Summary
Human peripheral blood NK cells may be divided into two main subsets: CD56brightCD16− and CD56dimCD16+. Since TGF-β is known to influence the development of many leukocyte lineages, its effects on NK cell differentiation either from human CD34+Lin− hematopoietic progenitor/stem cells in vitro or from peripheral blood NK cells were investigated. TGF-β represses development of NK cells from CD34+ progenitors and inhibits differentiation of CD16+ NK cells. Moreover, TGF-β also results in conversion of a minor fraction of CD56brightCD16+ cells found in peripheral blood into CD56brightCD16− cells, highlighting a possible role of the former as a developmental intermediate and of TGF-β in influencing the genesis of NK subsets found in blood.
Keywords: NK cells, Cell differentiation, Innate immunity
Introduction
Two main subsets of NK cells are present in human peripheral blood, termed CD56dimCD16+ and CD56brightCD16− [1, 2]. CD56dimCD16+ NK cells make up the vast majority of peripheral blood NK cells (>90%). They are highly cytotoxic towards tumors and some virus infected cells, but produce only low levels of cytokines [2, 3]. In contrast, CD56brightCD16− NK cells, representing <10% of blood NK cells, display less cytotoxic activity ex vivo, but efficiently produce IFN-γ, TNF-α, LTα, IL-10, and GM-CSF in response to non-specific stimulation or monokines [2, 3]. These two subsets also differ in the expression of many other cell surface receptors, including killer-cell immunoglobulin-like receptors (KIR) and CD94/NKG2A heterodimers, which are preferentially found on CD56dimCD16+ and CD56brightCD16− NK cells, respectively. Microarray comparisons of the two subsets have identified numerous additional genes that are differentially expressed [4, 5]. By contrast, most NK cells in lymph nodes have a CD56brightCD16− phenotype [6, 7]. Recently, several groups have identified an additional subset of NK-like cells in mucosa-associated lymphoid tissue that secrete IL-22 [8, 9]. In mice, these NK-22 cells are non-cytotoxic and do not produce IFNγ, but are characterized by expression of the RORγt transcription factor and IL-7Rα (CD127) [8, 9]. Decidual NK cells with a distinct phenotype are also found in abundance at the maternal-fetal interface during human pregnancy [4, 10].
TGF-β1,2,3 are multi-functional pleiotropic cytokines, playing significant roles in embryogenesis, development, tissue renewal, and regulation of the immune system. TGF-β1 is the predominant form expressed by immune cells [11]. TGF-β is known to regulate many branches of hematopoiesis, affecting differentiation and proliferation of hematopoietic stem cells as well as progenitor cells of erythrocytes, granulocytes, monocytes/macrophages, dendritic cells, megakaryocytes, and other lineages [12]. The effects of TGF-β are often very dependent on the developmental stage and in vivo environmental context, with different (and often opposing) effects at different stages [12]. TGF-β also influences T cells at several different phases of development/differentiation, including controlling formation of both inflammatory Th17 cells and Foxp3+ regulatory T cells [11].
To date, TGF-β has been shown to exert many effects on NK cells, including inhibition of proliferation, cytotoxicity, and IFNγ production, and down-regulation of activating receptors such as NKG2D and NKp30 [13–16]. In the present paper, its effect on NK cell development and differentiation has been explored, from both immature progenitors and from mature peripheral blood NK cells.
