Abstract
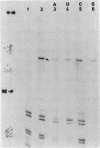
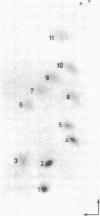
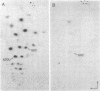
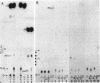
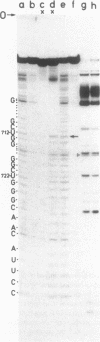
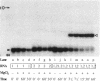
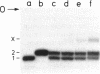
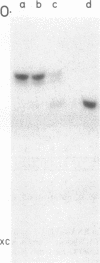
Genomic RNA of the hepatitis delta agent has a highly conserved element of local tertiary structure. This element contains two nucleotides which become covalently crosslinked to each other upon irradiation with UV light. Using direct RNA analysis, we now identify the two nucleotides as U-712 and U-865 and show that the UV-induced crosslink can be broken by re-exposure to a 254 nm peak UV light source. In the rod-like secondary structural model of delta RNA, nucleotides U-712 and U-865 are off-set from each other by 5-6 bases, a distance too great to permit crosslinking. This model needs to be modified. Our data indicate that bases U-712 and U-865 closely approximate each other and suggest that the smooth helical contour proposed for delta RNA is interrupted by the UV-sensitive element. The nucleotide sequence shows that the UV-sensitive site does not have a particularly high density of conventional Watson-Crick base pairs compared to the rest of the genome. However, this element may have a number of non-Watson-Crick bonds which confer stability. Following UV-crosslinking and digestion with 1 mg/ml of RNase T1 at 37 degrees C for 45 min in 10 mM Tris-HCl, 1 mM EDTA (conditions expected to give complete digestion), this element can be isolated as part of a 54 nucleotide long partial digestion product containing at least 16 internal G residues. UV-crosslinking analysis shows that this unusual tertiary structural element can form in a bimolecular complex.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Romaniuk P. J., Bakken A. H. RNA-protein interactions of stored 5S RNA with TFIIIA and ribosomal protein L5 during Xenopus oogenesis. Dev Biol. 1991 Mar;144(1):129–144. doi: 10.1016/0012-1606(91)90485-l. [DOI] [PubMed] [Google Scholar]
- Atmadja J., Brimacombe R., Blöcker H., Frank R. Investigation of the tertiary folding of Escherichia coli 16S RNA by in situ intra-RNA cross-linking within 30S ribosomal subunits. Nucleic Acids Res. 1985 Oct 11;13(19):6919–6936. doi: 10.1093/nar/13.19.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlen L. S., Sampson J. R., Uhlenbeck O. C. An ultraviolet light-induced crosslink in yeast tRNA(Phe). Nucleic Acids Res. 1992 Aug 11;20(15):4055–4059. doi: 10.1093/nar/20.15.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Baroudy B. M., Wells F. V., Gerin J. L., Robertson H. D. An ultraviolet-sensitive RNA structural element in a viroid-like domain of the hepatitis delta virus. Science. 1989 Feb 3;243(4891):649–652. doi: 10.1126/science.2492676. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Paul C. P., Robertson H. D. Analysis of ultraviolet-induced RNA-RNA cross-links: a means for probing RNA structure-function relationships. Methods Enzymol. 1989;180:418–442. doi: 10.1016/0076-6879(89)80115-6. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Robertson H. D. RNA fingerprinting. Methods Enzymol. 1989;180:130–154. doi: 10.1016/0076-6879(89)80098-9. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Benenfeld B. J., Robertson H. D. Ultraviolet light-induced crosslinking reveals a unique region of local tertiary structure in potato spindle tuber viroid and HeLa 5S RNA. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6590–6594. doi: 10.1073/pnas.82.19.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch A. D., Lee S. E., Neel O. D., Robertson H. D. Prominent polypurine and polypyrimidine tracts in plant viroids and in RNA of the human hepatitis delta agent. Nucleic Acids Res. 1993 Jul 25;21(15):3529–3535. doi: 10.1093/nar/21.15.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch A. D., Robertson H. D. A replication cycle for viroids and other small infectious RNA's. Science. 1984 Feb 3;223(4635):450–455. doi: 10.1126/science.6197756. [DOI] [PubMed] [Google Scholar]
