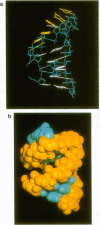
Abstract
A new modeling technique for arriving at the three dimensional (3-D) structure of an RNA stem-loop has been developed based on a conformational search by a genetic algorithm and the following refinement by energy minimization. The genetic algorithm simultaneously optimizes a population of conformations in the predefined conformational space and generates 3-D models of RNA. The fitness function to be optimized by the algorithm has been defined to reflect the satisfaction of known conformational constraints. In addition to a term for distance constraints, the fitness function contains a term to constrain each local conformation near to a prepared template conformation. The technique has been applied to the two loops of tRNA, the anticodon loop and the T-loop, and has found good models with small root mean square deviations from the crystal structure. Slightly different models have also been found for the anticodon loop. The analysis of a collection of alternative models obtained has revealed statistical features of local variations at each base position.
Full text
PDF
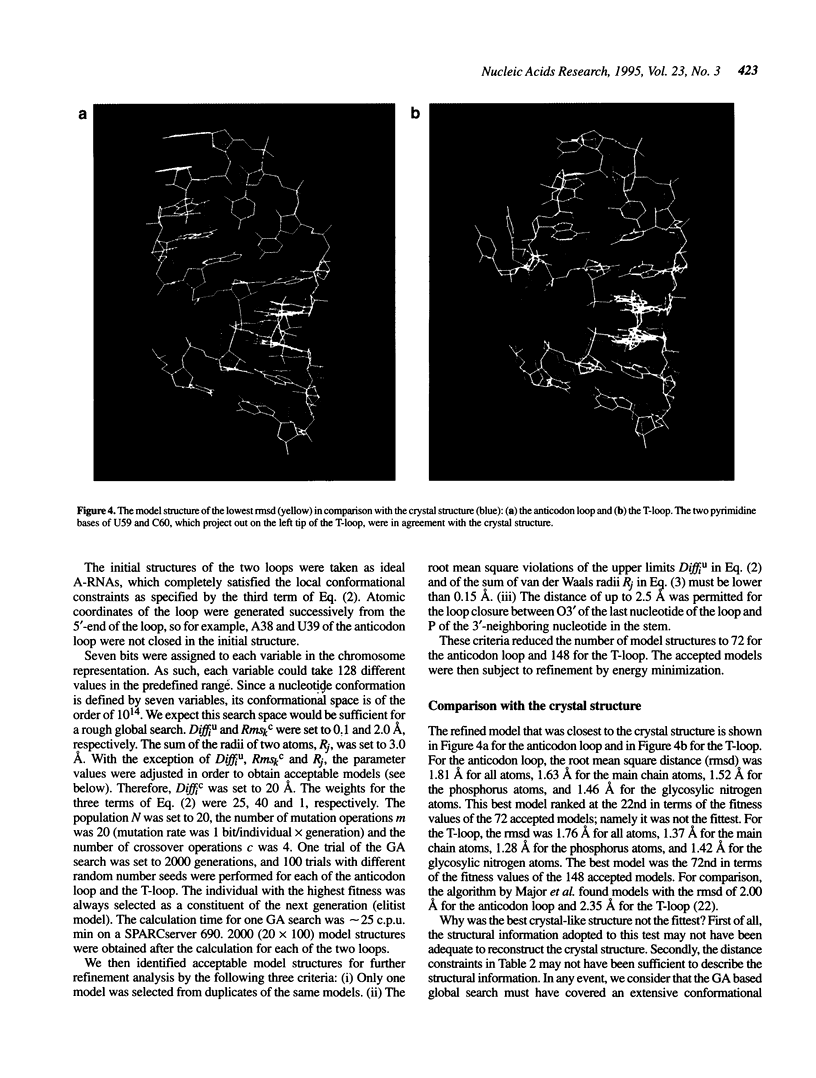
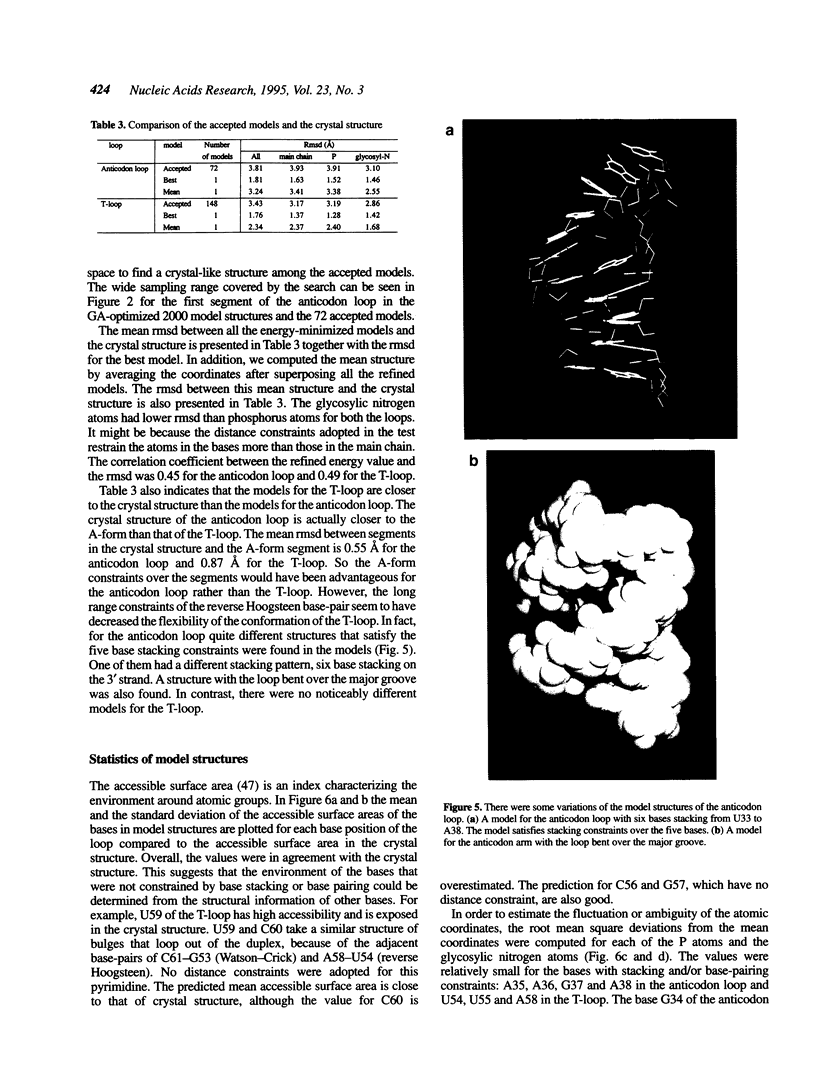
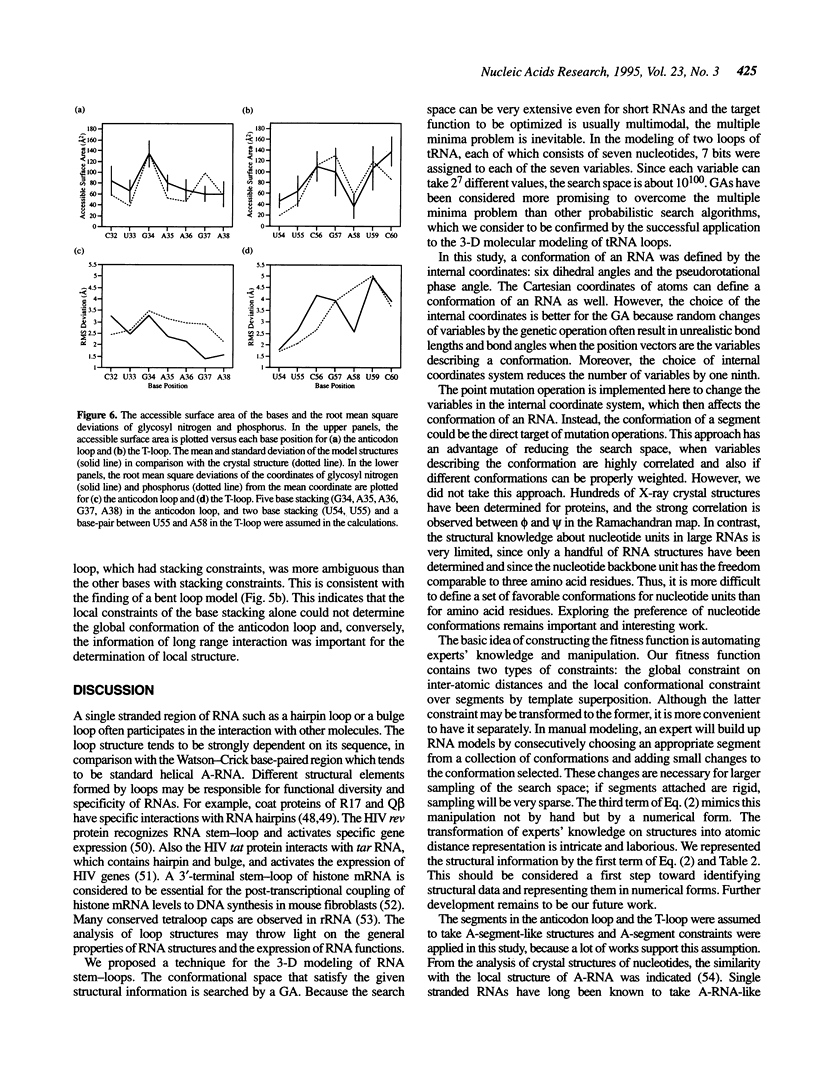

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achter E. K., Felsenfeld G. The conformation of single-strand polynucleotides in solution: sedimentation studies of apurinic acid. Biopolymers. 1971;10(9):1625–1634. doi: 10.1002/bip.360100916. [DOI] [PubMed] [Google Scholar]
- Amano M., Kawakami M. Assignment of the magnetic resonances of the imino protons and methyl protons of Bombyx mori tRNA(GlyGCC) and the effect of ion binding on its structure. Eur J Biochem. 1992 Dec 15;210(3):671–681. doi: 10.1111/j.1432-1033.1992.tb17468.x. [DOI] [PubMed] [Google Scholar]