Results and Discussion
TGF-β affects the numbers and phenotype of NK cells developing from human CD34+ progenitor cells
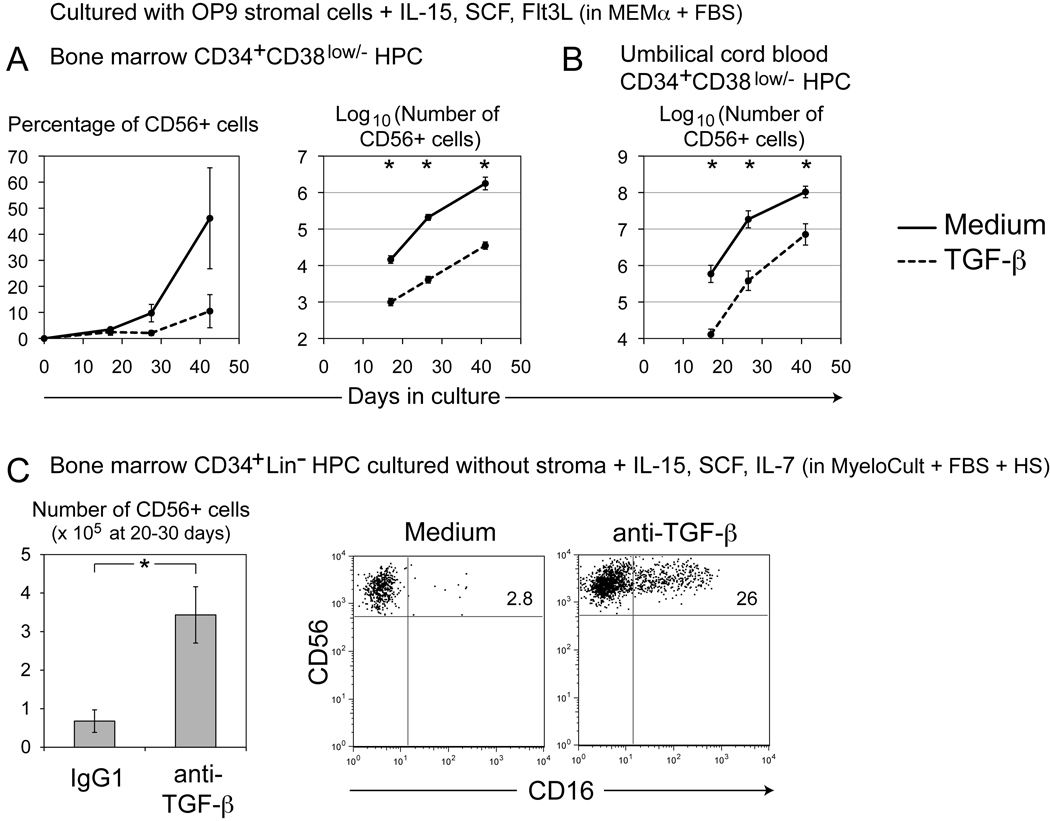
To investigate the effects of TGF-β on NK cell development, CD34+CD38low/− hematopoietic progenitor/stem cells (HPC) from human bone marrow were cultured in the presence of OP9 stromal cells with IL-15, SCF, and Flt3L, cytokines known to facilitate NK cell development. Supplementation of the cultures with TGF-β1 (2ng/ml) resulted in lower percentages and dramatically reduced numbers of CD56+ NK cells (Figure 1a). In similar cultures using CD34+CD38low/− HPC isolated from human umbilical cord blood, TGF-β again repressed the numbers of NK cells that developed (Figure 1b). TGF-β also appeared to inhibit or delay the acquisition of markers of NK cell maturation or subset formation such as CD94, CD16, and KIR (Supporting Information Figure 1). TGF-β similarly inhibited or delayed the ability of developing NK cells to lyse the OP9 stromal cell monolayer.
Figure 1. TGF-β affects the number of NK cells and the percentage of CD16+ NK cells developing from CD34+ HPC in vitro.
(A) Approximately 5000 human CD34+CD38low/−CD56− HPC were sorted from enriched bone marrow CD34+ cells, and cultured on OP9 stromal cells with IL-15 (20ng/ml), SCF (20ng/ml), and Flt3L (5ng/ml) in MEMα medium plus 20% heat-inactivated FBS. TGF-β1 (2ng/ml) was added to some cultures. Numbers of NK cells that developed were calculated as described in the Materials and methods. Plots represent the means of three experiments from two donors. (B) Similar cultures were initiated using approximately 5000 CD34+CD38low/−CD56− HPC sorted from umbilical cord blood (Mean of three experiments from three donors). (C) 5000 to 12000 human CD34+Lin− HPC were sorted from bone marrow aspirates, then cultured with IL-15 (20ng/ml), SCF (20ng/ml), and IL-7 (20ng/ml) in MyeloCult medium plus 10% heat-inactivated human serum and 5% heat-inactivated FBS. Cultures were additionally supplemented with IgG1 control or anti-TGF-β neutralizing mAb (clone 1D11) (10µg/ml). Mean numbers of developing NK cells were calculated for 8 experiments from 4 donors. Flow cytometry dot-plots were gated on resulting CD56+ cells. No CD3+ cells developed in these cultures. In (A), (B), and (C), data show mean ± SEM. *p<0.05, two-tailed Student’s t test comparing LOG10-transformed values.
