Abstract
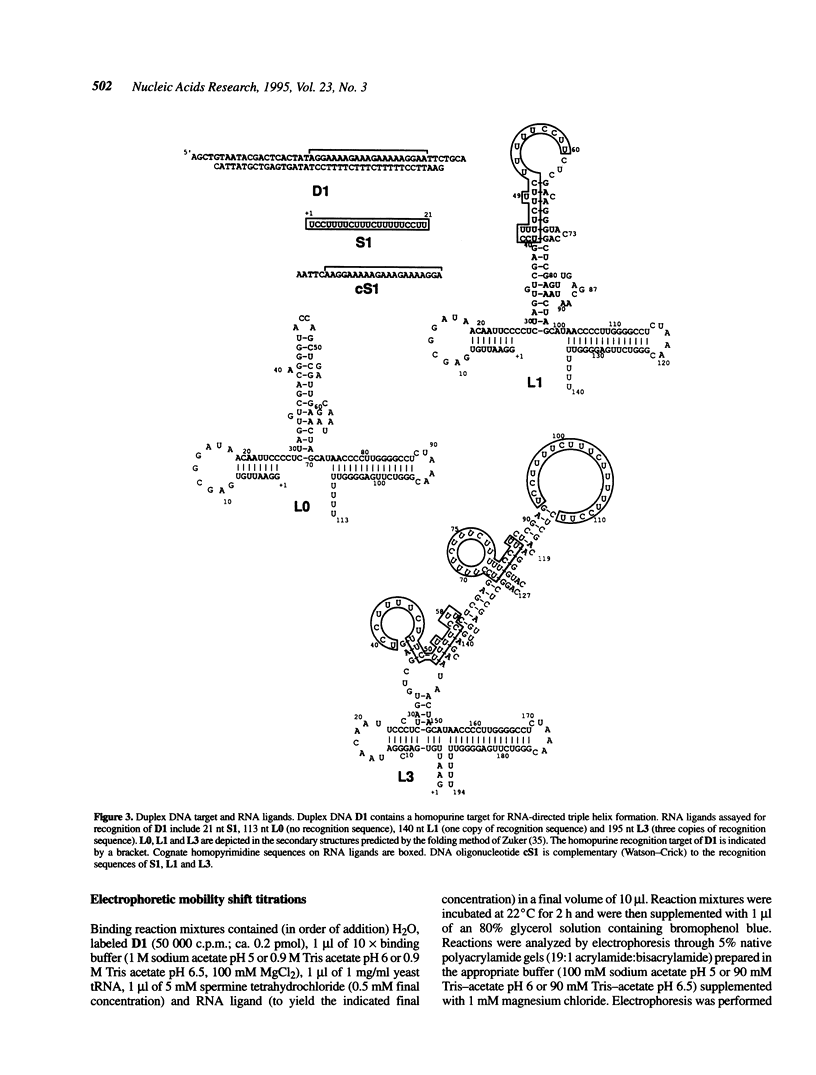
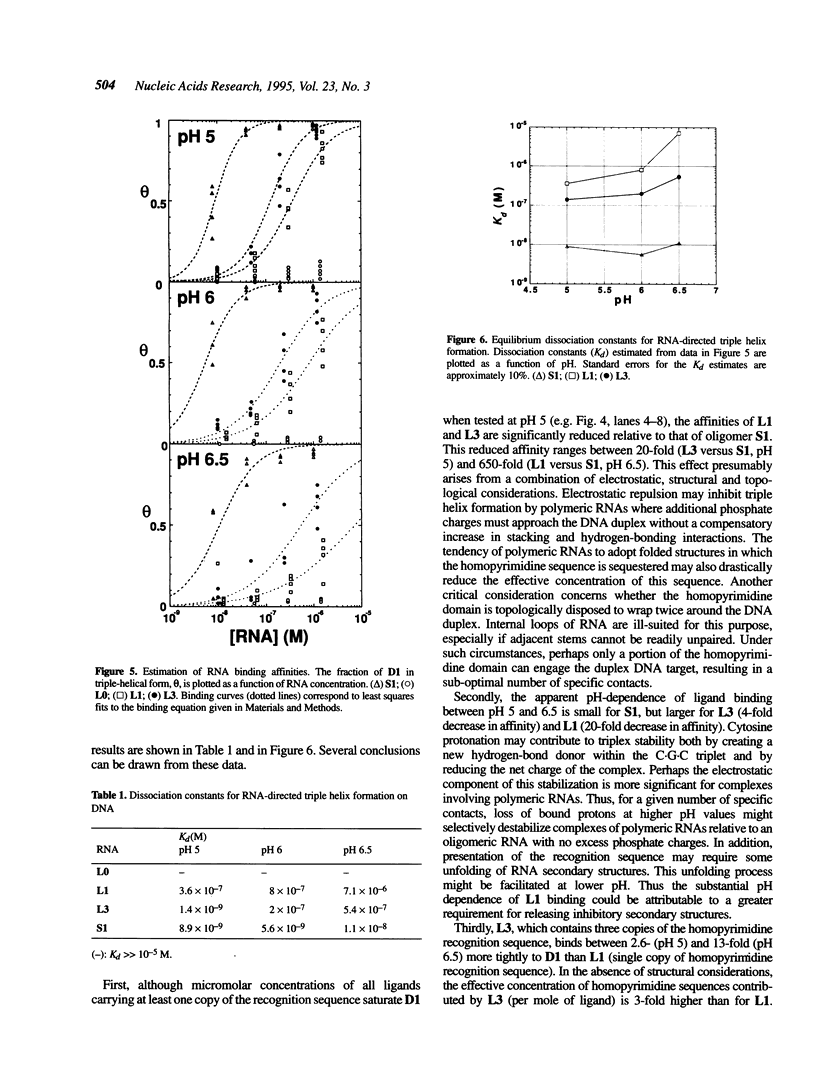
We are interested in creating artificial gene repressors based on duplex DNA recognition by nucleic acids. Homopyrimidine RNA oligonucleotides bind to duplex DNA at homopurine/homopyrimidine sequences under slightly acidic conditions. Recognition is sequence-specific, involving rU.dA.dT and rC+.dG.dC base triplets. Affinities were determined for folded polymeric RNAs (ca. 100-200 nt) containing 0, 1 or 3 copies of a 21 nt RNA sequence that binds duplex DNA by triple helix formation. When this recognition sequence was inserted into the larger folded RNAs, micromolar concentrations of the resulting RNA ligands bound a duplex DNA target at pH 5. However, these binding affinities were at least 20-fold lower than the affinity of an RNA oligonucleotide containing only the recognition sequence. Enzymatic probing of folded RNAs suggests that reduced affinity arises from unfavorable electrostatic, structural and topological considerations. The affinity of a polymeric RNA with three copies of the recognition sequence was greater than that of a polymeric RNA with a single copy of the sequence. This affinity difference ranged from 2.6- to 13-fold, depending on pH. Binding of duplex DNA by polymeric RNA might be improved by optimizing the RNA structure to efficiently present the recognition sequence.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beal P. A., Dervan P. B. Second structural motif for recognition of DNA by oligonucleotide-directed triple-helix formation. Science. 1991 Mar 15;251(4999):1360–1363. doi: 10.1126/science.2003222. [DOI] [PubMed] [Google Scholar]
- Beru N., Smith D., Goldwasser E. Evidence suggesting negative regulation of the erythropoietin gene by ribonucleoprotein. J Biol Chem. 1990 Aug 25;265(24):14100–14104. [PubMed] [Google Scholar]
- Birg F., Praseuth D., Zerial A., Thuong N. T., Asseline U., Le Doan T., Hélène C. Inhibition of simian virus 40 DNA replication in CV-1 cells by an oligodeoxynucleotide covalently linked to an intercalating agent. Nucleic Acids Res. 1990 May 25;18(10):2901–2908. doi: 10.1093/nar/18.10.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney M., Czernuszewicz G., Postel E. H., Flint S. J., Hogan M. E. Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science. 1988 Jul 22;241(4864):456–459. doi: 10.1126/science.3293213. [DOI] [PubMed] [Google Scholar]
- Davis T. L., Firulli A. B., Kinniburgh A. J. Ribonucleoprotein and protein factors bind to an H-DNA-forming c-myc DNA element: possible regulators of the c-myc gene. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9682–9686. doi: 10.1073/pnas.86.24.9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durland R. H., Kessler D. J., Gunnell S., Duvic M., Pettitt B. M., Hogan M. E. Binding of triple helix forming oligonucleotides to sites in gene promoters. Biochemistry. 1991 Sep 24;30(38):9246–9255. doi: 10.1021/bi00102a017. [DOI] [PubMed] [Google Scholar]