- Benedetti G., Morosetti S. Three-dimensional folding of Tetrahymena thermophila rRNA IVS sequence: a proposal. J Biomol Struct Dyn. 1991 Apr;8(5):1045–1055. doi: 10.1080/07391102.1991.10507864. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Brimacombe R., Atmadja J., Stiege W., Schüler D. A detailed model of the three-dimensional structure of Escherichia coli 16 S ribosomal RNA in situ in the 30 S subunit. J Mol Biol. 1988 Jan 5;199(1):115–136. doi: 10.1016/0022-2836(88)90383-x. [DOI] [PubMed] [Google Scholar]
- Brunel C., Romby P., Westhof E., Ehresmann C., Ehresmann B. Three-dimensional model of Escherichia coli ribosomal 5 S RNA as deduced from structure probing in solution and computer modeling. J Mol Biol. 1991 Sep 5;221(1):293–308. doi: 10.1016/0022-2836(91)80220-o. [DOI] [PubMed] [Google Scholar]
- Cheong C., Varani G., Tinoco I., Jr Solution structure of an unusually stable RNA hairpin, 5'GGAC(UUCG)GUCC. Nature. 1990 Aug 16;346(6285):680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- Chu W. C., Kintanar A., Horowitz J. Correlations between fluorine-19 nuclear magnetic resonance chemical shift and the secondary and tertiary structure of 5-fluorouracil-substituted tRNA. J Mol Biol. 1992 Oct 20;227(4):1173–1181. doi: 10.1016/0022-2836(92)90529-s. [DOI] [PubMed] [Google Scholar]
- Dandekar T., Argos P. Potential of genetic algorithms in protein folding and protein engineering simulations. Protein Eng. 1992 Oct;5(7):637–645. doi: 10.1093/protein/5.7.637. [DOI] [PubMed] [Google Scholar]
- Davis P. W., Thurmes W., Tinoco I., Jr Structure of a small RNA hairpin. Nucleic Acids Res. 1993 Feb 11;21(3):537–545. doi: 10.1093/nar/21.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dock-Bregeon A. C., Westhof E., Giegé R., Moras D. Solution structure of a tRNA with a large variable region: yeast tRNASer. J Mol Biol. 1989 Apr 20;206(4):707–722. doi: 10.1016/0022-2836(89)90578-0. [DOI] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Fuller W., Hodgson A. Conformation of the anticodon loop intRNA. Nature. 1967 Aug 19;215(5103):817–821. doi: 10.1038/215817a0. [DOI] [PubMed] [Google Scholar]
