Abstract
Despite partial sequence identity and structural similarity, human β-defensin 3 (HBD3) kills Staphylococcus aureus with a 4- to 8-fold higher efficiency than human β-defensin 2 (HBD2), whereas the activities against Escherichia coli are identical. The design and characterization of HBD2/HBD3 chimeric peptides revealed that distinct molecular regions are responsible for their divergent killing properties. Two of the chimeras killed both E. coli and S. aureus with an even higher efficacy than the wild-type molecules. Moreover, one of these two chimeras maintained its high killing activities in the presence of physiologic salt concentrations. Due to the broad spectrum of their antimicrobial activities against many human multidrug-resistant pathogens, these two designer peptides of human origin represent promising templates for a new class of antibiotics.
Antimicrobial peptides have been identified and isolated from a wide variety of organisms, including humans, where they mostly participate in the first line of defense against pathogens (9). Among them are the defensins, a family of small antimicrobial peptides ranging from about 2 to 6 kDa, which are rich in cationic amino acid and cysteine residues (1, 3, 21, 23, 26). Their abundance in humans and other vertebrates, along with their broad microbicidal activity, made them important effector molecules of neutrophils, mucosal surfaces, skin, and epithelia (13).
The human defensins are classified into two subfamilies, α- and β-defensins, which are distinguished by the localization and pairing of six cysteine residues (2, 22, 24). To date, 40 potential coding regions for human β-defensins have been found (15). The expression of many of these defensins was confirmed at the mRNA level, but only HBD1 to HBD4 have been characterized explicitly and are predominantly expressed in various epithelial tissues. The human β-defensins HBD2 and HBD3 were isolated initially from keratinocytes of patients with psoriasis (5, 6). Both molecules are inducible by bacterial challenge and proinflammatory cytokines, including tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), and interleukin-1β (IL-1β) (4, 6, 7, 10, 19, 25). Interestingly, human β-defensins themselves can act as chemokines by stimulating the adaptive immune response. HBD2 is known to mobilize memory T cells and immature dendritic cells and was shown to be mediated by human chemokine receptor 6 (CCR6) (29). HBD3 recruited monocytes; it was dependent on disulfide bond connectivity (28).
HBD2 is highly active against Gram-negative Escherichia coli, but it was found to be bacteriostatic only against Gram-positive Staphylococcus aureus even at comparatively very high protein concentrations (6). However, the antimicrobial activity of HBD2 against E. coli is strongly decreased in the presence of physiological salt concentrations (25). In contrast, HBD3 possesses bactericidal activity at low concentrations not only against E. coli but also against S. aureus (4, 5). Moreover, the bactericidal activity of the more positively charged HBD3 against S. aureus is described to be not sensitive to physiological salt concentration (5).
HBD2 and HBD3 show remarkable differences in their antimicrobial activities against Gram-positive and Gram-negative bacteria and in their sensitivity to physiological salt concentration (15). Therefore, we aim to identify the determinants responsible for these activities. Based on the three-dimensional structure, we designed six chimeric HBD2/HBD3 molecules and studied their antimicrobial activity against E. coli and S. aureus strains. Two of these chimeras showed an enhanced activity compared to that of their parent molecules and therefore were tested further against a broad range of multidrug-resistant pathogens. The high activity against these pathogens makes these molecules promising candidates for clinical studies.
MATERIALS AND METHODS
Generation of chimeric HBD2 and HBD3 constructs.
The nucleotide sequences encoding HBD2 and HBD3 chimeric peptides were codon optimized for bacterial expression and synthesized by GENEART (Regensburg, Germany). They were cloned into the expression vector pET-30a (Novagen) using KspI and XhoI cleavage sites to generate a fusion protein with an N-terminal His6 tag and an enterokinase or a factor Xa cleavage site. The resultant protein sequences of chimeric peptides are the following (underlined sequences belong to HBD2/HBD3 chimeras): C1, MHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTDDDDKGIGDPVTCLKSGAICHPVFCPRRYKQIGKCSTRGRKCCRRKK; C2, MHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTDDDDKGIGDPVTCLKSGGRCAVLSCLPKEEQIGTCGLPGTKCCKKP; C3, MHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTIEGRGIINTLQKYYCRVRGAICHPVFCPRRYKQIGTCGLPGTKCCKKP; C4, MHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTIEGRGIINTLQKYYCRVRGGRCAVLSCLPKEEQIGTCGLPGTKCCKKP; C5, MHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTIEGRGIINTLQKYYCRVRGAICHPVFCPRRYKQIGKCSTRGRKCCRRKK; and C6, MHHHHHHSSGLVPRGSGMKETAAAKFERQHMDSPDLGTDDDDKGIGDPVTCLKSGGRCAVLSCLPKEEQIGKCSTRGRKCCRRKK.
Protein expression and synthesis.
HBD3 was expressed and purified as described previously (5); the same procedure was used for HBD2. The chimeras were expressed in the following E. coli strains: BL21(DE3) pLysS (C1, C2, and C6), BL21(DE3) (C4), and BLR(DE3) (C3). All strains were cultured in LB medium with 50 μg/ml kanamycin at 37°C. When culture broths reached an optical density at 600 nm of ∼0.6, isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 1 or 0.3 mM (C2) to induce protein expression. The cell growth was continued at 37°C for 3 h, with the exception of C2, which was incubated overnight, and of C1, which was incubated at 25°C.
Due to the low yield of the recombinant expression of peptides C3, C5, and C6, these peptides were chemically synthesized by Biosyntan GmbH (Berlin, Germany) using standard Fmoc-based solid-phase synthesis. The synthetic peptides were generated without the N-terminal fusion part, as is the case for the recombinantly produced chimeras.
Purification and renaturation of recombinantly expressed and synthesized peptides.