Parallel results were observed in another experimental system. Bone marrow CD34+Lin− HPC were grown without stromal cells in a different culture medium with IL-15, SCF, and IL-7. Under these conditions, when the activity of TGF-β was blocked with neutralizing anti-TGF-β1,2,3 monoclonal antibody (mAb), increased numbers of NK cells were observed at approximately 30 days of culture (Figure 1c). Larger numbers of other cell types were also observed, including adherent CD56− MHC class II+ CD11c+ cells. Blockade of TGF-β activity also affected the phenotype of NK cells that developed from HPC in these cultures. In the presence of anti-TGF-β mAb, greater percentages of NK cells expressed CD16 (Figure 1c; Supporting Information Figure 2). An increase in the number of KIR+ NK cells was also observed upon neutralization of TGF-β (Supporting Information Figure 2). Both CD16 and KIR are characteristic of the CD56dimCD16+ peripheral blood NK cell subset. The TGF-β that mediated these effects may have been derived from the NK cells themselves or from other cell types that also developed in these cultures. However, it is likely that the TGF-β was derived from the serum in the culture system, since MyeloCult medium contains horse serum and FBS, and culture medium was additionally supplemented with FBS and human serum (HS). This medium showed similar levels of total TGF-β1 before and after culture of developing NK cells (~ 50pg/ml) (measured by Biosource multispecies ELISA). It should be noted that addition of anti-TGF-β mAb did not augment NK cell numbers in OP9 stromal cell co-cultures (such as those depicted in Figure 1a–b). This discrepancy in the activity of anti-TGF-β mAb may have been due to: (i) differences in the quantity of endogenous serum TGF-β between the two varieties of culture medium, (ii) differences in cell number in culture that might overwhelm the 10µg/ml concentration of mAb or (iii) production or depletion of TGF-β by OP9 stromal cells. However, in clinical protocols for amplification of NK cells, it may be important to control the levels of TGF-β in culture to achieve maximal yields of NK cells. Effects of TGF-β on numbers of NK cells have also been observed in murine transgenic models [17].
TGF-β directly inhibits development/differentiation of NK cell subsets
In the preceding experiments, other cell types also developed in these cultures in vitro. Thus, the observed effects may have been indirect, with TGF-β influencing other cell lineages or stromal cells which, in turn, affected NK cell development. In addition, TGF-β has previously been shown to affect proliferation and differentiation of HPC [12]. Therefore, the effect of TGF-β on developing NK cells was investigated directly. CD161 (NKR-P1A) is one of the earliest markers to be expressed during human NK cell development [18]. Therefore, CD161+ cells (that had not yet acquired CD94 or CD16 expression) were sorted from cultures undergoing NK development from umbilical cord blood HPC (as Figure 1b above). When placed back into medium with cytokines but without stromal cells, a proportion of these cells acquired CD94 and CD16 expression (Figure 2). However, addition of 2ng/ml TGF-β1 completely blocked this developmental progression/differentiation and greatly reduced proliferation of the cells (Figure 2). Analogous results were observed with sorted CD56+CD161+CD94−CD16− cells (not shown).
Figure 2. TGF-β inhibits acquisition of CD94 and CD16 on developing NK cells.
