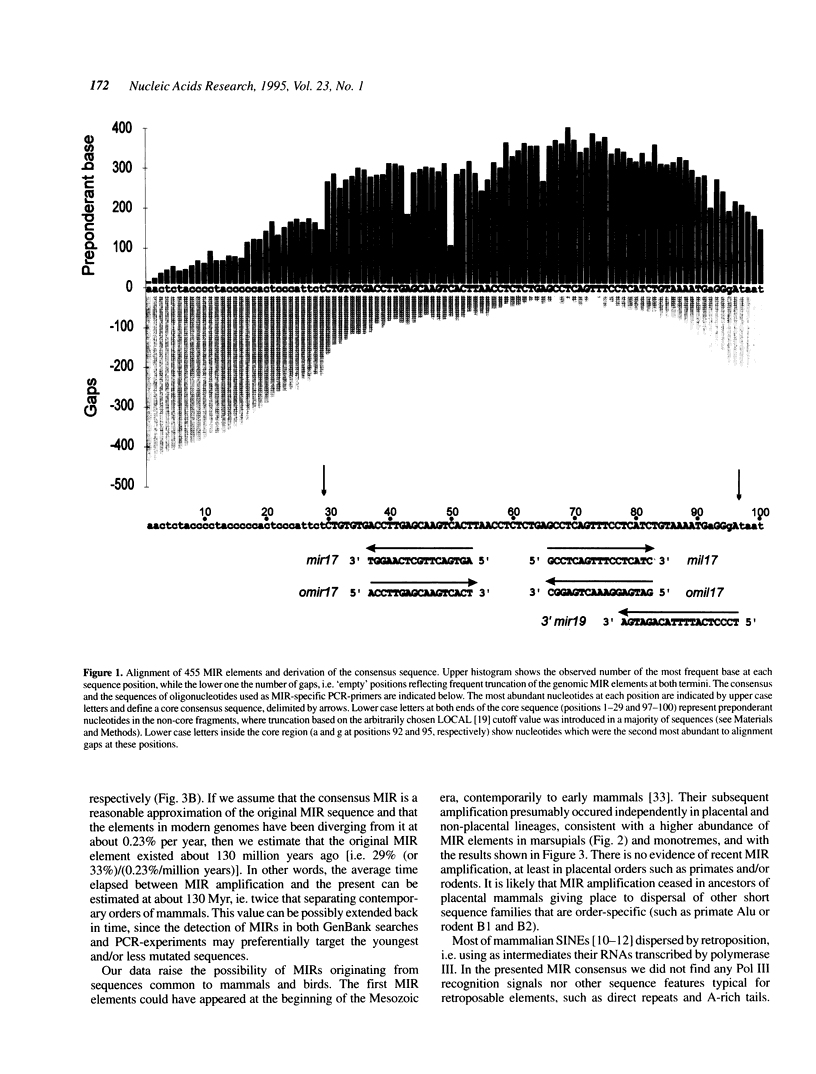
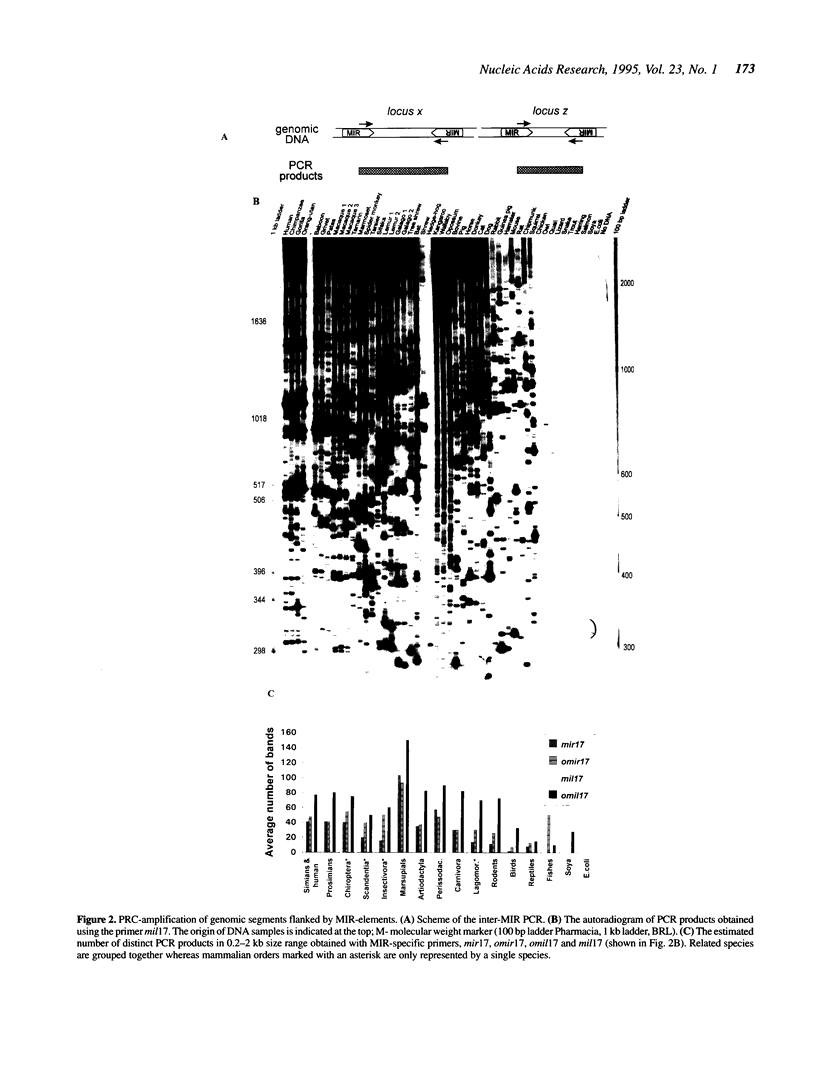
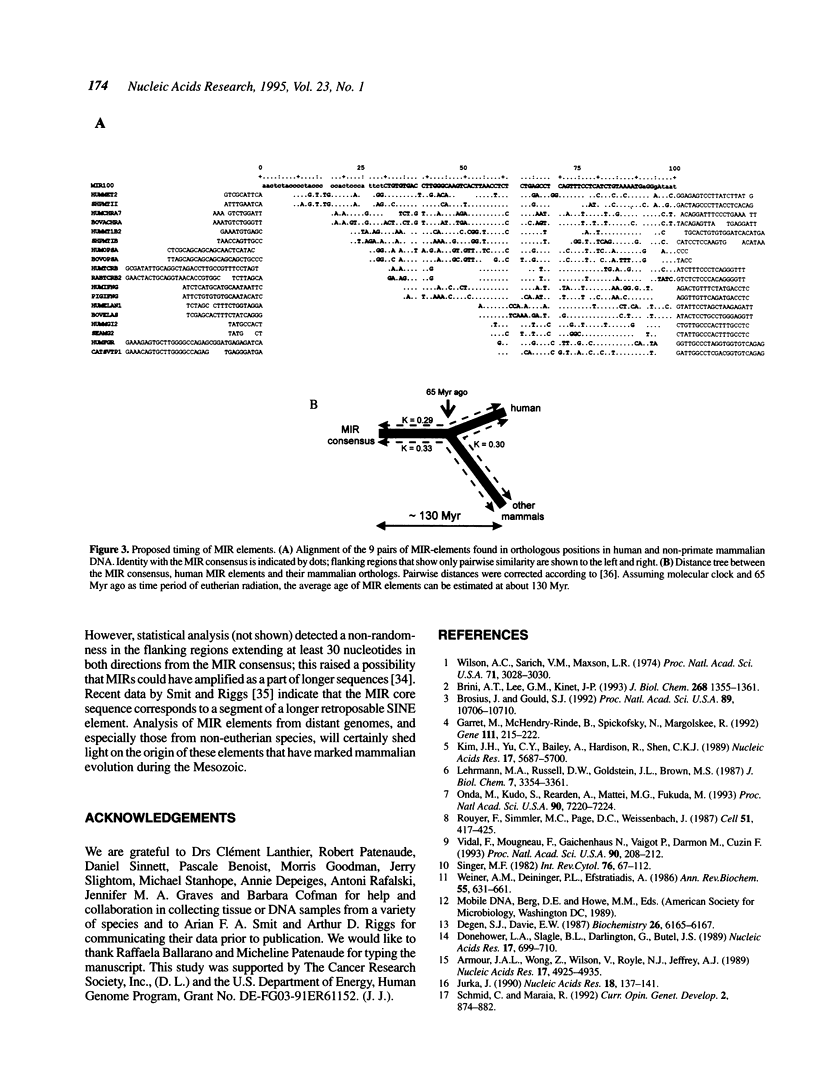
Short interspersed elements (SINEs) are ubiquitous in mammalian genomes. Remarkable variety of these repeats among placental orders indicates that most of them amplified in each lineage independently, following mammalian radiation. Here, we present an ancient family of repeats, whose sequence divergence and common occurrence among placental mammals, marsupials and monotremes indicate their amplification during the Mesozoic era. They are called MIRs for abundant Mammalian-wide Interspersed Repeats. With approximately 120,000 copies still detectable in the human genome (0.2-0.3% DNA), MIRs represent a 'fossilized' record of a major genetic event preceding the radiation of placental orders.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adey N. B., Tollefsbol T. O., Sparks A. B., Edgell M. H., Hutchison C. A., 3rd Molecular resurrection of an extinct ancestral promoter for mouse L1. Proc Natl Acad Sci U S A. 1994 Feb 15;91(4):1569–1573. doi: 10.1073/pnas.91.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armour J. A., Wong Z., Wilson V., Royle N. J., Jeffreys A. J. Sequences flanking the repeat arrays of human minisatellites: association with tandem and dispersed repeat elements. Nucleic Acids Res. 1989 Jul 11;17(13):4925–4935. doi: 10.1093/nar/17.13.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brini A. T., Lee G. M., Kinet J. P. Involvement of Alu sequences in the cell-specific regulation of transcription of the gamma chain of Fc and T cell receptors. J Biol Chem. 1993 Jan 15;268(2):1355–1361. [PubMed] [Google Scholar]
- Brosius J., Gould S. J. On "genomenclature": a comprehensive (and respectful) taxonomy for pseudogenes and other "junk DNA". Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degen S. J., Davie E. W. Nucleotide sequence of the gene for human prothrombin. Biochemistry. 1987 Sep 22;26(19):6165–6177. doi: 10.1021/bi00393a033. [DOI] [PubMed] [Google Scholar]
