Abstract
DNA methylation of cytosine residues is a widespread phenomenon and has been implicated in a number of biological processes in both prokaryotes and eukaryotes. This methylation occurs at the 5-position of cytosine and is catalyzed by a distinct family of conserved enzymes, the cytosine-5 methyltransferases (m5C-MTases). We have cloned a fission yeast gene pmt1+ (pombe methyltransferase) which encodes a protein that shares significant homology with both prokaryotic and eukaryotic m5C-MTases. All 10 conserved domains found in these enzymes are present in the pmt1 protein. This is the first m5C-MTase homologue cloned from a fungal species. Its presence is surprising, given the inability to detect DNA methylation in yeasts. Haploid cells lacking the pmt1+ gene are viable, indicating that pmt1+ is not an essential gene. Purified, bacterially produced pmt1 protein does not possess obvious methyltransferase activity in vitro. Thus the biological significance of the m5C-MTase homologue in fission yeast is currently unclear.
Full text
PDF




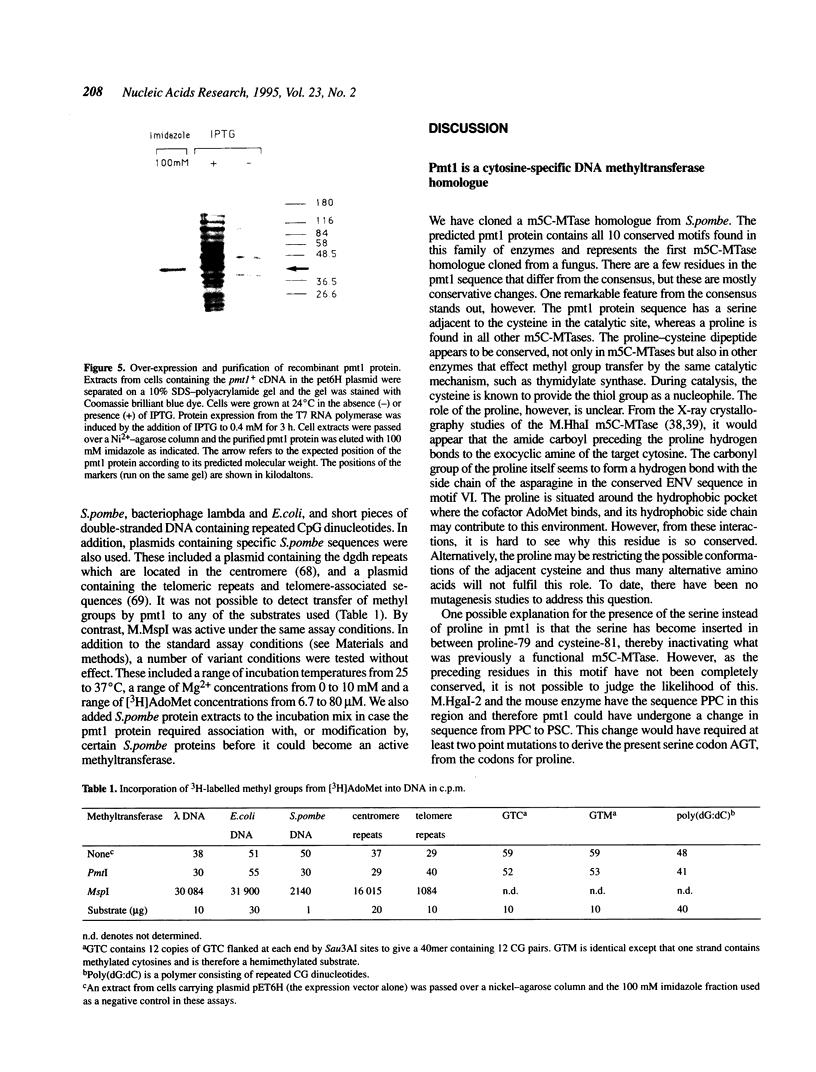


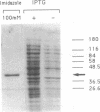
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Tamame M., Vilanueva J. R., Santos T. Developmental modulation of DNA methylation in the fungus Phycomyces blakesleeanus. Nucleic Acids Res. 1985 Sep 25;13(18):6545–6558. doi: 10.1093/nar/13.18.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F., Tamame M., Villanueva J. R., Santos T. DNA methylation in the fungi. J Biol Chem. 1984 Jul 10;259(13):8033–8036. [PubMed] [Google Scholar]
- Bestor T. H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992 Jul;11(7):2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988 Oct 20;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Caserta M., Zacharias W., Nwankwo D., Wilson G. G., Wells R. D. Cloning, sequencing, in vivo promoter mapping, and expression in Escherichia coli of the gene for the HhaI methyltransferase. J Biol Chem. 1987 Apr 5;262(10):4770–4777. [PubMed] [Google Scholar]
- Cedar H. DNA methylation and gene activity. Cell. 1988 Apr 8;53(1):3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- Chen L., MacMillan A. M., Chang W., Ezaz-Nikpay K., Lane W. S., Verdine G. L. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991 Nov 19;30(46):11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Crain P. F., McCloskey J. A. Analysis of modified bases in DNA by stable isotope dilution gas chromatography-mass spectrometry: 5-methylcytosine. Anal Biochem. 1983 Jul 1;132(1):124–131. doi: 10.1016/0003-2697(83)90434-7. [DOI] [PubMed] [Google Scholar]
- Cross S. H., Charlton J. A., Nan X., Bird A. P. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994 Mar;6(3):236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- Cross S., Kovarik P., Schmidtke J., Bird A. Non-methylated islands in fish genomes are GC-poor. Nucleic Acids Res. 1991 Apr 11;19(7):1469–1474. doi: 10.1093/nar/19.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S., Cedar H. Role of DNA methylation in the regulation of transcription. Curr Opin Genet Dev. 1994 Apr;4(2):255–259. doi: 10.1016/s0959-437x(05)80052-8. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A. Parental imprinting of autosomal mammalian genes. Curr Opin Genet Dev. 1994 Apr;4(2):265–280. doi: 10.1016/s0959-437x(05)80054-1. [DOI] [PubMed] [Google Scholar]