Co-cultures of OP9 stromal cells and HPC isolated from human umbilical cord blood were initiated as described in Figure 1a–b. At day 17, CD161+CD94−CD16− cells were sorted to high purity and placed back into culture with IL-15 (20ng/ml), SCF (20ng/ml), and Flt3L (5ng/ml) without stromal cells in MEMα medium plus 20% FBS. TGF-β1 (2ng/ml) was added to some cultures. After 24 additional days, resulting cells were counted and stained for flow cytometry. Plots were gated on live cells by FSC/SSC/PI. Numbers indicate the percentage of cells in each quadrant. Mean fold change in cell number ± SEM from three similar independent experiments is also shown. *p<0.05 in Student’s t test.
TGF-β inhibits and down-regulates CD16 expression on CD56bright NK cells from peripheral blood
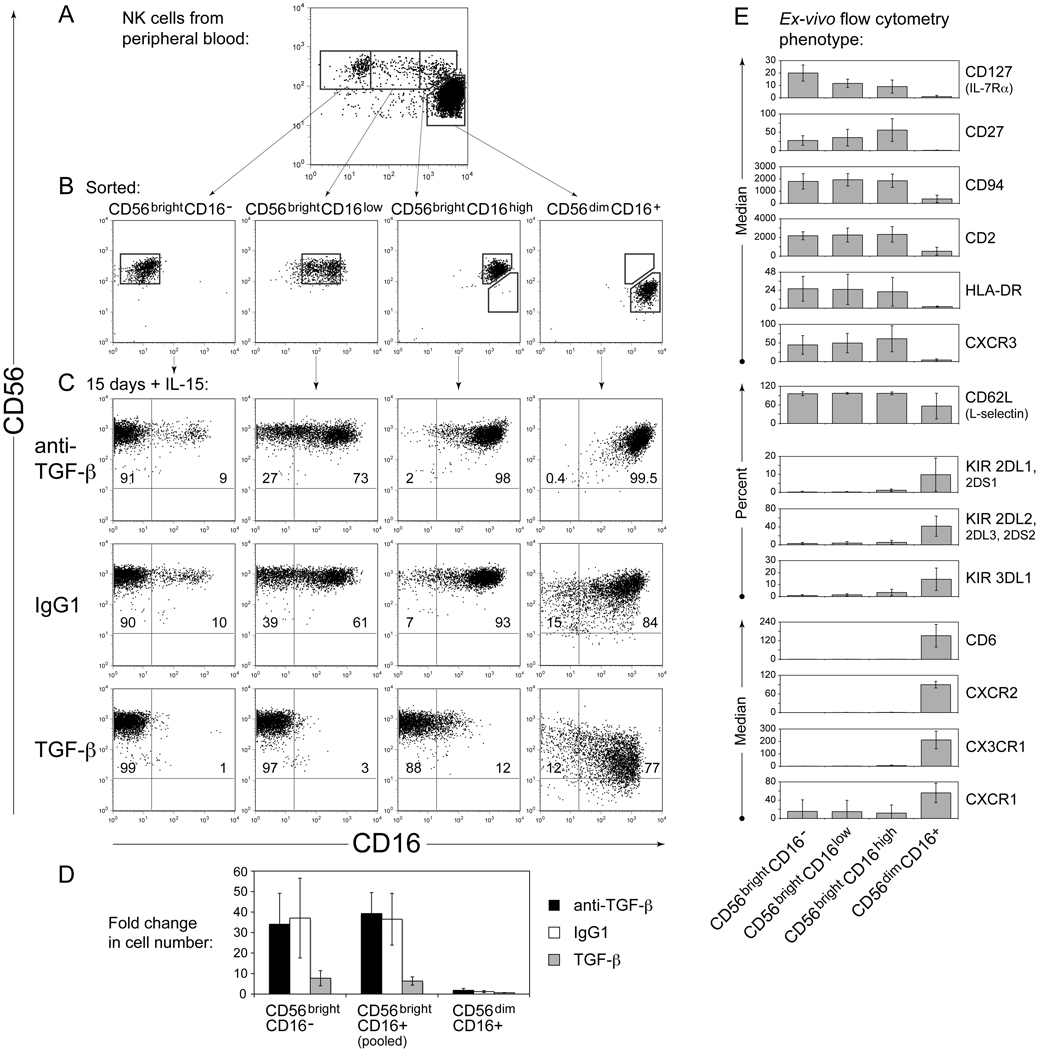
Several recent studies have provided insights into the developmental relationship between human NK cell subsets. Culture of CD56brightCD16− NK cells with synovial or dermal fibroblasts was reported to cause differentiation into CD56dimCD16+KIR+ cells exhibiting many characteristics of ex-vivo CD56dimCD16+ NK cells [19]. Related results were obtained upon culture with IL-2 [20, 21], or after transfer of human NK cells into mouse models [19, 22]. Similarly, when CD56brightCD16− NK cells were sorted from human peripheral blood and cultured with IL-15 for 15 days, appearance of some CD16+ cells was observed (Figure 3c column 1, Supporting Information Figure 3). However, percentages of KIR (or other features of CD56dim NK cells) did not increase substantially (not shown and Supporting Information Figure 4), suggesting that stromal cell factors or IL-2 (as opposed to IL-15) are required for complete transition to a CD56dimCD16+ phenotype [19, 20]. Neutralization of TGF-β activity with mAb did not affect, or increased, CD16 expression (Figure 3c column 1, Supporting Information Figure 3). On the other hand, addition of 2ng/ml TGF-β1 largely blocked the appearance of CD16+ cells (Figure 3c column 1, Supporting Information Figure 3). Here, sorted NK cells were highly pure, precluding indirect effects of other cell types. Thus, TGF-β inhibited the acquisition of CD16 on NK cells from peripheral blood, as well as on NK cells derived from bone marrow progenitors.
Figure 3. TGF-β inhibits and down-regulates CD16 expression on CD56bright NK cells from blood.
(A) NK cells were isolated from human peripheral blood by depletion of other cell lineages (CD3+, CD4+, CD19+, CD36+ or CD66b+), then stained with anti-CD56 and anti-CD16 mAb, and sorted into sub-populations as indicated. (B) Purity of cells immediately after sorting. (C) Phenotype of sorted cells after culture for 15 days with IL-15 (20ng/ml) plus IgG1, or anti-TGF-β 1D11 mAb (10µg/ml), or added TGF-β1 (2ng/ml). MyeloCult medium was used (plus 10% HS and 5% FBS). Numbers indicate the percentage of cells in each quadrant. Only CD56+ cells are shown in A; all live cells are shown in B and C (gated by FSC/SSC). Data in A/B versus C were collected with different staining and instrument protocols, so staining intensities are not directly comparable. (D) Changes in cell number in these experiments; data show mean ± SEM from three independent experiments. Results from CD56brightCD16low and CD56brightCD16high populations were pooled for analysis and labeled CD56brightCD16+. (E) Ex-vivo flow cytometry staining of human peripheral blood NK cell populations isolated, stained, and gated as depicted in (A). Data show median fluorescence intensity (minus median fluorescence of IgG control) or percentage of cells staining positively for each indicated mAb ± SD from 3–6 donors.