- Gabb H. A., Harris M. E., Pandey N. B., Marzluff W. F., Harvey S. C. Molecular modeling to predict the structural and biological effects of mutations in a highly conserved histone mRNA loop sequence. J Biomol Struct Dyn. 1992 Jun;9(6):1119–1130. doi: 10.1080/07391102.1992.10507983. [DOI] [PubMed] [Google Scholar]
- Gautheret D., Cedergren R. Modeling the three-dimensional structure of RNA. FASEB J. 1993 Jan;7(1):97–105. doi: 10.1096/fasebj.7.1.7678567. [DOI] [PubMed] [Google Scholar]
- Gautheret D., Major F., Cedergren R. Modeling the three-dimensional structure of RNA using discrete nucleotide conformational sets. J Mol Biol. 1993 Feb 20;229(4):1049–1064. doi: 10.1006/jmbi.1993.1104. [DOI] [PubMed] [Google Scholar]
- Gulik A., Inoue H., Luzzati V. Conformation of single-stranded polynucleotides: small-angle x-ray scattering and spectroscopic study of polyribocytidylic acid in water and in water-alcohol solutions. J Mol Biol. 1970 Oct 28;53(2):221–238. doi: 10.1016/0022-2836(70)90296-2. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Hubbard J. M., Hearst J. E. Computer modeling 16 S ribosomal RNA. J Mol Biol. 1991 Oct 5;221(3):889–907. doi: 10.1016/0022-2836(91)80182-t. [DOI] [PubMed] [Google Scholar]
- Hubbard J. M., Hearst J. E. Predicting the three-dimensional folding of transfer RNA with a computer modeling protocol. Biochemistry. 1991 Jun 4;30(22):5458–5465. doi: 10.1021/bi00236a019. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Westhof E., Michel F. Function of P11, a tertiary base pairing in self-splicing introns of subgroup IA. J Mol Biol. 1991 Oct 20;221(4):1153–1164. doi: 10.1016/0022-2836(91)90925-v. [DOI] [PubMed] [Google Scholar]
- Kajava A., Rüterjans H. Molecular modelling of the 3-D structure of RNA tetraloops with different nucleotide sequences. Nucleic Acids Res. 1993 Sep 25;21(19):4556–4562. doi: 10.1093/nar/21.19.4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Quigley G. J., Suddath F. L., McPherson A., Sneden D., Kim J. J., Weinzierl J., Rich A. Three-dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science. 1973 Jan 19;179(4070):285–288. doi: 10.1126/science.179.4070.285. [DOI] [PubMed] [Google Scholar]