- Faugeron G., Rhounim L., Rossignol J. L. How does the cell count the number of ectopic copies of a gene in the premeiotic inactivation process acting in Ascobolus immersus? Genetics. 1990 Mar;124(3):585–591. doi: 10.1093/genetics/124.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fikes J. D., Becker D. M., Winston F., Guarente L. Striking conservation of TFIID in Schizosaccharomyces pombe and Saccharomyces cerevisiae. Nature. 1990 Jul 19;346(6281):291–294. doi: 10.1038/346291a0. [DOI] [PubMed] [Google Scholar]
- Finnegan E. J., Dennis E. S. Isolation and identification by sequence homology of a putative cytosine methyltransferase from Arabidopsis thaliana. Nucleic Acids Res. 1993 May 25;21(10):2383–2388. doi: 10.1093/nar/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foss H. M., Roberts C. J., Claeys K. M., Selker E. U. Abnormal chromosome behavior in Neurospora mutants defective in DNA methylation. Science. 1993 Dec 10;262(5140):1737–1741. doi: 10.1126/science.7505062. [DOI] [PubMed] [Google Scholar]
- Friedman S., Ansari N. Binding of the EcoRII methyltransferase to 5-fluorocytosine-containing DNA. Isolation of a bound peptide. Nucleic Acids Res. 1992 Jun 25;20(12):3241–3248. doi: 10.1093/nar/20.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Faugeron G. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol Cell Biol. 1989 Jul;9(7):2818–2827. doi: 10.1128/mcb.9.7.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C., Nogueira T. I., Faugeron G. Perpetuation of cytosine methylation in Ascobolus immersus implies a novel type of maintenance methylase. J Mol Biol. 1994 Jul 1;240(1):42–51. doi: 10.1006/jmbi.1994.1416. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985 Feb;40(2):381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hoheisel J. D., Maier E., Mott R., McCarthy L., Grigoriev A. V., Schalkwyk L. C., Nizetic D., Francis F., Lehrach H. High resolution cosmid and P1 maps spanning the 14 Mb genome of the fission yeast S. pombe. Cell. 1993 Apr 9;73(1):109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- Ingrosso D., Fowler A. V., Bleibaum J., Clarke S. Sequence of the D-aspartyl/L-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J Biol Chem. 1989 Nov 25;264(33):20131–20139. [PubMed] [Google Scholar]
- Kinsey J. A., Helber J. Isolation of a transposable element from Neurospora crassa. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1929–1933. doi: 10.1073/pnas.86.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A. J. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature. 1987 Apr 2;326(6112):466–470. doi: 10.1038/326466a0. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Nelson J. L., Roberts R. J. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 1991 Nov 25;19(22):6183–6190. doi: 10.1093/nar/19.22.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimasauskas S., Timinskas A., Menkevicius S., Butkienè D., Butkus V., Janulaitis A. Sequence motifs characteristic of DNA[cytosine-N4]methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989 Dec 11;17(23):9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käufer N. F., Simanis V., Nurse P. Fission yeast Schizosaccharomyces pombe correctly excises a mammalian RNA transcript intervening sequence. Nature. 1985 Nov 7;318(6041):78–80. doi: 10.1038/318078a0. [DOI] [PubMed] [Google Scholar]