- Donehower L. A., Slagle B. L., Wilde M., Darlington G., Butel J. S. Identification of a conserved sequence in the non-coding regions of many human genes. Nucleic Acids Res. 1989 Jan 25;17(2):699–710. doi: 10.1093/nar/17.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner D. V., Jurka J. Multiple aligned sequence editor (MASE). Trends Biochem Sci. 1988 Aug;13(8):321–322. doi: 10.1016/0968-0004(88)90129-6. [DOI] [PubMed] [Google Scholar]
- Garret M., McHendry-Rinde B., Spickofsky N., Margolskee R. F. Isolation of a clone which induces expression of the gene encoding the human tumor necrosis factor receptor. Gene. 1992 Feb 15;111(2):215–222. doi: 10.1016/0378-1119(92)90689-m. [DOI] [PubMed] [Google Scholar]
- Hwu H. R., Roberts J. W., Davidson E. H., Britten R. J. Insertion and/or deletion of many repeated DNA sequences in human and higher ape evolution. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3875–3879. doi: 10.1073/pnas.83.11.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kaplan D. J., Duncan C. H., Walichiewicz J., Milosavljevic A., Murali G., Solus J. F. Identification and characterization of new human medium reiteration frequency repeats. Nucleic Acids Res. 1993 Mar 11;21(5):1273–1279. doi: 10.1093/nar/21.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Novel families of interspersed repetitive elements from the human genome. Nucleic Acids Res. 1990 Jan 11;18(1):137–141. doi: 10.1093/nar/18.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido Y., Aono M., Yamaki T., Matsumoto K., Murata S., Saneyoshi M., Okada N. Shaping and reshaping of salmonid genomes by amplification of tRNA-derived retroposons during evolution. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2326–2330. doi: 10.1073/pnas.88.6.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Yu C. Y., Bailey A., Hardison R., Shen C. K. Unique sequence organization and erythroid cell-specific nuclear factor-binding of mammalian theta 1 globin promoters. Nucleic Acids Res. 1989 Jul 25;17(14):5687–5700. doi: 10.1093/nar/17.14.