Bacteria were harvested by centrifugation and resuspended in 50 mM Tris-HCl, 150 mM NaCl, pH 7.5 (C1, C2, and C4) or in 100 mM Tris-HCl, 6 M guanidine hydrochloride, pH 8 (C3 and C6). The bacteria suspension was sonicated for 2 min and centrifuged. The supernatants containing soluble expressed peptide C1, C2, or C4 were loaded onto Ni2+-nitrilotriacetic acid (NTA) agarose (Qiagen, Hilden, Germany) columns equilibrated with 50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, pH 8.0. The column was washed with 50 mM sodium phosphate, 300 mM NaCl, 40 mM imidazole, pH 8. Chimeric peptides were eluted using 50 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole, pH 8.0. Supernatants containing denatured peptides C3 and C6 were loaded onto Ni2+-NTA agarose columns (Qiagen, Hilden, Germany) equilibrated with 100 mM Tris-HCl, 6 M guanidine hydrochloride, pH 8. After the column was washed with the sample's buffer, the peptides were eluted with 50 mM sodium acetate, 6 M guanidine hydrochloride, pH 4. The elutions were subjected to refolding by following the method of Jung et al. (11) except for final dialysis, which was performed against 50 mM sodium acetate, pH 5.
The synthesized peptides C3, C5, and C6 have been delivered as lyophilized materials that were reconstituted in 10 mM HCl with a final peptide concentration of 1 mg/ml. Samples were diluted 1:40 in 50 mM Tris-HCl, 1 M guanidine hydrochloride, 4 mM reduced glutathione, and 0.4 mM oxidized glutathione, pH 8.5, and were incubated at 37°C overnight.
After incubation, the solvent of the samples was exchanged to 50 mM sodium phosphate, pH 6, by dialysis.
Cleavage of recombinantly expressed peptides by enterokinase or factor Xa.
The buffers of both the soluble expressed and renatured peptides were exchanged to 50 mM sodium phosphate, pH 8.0, for cleavage with EnterokinaseMax (Invitrogen) or to 50 mM Tris-HCl, 1 mM CaCl2, pH 8.0, for cleavage with factor Xa (Roche). Cleavage was performed at room temperature for 24 to 96 h.
Purification of mature peptides.
After the cleavage of the recombinant peptides and the renaturation of the synthesized peptides, respectively, all samples were purified by reverse-phase high-performance liquid chromatography (RP-HPLC) using C18 columns (Macherey-Nagel, Düren, Germany) and acetonitrile-trifluoroacetic acid in the mobile phase. The average masses of the purified peptides were confirmed by mass spectrometry in linear mode using a 4700 proteomics analyzer matrix-assisted laser desorption-ionization time-of-flight/time-of-flight (MALDI-TOF/TOF) mass spectrometer (Applied Biosystems, Framingham, MA).
Determination of antimicrobial activity.
Susceptibility to the chimeras was determined in a modified microdilution assay as described previously (18). Briefly, after a 2- to 3-h growth period in brain heart infusion broth at 37°C, the test strains were adjusted to 104 to 105 bacteria per ml in 10 mM sodium phosphate, pH 7.2. Ten μl of the chimera solutions (range of final concentrations tested, 0.0125 to 100 μg/ml) was added to 100 μl of the bacterial suspension and incubated at 37°C for 2 h before CFU numbers were determined. Phosphate buffer as well as the chimera solvent (0.01% acetic acid) served as negative controls. The antibacterial activities of the peptides were given either as LD90 (≥90% lethal dose) or as minimal bactericidal concentrations (MBCs) (≥99.9% killing). Arbitrarily, a strain was defined as susceptible to a peptide if it had an MBC or LD90 of ≤12,5 μg/ml, as intermediately susceptible if it had an MBC of 25 to 100 μg/ml, or as resistant if it had an MBC or LD90 of >100 μg/ml. E. coli ATCC 35218 served as the control strain.
For the susceptibility testing of anaerobic bacteria, the strains were incubated under anaerobic conditions using prereduced media. E. coli ATCC 35218 subjected to these conditions served as the control for unimpaired chimera activity under anaerobic conditions.
As the sensitivity testing of yeasts in 10 mM phosphate buffer resulted in the significant unspecific killing of the cells, the phosphate buffer was supplemented with 10% brain heart infusion and 2% glucose. However, under these conditions the activity of C5 and C5 was somewhat reduced, as shown by control experiments with E. coli strain ATCC 35218.
The salt sensitivity of the chimeras C3 and C5 was determined in control experiments with 10 mM phosphate buffer supplemented with sodium chloride at final concentrations of 50 to 150 mM. Salt sensitivity was tested using strains E. coli ATCC 35218 and S. aureus ATCC 12600.
Hemolysis.
Peptides from stock solutions (C5 and melittin from bee venom [Sigma]) were 2-fold serially diluted in 50 mM sodium phosphate, 50 mM NaCl, pH 7.4, in a V-bottom 96-well microtiter plate (Sarstedt, Germany) after being coated with 0.1% bovine serum albumin (BSA) for 15 min. Human red blood cells (blood group A, rh+) were washed and resuspended in the same buffer to a hematocrit of 12.5%. The erythrocyte suspension (20 μl) was added to the diluted peptides (80 μl), resulting in a final hematocrit of 2.5%. The experiments were performed in quadruplicate. For controls, erythrocytes were incubated in the same volume of distilled water (maximal lysis) or in buffer supplemented with the corresponding volume of peptide solvent (spontaneous lysis). The plate subsequently was incubated at 37°C for 1 h. After gentle centrifugation, 20 μl of the supernatant was transferred to a flat-bottom 96-well microtiter plate (Sarstedt, Germany) and 10-fold diluted with 50 mM sodium phosphate, 150 mM NaCl, pH 7.4. The absorbance of the diluted supernatants was measured at 405 nm with a microtiter plate reader (Titertek Multiskan MCC/340 MKII; Flow Laboratories). Percent lysis was defined as (experimental lysis − spontaneous lysis)/(maximal lysis − spontaneous lysis) × 100 and plotted against micromolar peptide concentrations, giving medians and ranges.