Many individuals possess a substantial population of CD56bright NK cells that also express CD16, appearing between CD56brightCD16− and CD56dimCD16+ cells in two-color flow cytometric analysis (Figure 3a) [1]. These cells were hypothesized to be either a developmental intermediate between the other two subsets, or a distinct functional subset [1]. Thus, the effects of TGF-β were examined on each. Four populations of cells were sorted from peripheral blood NK cells (CD56brightCD16−, CD56brightCD16low, CD56brightCD16high, and CD56dimCD16+ (Figure 3a,b)), then cultured for approximately two weeks with IL-15. Upon culture with anti-TGF-β neutralizing mAb, the CD16+ subsets largely retained their CD16 expression (Figure 3c, Supporting Information Figure 3). In contrast, upon addition of TGF-β1, both CD56brightCD16low and CD56brightCD16high cells robustly lost CD16 expression to become CD56brightCD16− (Figure 3c, Supporting Information Figure 3). CD56dimCD16+ NK cells showed a reduced tendency to down-regulate CD16 expression upon culture with TGF-β (Figure 3c, Supporting Information Figure 3). These results are consistent with a previously published observation, in which total CD16+ blood NK cells sorted and cultured with TGF-β1 gave rise to a subset that lost CD16 expression, and another subset that remained CD16+ [23]. Here, the CD56brightCD16+ NK cell subset was identified to be most responsive to TGF-β in terms of CD16 down-regulation. This loss of CD16 expression was also accompanied by proliferation [23] (Figure 3d). CD56brightCD16+ and CD56brightCD16− NK cells showed similar average levels of proliferation in culture (Figure 3d), implying that these results were not due to outgrowth of rare sort contaminants. TGF-β inhibited proliferation of all subsets (Figure 3d). In these experiments with peripheral blood NK cells, effects of anti-TGF-β mAb were more limited, perhaps for the reasons mentioned above, or perhaps due to differential sensitivities or different responses of progenitors to TGF-β versus mature NK cells.
Therefore, for CD56bright NK cells, 2-week exposure to TGF-β inhibited acquisition of CD16 and down-regulated pre-existing CD16 expression. The majority of CD56dimCD16+ cells showed different responses to TGF-β (such as down-regulation of CD56 [Supporting Information Figure 3] and CX3CR1 chemokine receptors [Supporting Information Figure 4]). It is possible that divergent effects of TGF-β may be related to differences in TGF-β receptor expression (Supporting Information Figure 5). Although these may simply reflect subset-specific effects of TGF-β (e.g. on CD16 and ADCC function), these results may have relevance to NK cell differentiation pathways:
In accordance with recent reports [19, 20, 22], human NK cells may differentiate along a pathway from CD56brightCD16− cells to CD56brightCD16+ intermediate cells, to fully cytotoxic CD56dimCD16+ NK cells. TGF-β may be able to block progression along this pathway, and is able to convert CD56brightCD16+ intermediate cells back to a CD16− phenotype. However, CD56dimCD16+ NK cells may be fully differentiated to a committed mature effector phenotype and thus refractory to these effects of TGF-β.
On the other hand, TGF-β may primarily generate decidual-like NK cells resembling those at the maternal-fetal interface [23]. TGF-β has been shown to up-regulate CD9 and CD103 (αE integrin) molecules found on decidual NK cells [23], and expression of CD9 is eliminated with addition of anti-TGF-β mAb on NK cells cultured in IL-15 (not shown).
For each of the three sorted CD56bright NK populations from peripheral blood, culture with TGF-β1 or anti-TGF-β did not markedly change percentages of KIR+ cells (not shown and Supporting Information Figure 4). Statistically significant changes in KIR percentage have previously been seen with TGF-β treatment of enriched total NK populations, likely attributed to survival/expansion of mature NK cells that had already acquired KIR [23] (Supporting Information Figure 4). TGF-β may differentially effect de novo acquisition of KIR on immature NK cells developing from HPC versus more mature circulating peripheral blood NK cells. Alternatively, in HPC cultures, adherent MHC class II+ CD11c+ cells, or other cells (that also developed), or OP9 stromal cells, may have provided additional signals necessary for KIR expression.