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrman M. A., Russell D. W., Goldstein J. L., Brown M. S. Alu-Alu recombination deletes splice acceptor sites and produces secreted low density lipoprotein receptor in a subject with familial hypercholesterolemia. J Biol Chem. 1987 Mar 5;262(7):3354–3361. [PubMed] [Google Scholar]
- Li W. H., Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987 Mar 5;326(6108):93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- Martin A. P., Palumbi S. R. Body size, metabolic rate, generation time, and the molecular clock. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyzis R. K., Torney D. C., Meyne J., Buckingham J. M., Wu J. R., Burks C., Sirotkin K. M., Goad W. B. The distribution of interspersed repetitive DNA sequences in the human genome. Genomics. 1989 Apr;4(3):273–289. doi: 10.1016/0888-7543(89)90331-5. [DOI] [PubMed] [Google Scholar]
- Novacek M. J. Mammalian phylogeny: shaking the tree. Nature. 1992 Mar 12;356(6365):121–125. doi: 10.1038/356121a0. [DOI] [PubMed] [Google Scholar]
- Onda M., Kudo S., Rearden A., Mattei M. G., Fukuda M. Identification of a precursor genomic segment that provided a sequence unique to glycophorin B and E genes. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7220–7224. doi: 10.1073/pnas.90.15.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart F. P., Ritch T. G., Deininger P. L., Schmid C. W. Renaturation rate studies of a single family of interspersed repeated sequences in human deoxyribonucleic acid. Biochemistry. 1981 May 26;20(11):3003–3010. doi: 10.1021/bi00514a003. [DOI] [PubMed] [Google Scholar]
- Rouyer F., Simmler M. C., Page D. C., Weissenbach J. A sex chromosome rearrangement in a human XX male caused by Alu-Alu recombination. Cell. 1987 Nov 6;51(3):417–425. doi: 10.1016/0092-8674(87)90637-4. [DOI] [PubMed] [Google Scholar]