Cytotoxicity.
HepG2 and MDCK cells suspended in Dulbecco's modified essential medium (DMEM)-high glucose culture medium (PAA Laboratories, Paschberg, Austria) supplemented with 10% (vol/vol) fetal calf serum, 1% (vol/vol) penicillin-streptomycin (PAA Laboratories, Paschberg, Austria) were seeded on a 96-well microtiter plate at a density of 1 × 104 cells per well. After incubation overnight at 37°C under 5% CO2 in a humidified atmosphere, cells were washed twice in 20 mM HEPES, 150 mM NaCl, pH 7.4. Peptides were diluted in the wash buffer (range of final concentrations tested, 2.5 to 250 μg/ml or 0.5 to 50 μM) and 100 μl was added per well. After an incubation of 2 h at 37°C under 5% CO2, 20 μl of CellTiter blue reagent (Promega GmbH, Mannheim, Germany) was added. During a further 2 h of incubation, viable cells metabolized the indicator dye, resulting in an increase of fluorescence that was measured on a microplate fluorescence reader (Lambda Fluoro 320; MWG Biotech, Ebersberg, Germany) using excitation and emission filters of 530 and 590 nm, respectively. Cells were incubated with 1% (vol/vol) Triton X-100 for maximum lysis (100% value) and with buffer for minimum lysis (0% value). Percent cytotoxicity was defined as (0% value − experimental value)/(0% value − 100% value) × 100 and plotted against the micromolar peptide concentration. Two independent experiments have been performed, each as triplicates, for both cell lines.
Molecular modeling.
Models of the three-dimensional structures of the HBD2/HBD3 chimeras were generated using the structures of HBD2 (Protein Databank [PDB] accession code 1FQQ) and HBD3 (PDB accession code 1kJ6) as templates. The positions of the six cysteine residues in each molecule were used to superimpose the two structures. To build the chimeric molecules, the relevant parts were taken from each structure and combined. Finally, these model structures were energy minimized using the steepest-descent algorithm implemented in the GROMOS force field (27).
RESULTS
Construction of the chimeras.
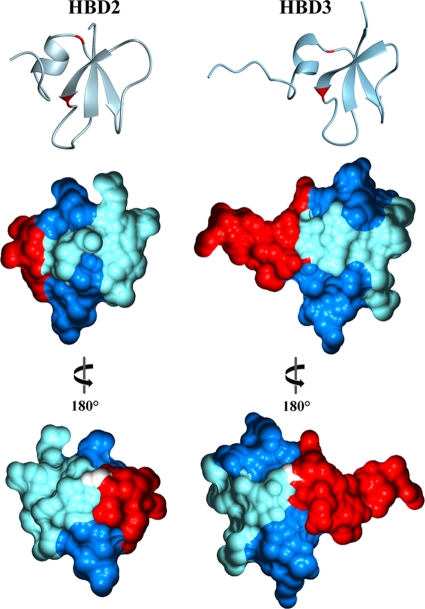
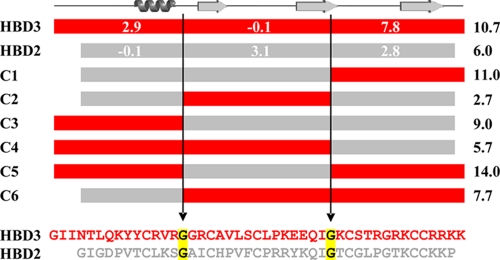
Besides the β-defensin typical cysteine residues, two glycine residues are conserved in HBD2 and HBD3. Using these glycines as boundaries, the resulting three sequence segments form four single, self-contained epitopes at the molecular surface (Fig. 1). Whereas the N-terminal and the middle segment form a single surface epitope each, the C-terminal segment is involved in two surface areas (Fig. 1). Since the surface properties of these molecules are responsible for the initial interaction with the bacterial membranes, the three sequence segments were used for the construction of six chimeras (Fig. 2).
FIG. 1.
Three-dimensional structures and molecular surfaces of HBD2 and HBD3. The positions of the two conserved glycines dividing the peptide surfaces into four single, self-containing epitopes are marked in red in the ribbon representations (upper panel). The molecular surfaces are depicted in the same orientation (middle panel) as the ribbon representation or rotated 180° (lower panel). The N-terminal epitope is colored red, the middle epitope is colored pale blue, and the C-terminal epitopes are colored blue. White surfaces depict the conserved glycines. Graphical representations were generated using the programs MOLMOL (12) and GRASP2 (16).
FIG. 2.
Schematic representation of the HBD2/HBD3 chimeric molecules (C1 to C6) and sequence alignment of HBD2 and HBD3. Sequences of HBD2 and HBD3 were divided into three regions, and the conserved glycines (marked in yellow) served as the internal borders for the construction of the chimeric proteins. Numbers represent the charge of the corresponding amino acid stretch (white) or the entire chimera (right column) at pH 7. Positions of α-helix and β-strands (arrows) according to the primary structure of the defensins are depicted on top.
Expression and purification.
Chimeras were recombinantly expressed in E. coli or chemically synthesized. After renaturation, the peptides were purified by RP-HPLC and analyzed by mass spectrometry. The analysis of the fractions revealed that the expected mass of chimera 1 was detected in seven distinct fractions (named C1.1 to C1.7). The different elution times might be due to a different combination of the six cysteine residues leading to the formation of disulfide bridge isomers. The elution times and measured and expected masses are summarized in Table S1 in the supplemental material. The chimeras C2, C3, C4, C5, and C6 each were eluted in a single fraction with the expected mass.