- Escudé C., François J. C., Sun J. S., Ott G., Sprinzl M., Garestier T., Hélène C. Stability of triple helices containing RNA and DNA strands: experimental and molecular modeling studies. Nucleic Acids Res. 1993 Dec 11;21(24):5547–5553. doi: 10.1093/nar/21.24.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François J. C., Saison-Behmoaras T., Thuong N. T., Hélène C. Inhibition of restriction endonuclease cleavage via triple helix formation by homopyrimidine oligonucleotides. Biochemistry. 1989 Dec 12;28(25):9617–9619. doi: 10.1021/bi00451a011. [DOI] [PubMed] [Google Scholar]
- Goddard J. P. The structures and functions of transfer RNA. Prog Biophys Mol Biol. 1977;32(3):233–308. [PubMed] [Google Scholar]
- Han H., Dervan P. B. Sequence-specific recognition of double helical RNA and RNA.DNA by triple helix formation. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3806–3810. doi: 10.1073/pnas.90.9.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanvey J. C., Shimizu M., Wells R. D. Site-specific inhibition of EcoRI restriction/modification enzymes by a DNA triple helix. Nucleic Acids Res. 1990 Jan 11;18(1):157–161. doi: 10.1093/nar/18.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hélène C. Rational design of sequence-specific oncogene inhibitors based on antisense and antigene oligonucleotides. Eur J Cancer. 1991;27(11):1466–1471. doi: 10.1016/0277-5379(91)90033-a. [DOI] [PubMed] [Google Scholar]
- Ing N. H., Beekman J. M., Kessler D. J., Murphy M., Jayaraman K., Zendegui J. G., Hogan M. E., O'Malley B. W., Tsai M. J. In vivo transcription of a progesterone-responsive gene is specifically inhibited by a triplex-forming oligonucleotide. Nucleic Acids Res. 1993 Jun 25;21(12):2789–2796. doi: 10.1093/nar/21.12.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohwi Y., Malkhosyan S. R., Kohwi-Shigematsu T. Intramolecular dG.dG.dC triplex detected in Escherichia coli cells. J Mol Biol. 1992 Feb 20;223(4):817–822. doi: 10.1016/0022-2836(92)90242-c. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd DNA triple-helix formation: an approach to artificial gene repressors? Bioessays. 1992 Dec;14(12):807–815. doi: 10.1002/bies.950141204. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Dervan P. B., Wold B. J. Kinetic analysis of oligodeoxyribonucleotide-directed triple-helix formation on DNA. Biochemistry. 1990 Sep 18;29(37):8820–8826. doi: 10.1021/bi00489a045. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Dervan P. B., Wold B. Analysis of promoter-specific repression by triple-helical DNA complexes in a eukaryotic cell-free transcription system. Biochemistry. 1992 Jan 14;31(1):70–81. doi: 10.1021/bi00116a012. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd Inhibition of T7 RNA polymerase initiation by triple-helical DNA complexes: a model for artificial gene repression. Biochemistry. 1992 Aug 25;31(33):7587–7594. doi: 10.1021/bi00148a021. [DOI] [PubMed] [Google Scholar]