- Branch A. D., Robertson H. D. Efficient trans cleavage and a common structural motif for the ribozymes of the human hepatitis delta agent. Proc Natl Acad Sci U S A. 1991 Nov 15;88(22):10163–10167. doi: 10.1073/pnas.88.22.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey J. L., Bergmann K. F., Brown T. L., Gerin J. L. Structural requirements for RNA editing in hepatitis delta virus: evidence for a uridine-to-cytidine editing mechanism. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):7149–7153. doi: 10.1073/pnas.89.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M. F., Baker S. C., Soe L. H., Kamahora T., Keck J. G., Makino S., Govindarajan S., Lai M. M. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J Virol. 1988 Jul;62(7):2403–2410. doi: 10.1128/jvi.62.7.2403-2410.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M., Hsieh S. Y., Taylor J. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J Virol. 1990 Oct;64(10):5066–5069. doi: 10.1128/jvi.64.10.5066-5069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M., Hsieh S. Y., Taylor J. The antigen of hepatitis delta virus: examination of in vitro RNA-binding specificity. J Virol. 1991 Aug;65(8):4057–4062. doi: 10.1128/jvi.65.8.4057-4062.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P. J., Kalpana G., Goldberg J., Mason W., Werner B., Gerin J., Taylor J. Structure and replication of the genome of the hepatitis delta virus. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8774–8778. doi: 10.1073/pnas.83.22.8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C., Moore P. B. Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures. Biochemistry. 1992 Sep 15;31(36):8406–8414. doi: 10.1021/bi00151a003. [DOI] [PubMed] [Google Scholar]
- Cruse W. B., Saludjian P., Biala E., Strazewski P., Prangé T., Kennard O. Structure of a mispaired RNA double helix at 1.6-A resolution and implications for the prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 1994 May 10;91(10):4160–4164. doi: 10.1073/pnas.91.10.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsillo P., Huber P. W. The use of chemical nucleases to analyze RNA-protein interactions. The TFIIIA-5 S rRNA complex. J Biol Chem. 1991 Nov 5;266(31):21075–21082. [PubMed] [Google Scholar]
- Doudna J. A., Grosshans C., Gooding A., Kundrot C. E. Crystallization of ribozymes and small RNA motifs by a sparse matrix approach. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7829–7833. doi: 10.1073/pnas.90.16.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs W. D., Cech T. R. An ultraviolet-inducible adenosine-adenosine cross-link reflects the catalytic structure of the Tetrahymena ribozyme. Biochemistry. 1990 Jun 12;29(23):5605–5613. doi: 10.1021/bi00475a027. [DOI] [PubMed] [Google Scholar]
- Ellington A. D. RNA ligands: out of shape but fir for recognition. Curr Biol. 1993 Jun 1;3(6):375–377. doi: 10.1016/0960-9822(93)90206-4. [DOI] [PubMed] [Google Scholar]
- Farci P., Mandas A., Coiana A., Lai M. E., Desmet V., Van Eyken P., Gibo Y., Caruso L., Scaccabarozzi S., Criscuolo D. Treatment of chronic hepatitis D with interferon alfa-2a. N Engl J Med. 1994 Jan 13;330(2):88–94. doi: 10.1056/NEJM199401133300202. [DOI] [PubMed] [Google Scholar]
- Feldstein P. A., Buzayan J. M., Bruening G. Two sequences participating in the autolytic processing of satellite tobacco ringspot virus complementary RNA. Gene. 1989 Oct 15;82(1):53–61. doi: 10.1016/0378-1119(89)90029-2. [DOI] [PubMed] [Google Scholar]
- Francki R. I. Plant virus satellites. Annu Rev Microbiol. 1985;39:151–174. doi: 10.1146/annurev.mi.39.100185.001055. [DOI] [PubMed] [Google Scholar]
- Gottlieb P. A., Prasad Y., Smith J. B., Williams A. P., Dinter-Gottlieb G. Evidence that alternate foldings of the hepatitis delta RNA confer varying rates of self-cleavage. Biochemistry. 1994 Mar 15;33(10):2802–2808. doi: 10.1021/bi00176a008. [DOI] [PubMed] [Google Scholar]