CD56brightCD16+ NK cells most closely resemble CD56brightCD16− NK cells
These experiments highlighted the CD56brightCD16+ NK cell subset as a potential intermediate in NK cell subset differentiation. Although the phenotype of CD56brightCD16+ NK cells from blood has been previously examined [1], many additional molecules have subsequently been shown to be expressed at higher levels and/or in greater percentages on either CD56brightCD16− NK cells or CD56dimCD16+ NK cells (listed in Figure 3e and references [2, 4, 24–26]. Importantly, expression of each of these molecules on the intermediate CD56brightCD16+ populations (both CD16low and CD16high) was similar to CD56brightCD16− NK cells, rather than to CD56dim NK cells (Figure 3e). Similarly, intracellular staining of perforin was lower in CD56brightCD16−, CD56brightCD16low, and CD56brightCD16high NK cells, while CD56dimCD16+ cells stained to a higher level (not shown). Thus, with the exception of CD16, for each of the markers examined, CD56brightCD16+ NK cells more closely resembled CD56brightCD16− NK cells than CD56dim NK cells. Effects of TGF-β on expression of these markers are depicted in Supporting Information Figure 4. Several of these molecules have been used to describe potentially analogous mouse NK cell subsets [27–29]. Another subset from human blood with intermediate phenotype, CD56dimCD94high, has recently been described [30], which could potentially be downstream of CD56brightCD16+ cells in NK differentiation. Interestingly, CD56brightCD16+ and CD56brightCD16− NK cells are more prevalent in the early period following bone marrow transplantation [31].
Concluding remarks
During the development of NK cells from bone marrow or umbilical cord blood CD34+ HPC in vitro, TGF-β was shown to influence the number of resulting NK cells (Figure 1). TGF-β also inhibited formation of CD16+ NK cells in sorted cultures of bone marrow HPC, immature CD161+CD94−CD16− NK cells, or peripheral blood CD56brightCD16− NK cells (Figures 1,2,3). Furthermore, addition of TGF-β to CD56brightCD16+ NK cells resulted in their conversion to CD56brightCD16− cells (Figure 3). Thus, levels of TGF-β at sites of NK development in vivo may influence the developmental progression and subset formation of NK cells.
Materials and methods
Cells
Bone marrow HPC were isolated from fresh normal whole human bone marrow (All Cells LLC., Emeryville, CA, USA) using RosetteSep human progenitor reagent (Stem Cell Technologies, Vancouver, BC, Canada), followed by flow cytometry sorting for CD34+Lin− (CD3−CD14−CD16−CD19−CD20−CD56−) cells (Figure 1c). Alternatively, CD34+CD38low/−CD56− bone marrow HPC were sorted from frozen enriched human bone marrow CD34+ progenitors (Lonza, Basel, Switzerland) (Figure 1a). In accordance with approved guidelines established by the Research Ethics Board of Sunnybrook Health Sciences Center, umbilical cord blood HPC were isolated as previously described [32] by negative selection using the StemSep Human Progenitor Enrichment Kit (StemCell Technologies), and flow cytometry sorting for CD34+CD38low/−CD56− cells. Peripheral blood NK cells were isolated from LeukoPaks (Massachusetts General Hospital Blood Apheresis Center, Boston, MA, USA) using RosetteSep human NK cell reagent (Stem Cell Technologies), then immediately analyzed or sorted by flow cytometry.
Cell culture conditions
Sorted cells were cultured in vitro in 24, 12, or 6 well plates with human cytokines (Peprotech, Rocky Hill, NJ, USA) in several culture media as indicated. MyeloCult medium was from Stem Cell Technologies. Anti-TGFβ (clone 1D11) mAb was obtained from R&D Systems (Minneapolis, MN, USA). Medium, cytokines and neutralizing mAb were refreshed every 3–5 days by changing all or half the culture volume. Where indicated, cells were co-cultured on confluent monolayers of non-irradiated OP9 cells. Every 3–11 days, co-cultured cells were disaggregated as described [32] and placed onto new monolayers of OP9 (more frequently after stromal cell lysis was observed). Numbers of NK cells that developed were calculated by multiplying the percentage of CD56+ cells (from flow cytometry) X the cell count X the factor by which the cells had been diluted during culture. Measurements taken at approximately the same duration from different experiments were averaged to single data points. OP9 cells support NK cell but not T cell development from HPC [32].
Flow cytometry
Samples were analyzed using FACSCalibur instruments (BD, Franklin Lakes, NJ, USA) and the following fluorescently conjugated mAb clones. From BD Pharmingen: CD56 (NCAM16.2, B159), CD16 (3G8), KIR (HP-3E4 ,CH-L, DX9), CD127 (hIL-7R-M21), CD27 (M-T271), CD94 (HP-3D9), CD2 (RPA-2.10), CD161 (DX12), HLA-DR (L243), CD62L (Dreg-56), CD25 (M-A251), CD122 (Mik-β2), CD34 (563, 581), CD38 (HIT2), and Lineage cocktail (340546). From R&D Systems: CXCR1 (42705), and CXCR3 (49801). From Biolegend (San Diego, CA, USA): CXCR2 (5E8), CD6 (BL-CD6), and CD44 (IM7). From MBL International (Woburn, MA, USA): CX3CR1 (2A9-1). From eBioscience (San Diego, CA, USA): CD16 (CB16).