- Maher L. J., 3rd, Wold B., Dervan P. B. Inhibition of DNA binding proteins by oligonucleotide-directed triple helix formation. Science. 1989 Aug 18;245(4919):725–730. doi: 10.1126/science.2549631. [DOI] [PubMed] [Google Scholar]
- Michel F., Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990 Dec 5;216(3):585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Sobell H. M. A molecular model for gene repression. Proc Natl Acad Sci U S A. 1966 May;55(5):1201–1205. doi: 10.1073/pnas.55.5.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton K. W. The triple helix: a potential mechanism for gene regulation. J Exp Pathol. 1985 Fall;2(3):135–148. [PubMed] [Google Scholar]
- Moser H. E., Dervan P. B. Sequence-specific cleavage of double helical DNA by triple helix formation. Science. 1987 Oct 30;238(4827):645–650. doi: 10.1126/science.3118463. [DOI] [PubMed] [Google Scholar]
- Orson F. M., Thomas D. W., McShan W. M., Kessler D. J., Hogan M. E. Oligonucleotide inhibition of IL2R alpha mRNA transcription by promoter region collinear triplex formation in lymphocytes. Nucleic Acids Res. 1991 Jun 25;19(12):3435–3441. doi: 10.1093/nar/19.12.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plum G. E., Park Y. W., Singleton S. F., Dervan P. B., Breslauer K. J. Thermodynamic characterization of the stability and the melting behavior of a DNA triplex: a spectroscopic and calorimetric study. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9436–9440. doi: 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel E. H., Flint S. J., Kessler D. J., Hogan M. E. Evidence that a triplex-forming oligodeoxyribonucleotide binds to the c-myc promoter in HeLa cells, thereby reducing c-myc mRNA levels. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8227–8231. doi: 10.1073/pnas.88.18.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. W., Crothers D. M. Stability and properties of double and triple helices: dramatic effects of RNA or DNA backbone composition. Science. 1992 Nov 27;258(5087):1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- Semerad C. L., Maher L. J., 3rd Exclusion of RNA strands from a purine motif triple helix. Nucleic Acids Res. 1994 Dec 11;22(24):5321–5325. doi: 10.1093/nar/22.24.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton S. F., Dervan P. B. Influence of pH on the equilibrium association constants for oligodeoxyribonucleotide-directed triple helix formation at single DNA sites. Biochemistry. 1992 Nov 17;31(45):10995–11003. doi: 10.1021/bi00160a008. [DOI] [PubMed] [Google Scholar]
- Skoog J. U., Maher L. J., 3rd Repression of bacteriophage promoters by DNA and RNA oligonucleotides. Nucleic Acids Res. 1993 May 11;21(9):2131–2138. doi: 10.1093/nar/21.9.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]