Antimicrobial activity against E. coli and S. aureus.
The antimicrobial activity of HBD2 and HBD3 was tested by a microdilution assay using different E. coli and S. aureus reference strains (Table 1). HBD2 and HBD3 exhibit the same activity against all E. coli reference strains, whereas HBD3 is four to eight times more effective in killing all S. aureus reference strains. The reproducibility of the microdilution assay was verified by determining the MBC and LD90 values in triplicate for E. coli ATCC 35218 (Table 2).
TABLE 1.
Antimicrobial activity of HBD2 and HBD3 against different reference strains of E. coli and S. aureusa
| Strain | HBD2 |
HBD3 |
||
|---|---|---|---|---|
| MBC (μg/ml) | LD90 (μg/ml) | MBC (μg/ml) | LD90 (μg/ml) | |
| E. coli ATCC 35218 | 0.78 | 0.2 | 0.78 | 0.2 |
| E. coli ATCC 25922 | 1.56 | 0.39 | 1.56 | 0.39 |
| E. coli ATCC 11775 | 0.78 | 0.2 | 0.78 | 0.39 |
| S. aureus ATCC 12600 | 12.5 | 6.25 | 1.56 | 0.39 |
| S. aureus ATCC 29213 | 12.5 | 6.25 | 3.125 | 0.39 |
| S. aureus ATCC 25923 | 25 | 12.5 | 3.125 | 0.78 |
Studies were performed in 9 mM sodium phosphate, 0.001 % (vol/vol) trifluoroacetic acid, pH 7.2.
TABLE 2.
Antimicrobial activity of HBD2, HBD3, and the chimeric molecules (C1 to C6)a
| Molecule |
E. coli ATCC 35218 |
S. aureus ATCC 12600 |
||
|---|---|---|---|---|
| MBC (μg/ml) | LD90 (μg/ml) | MBC (μg/ml) | LD90 (μg/ml) | |
| HBD2 | 0.39/0.78/0.78 | 0.1/0.2/0.2 | 12.5 | 1.56 |
| HBD3 | 1.56/1.56/3.125 | 0.39/0.39/0.39 | 3.13 | 0.39 |
| C1.1 | 0.2 | 0.1 | 6.25 | 0.78 |
| C1.2 | 0.1 | 0.025 | 3.13 | 0.39 |
| C1.3 | 0.1 | 0.025 | 1.56 | 0.39 |
| C1.4 | 0.2 | 0.025 | 3.13 | 0.78 |
| C1.5 | 0.1 | 0.025 | 6.25 | 0.39 |
| C1.6 | 0.2 | 0.025 | 6.25 | 3.13 |
| C1.7 | 0.2 | 0.05 | 6.25 | 0.39 |
| C2 | ND | 0.39 | >100 | >100 |
| C3 | 0.1 | 0.025 | 0.78 | 0.1 |
| C4 | 6.25 | 1.56 | 3.13 | 0.39 |
| C5 | 0.39 | 0.1 | 0.39 | 0.1 |
| C6 | >100 | ND | >100 | >100 |
Studies were performed in 9 mM sodium phosphate, 0.001 % (vol/vol) trifluoroacetic acid, pH 7.2. ND, not determinable.
In chimeras C1.1 to C1.7, C2, and C3, which each contain a particular molecular region of HBD3, C3 exhibits the highest activity against S. aureus. The efficiency of killing was four times increased compared to that of HBD3. The MBCs of the C1 variants were similar to that of HBD3, whereas C2 showed a clearly reduced activity. Therefore, the middle molecular region of HBD3 seems to be dispensable for S. aureus killing. Of the chimeras C4 and C6, which contain either the N-terminal or the C-terminal region of HBD3, only C4 exhibits a high activity against S. aureus. Therefore, only the N-terminal region contains the molecular region critical for activity against S. aureus. Interestingly, the same tendency holds true for the activity of HBD3 against E. coli.
The bactericidal activity against S. aureus of the C1 variants that do not contain the N-terminal region of HBD3 must originate from the middle region of HBD2, which also was revealed to be responsible for the killing of E. coli. In chimeras C4, C5, and C6, which all comprise merely one of the HBD2 regions, only C5 showed an activity against E. coli as high as that of HBD2 (Table 2). The activity of C4 was at least eight times reduced compared to that of HBD2, whereas C6 showed an even more drastically reduced activity. Therefore, the middle region of HBD2 appears to be the determinant of its activity against E. coli. This is emphasized by comparing the MBCs for chimeras C1 and C2, which contain either the middle or the C-terminal region of HBD2. Only C1 exhibited a high activity against E. coli. In contrast, the N-terminal region of HBD2 seems to be dispensable for the activity against E. coli.
In summary, C3 and C5 contain the N-terminal region of HBD3 as well as the middle region of HBD2 and differ only in their C-terminal region. Both peptides exhibited the same or an increased activity against S. aureus and E. coli (Table 2). In contrast, the chimeras C2 and C6 contain neither the N-terminal region of HBD3 nor the middle region of HBD2 and showed drastically reduced killing efficiencies. This confirms the importance of the N-terminal region of HBD3 and the middle region of HBD2 as determinants for their antimicrobial activities.
In addition to their antimicrobial activity, human β-defensins can act as chemokines, and HBD2 and HBD3 induce chemotaxis through interaction with CCR2 on monocytes (17). Therefore, we tested chimeras C3 and C5 with respect to their chemoattractant properties for peripheral blood monocytes accordingly. Compared to the positive control MCP-1, a chemotactic activity of these molecules could not be observed (data not shown).
Antimicrobial activity in the presence of high salt concentrations.