- Schmid C., Maraia R. Transcriptional regulation and transpositional selection of active SINE sequences. Curr Opin Genet Dev. 1992 Dec;2(6):874–882. doi: 10.1016/s0959-437x(05)80110-8. [DOI] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Sinnett D., Deragon J. M., Simard L. R., Labuda D. Alumorphs--human DNA polymorphisms detected by polymerase chain reaction using Alu-specific primers. Genomics. 1990 Jul;7(3):331–334. doi: 10.1016/0888-7543(90)90166-r. [DOI] [PubMed] [Google Scholar]
- Smit A. F. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993 Apr 25;21(8):1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A. F., Riggs A. D. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 1995 Jan 11;23(1):98–102. doi: 10.1093/nar/23.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. F., Waterman M. S. Identification of common molecular subsequences. J Mol Biol. 1981 Mar 25;147(1):195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- Vidal F., Mougneau E., Glaichenhaus N., Vaigot P., Darmon M., Cuzin F. Coordinated posttranscriptional control of gene expression by modular elements including Alu-like repetitive sequences. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):208–212. doi: 10.1073/pnas.90.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Wilson A. C., Sarich V. M., Maxson L. R. The importance of gene rearrangement in evolution: evidence from studies on rates of chromosomal, protein, and anatomical evolution. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3028–3030. doi: 10.1073/pnas.71.8.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz E., Labuda M., Sinnett D., Glorieux F. H., Labuda D. Linkage mapping by simultaneous screening of multiple polymorphic loci using Alu oligonucleotide-directed PCR. Proc Natl Acad Sci U S A. 1992 Sep 15;89(18):8448–8451. doi: 10.1073/pnas.89.18.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietkiewicz E., Rafalski A., Labuda D. Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics. 1994 Mar 15;20(2):176–183. doi: 10.1006/geno.1994.1151. [DOI] [PubMed] [Google Scholar]