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grant AI050207 to JLS, and Canadian Institutes of Health Research (CIHR) Grant 74754 to JRC. We thank Brian Tilton, Patricia Rogers, Gisele Knowles and Arian Khandani for assistance with flow cytometry sorting. We are grateful to Dr. Elaine Herer and the staff of the Perinatal and Gynaecology Program of Women’s College Hospital and Sunnybrook HSC. We also thank Aruz Mesci, Jason Fine, Peter Chen, Mobin Karimi, Oscar Aguilar, Christina Kirkham, and Muhammad Ali Akbar for assistance or helpful discussions. JRC is supported by a CIHR New Investigator Award and an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. JCZP is a Canada Research Chair in Developmental Immunology.
Abbreviations
- KIR
killer-cell immunoglobulin-like receptors
- HPC
hematopoietic progenitor/stem cells
- HS
human serum
Footnotes
Conflict of Interest:
The authors declare no financial or commercial conflict of interest.
References
- 1.Nagler A, Lanier LL, Cwirla S, Phillips JH. Comparative studies of human FcRIII-positive and negative natural killer cells. J Immunol. 1989;143:3183–3191. [PubMed] [Google Scholar]
- 2.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 3.Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, Carson WE, Caligiuri MA. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood. 2001;97:3146–3151. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 4.Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, Masch R, et al. Human decidual natural killer cells are a unique NK cell subset with immunomodulatory potential. J Exp Med. 2003;198:1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanna J, Bechtel P, Zhai Y, Youssef F, McLachlan K, Mandelboim O. Novel insights on human NK cells' immunological modalities revealed by gene expression profiling. J Immunol. 2004;173:6547–6563. doi: 10.4049/jimmunol.173.11.6547. [DOI] [PubMed] [Google Scholar]
- 6.Fehniger TA, Cooper MA, Nuovo GJ, Cella M, Facchetti F, Colonna M, Caligiuri MA. CD56bright natural killer cells are present in human lymph nodes and are activated by T cell-derived IL-2: a potential new link between adaptive and innate immunity. Blood. 2003;101:3052–3057. doi: 10.1182/blood-2002-09-2876. [DOI] [PubMed] [Google Scholar]
- 7.Ferlazzo G, Thomas D, Lin SL, Goodman K, Morandi B, Muller WA, Moretta A, Munz C. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J Immunol. 2004;172:1455–1462. doi: 10.4049/jimmunol.172.3.1455. [DOI] [PubMed] [Google Scholar]
- 8.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, Doherty JM, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vivier E, Spits H, Cupedo T. Interleukin-22-producing innate immune cells: new players in mucosal immunity and tissue repair? Nat Rev Immunol. 2009;9:229–234. doi: 10.1038/nri2522. [DOI] [PubMed] [Google Scholar]
- 10.Moffett-King A. Natural killer cells and pregnancy. Nat Rev Immunol. 2002;2:656–663. doi: 10.1038/nri886. [DOI] [PubMed] [Google Scholar]
- 11.Li MO, Flavell RA. TGF-beta: a master of all T cell trades. Cell. 2008;134:392–404. doi: 10.1016/j.cell.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fortunel NO, Hatzfeld A, Hatzfeld JA. Transforming growth factor-beta: pleiotropic role in the regulation of hematopoiesis. Blood. 2000;96:2022–2036. [PubMed] [Google Scholar]
- 13.Bellone G, Aste-Amezaga M, Trinchieri G, Rodeck U. Regulation of NK cell functions by TGF-beta 1. J Immunol. 1995;155:1066–1073. [PubMed] [Google Scholar]
- 14.Castriconi R, Cantoni C, Della Chiesa M, Vitale M, Marcenaro E, Conte R, Biassoni R, et al. Transforming growth factor beta 1 inhibits expression of NKp30 and NKG2D receptors: consequences for the NK-mediated killing of dendritic cells. Proc Natl Acad Sci USA. 2003;100:4120–4125. doi: 10.1073/pnas.0730640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eriksson M, Meadows SK, Wira CR, Sentman CL. Unique phenotype of human uterine NK cells and their regulation by endogenous TGF-beta. J Leukoc Biol. 2004;76:667–675. doi: 10.1189/jlb.0204090. [DOI] [PubMed] [Google Scholar]