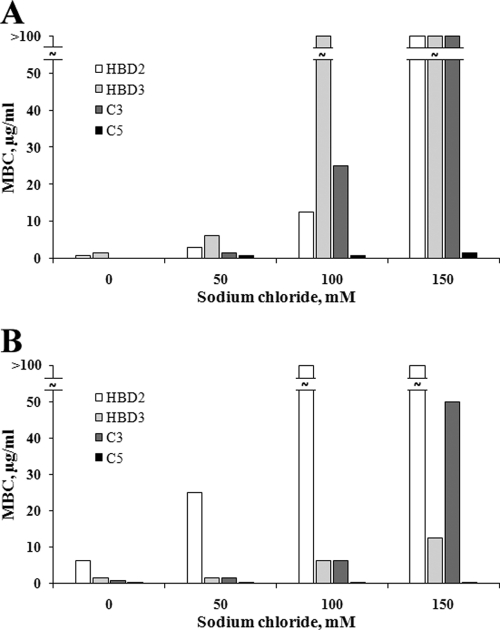
HBD2 and HBD3 exhibited a clear reduction of their antimicrobial activities at high sodium chloride concentrations (Fig. 3, Table 3). In the case of E. coli, HBD2 was less sensitive to high salt concentrations and maintained a considerably decreased but still remarkable activity (MBC, 12.5 μg/ml) in the presence of 100 mM sodium chloride, whereas HBD3 was almost inactive (MBC, >100 μg/ml) (Fig. 3A). In contrast, HBD3 showed considerably diminished but still substantial activity (MBC, 12.5 μg/ml) against S. aureus even in the presence of 150 mM sodium chloride, whereas HBD2 was almost inactive (MBC > 100 μg/ml already at 100 mM sodium chloride) (Fig. 3B). The chimera C3 behaved like HBD2 in the case of E. coli (Fig. 3A) and in principal like HBD3 in the case of S. aureus concerning its salt sensitivity (Fig. 3B). Most interestingly, C5 showed only a slight, if any, sensitivity to increasing concentrations of sodium chloride in cases of both microbes and was highly active even at 150 mM sodium chloride. C5 depicted the same antimicrobial activity against E. coli as HBD3 in the absence of sodium chloride (Fig. 3A and B).
FIG. 3.
Salt sensitivity of the antimicrobial activities of HBD2, HBD3, and chimeras C3 and C5. The salt sensitivity was tested against E. coli ATCC35218 (A) and S. aureus ATCC12600 (B) by measuring the MBCs in the presence of 50, 100, and 150 mM sodium chloride.
TABLE 3.
Salt sensitivity of the antimicrobial activities of HBD2, HBD3, and chimeras C3 and C5a
| Molecule and NaCl concn (mM) |
E. coli ATCC 35218 |
S. aureus ATCC 12600 |
||
|---|---|---|---|---|
| MBC (μg/ml) | LD90 (μg/ml) | MBC (μg/ml) | LD90 (μg/ml) | |
| HBD2 | ||||
| 0 | 0.78 | 0.2 | 6.25 | 1.56 |
| 50 | 3.13 | 0.78 | 25 | 25 |
| 100 | 12.5 | 3.13 | >100 | 50 |
| 150 | >100 | 25 | >100 | >100 |
| HBD3 | ||||
| 0 | 1.56 | 0.39 | 1.56 | 0.78 |
| 50 | 6.25 | 1.56 | 1.56 | 0.78 |
| 100 | >100 | 12.5 | 6.25 | 1.56 |
| 150 | >100 | >100 | 12.5 | 6.25 |
| Chimera C3 | ||||
| 0 | 0.2 | 0.1 | 0.78 | 0.2 |
| 50 | 1.56 | 0.1 | 1.56 | 0.78 |
| 100 | 25 | 6.25 | 6.25 | 3.13 |
| 150 | >100 | 12.5 | 50 | 12.5 |
| Chimera C5 | ||||
| 0 | 0.2 | 0.05 | 0.39 | 0.1 |
| 50 | 0.78 | 0.1 | 0.39 | 0.1 |
| 100 | 0.78 | 0.2 | 0.39 | 0.2 |
| 150 | 1.56 | 0.2 | 0.39 | 0.1 |
The salt sensitivity was tested against E. coli ATCC 35218 and S. aureus ATCC 12600 by measuring the MBC and LD90 values in the presence of 50, 100, and 150 mM sodium chloride.
Antimicrobial activity against multiresistant human pathogens.
Due to the exceptional antimicrobial activity and remarkable salt tolerance of chimeras C3 and C5, a broad range of human Gram-negative and Gram-positive pathogens and Candida strains were tested for their susceptibility to these peptides (Table 4). Both chimeras possess broad-spectrum antimicrobial activity and kill bacteria, including multiresistant strains (extended-spectrum β-lactamase, vancomycin-resistant enterococci, and methicillin-resistant S. aureus), with very high efficacy. In most cases the MBCs of C5 were below 1 μg/ml, whereas the MBCs of C3 slightly exceeded 1 μg/ml. However, independently of the bacterial strain or species both chimeras killed efficiently at very low concentrations.
TABLE 4.