- Green M. R. RNA bent for recognition. Curr Biol. 1991 Aug;1(4):245–247. doi: 10.1016/0960-9822(91)90071-4. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Lumelsky N., Altman S. Specific interactions in RNA enzyme-substrate complexes. Science. 1989 Dec 22;246(4937):1578–1584. doi: 10.1126/science.2480641. [DOI] [PubMed] [Google Scholar]
- Hannon G. J., Maroney P. A., Branch A., Benenfield B. J., Robertson H. D., Nilsen T. W. Accurate processing of human pre-rRNA in vitro. Mol Cell Biol. 1989 Oct;9(10):4422–4431. doi: 10.1128/mcb.9.10.4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holbrook S. R., Cheong C., Tinoco I., Jr, Kim S. H. Crystal structure of an RNA double helix incorporating a track of non-Watson-Crick base pairs. Nature. 1991 Oct 10;353(6344):579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- Hutchins C. J., Rathjen P. D., Forster A. C., Symons R. H. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986 May 12;14(9):3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J. A., SantaLucia J., Jr, Tinoco I., Jr Determination of RNA structure and thermodynamics. Annu Rev Biochem. 1993;62:255–287. doi: 10.1146/annurev.bi.62.070193.001351. [DOI] [PubMed] [Google Scholar]
- Kos A., Dijkema R., Arnberg A. C., van der Meide P. H., Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986 Oct 9;323(6088):558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- Kuo M. Y., Sharmeen L., Dinter-Gottlieb G., Taylor J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J Virol. 1988 Dec;62(12):4439–4444. doi: 10.1128/jvi.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Z., Lin J. H., Chao M., McKnight K., Lai M. M. RNA-binding activity of hepatitis delta antigen involves two arginine-rich motifs and is required for hepatitis delta virus RNA replication. J Virol. 1993 Apr;67(4):2221–2227. doi: 10.1128/jvi.67.4.2221-2227.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. H., Chang M. F., Baker S. C., Govindarajan S., Lai M. M. Characterization of hepatitis delta antigen: specific binding to hepatitis delta virus RNA. J Virol. 1990 Sep;64(9):4051–4058. doi: 10.1128/jvi.64.9.4051-4058.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G. X., Chao M., Hsieh S. Y., Sureau C., Nishikura K., Taylor J. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol. 1990 Mar;64(3):1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J., Favre A., Yaniv M. Molecular model for transfer RNA. Nature. 1969 Sep 27;223(5213):1333–1335. doi: 10.1038/2231333a0. [DOI] [PubMed] [Google Scholar]
- Ofengand J., Liou R. Evidence for pyrimidine-pyrimidine cyclobutane dimer formation in the covalent cross-linking between transfer ribonucleic acid and 16S ribonucleic acid at the ribosomal P site. Biochemistry. 1980 Oct 14;19(21):4814–4822. doi: 10.1021/bi00562a016. [DOI] [PubMed] [Google Scholar]
- Paul C. P., Levine B. J., Robertson H. D., Branch A. D. Transcripts of the viroid central conserved region contain the local tertiary structural element found in full-length viroid. FEBS Lett. 1992 Jun 22;305(1):9–14. doi: 10.1016/0014-5793(92)80644-v. [DOI] [PubMed] [Google Scholar]
- Perrotta A. T., Been M. D. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature. 1991 Apr 4;350(6317):434–436. doi: 10.1038/350434a0. [DOI] [PubMed] [Google Scholar]
- Polish L. B., Gallagher M., Fields H. A., Hadler S. C. Delta hepatitis: molecular biology and clinical and epidemiological features. Clin Microbiol Rev. 1993 Jul;6(3):211–229. doi: 10.1128/cmr.6.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzetto A., Hoyer B. H., Popper H., Engle R., Purcell R. H., Gerin J. L. Titration of the infectivity of hepatitis D virus in chimpanzees. J Infect Dis. 1987 Jan;155(1):72–78. doi: 10.1093/infdis/155.1.72. [DOI] [PubMed] [Google Scholar]
- Prince J. B., Taylor B. H., Thurlow D. L., Ofengand J., Zimmermann R. A. Covalent crosslinking of tRNA1Val to 16S RNA at the ribosomal P site: identification of crosslinked residues. Proc Natl Acad Sci U S A. 1982 Sep;79(18):5450–5454. doi: 10.1073/pnas.79.18.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prody G. A., Bakos J. T., Buzayan J. M., Schneider I. R., Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986 Mar 28;231(4745):1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- Riesner D., Henco K., Rokohl U., Klotz G., Kleinschmidt A. K., Domdey H., Jank P., Gross H. J., Sänger H. L. Structure and structure formation of viroids. J Mol Biol. 1979 Sep 5;133(1):85–115. doi: 10.1016/0022-2836(79)90252-3. [DOI] [PubMed] [Google Scholar]