- 16.Trotta R, Col JD, Yu J, Ciarlariello D, Thomas B, Zhang X, Allard J, et al. TGF-beta utilizes SMAD3 to inhibit CD16-mediated IFN-gamma production and antibody-dependent cellular cytotoxicity in human NK cells. J Immunol. 2008;181:3784–3792. doi: 10.4049/jimmunol.181.6.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laouar Y, Sutterwala FS, Gorelik L, Flavell RA. Transforming growth factor-beta controls T helper type 1 cell development through regulation of natural killer cell interferon-gamma. Nat Immunol. 2005;6:600–607. doi: 10.1038/ni1197. [DOI] [PubMed] [Google Scholar]
- 18.Blom B, Spits H. Development of human lymphoid cells. Annu Rev Immunol. 2006;24:287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- 19.Chan A, Hong DL, Atzberger A, Kollnberger S, Filer AD, Buckley CD, McMichael A, et al. CD56bright human NK cells differentiate into CD56dim cells: role of contact with peripheral fibroblasts. J Immunol. 2007;179:89–94. doi: 10.4049/jimmunol.179.1.89. [DOI] [PubMed] [Google Scholar]
- 20.Romagnani C, Juelke K, Falco M, Morandi B, D'Agostino A, Costa R, Ratto G, et al. CD56brightCD16− killer Ig-like receptor- NK cells display longer telomeres and acquire features of CD56dim NK cells upon activation. J Immunol. 2007;178:4947–4955. doi: 10.4049/jimmunol.178.8.4947. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi E, Kuranaga N, Satoh K, Habu Y, Shinomiya N, Asano T, Seki S, Hayakawa M. Induction of CD16+ CD56bright NK cells with antitumour cytotoxicity not only from CD16− CD56bright NK Cells but also from CD16− CD56dim NK cells. Scand J Immunol. 2007;65:126–138. doi: 10.1111/j.1365-3083.2006.01883.x. [DOI] [PubMed] [Google Scholar]
- 22.Huntington ND, Legrand N, Alves NL, Jaron B, Weijer K, Plet A, Corcuff E, et al. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J Exp Med. 2009;206:25–34. doi: 10.1084/jem.20082013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keskin DB, Allan DS, Rybalov B, Andzelm MM, Stern JN, Kopcow HD, Koopman LA, Strominger JL. TGFbeta promotes conversion of CD16+ peripheral blood NK cells into CD16− NK cells with similarities to decidual NK cells. Proc Natl Acad Sci USA. 2007;104:3378–3383. doi: 10.1073/pnas.0611098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vosshenrich CA, Garcia-Ojeda ME, Samson-Villeger SI, Pasqualetto V, Enault L, Richard-Le Goff O, Corcuff E, et al. A thymic pathway of mouse natural killer cell development characterized by expression of GATA-3 and CD127. Nat Immunol. 2006;7:1217–1224. doi: 10.1038/ni1395. [DOI] [PubMed] [Google Scholar]
- 25.Silva A, Andrews DM, Brooks AG, Smyth MJ, Hayakawa Y. Application of CD27 as a marker for distinguishing human NK cell subsets. Int Immunol. 2008;20:625–630. doi: 10.1093/intimm/dxn022. [DOI] [PubMed] [Google Scholar]
- 26.Poli A, Michel T, Theresine M, Andres E, Hentges F, Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology. 2009;126:458–465. doi: 10.1111/j.1365-2567.2008.03027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayakawa Y, Huntington ND, Nutt SL, Smyth MJ. Functional subsets of mouse natural killer cells. Immunol Rev. 2006;214:47–55. doi: 10.1111/j.1600-065X.2006.00454.x. [DOI] [PubMed] [Google Scholar]
- 28.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 29.Gregoire C, Chasson L, Luci C, Tomasello E, Geissmann F, Vivier E, Walzer T. The trafficking of natural killer cells. Immunol Rev. 2007;220:169–182. doi: 10.1111/j.1600-065X.2007.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Mao HC, Wei M, Hughes T, Zhang J, Park IK, Liu S, et al. CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK-cell subsets. Blood. 2010;115:274–281. doi: 10.1182/blood-2009-04-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dulphy N, Haas P, Busson M, Belhadj S, Peffault de Latour R, Robin M, Carmagnat M, et al. An unusual CD56(bright) CD16(low) NK cell subset dominates the early posttransplant period following HLA-matched hematopoietic stem cell transplantation. J Immunol. 2008;181:2227–2237. doi: 10.4049/jimmunol.181.3.2227. [DOI] [PubMed] [Google Scholar]
- 32.La Motte-Mohs RN, Herer E, Zuniga-Pflucker JC. Induction of T-cell development from human cord blood hematopoietic stem cells by Delta-like 1 in vitro. Blood. 2005;105:1431–1439. doi: 10.1182/blood-2004-04-1293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.