Antimicrobial activity of chimeras C3 and C5a
| Strain | C3 |
C5 |
||
|---|---|---|---|---|
| MBC (μg/ml) | LD90 (μg/ml) | MBC (μg/ml) | LD90 (μg/ml) | |
| Gram negative | ||||
| E. coli E4 ESBL (ci) | 0.78 | 0.78 | 0.39 | 0.1 |
| E. coli E9 ESBL (ci) | 0.78 | 0.78 | 0.39 | 0.1 |
| Klebsiella pneumoniae ATCC 700603 ESBL | 0.39 | 0.2 | 0.39 | 0.2 |
| K. pneumoniae CF1 ESBL (ci) | 3.13 | 1.56 | 1.56 | 0.78 |
| K. pneumoniae CF7 ESBL (ci) | 1.56 | 0.78 | 0.39 | 0.2 |
| Pseudomonas aeruginosa CF 453 mr (ci) | 1.56 | 1.56 | 0.39 | 0.2 |
| P. aeruginosa CF 479 mr (ci) | 1.56 | 0.78 | 0.78 | 0.2 |
| P. aeruginosa CF 509 mr (ci) | 1.56 | 0.78 | 0.78 | 0.2 |
| P. aeruginosa CF 629 mr (ci) | 1.56 | 0.78 | 0.78 | 0.2 |
| P. aeruginosa CF 640 mr (ci) | 1.56 | 0.78 | 0.39 | 0.2 |
| Gram positive | ||||
| E. faecalis ATCC 51299 VRE | 0.78 | 0.2 | 1.56 | 0.39 |
| E. faecium 354 VRE (ci) | 1.56 | 0.78 | 0.39 | 0.2 |
| E. faecium 356 VRE (ci) | 0.78 | 0.39 | 0.39 | 0.2 |
| E. faecium G 70 VRE (ci) | 1.56 | 0.78 | 1.56 | 0.39 |
| E. faecium G 71 VRE (ci) | 1.56 | 0.78 | 0.78 | 0.2 |
| S. aureus ATCC 33393 MRSA | 0.78 | 0.39 | 0.39 | 0.2 |
| S. aureus ATCC 43300 MRSA | 0.78 | 0.39 | 0.78 | 0.2 |
| S. aureus MRSA 344 (ci) | 0.78 | 0.39 | 0.39 | 0.2 |
| S. aureus MRSA 355 (ci) | 0.78 | 0.39 | 0.39 | 0.2 |
| S. aureus MRSA 358 (ci) | 0.78 | 0.39 | 0.39 | 0.2 |
| Fungi | ||||
| Candida albicans ATCC 24433 | 12.5 | 6.25 | 100 | 25 |
| C. albicans ATCC 36801 | 50 | 12.5 | >100 | 50 |
| C. albicans ATCC 44373 | 25 | 12.5 | >100 | 25 |
| C. albicans ATCC 76615 | 50 | 12.5 | >100 | 50 |
| C. albicans ATCC 90028 | 12.5 | 6.25 | >100 | 25 |
| C. albicans DSMZ 11949 | 50 | 12.5 | >100 | 25 |
| Anaerobic bacteria | ||||
| Bacteroides fragilis ATCC 25285 | 6.25 | 0.2 | 50 | 1.56 |
| Clostridium perfringens ATCC 13124 | >100 | >100 | 6.25 | 1.56 |
| Peptostreptococcus anaerobius ATCC 27337 | 50 | 0.78 | 25 | 0.39 |
| Propionibacterium acnes ATCC 6919 | 1.56 | 0.39 | 3.125 | 0.78 |
| Prevotella oralis ATCC 33321 | >100 | 12.5 | >100 | 50 |
ci, clinical isolate; mr, multiresistant. Studies were performed in 9 mM sodium phosphate, 0.001 % (vol/vol) trifluoroacetic acid, pH 7.2.
Cytotoxicity.
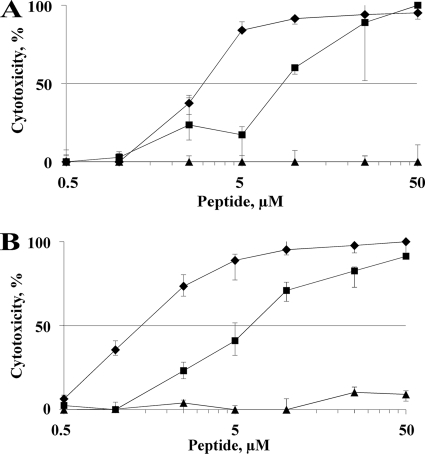
Chimera C5 retained its high antibacterial activity at physiologic salt concentrations, making it a tempting candidate for therapeutic use. Therefore, together with HBD3 and HBD2 it was tested for its cytotoxic effect on nucleated, metabolically active human cells. The hepatic HepG2 and the renal MDCK cell lines have been used here as models. C5 showed cytotoxicity at concentrations higher than 1 μM (5 μg/ml) in the case of HepG2 cells and at even lower concentrations in the case of MDCK cells (Fig. 4). At a concentration between 1 and 3 μM (5 to 15 μg/ml), C5 reached 50% cytotoxicity in these cell lines. Whereas HBD2 did not show any significant cytotoxic effect, even at 50 μM (250 μg/ml), HBD3 revealed cytotoxicity against HepG2 and MDCK cells starting from a concentration higher than 1 μM (5 μg/ml), reaching the 50% value between 5 and 10 μM (25 to 50 μg/ml). C5 reached almost 100% cytotoxicity at a concentration of 10 μM (50 μg/ml), whereas HBD3 amounted to 50 μM (250 μg/ml).
FIG. 4.
Cytotoxic activity of HBD2, HBD3, and C5. HepG2 (A) and MDCK (B) cells were incubated with increasing amounts of HBD2 (▴), HBD3 (▪), and C5 (⧫). Cytotoxicity was measured fluorometrically. Metabolically inactive target cells showed a decrease of fluorescence compared to that of viable, metabolically active cells that is expressed in percent cytotoxicity.
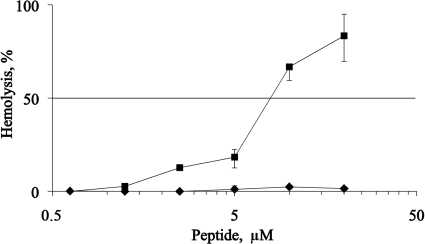
Hemolysis.
C5 peptide did not exhibit hemolytic activity against fresh human red blood cells at the highest concentration tested, i.e., 20 μM. For comparison, the hemolytic activity of the potent peptide melittin is almost 100% at the same concentration (Fig. 5).