- Levin H. L., Weaver D. C., Boeke J. D. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol Cell Biol. 1990 Dec;10(12):6791–6798. doi: 10.1128/mcb.10.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. D., Meehan R. R., Henzel W. J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992 Jun 12;69(6):905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- Lewis J., Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991 Jul 22;285(2):155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- Li E., Bestor T. H., Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992 Jun 12;69(6):915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lin P. M., Lee C. H., Roberts R. J. Cloning and characterization of the genes encoding the MspI restriction modification system. Nucleic Acids Res. 1989 Apr 25;17(8):3001–3011. doi: 10.1093/nar/17.8.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man. Clustering in the conserved core of molecule. J Mol Biol. 1988 May 20;201(2):289–314. doi: 10.1016/0022-2836(88)90139-8. [DOI] [PubMed] [Google Scholar]
- Maier E., Hoheisel J. D., McCarthy L., Mott R., Grigoriev A. V., Monaco A. P., Larin Z., Lehrach H. Complete coverage of the Schizosaccharomyces pombe genome in yeast artificial chromosomes. Nat Genet. 1992 Jul;1(4):273–277. doi: 10.1038/ng0792-273. [DOI] [PubMed] [Google Scholar]
- Mi S., Roberts R. J. The DNA binding affinity of HhaI methylase is increased by a single amino acid substitution in the catalytic center. Nucleic Acids Res. 1993 May 25;21(10):2459–2464. doi: 10.1093/nar/21.10.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Murakami S., Matsumoto T., Niwa O., Yanagida M. Structure of the fission yeast centromere cen3: direct analysis of the reiterated inverted region. Chromosoma. 1991 Dec;101(4):214–221. doi: 10.1007/BF00365153. [DOI] [PubMed] [Google Scholar]
- Nicolas A., Hamza H., Mekki-Berrada A., Kalogeropoulos A., Rossignol J. L. Premeiotic and Meiotic Instability Generates Numerous b2 Mutation Derivatives in Ascobolus. Genetics. 1987 May;116(1):33–43. doi: 10.1093/genetics/116.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Osterman D. G., DePillis G. D., Wu J. C., Matsuda A., Santi D. V. 5-Fluorocytosine in DNA is a mechanism-based inhibitor of HhaI methylase. Biochemistry. 1988 Jul 12;27(14):5204–5210. doi: 10.1021/bi00414a039. [DOI] [PubMed] [Google Scholar]
- Prentice H. L. High efficiency transformation of Schizosaccharomyces pombe by electroporation. Nucleic Acids Res. 1992 Feb 11;20(3):621–621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proffitt J. H., Davie J. R., Swinton D., Hattman S. 5-Methylcytosine is not detectable in Saccharomyces cerevisiae DNA. Mol Cell Biol. 1984 May;4(5):985–988. doi: 10.1128/mcb.4.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renbaum P., Abrahamove D., Fainsod A., Wilson G. G., Rottem S., Razin A. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI). Nucleic Acids Res. 1990 Mar 11;18(5):1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Pfeifer G. P. X-chromosome inactivation and cell memory. Trends Genet. 