- Rizzetto M., Canese M. G., Aricò S., Crivelli O., Trepo C., Bonino F., Verme G. Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers. Gut. 1977 Dec;18(12):997–1003. doi: 10.1136/gut.18.12.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaniuk P. J. The role of highly conserved single-stranded nucleotides of Xenopus 5S RNA in the binding of transcription factor IIIA. Biochemistry. 1989 Feb 7;28(3):1388–1395. doi: 10.1021/bi00429a067. [DOI] [PubMed] [Google Scholar]
- Sharmeen L., Kuo M. Y., Dinter-Gottlieb G., Taylor J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J Virol. 1988 Aug;62(8):2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y. A., Kumar P. K., Kawakami J., Nishikawa F., Taira K., Nishikawa S. Systematic substitution of individual bases in two important single-stranded regions of the HDV ribozyme for evaluation of the role of specific bases. FEBS Lett. 1993 Jul 12;326(1-3):158–162. doi: 10.1016/0014-5793(93)81782-u. [DOI] [PubMed] [Google Scholar]
- Symons R. H. The intriguing viroids and virusoids: what is their information content and how did they evolve? Mol Plant Microbe Interact. 1991 Mar-Apr;4(2):111–121. doi: 10.1094/mpmi-4-111. [DOI] [PubMed] [Google Scholar]
- Tabler M., Schnölzer M., Sänger H. L. Molecular cloning of potato spindle tuber viroid (PSTV) cDNA synthesized by enzymatic elongation of PSTV-specific DNA primers: a general strategy for viroid cloning. Biosci Rep. 1985 Feb;5(2):143–158. doi: 10.1007/BF01117061. [DOI] [PubMed] [Google Scholar]
- Tanner N. K., Schaff S., Thill G., Petit-Koskas E., Crain-Denoyelle A. M., Westhof E. A three-dimensional model of hepatitis delta virus ribozyme based on biochemical and mutational analyses. Curr Biol. 1994 Jun 1;4(6):488–498. doi: 10.1016/s0960-9822(00)00109-3. [DOI] [PubMed] [Google Scholar]
- Taylor J. M. The structure and replication of hepatitis delta virus. Annu Rev Microbiol. 1992;46:253–276. doi: 10.1146/annurev.mi.46.100192.001345. [DOI] [PubMed] [Google Scholar]
- Wang K. S., Choo Q. L., Weiner A. J., Ou J. H., Najarian R. C., Thayer R. M., Mullenbach G. T., Denniston K. J., Gerin J. L., Houghton M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature. 1986 Oct 9;323(6088):508–514. doi: 10.1038/323508a0. [DOI] [PubMed] [Google Scholar]
- Weiner A. J., Choo Q. L., Wang K. S., Govindarajan S., Redeker A. G., Gerin J. L., Houghton M. A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta. J Virol. 1988 Feb;62(2):594–599. doi: 10.1128/jvi.62.2.594-599.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly B., Varani G., Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993 Feb 2;32(4):1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- Wu H. N., Lin Y. J., Lin F. P., Makino S., Chang M. F., Lai M. M. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv M., Favre A., Barrell B. G. Structure of transfer RNA. Evidence for interaction between two non-adjacent nucleotide residues in tRNA from Escherichia coli. Nature. 1969 Sep 27;223(5213):1331–1333. doi: 10.1038/2231331a0. [DOI] [PubMed] [Google Scholar]
- Zheng H., Fu T. B., Lazinski D., Taylor J. Editing on the genomic RNA of human hepatitis delta virus. J Virol. 1992 Aug;66(8):4693–4697. doi: 10.1128/jvi.66.8.4693-4697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Schüler D. Low resolution three-dimensional models of the 7SL RNA of the signal recognition particle, based on an intramolecular cross-link introduced by mild irradiation with ultraviolet light. Biochem Cell Biol. 1989 Aug;67(8):434–442. doi: 10.1139/o89-069. [DOI] [PubMed] [Google Scholar]
- di Bisceglie A. M. Interferon therapy for chronic viral hepatitis. N Engl J Med. 1994 Jan 13;330(2):137–138. doi: 10.1056/NEJM199401133300211. [DOI] [PubMed] [Google Scholar]