FIG. 5.
Hemolytic activity of bee venom melittin (▪) and chimera C5 (⧫) against human red blood cells. At pH 7.4 and in the presence of 150 mM NaCl, no hemolytic activity of C5 was detected at up to 20 μM peptide. For a control, melittin was used as a potent hemolytic agent.
DISCUSSION
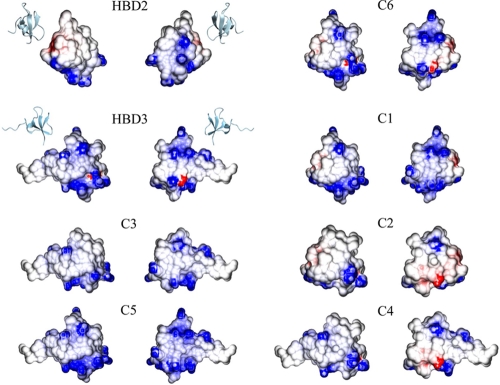
A common feature of most antimicrobial peptides is their net-positive charge, causing the initial attraction to the target membrane via interaction with the negative charges of the phospholipids (9). Actually, only in chimeras 3 and 5 were the negatively charged amino acid residues from the native proteins completely removed, resulting in higher net-positive charges than those of HBD2 or HBD3, respectively (Fig. 6). Nevertheless, the HBD2/HBD3 chimeric peptides show a clear variation in their antimicrobial activity that does not depend on the overall net-positive charge of the molecules. More important for the antimicrobial activity is the charge distribution or regional charge density on the molecule's surface. For example, HBD2 compared to chimera 4 and HBD3 compared to chimera 1 show an equal net-positive charge but strongly differ in their killing efficacy. Furthermore, the corresponding high-activity determinants of HBD2 and HBD3 both possess a similar positive net charge, whereas the counterparts are uncharged (Fig. 2). However, the additional positive charges in the C terminus of chimera 5 compared to that of chimera 3 apparently mediate its high antimicrobial activity under physiologic salt concentrations. This is in agreement with the results of Scudiero et al., who investigated chimeric peptides of HBD1 and HBD3 and stated that the C terminus of HBD3 is fundamental for antibacterial activity at high ionic strength (20). Recently, Scudiero et al. also linked the first six N-terminal amino acids, which do not have an effect on the net positive charge, to the salt tolerance of HBD3, confirming that the charge alone is not pivotal for the antimicrobial activity (20).
FIG. 6.
Electrostatic surface potentials of HBD2, HBD3, and the chimeras. Ribbon representations of HBD2 (PDB accession code 1FQQ) and HBD3 (PDB accession code 1kJ6) depict the orientations of the molecules, each presented in front and back views (180° rotated). Structures of the chimeras (C1 to C6) were generated by molecular modeling using the structures of HBD2 and HBD3 as templates. Negatively charged regions are colored red, and positively charged regions are colored blue. Graphical representations were generated using the programs MOLMOL (12) and GRASP2 (16).
Our results confirm and contradict the investigations of Hoover et al., who studied derivatives of HBD3 and linked the activity against S. aureus to the N terminus (as is done in the present work) and the activity against E. coli to the C terminus (8). The latter statement is in contrast to the strongly inactive chimera C6, which comprises the C-terminal region of HBD3. However, the molecular regions themselves apparently are sufficient to exert the antimicrobial activities of the β-defensins. All chimeric peptides that contain the middle region of HBD2 possess at least the same efficacy against E. coli. The same holds true for chimeras harboring the N-terminal region of HBD3. Moreover, the combination of the molecular regions of HBD2 and HBD3 responsible for killing E. coli and S. aureus led to not only a combination of the activities but also to an enhancement. Hence, there might exist a synergism between the peptide regions.
Chimeras C3 and C5 unify the molecular regions from HBD2 and HBD3 that are responsible for the high killing activities against E. coli and S. aureus. Unlike the native HBD2 or HBD3, they allow the efficient killing of both bacterial strains and possess even lower MBCs at physiologic salt concentrations. Their activity against a broad range of microbes, including many multiresistant pathogens, makes them promising candidates for the development of therapeutics, especially chimera C5. In this context, cytotoxic side effects against eukaryotic cells must be excluded or at least must be in an acceptable ratio to the cure. The most attractive application of antimicrobial proteins is a systemic administration so that the highest concentration of protein will meet the blood cells. Our results show that concentrations of even 20 μM (100 μg/ml) do not harm human erythrocytes. Therefore, the more-than 250 times lower concentration (0.39 μg/ml) needed to kill 99.9% of S. aureus under physiologic concentrations opens a reasonable therapeutic window. Since all systemically administered substances will be cleared in the liver and/or kidney, the effect on hepatic HepG2 and renal MDCK cells has been studied as well. The MBC of chimera C5 against S. aureus under physiologic salt concentrations (0.39 μg/ml) is more than six times lower than the concentration at which C5 becomes cytotoxic (2.5 μg/ml, or 0.5 μM) against MDCK cells. This suggests that 99.9% of bacteria are killed at less than a sixth of the concentration that might become harmful to liver cells. C5 is only 2 to 10 times more cytotoxic (50% values) than wild-type HBD3, which is naturally expressed in many human tissues, but the MBC of C5 for E. coli is more than 60 times lower and for S. aureus more than 30 times lower than those for HBD3 (Tab. 3). This suggests that cytotoxic side effects against human host cells are at least in a reasonable ratio to the cure even if they are not totally excludable. This is supported by the work of Nishimura et al., whose investigations did not reveal any cytotoxic effects of HBD1, HBD2, and HBD3 on other cell types, including primary epithelial cells and fibroblasts (14). The noncytotoxic appearance of the native β-defensins 2 and 3 is a good basis for the development of therapeutic drugs. The presented work emphasizes that designer peptides with high antimicrobial activity may contribute to the search and development of new antibiotics against the rising number of multiresistant pathogens.