1992 May;8(5):169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- Rothnie H. M., McCurrach K. J., Glover L. A., Hardman N. Retrotransposon-like nature of Tp1 elements: implications for the organisation of highly repetitive, hypermethylated DNA in the genome of Physarum polycephalum. Nucleic Acids Res. 1991 Jan 25;19(2):279–286. doi: 10.1093/nar/19.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. J., Rodland K. D., Rachlin E. M., McCloskey J. A. Differential DNA methylation during the vegetative life cycle of Neurospora crassa. J Bacteriol. 1987 Jun;169(6):2902–2905. doi: 10.1128/jb.169.6.2902-2905.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P. J., Welsch J. A., Rachlin E. M., McCloskey J. A. Different levels of DNA methylation in yeast and mycelial forms of Candida albicans. J Bacteriol. 1987 Sep;169(9):4393–4395. doi: 10.1128/jb.169.9.4393-4395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi D. V., Hardy L. W. Catalytic mechanism and inhibition of tRNA (uracil-5-)methyltransferase: evidence for covalent catalysis. Biochemistry. 1987 Dec 29;26(26):8599–8606. doi: 10.1021/bi00400a016. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Cambareri E. B., Jensen B. C., Haack K. R. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987 Dec 4;51(5):741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Garrett P. W. DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6870–6874. doi: 10.1073/pnas.85.18.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. S., Kaplan B. E., Sowers L. C., Newman E. M. Mechanism of human methyl-directed DNA methyltransferase and the fidelity of cytosine methylation. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Som S., Bhagwat A. S., Friedman S. Nucleotide sequence and expression of the gene encoding the EcoRII modification enzyme. Nucleic Acids Res. 1987 Jan 12;15(1):313–332. doi: 10.1093/nar/15.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. L., Stuhlmann H., Jähner D., Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Taylor C., Ford K., Connolly B. A., Hornby D. P. Determination of the order of substrate addition to MspI DNA methyltransferase using a novel mechanism-based inhibitor. Biochem J. 1993 Apr 15;291(Pt 2):493–504. doi: 10.1042/bj2910493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum Y., Sedat J., Razin A. The absence of detectable methylated bases in Drosophila melanogaster DNA. FEBS Lett. 1982 Sep 6;146(1):148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- Wilke K., Rauhut E., Noyer-Weidner M., Lauster R., Pawlek B., Behrens B., Trautner T. A. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 1988 Aug;7(8):2601–2609. doi: 10.1002/j.1460-2075.1988.tb03110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- Wyszynski M. W., Gabbara S., Bhagwat A. S. Substitutions of a cysteine conserved among DNA cytosine methylases result in a variety of phenotypes. Nucleic Acids Res. 1992 Jan 25;20(2):319–326. doi: 10.1093/nar/20.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen R. W., Vertino P. M., Nelkin B. D., Yu J. J., el-Deiry W., Cumaraswamy A., Lennon G. G., Trask B. J., Celano P., Baylin S. B. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992 May 11;20(9):2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]