- Krol A., Westhof E., Bach M., Lührmann R., Ebel J. P., Carbon P. Solution structure of human U1 snRNA. Derivation of a possible three-dimensional model. Nucleic Acids Res. 1990 Jul 11;18(13):3803–3811. doi: 10.1093/nar/18.13.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. J., Chodchoy N., Marzluff W. F., Skoultchi A. I. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem-loop sequence at the 3' end of the mRNA. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major F., Gautheret D., Cedergren R. Reproducing the three-dimensional structure of a tRNA molecule from structural constraints. Proc Natl Acad Sci U S A. 1993 Oct 15;90(20):9408–9412. doi: 10.1073/pnas.90.20.9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major F., Turcotte M., Gautheret D., Lapalme G., Fillion E., Cedergren R. The combination of symbolic and numerical computation for three-dimensional modeling of RNA. Science. 1991 Sep 13;253(5025):1255–1260. doi: 10.1126/science.1716375. [DOI] [PubMed] [Google Scholar]
- Malhotra A., Tan R. K., Harvey S. C. Prediction of the three-dimensional structure of Escherichia coli 30S ribosomal subunit: a molecular mechanics approach. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1950–1954. doi: 10.1073/pnas.87.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei H. Y., Kaaret T. W., Bruice T. C. A computational approach to the mechanism of self-cleavage of hammerhead RNA. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9727–9731. doi: 10.1073/pnas.86.24.9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Moodie S. L., Thornton J. M. A study into the effects of protein binding on nucleotide conformation. Nucleic Acids Res. 1993 Mar 25;21(6):1369–1380. doi: 10.1093/nar/21.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Nikonowicz E. P., Pardi A. Three-dimensional heteronuclear NMR studies of RNA. Nature. 1992 Jan 9;355(6356):184–186. doi: 10.1038/355184a0. [DOI] [PubMed] [Google Scholar]