Supplementary Material
Acknowledgments
We thank Britta Hansen, Sonja Hollmer, Ursula Mundt, Silvia Voss, Maren Hartelt, and Björn Ahrens for their excellent technical assistance.
This study has been supported by the Deutsche Forschungsgemeinschaft (SFB 617, A9, A18, A26, Z1, and Z2).
Footnotes
Published ahead of print on 28 December 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Diamond, G., et al. 1991. Tracheal antimicrobial peptide, a cysteine-rich peptide from mammalian tracheal mucosa: peptide isolation and cloning of a cDNA. Proc. Natl. Acad. Sci. U. S. A. 88:3952-3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganz, T. 2005. Defensins and other antimicrobial peptides: a historical perspective and an update. Comb Chem. High Throughput Screen. 8:209-217. [DOI] [PubMed] [Google Scholar]
- 3.Ganz, T., et al. 1985. Defensins. Natural peptide antibiotics of human neutrophils. J. Clin. Investig. 76:1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia, J. R., et al. 2001. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 306:257-264. [DOI] [PubMed] [Google Scholar]
- 5.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 2001. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 6.Harder, J., J. Bartels, E. Christophers, and J. M. Schröder. 1997. A peptide antibiotic from human skin. Nature 387:861. [DOI] [PubMed] [Google Scholar]
- 7.Harder, J., et al. 2000. Mucoid Pseudomonas aeruginosa, TNF-alpha, and IL-1beta, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am. J. Respir. Cell Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 8.Hoover, D. M., Z. Wu, K. Tucker, W. Lu, and J. Lubkowski. 2003. Antimicrobial characterization of human beta-defensin 3 derivatives. Antimicrob. Agents Chemother. 47:2804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenssen, H., P. Hamill, and R. E. Hancock. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jia, H. P., et al. 2001. Discovery of new human beta-defensins using a genomics-based approach. Gene 263:211-218. [DOI] [PubMed] [Google Scholar]
- 11.Jung, S., et al. 2009. Hydramacin-1, structure and antibacterial activity of a protein from the basal metazoan hydra. J. Biol. Chem. 284:1896-1905. [DOI] [PubMed] [Google Scholar]
- 12.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51-55. [DOI] [PubMed] [Google Scholar]
- 13.Lehrer, R. I., and T. Ganz. 1996. Endogenous vertebrate antibiotics. Defensins, protegrins, and other cysteine-rich antimicrobial peptides. Ann. N. Y. Acad. Sci. 797:228-239. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura, M., et al. 2004. Effect of defensin peptides on eukaryotic cells: primary epithelial cells, fibroblasts and squamous cell carcinoma cell lines. J. Dermatol. Sci. 36:87-95. [DOI] [PubMed] [Google Scholar]
- 15.Pazgier, M., D. M. Hoover, D. Yang, W. Lu, and J. Lubkowski. 2006. Human beta-defensins. Cell. Mol. Life Sci. 63:1294-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrey, D., and B. Honig. 2003. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 374:492-509. [DOI] [PubMed] [Google Scholar]
- 17.Röhrl, J., D. Yang, J. J. Oppenheim, and T. Hehlgans. 2010. Human beta-defensin 2 and 3 and their mouse orthologs induce chemotaxis through interaction with CCR2. J. Immunol. 184:6688-6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sahly, H., et al. 2003. Burkholderia is highly resistant to human beta-defensin 3. Antimicrob. Agents Chemother. 47:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schröder, J. M. 1999. Epithelial antimicrobial peptides: innate local host response elements. Cell. Mol. Life Sci. 56:32-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scudiero, O., et al. 2010. Novel synthetic, salt-resistant analogs of human beta-defensins 1 and 3 endowed with enhanced antimicrobial activity. Antimicrob. Agents Chemother. 54:2312-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selsted, M. E., D. M. Brown, R. J. DeLange, S. S. Harwig, and R. I. Lehrer. 1985. Primary structures of six antimicrobial peptides of rabbit peritoneal neutrophils. J. Biol. Chem. 260:4579-4584. [PubMed] [Google Scholar]
- 22.Selsted, M. E., and S. S. Harwig. 1989. Determination of the disulfide array in the human defensin HNP-2. A covalently cyclized peptide. J. Biol. Chem. 264:4003-4007. [PubMed] [Google Scholar]
- 23.Selsted, M. E., S. S. Harwig, T. Ganz, J. W. Schilling, and R. I. Lehrer. 1985. Primary structures of three human neutrophil defensins. J. Clin. Investig. 76:1436-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selsted, M. E., et al. 1993. Purification, primary structures, and antibacterial activities of beta-defensins, a new family of antimicrobial peptides from bovine neutrophils. J. Biol. Chem. 268:6641-6648. [PubMed] [Google Scholar]
- 25.Singh, P. K., et al. 1998. Production of beta-defensins by human airway epithelia. Proc. Natl. Acad. Sci. U. S. A. 95:14961-14966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang, Y. Q., et al. 1999. A cyclic antimicrobial peptide produced in primate leukocytes by the ligation of two truncated alpha-defensins. Science 286:498-502. [DOI] [PubMed] [Google Scholar]
- 27.van Gunsteren, W. F., et al. 1996. Biomolecular simulation: the GROMOS96 manual and user guide. vdf Hochschulverlag AG, Zürich, Switzerland.
- 28.Wu, Z., et al. 2003. Engineering disulfide bridges to dissect antimicrobial and chemotactic activities of human beta-defensin 3. Proc. Natl. Acad. Sci. U. S. A. 100:8880-8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang, D., et al. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.