- Olsen H. S., Nelbock P., Cochrane A. W., Rosen C. A. Secondary structure is the major determinant for interaction of HIV rev protein with RNA. Science. 1990 Feb 16;247(4944):845–848. doi: 10.1126/science.2406903. [DOI] [PubMed] [Google Scholar]
- Richmond T. J. Solvent accessible surface area and excluded volume in proteins. Analytical equations for overlapping spheres and implications for the hydrophobic effect. J Mol Biol. 1984 Sep 5;178(1):63–89. doi: 10.1016/0022-2836(84)90231-6. [DOI] [PubMed] [Google Scholar]
- Romaniuk P. J., Lowary P., Wu H. N., Stormo G., Uhlenbeck O. C. RNA binding site of R17 coat protein. Biochemistry. 1987 Mar 24;26(6):1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Sun S. Reduced representation model of protein structure prediction: statistical potential and genetic algorithms. Protein Sci. 1993 May;2(5):762–785. doi: 10.1002/pro.5560020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unger R., Moult J. Genetic algorithms for protein folding simulations. J Mol Biol. 1993 May 5;231(1):75–81. doi: 10.1006/jmbi.1993.1258. [DOI] [PubMed] [Google Scholar]
- Veal J. M., Wilson W. D. Modeling of nucleic acid complexes with cationic ligands: a specialized molecular mechanics force field and its application. J Biomol Struct Dyn. 1991 Jun;8(6):1119–1145. doi: 10.1080/07391102.1991.10507875. [DOI] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- Witherell G. W., Uhlenbeck O. C. Specific RNA binding by Q beta coat protein. Biochemistry. 1989 Jan 10;28(1):71–76. doi: 10.1021/bi00427a011. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Winker S., Gutell R. R. Architecture of ribosomal RNA: constraints on the sequence of "tetra-loops". Proc Natl Acad Sci U S A. 1990 Nov;87(21):8467–8471. doi: 10.1073/pnas.87.21.8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Yao S., Wilson W. D. A molecular mechanics investigation of RNA complexes. I. Ethidium intercalation in an HIV-1 TAR RNA sequence with an unpaired adenosine. J Biomol Struct Dyn. 1992 Oct;10(2):367–387. doi: 10.1080/07391102.1992.10508653. [DOI] [PubMed] [Google Scholar]